The Translesion DNA Polymerase ζ Plays a Major Role in Ig and bcl-6 Somatic Hypermutation (original) (raw)
. Author manuscript; available in PMC: 2015 Nov 4.
Summary
Ig somatic mutations would be introduced by a polymerase (pol) while repairing DNA outside main DNA replication. We show that human B cells constitutively express the translesion pol ζ, which effectively extends DNA past mismatched bases (mispair extender), and pol η, which bypasses DNA lesions in an error-free fashion. Upon B cell receptor (BCR) engagement and coculture with activated CD4+ T cells, these lymphocytes upregulated pol ζ, downregulated pol η, and mu tated the Ig and bcl-6 genes. Inhibition of the pol ζ REV3 catalytic subunit by specific phosphorothioate-modified oligonucleotides impaired Ig and bcl-6 hypermutation and UV damage-induced DNA mutagenesis, without affecting cell cycle or viability. Thus, pol ζ plays a critical role in Ig and bcl-6 hypermutation, perhaps facilitated by the downregulation of pol η.
Introduction
Somatic hypermutation underlies the affinity maturation of the antibody response (McKean et al., 1984; Ueki et al., 1990; Ikematsu et al., 1993; Chang and Casali, 1994; Ikematsu et al., 1998; Neuberger et al., 1998). It targets both the Ig and the bcl-6 loci and introduces single base substitutions, with rare deletions or insertions (Pasqualucci et al., 1998; Shen et al., 1998; Zan et al., 1999, 2000a). Somatic Ig and bcl-6 point mutations accumulate at a rate of 10−3 to 10−4 per base per cell generation and extend 1.5–2.0 kb downstream of the transcription initiation site, with preference for certain hot spots (Peters and Storb, 1996; Fukita et al., 1998; Neuberger et al., 1998; Shen et al., 1998; Storb et al., 1998a; Zan et al., 1999, 2000a). They favor transitions over transversions and display strand polarity, as inferred from the A over T bias in murine Ig gene V sequences (Smith et al., 1996; Neuberger et al., 1998; Storb et al., 1998a) and G over C bias in human Ig V(D)J and bcl-6 sequences (Chang and Casali, 1994; Zan et al., 1999, 2000a).
The mechanism that underlies somatic hypermutation remains speculative, but DNA breaks have been identified in Ig V(D)J DNA regions of hypermutating B cells, suggesting a role of these lesions in hypermutation (Sale and Neuberger, 1998; Bross et al., 2000; Papavasiliou and Schatz, 2000). Mutations would be introduced as mismatched nucleotides by a DNA polymerase while repairing a single-strand DNA gap (Bertocci et al., 1998; Diaz and Flajnik, 1998; Storb et al., 1998b; Zan et al., 2000b) or double-strand DNA breaks (DSBs) through homologous recombination, possibly in concert with another polymerase(s), with low processivity and error prone (Papavasiliou and Schatz, 2000; Poltoratsky et al., 2000). After their introduction, the mutations would be “fixed” by the cellular mismatch repair complex and passed on to the progeny B cells (Rada et al., 1998; Shannon and Weigert, 1998).
Knowledge of DNA synthesis and repair in eukaryotes has increased considerably, leading to the identification of a dozen DNA polymerases. Pol α, δ, and ε are involved mainly in DNA replication and are expressed prevalently in the S phase of the cell cycle (Burgers, 1998). Pol γ specifically replicates mitochondrial DNA (Weissbach, 1979), and pol θ is restricted to DNA interstrand crosslink repair (Johnson et al., 2000a). Because of their specialized function, these DNA polymerases are unlikely to be involved in the hypermutation process. Pol β is essential for base excision repair and is error prone (Prasad et al., 1996; Sobol et al., 1996) but is not involved in hypermutation, as immune-incompetent mice reconstituted with pol β-deficient fetal liver cells mutate their Ig genes normally upon antigenic challenge (Esposito et al., 2000b). Both pol ζ and pol η (Rad30A) are efficient translesion polymerases (i.e., they carry out lesion bypass DNA synthesis) and could be involved in hypermutation. Pol ζ is responsible for most damage-induced and spontaneous DNA mutagenesis (mutagenic DNA repair) (Morrison et al., 1989; Lawrence and Hinkle, 1996; Nelson et al., 1996; Holbeck and Strathern, 1997; Gibbs et al., 1998, 2000; Lin et al., 1999; Murakumo et al., 2000), and pol η is the defective polymerase in patients with the variant form of Xeroderma pigmentosum (XP-V) (Johnson et al., 1999b; Masutani et al., 1999b). In spite of its highly distributive nature and intrinsic lack of proofreading capacity, pol η bypasses UV- and chemical-induced lesions by inserting deoxynucleotides mostly in an error-free fashion in yeast, mouse, and human cells, as it favors the intervention of exogenous 3′ → 5′ exonucleases (Johnson et al., 1999a, 1999c, 2000c; Washington et al., 1999; Masutani et al., 1999a; Haracska et al., 2000; Yamada et al., 2000).
As we (Diaz and Flajnik, 1998; Zan et al., 2000b) and others (Papavasiliou and Schatz, 2000; Poltoratsky et al., 2000) have suggested, the modalities of Ig and bcl-6 somatic mutations seem to best reflect the functional features of pol ζ (Papavasiliou and Schatz, 2000; Poltoratsky et al., 2000). This polymerase effectively extends DNA past a lesion (mispair extender) with relative high fidelity, possibly after the insertion of one or two mismatched nucleotides by a low processivity, error-prone polymerase (mispair inserter), such as the recently identified pol ι (Rad30B) (Tissier et al., 2000), pol κ (Ohashi et al., 2000), pol λ (Aoufouchi et al., 2000; Garcia-Diaz et al., 2000), or, perhaps, pol μ (Aoufouchi et al., 2000; Dominguez et al., 2000). Disruption of the pol ζ REV3 gene in mice results in early embryonic lethality (Bemark et al., 2000; Esposito et al., 2000a). However, human fibroblasts in which REV3 gene expression is inhibited by an antisense REV3 fragment display impaired damage-induced mutagenesis but normal growth (Gibbs et al., 1998). We therefore addressed the role of pol ζ in hypermutation using human B cells in vitro. In this study, we show that pol ζ plays a critical role in hypermutation, as indicated by the significant reduction of somatic Ig and bcl-6 hypermutation in B cells in which pol ζ mRNA transcripts were inhibited by specific oligonucleotides. We also show that pol ζ is upregulated by B cell receptor (BCR) engagement, which simultaneously downregulates pol η, thereby making pol ζ mispair extender activity abundantly available to possibly mutate Ig and bcl-6 DNA.
Results
Pol ζ Is Upregulated and pol η Is Downregulated in Mutating Human B Cells
Ig V(D)J and bcl-6 hypermutation occurs in germinal centers of secondary lymphoid organs, such as tonsils and lymph nodes, during the transition from IgD+CD38− naive B cell to IgD+CD38+ early centroblast and IgD−CD38+ full-blown centroblast, with some IgD+CD38+ B cells displaying an uncommonly high frequency of somatic mutations, possibly as a result of the sequestration of this subpopulation in the germinal center environment (Pascual et al., 1994; Liu et al., 1996; Wilson et al., 2000). In human tonsils, highly mutated V(D)J and bcl-6 DNA sequences are found in CD38+ germinal center and CD38− memory but not in CD38− naive B cells (Pascual et al., 1994; Liu et al., 1996; Pasqualucci et al., 1998; Shen et al., 1998; Wilson et al., 2000).
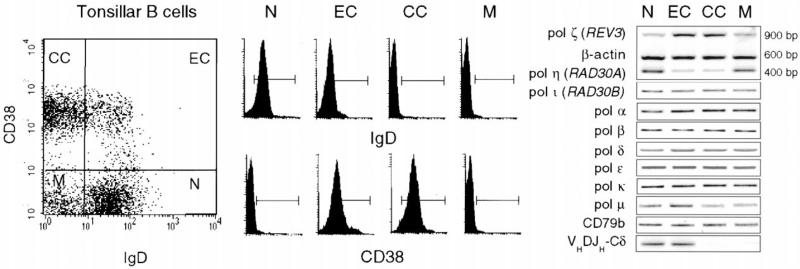
To analyze the involvement of DNA polymerases in somatic hypermutation, we sorted tonsillar B lymphocytes into IgD+CD38− (naive B cells), IgD+CD38+ (early centroblasts), IgD−CD38+ (germinal center centroblasts and cen trocytes), and IgD−CD38− (memory B cells) fractions and measured the expression of DNA pol ζ REV3 and pol η (RAD30A), as well as pol ι (RAD30B), pol α, pol β, pol δ, pol ε, pol κ, and pol μ by RT-PCR (Figure 1). The sizes of all amplified cDNAs were as predicted, and, in each case, the cDNA was cloned and found to contain the correct coding sequence (data not shown). Pol α, pol β, pol δ, pol ε, and the error-prone pol ι and pol κ were expressed at comparable levels in IgD+CD38−, IgD+CD38+, IgD−CD38+, and IgD−CD38− B cells. In contrast, the gene encoding the catalytic subunit of the translesion pol ζ REV3 was expressed at significantly higher levels in the mutating IgD+CD38+ and IgD−CD38+ B cells than in the nonmutating IgD+CD38− and IgD−CD38− B cells (Pascual et al., 1994; Liu et al., 1996; Shen et al., 1998; Wilson et al., 2000). The error-prone pol μ was equally expressed in IgD+CD38−, IgD−CD38+, and IgD−CD38− B cells but slightly upregulated in IgD+CD38+ cells. Pol η, which can perform error-free translesion DNA synthesis, was expressed in IgD+CD38− and slightly decreased in IgD−CD38− B cells but significantly downregulated in IgD+CD38+ and IgD−CD38+ B cells. The lack of VHDJH-Cδ transcripts in IgD−CD38+ and IgD−CD38− lymphocytes confirmed the (centroblastic and centrocytic) germinal center and memory phenotype, respectively, of these B cells, while the comparable amounts of amplified CD79b and β-actin cDNA indicated that comparable numbers of cells were analyzed for each fraction. The differential expression of DNA pol ζ and pol η in the tonsillar B cell subsets was emphasized by the amplification of these polymerases in the same vessel, together with β-actin (Figure 1), and was confirmed by multiple RT-PCRs using serially diluted cDNAs (see Supplemental Figure S1 at http://www.immunity.com/cgi/content/full/14/5/645/DC1) (data not shown). These ex vivo experiments show that human tonsillar IgD+CD38+ and IgD−CD38+ B cells, which undergo intensive Ig hypermutation in response to antigen binding and T cell help, display a significant alteration of the expression of the translesion DNA pol ζ and pol η.
Figure 1. Somatic Hypermutating Human Tonsillar B Cells Upregulate DNA pol ζ and Downregulate pol η.
Mutating IgD+CD38+ early centroblasts (EC) and IgD−CD38+ centroblasts/centrocytes (CC) and unmutating IgD+CD38− naive (N) and IgD−CD38− memory (M) B cells were isolated from human tonsillar B cells and analyzed for the expression of pol ζ REV3, pol η (RAD30A), pol ι (RAD30B), pol α, pol β, pol δ, pol ε, pol κ, and pol μ, using RT-PCR. Pol ζ, pol η, and β-actin were amplified in the same vessel. These findings were derived from one of three experiments yielding comparable results.
BCR Induces Upregulation of pol ζ, Downregulation of pol η, and Ig VHDJH Hypermutation
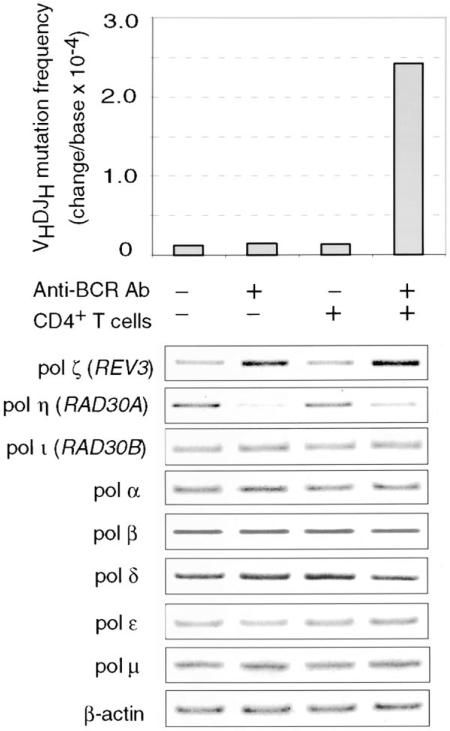
As we have shown in human IgM+IgD+ B cells, contact with activated CD4+ T cells is sufficient to induce Ig class switching and germinal center-like differentiation (Zan et al., 1998; Cerutti et al., 1998a, 1998b, 2000; Schaffer et al., 1999), but induction of somatic hypermutation requires BCR engagement (Zan et al., 1999, 2000a). We hypothesized that the pol ζ and pol η up- and downregulation found in mutating B cells was induced by the antigen-mediated crosslinking of the BCR on these germinal center lymphocytes. We used our human monoclonal CL-01 B cell line to prove that BCR engagement modulates pol ζ and pol η levels and that this DNA polymerase modulation is associated with somatic hypermutation. CL-01 B cells are IgM+IgD+ and, upon engagement of BCR and contact with activated CD4+ T cells, undergo Ig class switching and effectively mutate the Ig V(D)J and bcl-6 genes at a rate and with modalities that mimic those of B cells in vivo (Zan et al., 1999, 2000a). In these B cells, BCR engagement increased the expression of the pol ζ REV3 catalytic subunit while virtually ablating pol η expression and, together with activated CD4+ T cells, induced Ig VHDJH gene hypermutation (28 nucleotide changes in 112.5 kb analyzed) (Figure 2). No alteration in the expression of pol ζ and pol η and no hypermutation occurred in similar B cells cultured with activated CD4+ T cells without BCR engagement (two changes in 94 kb) or cultured with nil, with (two changes in 75 kb) or without (one change in 56 kb) reaction with anti-BCR Ab. Consistent with our previous findings (Zan et al., 1999, 2000a, 2000b), the B cells cultured with activated CD4+ T cells without BCR engagement underwent Ig class switching and germinal center phenotypic differentiation (data not shown). The specificity of the reciprocal modulation of DNA pol ζ and pol η was further emphasized by the relatively constant levels of pol ι, pol α, pol β, pol δ, pol ε, and pol μ in B cells reacted or not reacted with anti-BCR Ab and cultured with or without activated CD4+ T cells, as well as by further analysis involving multiple RT-PCRs using serially diluted cDNAs (data not shown). These in vitro experiments show that BCR engagement effectively modulates pol ζ and pol η expression and induces Ig hypermutation in a fashion that is consistent with the findings in ex vivo tonsillar B cells.
Figure 2. Upregulation of pol ζ and Downregulation of pol η Are Induced by BCR Engagement and Are Associated with Somatic Hypermutation.
Human IgM+IgD+ CL-01 cells were cultured in the presence or absence of activated CD4+ T cells with or without treatment with anti-BCR Ab. After 12 hr, 1, 2, 5, 7, and 9 days, 0.5 × 106 B cells from each culture were harvested. RNA was extracted and used to analyze the expression level of pol ζ REV3, pol η, pol ι, pol α, pol β, pol δ, pol ε, and pol μ by RT-PCR. Shown are the levels of the DNA polymerases after 12 hr of culture. Comparable levels were detected at day 1, 2, 5, 7, and 9. After 14 days, the cultures were terminated to identify and analyze the Ig VHDJH mutated transcripts. Histograms depict the findings derived from one of three independent experiments yielding comparable results.
BCR-Induced pol ζ and pol η Regulation Is Dose and Time Dependent
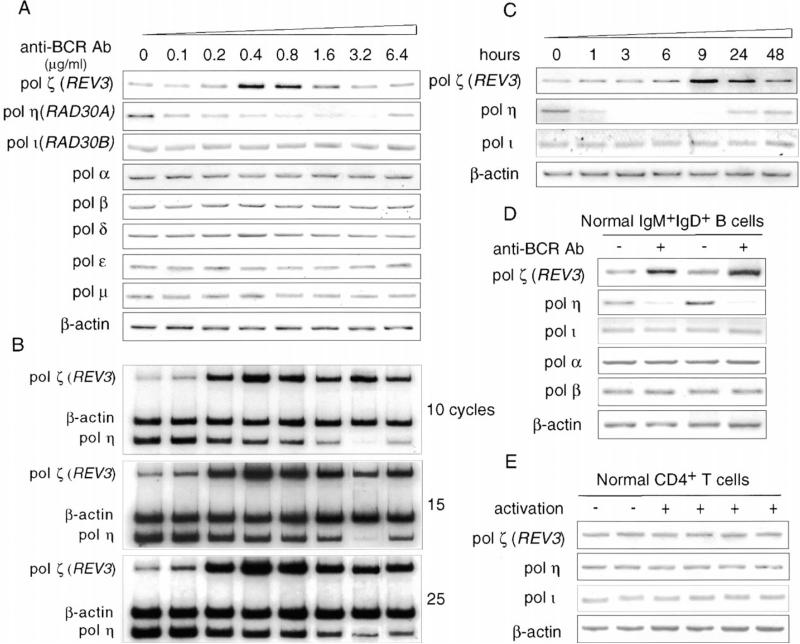
To assess the specificity of the BCR-induced modulation of DNA polymerase expression, we treated human CL-01 IgM+IgD+ B cells with increasing amounts of anti-BCR Ab and measured by RT-PCR the levels of pol ζ, pol η, pol ι, pol α, pol δ, pol ε, pol κ, and pol μ in these cells after 12 hr of culture (Figure 3). Consistent with the results of the ex vivo and in vitro experiments (Figures 1 and 2), these DNA polymerases were all constitutively expressed in the B cells (Figures 3A and 3B). Pol ζ REV3 was upregulated in a dose-dependent fashion by BCR engagement, which induced a concomitant downregulation of pol η but no significant changes in pol ι, pol α, pol δ, pol ε, pol κ, and pol μ. The reciprocal modulation of pol ζ and pol η began as early as 1 hr after BCR engagement, peaked within 9–24 hr, and lasted as long as 48 hr after a single treatment of B cells with anti-BCR Ab (Figure 3C). It was reproduced in freshly isolated normal human IgM+IgD+ B cells (Figure 3D) and confirmed by multiple RT-PCRs using serially diluted cDNAs (Supplemental Figure S1 and data not shown). It was not due to lymphocyte proliferation or differentiation, as indicated by the demonstration that CD154 or activated CD4+ T cells, which provide strong B cell proliferation and differentiation stimuli, did not influence the expression of pol ζ, pol η, pol ι, pol α, pol δ, pol ε, pol κ, or pol μ in the absence of BCR engagement (Figure 2 and data not shown). Finally, consistent with the lack of hypermutation in T cells (Bachl and Wabl, 1995), TCR complex crosslinking by an anti-CD3 Ab did not modulate the expression of pol ζ or pol η in freshly isolated normal human T cells (Figure 3E).
Figure 3. Pol ζ Is Upregulated and pol η Is Downregulated by BCR but Not TCR in a Dose- and Time-Dependent Fashion in Human Lymphocytes.
(A) Human CL-01 cells were reacted with increasing amounts of anti-BCR Ab for 1 hr at 4°C, washed, and cultured in FCS-RPMI for 12 hr. The level of pol ζ REV3, pol η, pol ι, pol α, pol β, pol δ, pol ε, and pol μ was measured by RT-PCR for each anti-BCR Ab concentration.
(B) Level of pol ζ REV3, pol η, and β-actin in the same B cells as in (A), as determined by single-vessel RT-PCR incorporating [α-32P]dCTP and consisting of 10, 15, or 25 cycles.
(C) Human CL-01 cells were reacted with anti-BCR Ab, washed, and cultured in FCS-RPMI. The level of pol ζ, pol η, and pol ι was measured at 1, 3, 6, 9, 24, and 48 hr.
(D) Freshly isolated normal human peripheral blood IgM+IgD+ B cells were reacted with anti-BCR Ab (1 μg/ml) for 1 hr at 4°C, washed, and cultured in FCS-RPMI for 12 hr before analysis of pol ζ, pol η, and pol ι transcripts.
(E) Normal human CD4+ normal T cells were cultured in FCS-RPMI in flat-bottom 96-well culture plates coated with 1:800 OKT3 anti-CD3 Ab for 12 hr and then analyzed for pol ζ, pol η, and pol ι transcripts.
Specific Inhibition of pol ζ Impairs Ig VHDJH and bcl-6 Somatic Hypermutation and Damage-Induced DNA Mutagenesis in Human B Cells
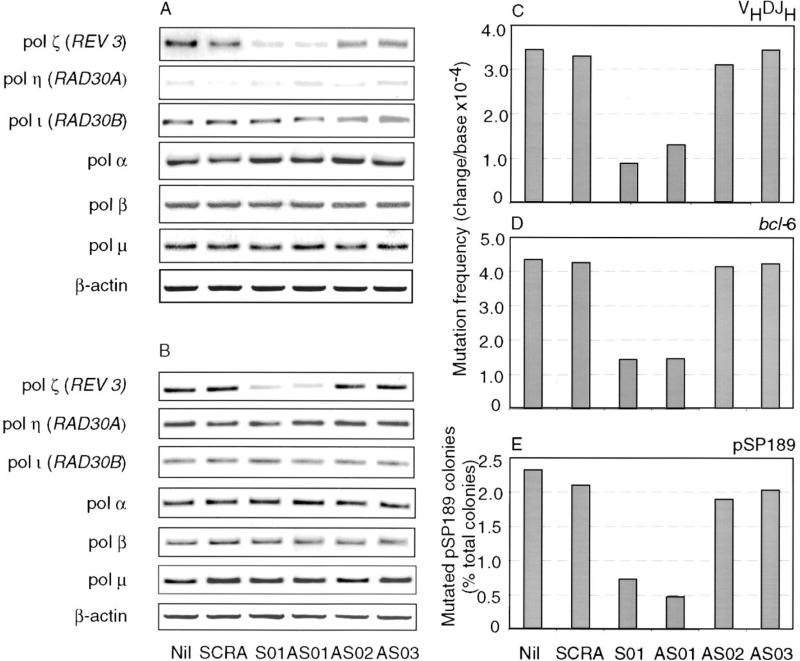
We used specific phosphorothioate oligonucleotides to inhibit the pol ζ REV3 catalytic subunit in human B cells induced by activated CD4+ T cells upon BCR engagement. The oligonucleotides were introduced into CL-01 cells, using the cationic lipid Cytofectin GSV (Lewis et al., 1996), which in our hands has yielded consistently more than 85% transfection efficiency, at a final concentration of 2 μM, starting at day –1 (a day prior to stimulation with anti-BCR Ab and activated CD4+ T cells) and every day thereafter. Pol ζ REV3 catalytic subunit, pol η, pol ι, pol α, pol β, and pol μ transcripts were determined by specific RT-PCR at days 1, 2, 5, 7, 9, and 14 of culture; mutated Ig VHDJH and bcl-6 transcripts were analyzed after 14 days of culture, as in the experiments of Figure 2. REV3 transcripts were significantly reduced by the specific sense S01 and antisense AS01 but not the scrambled (SCRA) or the antisense AS02 and AS03 oligonucleotides, as early as 36 hr after the initiation of the culture, and remained such throughout day 14 (Figure 4A and data not shown). None of the oligonucleotides inhibited pol ι, pol α, pol β, or pol μ expression. The pol ζ inhibition was confirmed by multiple RT-PCRs, using different numbers of cycles and serially diluted cDNAs (Supplemental Figure S1 and data not shown). An overall comparable pattern of DNA polymerase modulations was observed in B cells cultured in the absence of activated CD4+ T cells and without BCR engagement (Figure 4B).
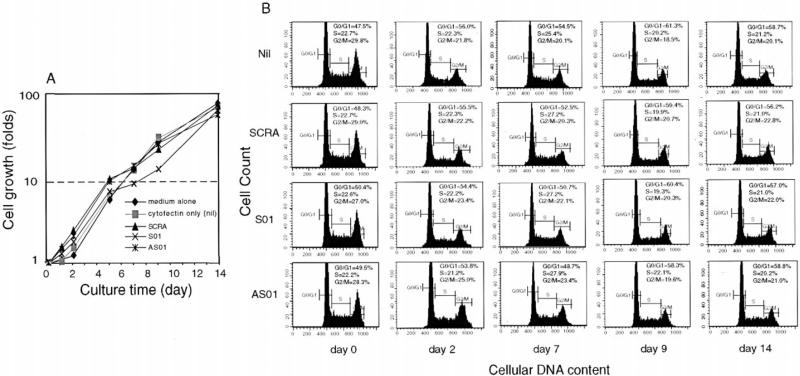
Figure 4. Specific Inhibition of pol ζ REV3 Expression Impairs Ig VHDJH and bcl-6 Somatic Hypermutation as well as Damage-Induced DNA Mutagenesis in CL-01 Cells Induced by BCR Engagement and Contact with Activated CD4+ T Cells.
(A) Human B cells cocultured with activated CD4+ T cells upon BCR engagement after daily treatment with Cytofectin GSV only (Nil) or Cytofectin GSV together with the specific pol ζ REV3 S01, AS01, AS02, AS03, or SCRA control oligonucleotide, from day –1 to day 13. B cells were harvested at days 1, 2, 5, 7, 9, and 14 by positive selection with a biotinylated anti-CD19 mAb and MiniMac System Streptavidin microbeads (Miltenyi Biotec, Inc.) for RNA extraction. Shown are the transcript levels of pol ζ REV3, pol η, pol ι, pol α, pol β, and pol μ, as measured by RT-PCR after 9 days of cultures (comparable pol ζ inhibition was observed at all other time points analyzed). These findings were derived from one of three independent experiments yielding comparable results (data are from the experiments of Table 1).
(B) Specific pol ζ REV3 oligonucleotides selectively inhibit the expression of pol ζ REV3 in CL-01 cells cultured in the absence of activated CD4+ T cells and without prior BCR engagement. All other experimental conditions and analyses are as in (A).
(C and D) Frequency of the mutations in the Ig VHDJH and bcl-6 transcripts in the B cells from the cultures of (A).
(E) CL-01 cells were treated with Cytofectin GSV only or Cytofectin GSV together with the pol ζ REV3 S01, AS01, SCRA, AS02, or AS03 oligonucleotides and then analyzed for the damage-induced mutagenesis using the UV-treated pSP189 shuttle vector. Histograms depict the frequency of 6-thioguanine-resistant mutants (white colonies out of about 4000 total colonies for each treatment).
The reduced pol ζ expression resulted in impaired Ig VHDJH and bcl-6 hypermutation. As we have shown (Zan et al., 2000a), bcl-6 mutates in response to the same stimuli and with the same modalities as Ig V(D)J genes. As determined by single-strand DNA conformation polymorphism (SSCP) analysis, CL-01 cells cocultured, upon BCR engagement, with activated CD4+ T cells for 14 days with daily treatment of the scrambled (SCRA) oligonucleotide, and Cytofectin GSV mutated as many as 12.31% and 9.62% of the 375 bp Ig VHDJH, and the 226 bp bcl-6 Exon I cDNA segments (Table 1), yielding a frequency of mutations of 3.28 × 10−4 and 4.25 × 10−4 change/base, respectively (Figure 4C). This frequency was comparable to that of similarly induced CL-01 cells treated with Cytofectin GSV alone (nil), AS02, or AS03. In contrast, CL-01 cells stimulated with anti-BCR Ab and activated CD4+ T cells but treated daily with the pol ζ _REV3_-specific S01 or AS01 oligonucleotide and Cytofectin GSV mutated as few as 3.34% and 3.14% of the expressed Ig VHDJH and bcl-6 transcripts, respectively, which corresponded to about 73% reduction (p<0.001) in mutation frequency, as compared to CL-01 cells treated with SCRA, AS02, or AS03 or Cytofectin GSV alone. This decrease in Ig and bcl-6 mutation frequency was not due to changes in cell cycle, viability, or proliferation, as these parameters were not significantly affected by Cytofectin GSV alone and/or the different phosphorothioate-modified oligonucleotides throughout the culture period (Figures 5A and 5B) but was associated with an impairment of the B cells to copy the UV-damaged shuttle pSP189 vector DNA (Figure 4D), a mutagenic function that reflects pol ζ Rev3 activity (Morrison et al., 1989; Lawrence and Hinkle, 1996; Nelson et al., 1996; Holbeck and Strathern, 1997; Gibbs et al., 1998, 2000; Lin et al., 1999; Murakumo et al., 2000), and increased cell death by UV light (data not shown).
Table 1.
Inhibition of Polymerase ζ (REV3) Expression Reduces Ig VHDJH and bcl-6 Somatic Hypermutation
| NIL | SCRA | S01 | AS01 | AS02 | AS03 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcripts | VHDJH | bcl-6 | VHDJH | bcl-6 | VHDJH | bcl-6 | VHDJH | bcl-6 | VHDJH | bcl-6 | VHDJH | bcl-6 |
| Experiment 1 | ||||||||||||
| Analyzed | 128 | ND | 109 | ND | 108 | ND | ND | ND | ND | ND | ND | ND |
| Mutated | 14 | ND | 13 | ND | 3 | ND | ND | ND | ND | ND | ND | ND |
| Experiment 2 | ||||||||||||
| Analyzed | 202 | 112 | 101 | 120 | 131 | 112 | 124 | 112 | 103 | 102 | 100 | 102 |
| Mutated | 31 | 12 | 15 | 12 | 6 | 4 | 8 | 4 | 10 | 9 | 10 | 9 |
| Experiment 3 | ||||||||||||
| Analyzed | 109 | 185 | 245 | 119 | 150 | 143 | 137 | 118 | 100 | 102 | 101 | 100 |
| Mutated | 12 | 17 | 28 | 11 | 4 | 4 | 5 | 4 | 11 | 11 | 11 | 10 |
| Total | ||||||||||||
| Analyzed | 439 | 297 | 455 | 239 | 389 | 255 | 261 | 230 | 203 | 204 | 201 | 202 |
| Mutated | 57 | 29 | 56 | 23 | 13 | 8 | 13 | 8 | 21 | 20 | 21 | 19 |
| Mutated (%) | 12.98 | 9.76 | 12.31 | 9.62 | 3.34 | 3.14 | 4.98 | 3.48 | 10.34 | 9.80 | 10.45 | 9.41 |
Figure 5. Cytofectin GSV and the pol ζ REV3 Phosphorothioate Oligonucleotides Do Not Significantly Affect B Cell Cycle or Proliferation.
(A) CD19+ cells from CL-01 cultures (same or similar to those of Figure 4) treated daily with Nil, Cytofectin GSV only, and Cytofectin GSV together with the SCRA, S01, or AS01 oligonucleotides, respectively, were harvested for counting and analysis of viability at days 0, 1, 2, 5, 7, 9, and 14 of culture. More than 90% of the B cells were viable at any time point.
(B) B cell (from [A]) cycle analysis by PI staining at days 0, 2, 7, 9, and 14 of culture.
The Residual Ig VHDJH and bcl-6 Mutations Found after Inhibition of pol ζ Do Not Differ from Those Introduced in the Presence of pol ζ
The residual Ig VHDJH and bcl-6 mutations in the CL-01 cells induced by activated CD4+ T cells upon BCR-engagement and treated with the _REV3_-specific oligonucleotides displayed features similar to those found in the CL-01 cells similarly induced but treated with the SCRA oligonucleotide or Cytofectin GSV alone. The 56 Ig VHDJH and 23 bcl-6 exon 1 mutated cDNA clones from the induced CL-01 cells treated with the SCRA oligonucleotide (Table 1) bore 58 and 27 independent point mutations, respectively (Table 2). Of the total 85 point mutations, 55 (65%) were transitions, while randomly occurring point mutations would be expected to be one-third transitions and two-thirds transversions. G nucleotides were mutated (49.4% of the total point mutations) at a frequency 68% higher than expected by chance alone (29.4%), after correcting for base composition, with G → A transitions accounting for 69.0% of the total G mutations and 52.7% of all transitions. The residual 26 Ig VHDJH and 16 bcl-6 exon 1 mutated cDNA clones from the induced CL-01 cells treated with the _REV3_-specific S01 and AS01 oligonucleotides (Table 1) bore 26 and 17 independent point mutations, respectively (Table 2). Of the total 43 point mutations, 29 (67.2%) were transitions. G nucleotides were mutated at a frequency (51.2% of the total point mutations) 74.2% higher than that expected by chance alone (29.4%), after correcting for base composition, with G → A transitions accounting and 58.6% for 77.3% of the total G mutations of all transitions.
Table 2.
Nature of the Base Substitutions in the Ig VHDJH and bcl-6 Genes of Induced CL-01 Cells Transfected by DNA Polymerase ζ (REV3)-Scrambled Oligonucleotides
| B Cells Transfected by DNA Polymerase ζ (REV3)-Scrambled Oligonucleotide | ||||
|---|---|---|---|---|
| Transitions | G → C | A → G | C → T | T → C |
| 55 | 29a | 5 | 17 | 4 |
| [28.3] | [8.3]b | [6.8] | [6.8] | [6.2] |
| Transversions | G → C | A → T | C → A | T → A |
| 11 | 5 | 1 | 4 | 1 |
| [28.3] | [8.2] | [6.8] | 6.8] | [6.2] |
| G → T | A → T | C → G | T → G | |
| 19 | 8 | 5 | 0 | 6 |
| [28.3] | [8.3] | [6.8] | [6.8] | [6.2] |
| Total Mutations | G → N | A → N | C → N | T → N |
| 85 | 42d | 11d | 21 | 11c |
| [85] | [25.0] | [20.5] | [20.5] | [18.7] |
| B Cells Transfected by DNA Polymerase ζ (REV3) → Specific Oligonucleotide | ||||
| Transitions | G → A | A → G | C → T | T → C |
| 29 | 17 | 2 | 7 | 3 |
| [14.3] | [4.2] | [3.5] | [3.5] | [3.2] |
| Transversions | G → C | A → C | C → A | T → A |
| 7 | 2 | 2 | 2 | 1 |
| [14.3] | [4.2] | [3.5] | [3.5] | [3.2] |
| G → T | A → T | C → G | T → G | |
| 7 | 3 | 0 | 1 | 3 |
| [14.3] | [4.2] | [3.5] | [3.5] | [3.2] |
| Total Mutations | G → N | A → N | C → N | T → N |
| 43 | 22d | 4d | 10 | 7 |
| [43] | [12.6] | [10.5] | [10.4] | [9.5] |
Discussion
As we showed, the induction of Ig and bcl-6 hypermutation (Zan et al., 1999, 2000a) but not that of class switch DNA recombination (Zan et al., 1998; Cerutti et al., 1998a, 1998b, 2000; Schaffer et al., 1999) requires, in addition to CD4+ T cell help, BCR engagement, thereby indicating that the function of the BCR goes beyond providing the structural correlate for the selection of higher affinity mutants. We show here that, in B cells, the expression of the translesion pol ζ is effectively modulated by BCR engagement and that the inhibition of this polymerase by specific oligonucleotides impairs Ig VHDJH and bcl-6 hypermutation and, to a comparable extent, DNA damage-induced mutagenesis without affecting cell cycle or growth. Thus, in human B cells, pol ζ is regulated by the BCR and plays a critical role in somatic hypermutation, perhaps facilitated by pol η downregulation, which is also BCR dependent. The modulation of the expression of these polymerases may be regulated not only at the transcriptional but also the posttranscriptional level.
B cells initiate somatic hypermutation upon exposure to antigen and activated CD4+ T cells in germinal centers of secondary lymphoid organs. The demonstration that BCR engagement signals in a dose-dependent fashion the upregulation of pol ζ with concomitant downregula tion of pol η, while inducing Ig and bcl-6 hypermutation, is highly consistent with the findings in tonsillar B cells: pol ζ was upregulated and pol η downregulated in IgD+CD38+ and IgD−CD38+ B cells, which include the majority of the highly hypermutating B lymphocytes (Pascual et al., 1994; Liu et al., 1996; Wilson et al., 2000), but not in IgD+CD38− naive B cells, which express unmutated Ig V(D)J and bcl-6 genes, and in IgD−CD38− memory B cells, which express mutated Ig V(D)J and bcl-6 genes, but in which the hypermutation process is extinct (Figure 1). The demonstration that BCR engagement increases pol ζ and decreases pol η but does not lead to somatic hypermutation unless activated T cells are also present (Figure 2) suggests that accessibility of the hypermutation machinery to the Ig and bcl-6 loci is important and likely requires multiple signals.
Our experiments show that pol ζ, which is essential for DNA damage-induced mutagenesis (Morrison et al., 1989; Lawrence and Hinkle, 1996; Nelson et al., 1996; Holbeck and Strathern, 1997; Gibbs et al., 1998, 2000; Lin et al., 1999; Murakumo et al., 2000), is also critical for the hypermutation of both the Ig and bcl-6 loci in human B cells. In spite of the lack of 3′ → 5′ exonuclease activity, pol ζ copies with high fidelity undamaged DNA template (Johnson et al., 2000b). It is very inefficient in inserting deoxynucleotides opposite DNA lesions but readily extends from such nucleotides once they have been inserted. Indeed, it extends damaged DNA efficiently, in concert with the error-prone, low processivity pol ι, after the incorporation of one or two mismatched deoxynucleotides opposite DNA lesions by this polymerase (Johnson et al., 2000b). Pol ι and pol ζ act sequentially: pol ι as a “mispair inserter” and pol ζ as “mispair extender.” The central role of pol ζ in damage-induced mutagenesis would be reflected in the central role of this polymerase in hypermutation. In Saccharomyces cerevisiae, pol ζ has been shown to be involved in the generation of most mutations associated with transcription (Datta and Jinks-Robertson, 1995) and, by engaging sites near DNA breaks, to increase the frequency of base substitutions in the vicinity of breaks by at least 100-fold (Strathern et al., 1995; Holbeck and Strathern, 1997). Both transcription and DNA breaks would be involved in the B cell somatic hypermutation process.
The meaning of the BCR-induced downregulation of pol η in human hypermutating B cells in vivo (Figure 1) and in vitro (Figures 2 and 4) is unclear. Pol η lacks proofreading 3′ → 5′ exonuclease activity, but its low processivity allows for extrinsic exonucleases to proofread the mismatched nucleotides it inserts (Bebenek et al., 2001). This, together with the inability of pol η to elongate a DNA chain after incorporating the incorrect nucleotide opposite a lesion, allows this polymerase to perform error-free translesion synthesis of damaged DNA (Johnson et al., 1999a, 1999a, 1999b, 2000a; Washington et al., 1999; Masutani et al., 2000; Matsuda et al., 2000). Our demonstration that BCR engagement upregulates pol ζ and profoundly downregulates pol η supports the speculation that the lack or scarcity of pol η facilitates the insertion of mismatched nucleotides (mutations) at the site of DNA breaks by another (error-prone) polymerase, such as pol ι or pol μ, which would then lead to the extension of the mutated DNA by the mispair extender pol ζ. Both pol ι and pol μ have been suggested to be preferentially expressed in secondary lymphoid tissues (Aoufouchi et al., 2000; Dominguez et al., 2000; Poltoratsky et al., 2000). Neither was downregulated by BCR engagement in vivo (Figure 1) or in vitro (Figures 2–4).
The involvement of pol ζ in the somatic hypermutation process was demonstrated using specific sense and antisense phosphorothioate-modified oligonucleotides, which effectively inhibited the expression of the REV3 gene without altering the expression of pol ι, pol α, pol β, pol μ, or β-actin (Figures 4A and 4B). The ability of S01 and AS01 but not AS02 and AS03 to actually inhibit pol ζ expression was highly consistent with the degree of effectiveness predicted for these oligonucleotides using the HYBsimulator (RNAture, Inc.). Antisense oligonucleotides would disrupt gene expression by providing a substrate for RNase H upon hybridization with mRNA (Gewirtz et al., 1998); sense oligonucleotides would impair gene transcription by hybridizing with double-strand DNA, within the major helix groove, to generate a triple-stranded molecule, thereby preventing duplex unwinding (Chan and Glazer, 1997; Gewirtz et al., 1998; Praseuth et al., 1999; Catapano et al., 2000; Faria et al., 2000).
The impairment of UV-induced DNA mutagenesis, as assessed using the pSP189 shuttle vector (Figure 4E), in the human B cells treated with the pol ζ REV3 S01 and AS01 but not the SCRA, AS02, or the AS03 phosphorothioate-modified oligonucleodites strengthened our findings on Ig and bcl-6 hypermutation. The normal cell cycle, viability, and proliferation of the human B cells in which pol ζ REV3 transcripts were inhibited by the specific oligonucleotides was evocative of the findings in human MSU-1.2-10A-42 fibroblasts, in which the expression of a high level of antisense REV3 mRNA significantly affected UV-induced mutagenesis but not cell division (Gibbs et al., 1998). The residual damage-induced mutagenic activity measured in the B cells treated with S01 and AS01 was consistent with the residual mutations detected in the Ig VHDJH and bcl-6 genes of the same cells (Figures 4C and 4D).
Recently, a potential RNA editing enzyme activation-induced (cytidine) deaminase (AID) has been found to play a crucial role in class switching and Ig somatic hypermutation (Muramatsu et al., 2000; Revy et al., 2000). In AID-deficient mice and patients with defective AID function, Ig somatic hypermutation appears to be defective (Muramatsu et al., 2000; Revy et al., 2000), but the role of AID in the somatic hypermutation process in unclear and possibly indirect. The most likely targets for AID could be any of the DNA enzymes that would be involved in somatic hypermutation, including pol ζ and perhaps an endonuclease that creates a single-strand nick or a double-strand cut. Thus, pol ζ and AID would represent different elements with different functions in the somatic hypermutation process, which would also include, as suggested by our experiments, a role for the modulation of pol η. Whether pol η overexpression affects Ig somatic hypermutation and whether AID expression is, like pol ζ and pol η, under the control of signals emanating from the BCR is currently being investigated in our laboratory.
Experimental Procedures
Lymphocytes
The phenotypic and functional features of the human monoclonal IgM+IgD+ CL-01 B cell line were reported (Zan et al., 1998, 1999, 2000a; Cerutti et al., 1998a, 1998b, 2000; Schaffer et al., 1999). To prepare sIgD+ B cells, human PBMCs were depleted of T cells (Cerutti et al., 1998b), reacted with a mouse mAb to human IgD (Southern Biotechnology Associates, Inc.), and then with magnetic Microbeads conjugated with goat anti-mouse IgG (Miltenyi Biotec, Inc.). The Microbead-treated sIgD+ B cells were separated using a MACS magnetic sorter (Miltenyi Biotec, Inc.). Tonsillar IgD+CD38−, IgD+CD38+, IgD−CD38+, and IgD−CD38− B cells were selected upon incubation of purified IgD+ B cells with MultiSort Release Reagent (Miltenyi Biotec, Inc.), FITC-conjugated mouse mAb to CD38 (PharMingen), and anti-FITC MicroBeads (Miltenyi Biotec, Inc.), as reported (Cerutti et al., 2000). The homogeneity of the separated cell fractions was assessed using a FACScalibur (Becton Dickinson). The human activated CD4+ T cells used for the hypermutation induction experiments were prepared as reported (Zan et al., 1999, 2000a).
Human DNA pol ζ, pol η, pol ι, pol α, pol β, pol δ, pol ε, pol κ, and pol μ Transcripts
The level of pol ζ, pol η (RAD30A), pol ι (RAD30B), pol α, pol β, pol δ, pol ε, pol κ, and pol μ was analyzed by RT-PCR. This was made semiquantitative by performing dilution analysis so that there was a linear relationship between the amount of cDNA used and the intensity of the PCR product. Each PCR consisted of 25 cycles, each of 1 min denaturation at 94°C, 1 min annealing at 60°C, and 1 min extension at 72°C. The amplifications were completed by an additional 10 min extension at 72°C. RT-PCR using [α-32P]dCTP consisted of 10, 15, and 25 cycles. RNA was extracted from 2 × 106 B cells, using the RNeasy Mini Kit (Qiagen, Inc.). The first-strand cDNAs were synthesized from equal amounts of total RNAs (4 μg), using the SuperScript Preamplification System (Life Technologies, Inc.). The different cDNAs were amplified using the same amount of first-strand cDNA template together with the following specific primers: pol ζ REV3, 5′-GAGATTCAGATGCATTCCTGG-3′ (forward), 5′-GTCTTGCTTGTAAGCCTTCAT-3′ (reverse), or 5′-GTTGCTTTGAT CGACACATCCC-3′ (for2), 5′-GAGGTTTTCAGTGCTCCTTCCAC-3′ (rev), 5′-ATGTGGACATCCTGCTCATCCG-3′ (rev2); pol η, 5′-CGAAATGATAATGACAGGGTAGCC-3′ (for), 5′-GGAGCAGTAAGAGATGAAAGCGAAG-3′ (rev); pol ι, 5′-TTGCTTTAGGGCTCAAGGGTG-3′ (for), 5′-TTCACATAAGTAAGGCTGGCGG-3′ (rev); pol α, 5′-AAGCACATCTTGACCCAGAGGAGC-3′ (for), 5′-CAGAGAACAGCAGAGGACAGA AGC-3′ (rev); pol β, 5′-AAGATTCGGCAGGATGATACGAG-3′ (for), 5′-AACACCCATGAACTTTGTCTCACC-3′ (rev); pol δ, 5′-GCCAGCAGGTCAAAGTCGTATC-3′ (for), 5′-GCACTGAGGTCTTCACAAACTCG-3′ (rev); pol ε, 5′-GATGAGGAGGAAAGAGATGGGG-3′ (for), 5′-GGGTTATTGAGCAGCAAGTGGG-3′ (rev); pol κ, 5′-TTGCTTTAGGGCTCAAGGGTG-3′ (for), 5′-TTCACATAAGTAAGGCTGGCGG-3′ (rev); pol μ, 5′-GCCTGTTCTCAGCACAATGTCTC-3′ (for), 5′-GTCTGGGAAATCCTCGCCTAAC-3′ (rev). In selected experiments, RT-PCR for pol ζ REV3, pol η, and β-actin were performed in the same vessel.
Induction of Somatic Hypermutation in Human B Cells
To induce Ig hypermutation, human IgM+IgD+ CL-01 B cells (0.5 × 106) were reacted for 1 hr at 4°C with IMMUNOBEAD-conjugated rabbit Ab to human Ig m chain and rabbit Ab to human Ig (H + L chain) (Irvine Scientific) (mixed 1:1 at 1 μg/ml) (anti-BCR Ab), and then washed with cold PBS before being placed in culture with activated CD4+ T cells. The activated CD4+ T cells were used at least 2 weeks from the last stimulation and were incubated for 6 hr with 20 ng/ml of 13-phorbol 12-myristate acetate (PMA) (Sigma Chemical Co.) and 500 ng/ml of ionomycin (Calbiochem-Novabiochem Co.) prior to coculture with B cells (Zan et al., 1999, 2000a). B cells were cocultured in the presence of 2.5 × 106 irradiated and activated CD4+ T cells or nil in a flat-bottom 6-well (5.0 ml FCS-RPMI volume) plate. After 7 days of culture during which fresh anti-BCR Ab was added every other day at a final concentration of 0.1 μg/ml, the lymphocytes were washed in fresh FCS-RPMI and then supplemented with freshly activated CD4+ T cells. After additional 7 days of similar treatment, the cultures were terminated. At 12 hr, 1 day, 2, 5, 7, 9, and 14 days from the initiation of the culture, the B cells were purified by positive selection with a biotinylated anti-CD19 mAb and MiniMac System Streptavidin microbeads (Miltenyi Biotec, Inc.) and used as a source of mRNA for the analysis of DNA polymerases and Ig V(D)J and bcl-6 mutations.
Analysis of Mutated Ig VHDJH and bcl-6 Transcripts
The VHDJH and bcl-6 transcripts were amplified by RT-PCR using the Pfu Turbo DNA polymerase (Stratagene) (Zan et al., 1999, 2000a). cDNAs were purified using the QIAquick PCR purification kit (Qiagen) and cloned into pCR-Blunt II TOPO (Zero Blunt TOPO PCR Cloning kit, Invitrogen). Mutated VHDJH transcripts were identified by SSCP analysis and then sequenced (Zan et al., 1999, 2000a). Identical somatic point mutations were counted only once, on the assumption that they were due to the same mutational events (shared mutations), although some of these mutations might have arisen independently. The expected frequency of mutations from one base to the other three was calculated by assuming randomness. The comparisons of the observed with the expected number of mutations were performed using a contingency table (χ2 test).
Inhibition of pol ζ Expression, Ig, and bcl-6 Somatic Hypermutation and UV Damage-Induced Mutagenesis by Specific pol ζ REV3 Oligonucleotides
The specific pol ζ REV3 phosphorothioate-modified oligodeoxynucleotides were designed by RNAture, Inc., using the proprietary HYBsimulator “Antisense Strategy,” and were synthesized by GenBase, Inc. S01 (5′-GACTTATGTCGGATG-3′) was identical to and AS01 (5′-CATCCGACATAAGTC-3′) was the reverse complement of the 15 nucleotides 7301–7315 of the human pol ζ REV3 mRNA sequence (GenBank number AF157476). SCRA (5′-CTACCGACTAATGAC-3′) oligodeoxynucleotide consisted of the same bases as AS01 but in a scrambled order. The AS02 (5′-ACTCTGTGGTTCAAC-3′) and AS03 (5′-TGACGTATGGCACTC-3′) were the reverse complements of the 15 nucleotides 713–727 and 8874–8888 residues of the human pol ζ REV3 mRNA sequence.
The phosphorothioate-modified oligodeoxynucleotides were introduced into CL-01 cells using Cytofectin GSV (Glen Research Corp.) (Lewis et al., 1996). Oligodeoxynucleotides (50 μM) and Cytofectin GSV (25 μg/ml) were allowed to associate in 250 μl Opti-MEM media (Life Technologies, Inc.) for 10 min at room temperature. The oligonucleotides/Cytofectin GSV mixture was supplemented with FCS-RPMI to yield a total volume of 2.5 ml. The resulting mixture was then added to 0.5 × 106 CL-01 cells in a flat-bottom 6-well plate which was incubated for 24 hr at 37°C. After incubation, the B cells were reacted with anti-BCR Ab and then cocultured with CD4+ T cells for a cycle of 14 days, as outlined above, with the oligonucleotides/Cytofectin GSV treatment being repeated every 24 hr. B cells were purified after 1, 2, 5, 7, 9, and 14 days by positive selection with a biotinylated anti-CD19 mAb and MiniMac System Streptavidin microbeads (Miltenyi Biotec, Inc.) for the the analysis of the DNA polymerases and the Ig VHDJH and bcl-6 transcripts. At the same time points, the same and similar cultures were analyzed for cell number, viability (trypan blue exclusion), and cycle, using propidium iodide (PI) and fluorescence flow cytometry (Noguchi, 1992).
Damage-induced DNA mutagenesis was analyzed essentially as reported by Seidman (Seidman, 1996). In brief, CL-01 cells were treated with Cytofectin GSV only or with Cytofectin GSV and the pol ζ REV3 S01, AS01, SCRA, AS02, and AS03 oligonucleotides daily for 3 days and transfected with the UV-irradiated (500 J/m2) pSP189 shuttle vector on day 4. After 2 more days of culture, the plasmid DNA was recovered, digested with Dpn I, and used to transform E. Coli MBM 7070 test strain. The frequency of white (mutated) colonies was taken as measure of B cell mutagenic activity.
Supplementary Material
Figure S1
Acknowledgments
We are grateful to Dr. William K. Holloman for helpful discussions and to Dr. Michael M. Seidman for the shuttle vector pSP189 and the test E. Coli strain MBM7070. We thank Shefali Shah and Pat Dramitinos for their excellent technical assistance. This work was supported by the National Institutes of Health grants AR 40908, AG 13910, and AI 45011 and by a Research Grant from the SLE Foundation, Inc. (New York) (to P.C.). A.S. was supported by a Cancer Research Institute (New York) fellowship.
References
- Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, Cocea L, Gupta N, Weill JC, Reynaud CA. Two novel human and mouse DNA polymerases of the pol X family. Nucleic Acids Res. 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachl J, Wabl M. Hypermutation in T cells questioned. Nature. 1995;375:285–286. doi: 10.1038/375285c0. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel TA. Proofreading of DNA polymerase η-dependent replication errors. J. Biol. Chem. 2001;276:2317–2320. doi: 10.1074/jbc.C000690200. [DOI] [PubMed] [Google Scholar]
- Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- Bertocci B, Quint L, Delbos F, Garcia C, Reynaud CA, Weill JC. Probing immunoglobulin gene hypermutation with microsatellites suggests a nonreplicative short patch DNA synthesis process. Immunity. 1998;9:257–265. doi: 10.1016/s1074-7613(00)80608-1. [DOI] [PubMed] [Google Scholar]
- Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- Burgers PM. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- Catapano CV, McGuffie EM, Pacheco D, Carbone GM. Inhibition of gene expression and cell proliferation by triple helix-forming oligonucleotides directed to the c-myc gene. Biochemistry. 2000;39:5126–5138. doi: 10.1021/bi992185w. [DOI] [PubMed] [Google Scholar]
- Cerutti A, Zan H, Schaffer A, Bergsagel L, Harindranath N, Max EE, Casali P. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plasmacytoid phenotypic differentiation in a human monoclonal IgM+ IgD+ B cell line. J. Immunol. 1998a;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Schaffer A, Shah S, Zan H, Liou HC, Goodwin RG, Casali P. CD30 is a CD40-inducible molecule that negatively regulates CD40-mediated immunoglobulin class switching in non-antigen-selected human B cells. Immunity. 1998b;9:247–256. doi: 10.1016/s1074-7613(00)80607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Schaffer A, Shah S, Goodwin RG, Zan H, Ely S, Casali P. Engagement of CD153 (CD30 ligand) by CD30+ T cells inhibits class switch DNA recombination and antibody production in human IgD+ IgM+ B cells. J. Immunol. 2000;165:786–794. doi: 10.4049/jimmunol.165.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Glazer PM. Triplex DNA: fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol. Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Diaz M, Flajnik M. Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol. Rev. 1998;162:13–24. doi: 10.1111/j.1600-065x.1998.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez-A C, Bernad A, Blanco L. DNA polymerase mu (Pol μ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Godindagger I, Klein U, Yaspo M, Cumano A, Rajewsky K. Disruption of the Rev3-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr. Biol. 2000a;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- Esposito G, Texido G, Betz UA, Gu H, Muller W, Klein U, Rajewsky K. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc. Natl. Acad. Sci. USA. 2000b;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria M, Wood CD, Perrouault L, Nelson JS, Winter A, White MR, Helene C, Giovannangeli C. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc. Natl. Acad. Sci. USA. 2000;97:3862–3867. doi: 10.1073/pnas.97.8.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, de Lera LT, Saniger ML, Ruiz JF, Parraga M, Garcia-Ortiz MJ, Kirchhoff T, del Mazo J, et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- Gewirtz AM, Sokol DL, Ratajczak MZ. Nucleic acid therapeutics: state of the art and future prospects. Blood. 1998;92:712–736. [PubMed] [Google Scholar]
- Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl. Acad. Sci. USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nature Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu H, Harindranath N, Ueki Y, Notkins AL, Casali P. Clonal analysis of a human antibody response. II. Sequences of the VH genes of human monoclonal IgM, IgG and IgA to rabies virus reveal preferential utilization of the VH3 segments and somatic hypermutation. J. Immunol. 1993;150:1325–1337. [PMC free article] [PubMed] [Google Scholar]
- Ikematsu W, Kobarg J, Ikematsu H, Ichiyoshi Y, Casali P. Clonal analysis of a human antibody response. III. Nucleotide sequences of human monoclonal IgM, IgG and IgA to rabies virus reveal Vκ gene utilization, junctional Vκ-Jκ and Vλ-Jλ heterogeneity, and somatic hypermutation. J. Immunol. 1998;161:2895–2905. [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc. Natl. Acad. Sci. USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of Xeroderma pigmentosum. Science. 1999a;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase η. Science. 1999b;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase θ. Proc. Natl. Acad. Sci. USA. 2000a;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000b;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J. Biol. Chem. 2000c;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Hinkle DC. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- Lewis JG, Lin KY, Kothavale A, Flanagan WM, Matteucci MD, DePrince RB, Mook RAJ, Hendren RW, Wagner RW. A serum-resistant cytofectin for cellular delivery of anti-sense oligodeoxynucleotides and plasmid DNA. Proc. Natl. Acad. Sci. USA. 1996;93:3176–3181. doi: 10.1073/pnas.93.8.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Hiaohua X, Wang Z. A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutation Research. 1999;433:89–99. doi: 10.1016/s0921-8777(98)00065-2. [DOI] [PubMed] [Google Scholar]
- Liu YJ, de Bouteiller O, Arpin C, Briere F, Galibert L, Ho S, Martinez-Valdez H, Banchereau J, Lebecque S. Normal human IgD+IgM– germinal center B cells can express up to 80 mutations in the variable region of their IgD transcripts. Immunity. 1996;4:603–613. doi: 10.1016/s1074-7613(00)80486-0. [DOI] [PubMed] [Google Scholar]
- Masutani C, Araki M, Yamada A, Kusumoto R, Nagimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymidine dimer by pass DNA polymerase activity. EMBO J. 1999a;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohamae N, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999b;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y, Roth T, Ishii H, Rasio D, Numata S, Croce CM, Fishel R. A human REV7 homolog that interacts with the polymerase zeta catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential rna editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Ehrenstein MR, Klix N, Jolly CJ, Yelamos J, Rada C, Milstein C. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol. Rev. 1998;162:107–116. doi: 10.1111/j.1600-065x.1998.tb01434.x. [DOI] [PubMed] [Google Scholar]
- Noguchi PD. Use of flow cytometry for DNA analysis. In: Coligan JE, Kruisbeek AM, Margulies DV, Shevach EM, editors. Current Protocols in Immunology. W. Strober. Greene Publishing Associates and Wiley-Interscience; New York: 1992. pp. 5.7.1–5.7.6. [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RSK, Klein U, Küppers R, Rajewsky K, Dalla-Favera R. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Poltoratsky V, Goodman MF, Scharff MD. Error-prone candidates vie for somatic mutation. J. Exp. Med. 2000;192:27–30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. Specific interaction of DNA polymerase and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- Praseuth D, Guieysse AL, Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochem. Biophys. Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Lagelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Sale JE, Neuberger MS. TdT-accessibe breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- Schaffer A, Cerutti A, Shah S, Zan H, Casali P. The evolutionary conserved sequence upstream of the human Ig Sγ3 region is an inducible promoter: synergistic activation by CD40 ligand and IL-4 via cooperative NF-κB and STAT-6 binding sites. J. Immunol. 1999;162:5327–5336. [PubMed] [Google Scholar]
- Seidman MM. Detection and characterization of mutations in mammalian cells with the pSP189 shuttle vector system. In: Pfeifer GP, editor. Technologies for Detection of DNA Damage and Mutations. Plenum Press; New York: 1996. pp. 373–390. [Google Scholar]
- Shannon M, Weigert M. Fixing mismatches. Science. 1998;279:1159–1160. doi: 10.1126/science.279.5354.1159. [DOI] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of the bcl-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Smith DS, Creadon G, Jena PK, Portanova JP, Kotzin BL, Wysocki LJ. Di- and trinucleotide target preferences of somatic mutagenesis in normal and autoreactive B cells. J. Immunol. 1996;156:2642–2652. [PubMed] [Google Scholar]
- Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- Storb U, Klotz EL, Hackett JJ, Kage K, Bozek G, Martin TE. A hypermutable insert in an immunoglobulin transgene contains hotspots of somatic mutation and sequences predicting highly stable structures in the RNA transcript. J. Exp. Med. 1998a;188:689–698. doi: 10.1084/jem.188.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U, Peters A, Klotz E, Kim N, Shen HM, Kage K, Rogerson B, Martin TE. Somatic hypermutation of immunoglobulin genes is linked to transcription. Cur. Top. Microb. Immunol. 1998c;229:11–20. doi: 10.1007/978-3-642-71984-4_2. [DOI] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R. pol ι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Goldfarb IS, Harindranath N, Gore M, Koprowski H, Notkins AL, Casali P. Clonal analysis of a human antibody response. Quantitation of precursors of antibody-producing cells and generation and characterization of monoclonal IgM, IgG, and IgA to rabies virus. J. Exp. Med. 1990;171:19–34. doi: 10.1084/jem.171.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase eta. J. Biol. Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- Weissbach A. The functional roles of mammalian DNA polymerase. Arch. Biochem. Biophys. 1979;198:386–396. doi: 10.1016/0003-9861(79)90511-3. [DOI] [PubMed] [Google Scholar]
- Wilson PC, Wilson K, Liu YJ, Banchereau J, Pascual V, Capra JD. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J. Exp. Med. 2000;91:1881–1894. doi: 10.1084/jem.191.11.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Masutani C, Iwai S, Hanaoka F. Complementation of defective translesion synthesis and UV light sensitivity in Xeroderma pigmentosum variant cells by human and mouse DNA polymerase eta. Nucleic Acids Res. 2000;28:2473–2480. doi: 10.1093/nar/28.13.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Schaffer A, Dramitinos P, Casali P. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β. Evidence for TGF-β but not IL-10-dependent direct Sμ→Sα and sequential Sμ→Sγ, Sγ→Sα DNA recombination. J. Immunol. 1998;161:5217–5225. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ cell line in vitro: definition of the requirements and the modalities of hypermutation. J. Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. BCR engagement and T cell contact induce bcl-6 hypermutation in human B cells: association with initiation of transcription and identity with Ig hypermutation. J. Immunol. 2000a;165:830–839. doi: 10.4049/jimmunol.165.2.830. [DOI] [PubMed] [Google Scholar]
- Zan H, Li Z, Gurrieri C, Yamaji K, Cerutti A, Flajnik MF, Diaz M, Casali P. BCR-engagement induces downregulation of the error-free DNA polymerase η in human B cells. Association with the introduction of Ig mutations by the translesion DNA polymerase ζ. FASEB J. 2000;14:A1043. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1