MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories (original) (raw)
Abstract
As molecular dynamics (MD) simulations continue to evolve into powerful computational tools for studying complex biomolecular systems, the necessity of flexible and easy-to-use software tools for the analysis of these simulations is growing. We have developed MDTraj, a modern, lightweight, and fast software package for analyzing MD simulations. MDTraj reads and writes trajectory data in a wide variety of commonly used formats. It provides a large number of trajectory analysis capabilities including minimal root-mean-square-deviation calculations, secondary structure assignment, and the extraction of common order parameters. The package has a strong focus on interoperability with the wider scientific Python ecosystem, bridging the gap between MD data and the rapidly growing collection of industry-standard statistical analysis and visualization tools in Python. MDTraj is a powerful and user-friendly software package that simplifies the analysis of MD data and connects these datasets with the modern interactive data science software ecosystem in Python.
Introduction
Molecular dynamics (MD) simulations yield a great deal of information about the structure, dynamics, and function of biological macromolecules by modeling the physical interactions between their atomic constituents. Modern MD simulations, often using distributed computing, graphics processing unit acceleration, or specialized hardware can generate large datasets containing hundreds of gigabytes or more of trajectory data tracking the positions of a system’s atoms over time (1). To use these vast and information-rich datasets to understand biomolecular systems and generate scientific insight, further computation, analysis, and visualization are required (2).
Within the last decade, the Python language (https://www.python.org/) has become a major hub for scientific computing. It features a wealth of high-quality open source packages, including those for interactive computing (3), machine learning (4), and visualization (5). This environment is ideal for both rapid development and high performance, as computational kernels can be implemented in the languages C, C++, and FORTRAN, but made available within a more user-friendly interactive environment.
In the MD community, the benefits of integration with such industry standard tools have not yet been fully realized because of a tradition of custom file formats and command-line analysis. To address this need, we have developed MDTraj, a modern, open, and lightweight Python library for analysis and manipulation of MD trajectories. The project has the following goals:
- To serve as a bridge between MD data and the modern statistical analysis and scientific visualization software ecosystem in Python.
- To support a wide range of MD data formats and computations.
- To run rapidly on modern hardware with efficient memory utilization, enabling the interactive analysis of large datasets.
Several other software packages for the analysis of MD trajectories exist, including the GROMACS tools (6), CPPTRAJ (7), VMD (8), MMTK (9), MDAnalysis (10), Bio3D (11), ST-Analyzer (12), LOOS (13), and Pteros (14). GROMACS and CPPTRAJ provide a broad range of functionality to users from the Unix command line, or with a simple interactive scripting environment. LOOS and Pteros are C++ toolkits that enable the construction of novel trajectory analysis programs, while VMD and ST-Analyzer provide convenient graphical interfaces. Like MDTraj, MMTK and MDAnalysis are written in Python while Bio3D is written in the statistical programming language R (https://www.r-project.org/). Each of these software packages has capabilities that have served to inform the development of MDTraj.
Materials and Methods
Capabilities and implementation
MDTraj is widely interoperable and extremely easy to use. First and foremost, MDTraj can load trajectory and/or topology data from the formats used by a broad range of MD packages, including AMBER (15), GROMACS (6), DESMOND (16), CHARMM (17), NAMD (18), TINKER (19), LAMMPS (20), OpenMM (21), ACEMD (22), and HOOMD-Blue (23); see Table 1 for a full list of supported file formats. This wide support enables consistent interfaces and reproducible analyses regardless of users’ preferred MD simulation packages.
Table 1.
List of supported file formats
| Package | File Formats |
|---|---|
| Many packages | pdb, xyz, dcd |
| Amber | prmtop, crd, netcdf, binpos, restrt |
| Gromacs | gro, xtc, trr |
| Desmond | dtr, stk |
| CHARMM | psf |
| LAMMPS | lammpstrj |
| TINKER | arc |
| HOOMD-Blue | xml |
| OpenMM | xml |
| TRIPOS | mol2 |
| MDTraj | hdf5 |
From its inception, MDTraj has been designed to work in concert with other packages for analysis and visualization. No single toolkit can provide all possible ways to analyze molecular simulations, especially given the rapid pace of development in statistics and data science. Rather than attempting to provide all conceivable functionality in one toolkit, MDTraj leverages Python and NumPy (http://www.numpy.org/) to empower users to connect their MD data with the large and rapidly growing ecosystem of data science tools available more broadly in the community.
MDTraj originated from the trajectory handling portions of MSMBuilder (24), where it now provides a stable base for handling trajectories, computing order parameters and projections, and providing the distance metrics—such as minimal root-mean-squared deviation (RMSD)—that are necessary for clustering. Additionally, it is now used inside tools that analyze data from the Folding@home distributed computing architecture (25), a structure-based virtual screening pipeline at Google Research, the PyEMMA Markov modeling package (26), the Ensembler and mBuild (27, 28) modeling tools, and countless individual analysis scripts. MDTraj is part of the Omnia consortium (http://omnia.md) suite of tools, which will be described in a later article.
Most data analyses for MD involve either extracting a vector of order parameters of each simulation snapshot or defining a distance metric between snapshots. MDTraj makes it very easy to rapidly extract these representations. It includes an extremely fast RMSD engine capable of operating near the machine floating point limit described in detail by Haque et al. (29), performing Theobald’s QCP algorithm (30) approximately three times faster than the original implementation. Functions for secondary-structure assignment (31), solvent-accessible surface area determination (32), hydrogen bond identification (33), residue-residue contact mapping, NMR scalar coupling constants (34), nematic order parameters (35), and the extraction of various internal degrees of freedom are similarly available. Where appropriate, these compute kernels are written in C or C++ and heavily optimized with vectorized instructions and multithreading. To enable interoperability, these data are returned to the user as multidimensional NumPy arrays, the standard numeric data storage format for the scientific Python ecosystem.
MDTraj also provides an atom selection language. Often, analysis functions are applied to a subset of atoms in the system. To generate arrays of these indices, the topology attribute and full Python grammar can be a powerful combination (i.e., Fig. 1, line 2). For users less familiar with Python or making the transition from other packages, a natural text-based selection syntax can be used as well (i.e., Fig. 1, line 3). These selection strings can be translated into standard Python syntax for pedagogical purposes or directly executed.
Figure 1.

The MDTraj atom selection language. Queries can be expressed using standard Python code (line 2), or an intuitive string-based syntax (line 3).
Ease-of-use is a central and deliberate goal at each level of the design and implementation of MDTraj. This starts with installation. Using the cross-platform Conda package manager, users can get started in seconds using the shell command conda install −c omnia mdtraj, which downloads and installs precompiled binaries of MDTraj (and all of its dependencies) on Windows, Linux, or Mac OS-X, without the requirement of administrator privileges.
The package has an extremely simple object model, which makes it very easy for new users to get started. Only a single class, Trajectory, needs to be mastered; it contains all relevant information about the MD trajectory, such as the atomic coordinates, unit cell dimensions, and simulation time. Loading files and performing analysis are generally done with functions (e.g., mdtraj.load, mdtraj.compute_) as opposed to classes to provide a simple and intuitive user experience that minimizes the need to remember complex object workflows.
MDTraj is extensively documented in a consistent format. The package itself contains over 9000 lines of Python docstrings that describe each function and class. The website, http://mdtraj.org, contains complete documentation, but more importantly contains 14 complete, executable example notebooks demonstrating topics including hydrogen-bond identification, Ramachandran plotting, and strategies for memory-limited computation on large datasets. These examples provide new users the patterns to get up and running with their own analyses immediately.
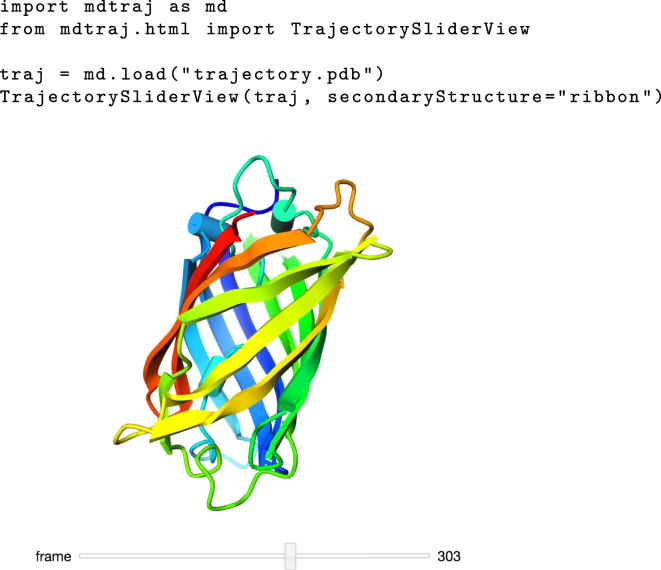
Furthermore, MDTraj includes a unique interactive WebGL-based three-dimensional structure viewer for the IPython notebook adapted from iview (36), shown in Fig. 2. Because it combines the analysis input code with results and plots into a single worksheet, the IPython notebook provides one of the most convenient user interfaces for interactive analysis. This convenience is further enhanced by MDTraj’s TrajectoryView widget, which runs inside the IPython notebook and provides a high-quality and fully interactive three-dimensional rendering of a trajectory. The viewer can save high-quality png images or STL three-dimensional models. MDTraj thus not only provides first-class scriptability but also high-quality three-dimensional visualization.
Figure 2.
MDTraj’s interactive WebGL-based protein and trajectory viewer. This feature requires a modern WebGL-enabled browser, and the IPython notebook that can be installed with Conda using the command conda install IPython-notebook. To see this figure in color, go online.
The development, engineering, and testing of MDTraj incorporates modern best practices for scientific computing (37). The package contains more than 1100 unit tests for individual components. These tests are continually run on each incremental contribution on both Windows and Linux, using multiple versions of Python and the required libraries. The project is hosted on GitHub, and development takes place fully openly and collaboratively. Users of MDTraj are often researchers who are interested in analyzing simulations in new ways, a task that involves not only MDTraj library functions but also writing new code. The simple coding style, open source licensing, GitHub pull-request-based development pattern (38), and active culture of collaborative code review enable these researchers to rapidly prototype new methods and extend MDTraj. This has been borne out by the MDTraj community, which comprises members from numerous academic and industrial research groups across the world that have contributed to the project over the past two years.
Results and Discussion
The capabilities of MDTraj serve as a bridge, connecting MD data with statistics and graphics libraries developed for general data science audiences. A key advantage of this design, for users and developers, is access to a much wider range of state-of-the-art analysis capabilities characterized by large feature sets, extensive documentation, and active user communities.
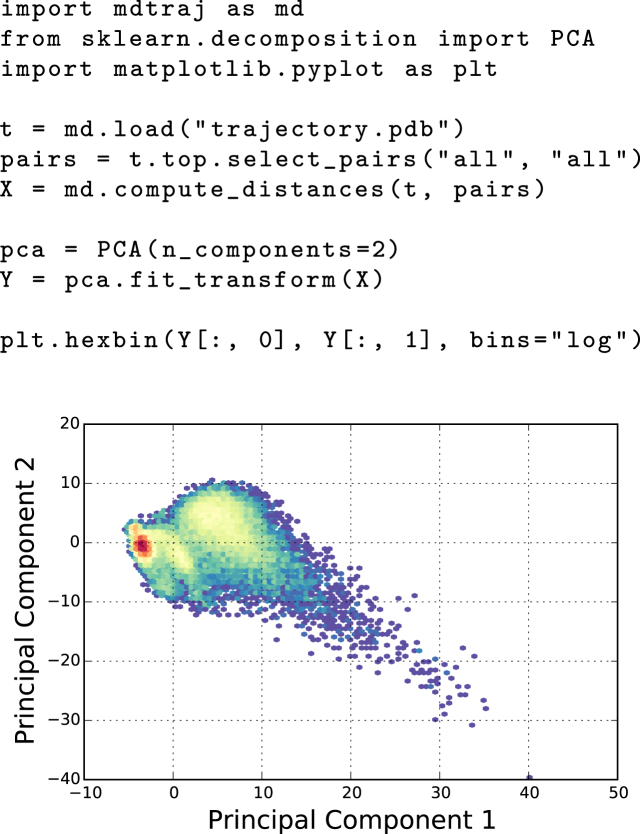
A demonstration of this integrative workflow is shown in Fig. 3, which combines MDTraj with the scikit-learn (4) for principal component analysis (PCA) and matplotlib (5) for visualization, to determine high-variance collective motions in a protein system. While PCA is a widely used method that is included in a variety of MD analysis packages, the advantage of integrating with the wider data science community is immediately evident when moving on to more complex statistical analysis. For example, a variety of sparse and kernelized PCA-like methods have been introduced into the machine learning community (39), and may be quite powerful for analyzing more complex protein systems. Because of its open and interoperable design, these cutting-edge statistical tools are readily available to MD researchers with MDTraj, without duplication of developer efforts and independent of the particular MD software used to perform the simulations.
Figure 3.
Demonstration of PCA with MDTraj, scikit-learn, and MATPLOTLIB. To see this figure in color, go online.
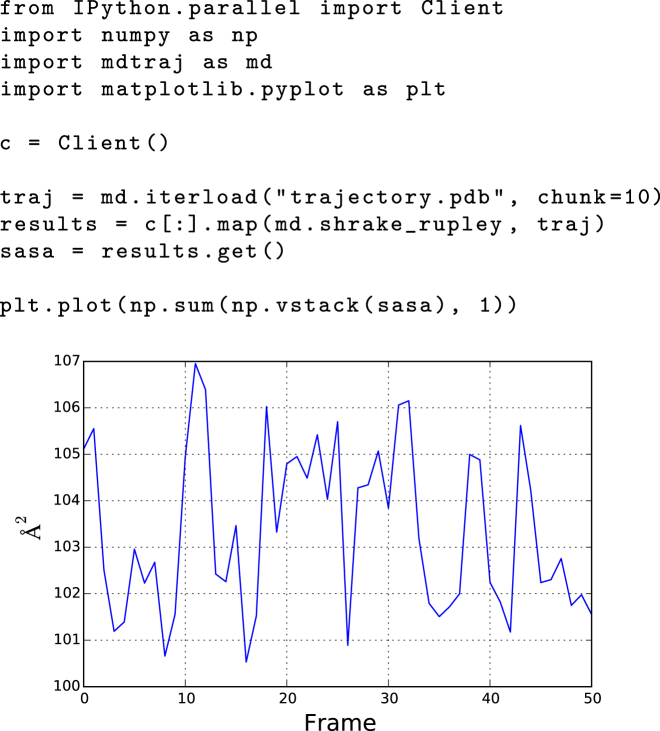
We generally find that file I/O and main memory are more limiting than raw CPU performance for MD analysis. For this reason, simple multinode parallelization, even over relatively slow interconnects, can often be extremely useful for accelerating calculations. As an example, Fig. 4 shows a demonstration of the use of MDTraj with the IPython parallel toolkit to parallelize the calculation of the solvent-accessible surface area of a trajectory over the individual snapshots of the trajectory. The code requires separately initializing an array of IPython engine processes on which the calculation is executed. These can be distributed over many nodes on a cluster or in the cloud and linked together by MPI or SSH. Because many simulation datasets contain many separate MD trajectories saved in separate files, a similar pattern can also be used to process individual files in parallel.
Figure 4.
Demonstration of solvent-accessible surface area calculation done in parallel with MDTraj and IPython. To see this figure in color, go online.
Conclusions
Within the field of trajectory analysis tools, MDTraj stands out due to its ease of use, flexibility, and Python-centric design, largely thanks to its organization around the intuitive Trajectory object in which data are stored as NumPy arrays. This design significantly enhances extensibility and gives users a great deal of latitude for freely accessing and manipulating the data according to the needs of their research. MDTraj speeds up analysis tasks by implementing computationally intensive operations (such as RMSD) using optimized low-level kernels written in C/C++. Furthermore, MDTraj can read and write a very wide range of trajectory file formats, ensuring interoperability across most MD software packages.
Software Availability
MDTraj is available under the GNU Lesser General Public License (LGPL), version 2.1 or later. Full documentation and examples are available at the project home page, http://mdtraj.org, and development is hosted on GitHub at http://github.com/mdtraj/mdtraj. The latest release, version 1.4.2, is archived at doi:10.5281/zenodo.18700.
Author Contributions
R.T.M., K.A.B., M.P.H., C.K., J.M.S., C.X.H., C.R.S., L.-P.W., and T.J.L. developed the software; R.T.M. drafted the article; R.T.M., C.X.H., M.P.H., L.-P.W., J.M.S., K.A.B., C.R.S., and T.J.L. edited the article; and all authors read and approved the final article.
Acknowledgments
We are grateful to the full team of MDTraj contributors: Patrick Riley, Teng Lin, Tim Moore, Ravi Ramanathan, Joshua Adelman, Chaya Stern, Gert Kiss, Muneeb Sultan, Yutong Zhao, Andrea Zonca, Ondrej Marsalek, Thomas Peulen, Anton Goloborodko, and Alexander Götz, as well as participants on the MDTraj discussion forum and issue tracker.
The authors acknowledge funding from the National Institutes of Health (grants No. R01-GM62868 and No. P30-CA008748) and National Science Foundation (grant No. MCB-0954714).
Editor: David Sept.
References
- 1.Klepeis J.L., Lindorff-Larsen K., Shaw D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Lane T.J., Shukla D., Pande V.S. To milliseconds and beyond: challenges in the simulation of protein folding. Curr. Opin. Struct. Biol. 2013;23:58–65. doi: 10.1016/j.sbi.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez F., Granger B.E. IPython: a system for interactive scientific computing. Comput. Sci. Eng. 2007;9:21–29. [Google Scholar]
- 4.Pedregosa F., Varoquaux G., Duchesnay E. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 5.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 6.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 7.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 8.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 9.Hinsen K. The molecular modeling toolkit: a new approach to molecular simulations. J. Comput. Chem. 2000;21:79–85. [Google Scholar]
- 10.Michaud-Agrawal N., Denning E.J., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant B.J., Rodrigues A.P.C., Caves L.S.D. Bio3D: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 12.Jeong J.C., Jo S., Im W. ST-Analyzer: a web-based user interface for simulation trajectory analysis. J. Comput. Chem. 2014;35:957–963. doi: 10.1002/jcc.23584. [DOI] [PubMed] [Google Scholar]
- 13.Romo T., Grossfield A. Engineering in Medicine and Biology Society, EMBC 2009. Annual International Conference of the IEEE. Institute of Electrical and Electronics Engineers; Piscataway, NJ: 2009. LOOS: an extensible platform for the structural analysis of simulations; pp. 2332–2335. [DOI] [PubMed] [Google Scholar]
- 14.Yesylevskyy S.O. Pteros: fast and easy to use open-source C++ library for molecular analysis. J. Comput. Chem. 2012;33:1632–1636. doi: 10.1002/jcc.22989. [DOI] [PubMed] [Google Scholar]
- 15.Case D., Darden T., Kollman P. University of California; San Francisco, CA: 2015. AMBER. [Google Scholar]
- 16.Bowers K., Chow E., Shaw D. ACM/IEEE SC 2006 Conference. Institute of Electrical and Electronics Engineers; New York: 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. 43–43. [Google Scholar]
- 17.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 18.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponder J.W., Richards F.M. An efficient Newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 1987;8:1016–1024. [Google Scholar]
- 20.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117:1–19. [Google Scholar]
- 21.Eastman P., Friedrichs M.S., Pande V.S. OpenMM 4: a reusable, extensible, hardware independent library for high performance molecular simulation. J. Chem. Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey M.J., Giupponi G., Fabritiis G.D. ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 23.Anderson J.A., Lorenz C.D., Travesset A. General purpose molecular dynamics simulations fully implemented on graphics processing units. J. Comput. Phys. 2008;227:5342–5359. [Google Scholar]
- 24.Beauchamp K.A., Bowman G.R., Pande V.S. MSMBuilder2: modeling conformational dynamics at the picosecond to millisecond scale. J. Chem. Theory Comput. 2011;7:3412–3419. doi: 10.1021/ct200463m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson S.M., Snow C.D., Pande V.S. Folding@home and Genome@home: using distributed computing to tackle previously intractable problems in computational biology. In: Grant R., editor. Computational Genomics: Theory and Applications. Horizon Scientific Press; Norfolk, VA: 2004. [Google Scholar]
- 26.Senne M., Trendelkamp-Schroer B., Noé F. EMMA: a software package for Markov model building and analysis. J. Chem. Theory Comput. 2012;8:2223–2238. doi: 10.1021/ct300274u. [DOI] [PubMed] [Google Scholar]
- 27.Parton, D. L., P. B. Grinaway, …, J. D. Chodera. 2015. Ensembler: enabling high-throughput molecular simulations at the superfamily scale. bioRxiv, 018036. [DOI] [PMC free article] [PubMed]
- 28.Klein C. mBuild: a component-based molecule builder tool that relies on equivalence relations for component composition. GitHub. 2014 http://imodels.github.io/mbuild/ [Google Scholar]
- 29.Haque, I. S., K. A. Beauchamp, and V. S. Pande. 2014. A fast 3 × N matrix multiply routine for calculation of protein RMSD. bioRxiv, 008631.
- 30.Theobald D.L. Rapid calculation of RMSDs using a quaternion-based characteristic polynomial. Acta Crystallogr. A. 2005;61:478–480. doi: 10.1107/S0108767305015266. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Shrake A., Rupley J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- 33.Baker E.N., Hubbard R.E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 34.Vögeli B., Ying J., Bax A. Limits on variations in protein backbone dynamics from precise measurements of scalar couplings. J. Am. Chem. Soc. 2007;129:9377–9385. doi: 10.1021/ja070324o. [DOI] [PubMed] [Google Scholar]
- 35.Allen M.P., Tildesley D.J. Computer Simulation of Liquids. Clarendon Press; Oxford, UK: 1989. Liquid crystals; pp. 300–305. [Google Scholar]
- 36.Li H., Leung K.-S., Wong M.-H. iview: an interactive WebGL visualizer for protein-ligand complex. BMC Bioinformatics. 2014;15:56. doi: 10.1186/1471-2105-15-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson G., Aruliah D.A., Wilson P. Best practices for scientific computing. PLoS Biol. 2014;12:e1001745. doi: 10.1371/journal.pbio.1001745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gousios G., Pinzger M., van Deursen A. Proceedings of the 36th International Conference on Software Engineering, ICSE 2014. Association for Computing Machinery (ACM); New York: 2014. An exploratory study of the pull-based software development model; pp. 345–355. [Google Scholar]
- 39.Burges C. Now Publishers; Boston, MA: 2010. Dimension Reduction: A Guided Tour, Foundations and Trends in Machine Learning. [Google Scholar]