Capturing Snapshots of APE1 Processing DNA Damage (original) (raw)
. Author manuscript; available in PMC: 2016 May 1.
Published in final edited form as: Nat Struct Mol Biol. 2015 Oct 12;22(11):924–931. doi: 10.1038/nsmb.3105
Abstract
DNA apurinic-apyrimidinic (AP) sites are prevalent non-coding threats to genomic stability and are processed by AP endonuclease 1 (APE1). APE1 incises the AP-site phosphodiester backbone, generating a DNA repair intermediate that is potentially cytotoxic. The molecular events of the incision reaction remain elusive due in part to limited structural information. We report multiple high-resolution human APE1:DNA structures that divulge novel features of the APE1 reaction, including the metal binding site, nucleophile, and arginine clamps that mediate product release. We also report APE1:DNA structures with a T:G mismatch 5′ to the AP-site, representing a clustered lesion occurring in methylated CpG dinucleotides. These reveal that APE1 molds the T:G mismatch into a unique Watson-Crick like geometry that distorts the active site reducing incision. These snapshots provide mechanistic clarity for APE1, while affording a rational framework to manipulate biological responses to DNA damage.
INTRODUCTION
Apurinic-apyrimidinic (AP) sites are a threat to genomic stability since they result in the loss of DNA coding potential, block replication forks, promote mutagenesis, and lead to DNA double strand breaks1–3. Spontaneous depurination can generate >10,000 AP-sites per cell per day4,5. Therefore, the cell has developed potent mechanisms for dealing with AP-sites during the process known as base excision repair (BER)6,7. Classical BER is initiated by a damage-specific DNA glycosylase that removes the damaged base generating an AP-site8. The phosphodiester bond 5′ to the AP-site is then incised by an AP endonuclease (APE), the major human form being APE1. Incision of the AP-site results in a single-nucleotide DNA gap with 3′-hydroxyl and 5′-deoxyribose phosphate termini. This potentially cytotoxic BER intermediate is the substrate for downstream BER processing by DNA polymerase β9,10.
Rational study of key mechanistic features of the APE1 reaction and its biology has been hampered due to the lack of high-resolution structures of APE1 in complex with DNA repair intermediates. Previous crystal structures of APE1:DNA complexes had suggested a metal-dependent mechanism where a protein side chain activates an attacking water molecule11. Importantly, this mechanism was inferred from structures that lacked key active site groups. The lack of structural detail has generated alternative mechanistic proposals for the role and number of divalent metals and for the identity and generation of the nucleophile12–14. Consequently, there is not a clear consensus for the APE1 mechanism15–19.
RESULTS
Capturing key structural snapshots facilitates mechanistic interpretation during nucleic acid enzymology20–22. Previous DNA bound APE1 substrate and product structures were determined to 2.95 and 2.65 Å, respectively, and provided clues into the mechanism of APE111. Yet, the resolution of these structures provided limited understanding of the molecular features of AP-site recognition and processing. To resolve mechanistic uncertainties, we have determined several high-resolution structures of APE1 substrate and product complexes.
High-resolution structure of APE1 bound to product DNA
The APE1 product complex was obtained in the presence of MgCl2 with a 21-mer dsDNA containing a centrally located AP-site analog (tetrahydrofuran, THF). The crystals diffracted to 1.57 Å (Table 1) and reveals APE1 flipping the AP-site out of the double helix into the active site binding pocket and kinking the DNA by ~35°. This places the AP-site in position for incision of the 5′-phosphate backbone and results in an orphan base opposite the AP-site (Fig. 1a). Overall, the protein and DNA structure is globally consistent with the previously reported 2.4 Å product structure (RMSD = 0.29 Å over 316 Cα atoms, Supplementary Fig. 1)18.
Table 1.
Data collection and refinement statistics of APE1:DNA co-complexes
| Product Complex | 2′OMePS substrate complex | 2′OMePS substrate complex (MnCl2) | E96Q D210N Mismatch Substrate | Mismatch Product | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P1 | P1 | P1 | P1 | P1 |
| Cell dimensions | |||||
| a, b, c (Å) | 44.5,61.7,72.1 | 44.4,60.8,73.2 | 44.3,60.6,73.3 | 44.5,60.7,73.2 | 44.4,61.6,72.3 |
| α, β, γ (°) | 83.9,78.8,88 | 83.0,80.5,89.1 | 83.1,80.6,89.0 | 82.8,80.3,89.2 | 83.8,78.7,88.2 |
| Resolution (Å) | 50-1.57 | 50-1.63 | 50-1.80 | 50-1.85 | 50-1.95 |
| _R_mergea | 11.5 (57.4) | 8.4 (54.1) | 6.9 (56.1) | 13.2 (43.0) | 5.8 (38.0) |
| I/σ_I_ | 16.37 (2.1) | 15.72 (2.0) | 18 (2.0) | 12.2 (2.7) | 20.8(2.8) |
| Completeness (%) | 99.6 (94.8) | 98.7 (88.3) | 99.8 (99.2) | 100 (99.9) | 96.1 (77.6) |
| Redundancy | 4.5 (2.5) | 4.1 (2.2) | 5.0 (3.0) | 4.3 (3.1) | 2.7 (1.6) |
| Refinement | |||||
| Resolution (Å) | 1.57 | 1.63 | 1.80 | 1.85 | 1.95 |
| No. reflections | 103855 | 137947 | 111181 | 58169 | 79881 |
| _R_work/_R_free | 0.18/0.21 | 0.17/0.20 | 0.18/0.22 | 0.17/0.21 | 0.19/0.23 |
| No. atoms | |||||
| Protein | 4335 | 4331 | 4353 | 4372 | 4309 |
| DNA | 855 | 915 | 879 | 896 | 858 |
| Water | 695 | 687 | 499 | 682 | 377 |
| B factors | |||||
| Protein | 19.0 | 17.9 | 20.1 | 21.6 | 31.5 |
| DNA/THF/metalb | 33.4/19.3/20 | 30/21.1/- | 36.1/23.4/32.1 | 35.8/25.0/- | 45/30/46 |
| Waterbulk/Waternucc | 33.1/- | 30.7/13.9 | 29.7/16.6 | 33.4/24.5 | 35.3/- |
| r.m.s. deviations | |||||
| Bond lengths (Å) | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| Bond angles (°) | 1.06 | 1.15 | 1.25 | 1.13 | 1.14 |
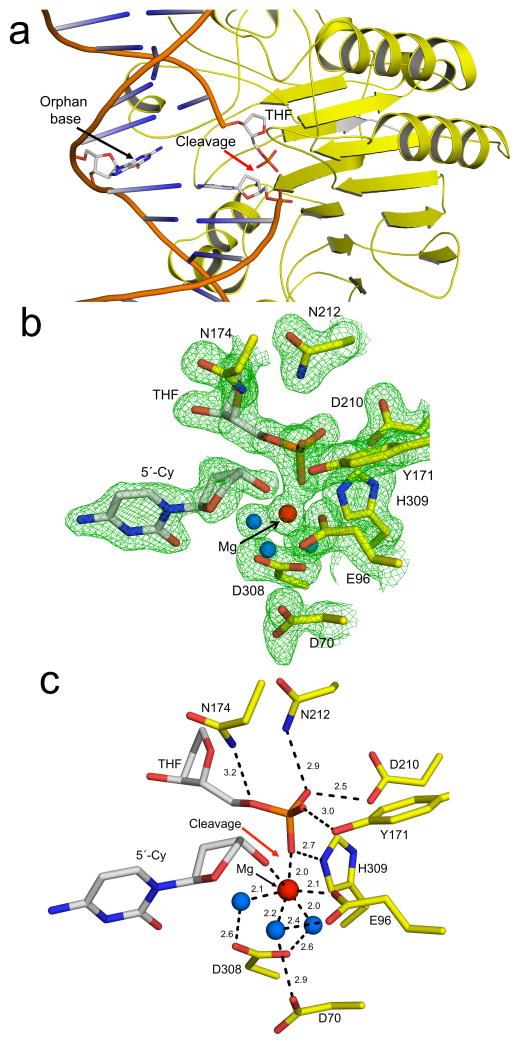
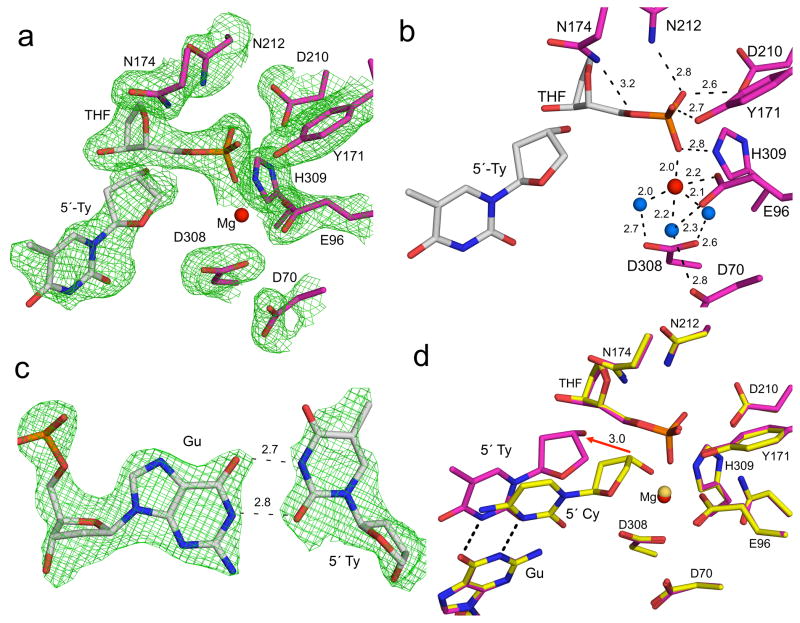
Figure 1. High-resolution APE1:DNA product complex.
(a) Overview of APE1:DNA product complex with APE1 shown in yellow and the 21-mer DNA shown in cartoon representations. The site of cleavage is indicated with a red arrow. The THF, 5′-Cy, and orphan base are shown in stick format (grey carbons). A focused view of the active site with (b) and without (c) density is shown with key residues and distances (Å) indicated. The protein side chains are shown in yellow and DNA residues in grey. Water molecules and Mg2+ are shown as blue and red spheres respectively. The omit map (green) is contoured at 3σ.
A detailed view of the active site shows a single Mg2+ (Fig. 1b, c). The Mg2+ coordinates E96 and three water molecules that are also coordinated to D70 and D308. Mutation of these aspartate residues has been shown to reduce activity23,24. Importantly, the Mg2+ in this product complex directly coordinates the oxygen of the 5′-phosphate and 3′-hydroxyl at the site of backbone cleavage; ligands that are generated following cleavage. The non-bridging oxygens of the 5′-phosphate interact with N212, D210, Y171, and H309 (Fig. 1c). The bridging oxygen of the cleaved AP-site is within hydrogen bonding distance to N174. Together these residues stabilize the cleaved product complex.
Arginine clamps swing to facilitate product binding
The pre-steady state kinetic description of APE1 is that rapid catalysis is followed by slow product release25. This feature may conceal cytotoxic incised DNA BER intermediates during DNA damage processing. In our product structure, we identified a unique contact with R181 not appreciated previously. Previously, R181 was observed to come within hydrogen bonding distance to E154 in the DNA bound and apoenzyme APE1 structures11,26. In our product complex, R181 has shifted 5.5 Å to come within hydrogen bonding distance of a backbone phosphate upstream (5′) of the AP-site (Fig. 2a). In this conformer, side-chain nitrogens of R181 are 2.7 and 2.8 Å away from a DNA backbone non-bridging oxygen. These contacts are only present after product formation as shown by an overlay with the apoenzyme and substrate complexes (Fig. 2a). This implies R181 facilitates product binding by shifting to clamp down on product DNA after catalysis.
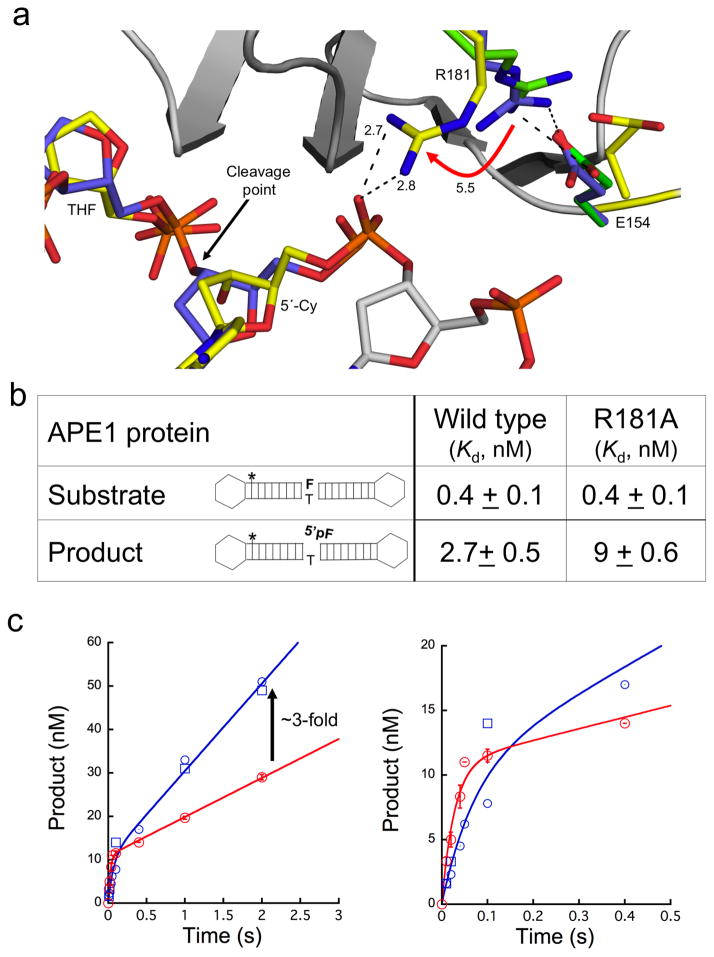
Figure 2. R181 swings to facilitate product stability.
(a) A focused view of R181 after overlaying our APE1 product complex (yellow) with the previous substrate (purple, PDB ID 1DE8) and apoenzyme (green, PDB ID 4QHE) structures. The cleavage point is indicated with a black arrow and the R181 shift (Å) is shown with a red arrow. Key distances and residues are indicated. (b) The binding affinities (nM) with the standard error for the substrate and product shown for the full length wild-type and R181A APE1. The hairpin for the product and substrate is shown for reference; the (*) indicates the location of the fluorescein label, ‘F’ is the THF moiety, and (p) is the 5′-phosphate group. (c) The wild-type (WT, red) APE1 or R181A mutant (blue) time-courses of product formation are shown with an initial rapid exponential phase followed by a linear phase. The time points for the wild-type enzyme are the mean and standard error of three independent experiments. The individual time points for two independent determinations for the mutant enzyme are shown (open blue squares and circles). The early time points are shown in the right panel. The line represents the best fit to an equation with rising exponential and linear terms. The arrow indicates the estimated increase in product dissociation for the mutant enzyme. See also Supplemental Table 1 for kinetic values.
To probe the role of R181, we performed protein-DNA binding studies using full-length wild-type or R181A mutant proteins and substrate or product DNA. The DNA was constructed to prevent end binding by utilizing hairpin oligonucleotides (Fig. 2b). The substrate DNA binding experiments where performed in the presence of 10 mM EDTA, to prevent catalysis, using fluorescence anisotropy. Figure 2b summarizes results with both wild-type and R181A APE1. These enzymes exhibited a low-affinity non-specific binding component and a high-affinity binding component. For the high-affinity component, both proteins bound to the substrate DNA with similar sub-nanomolar affinity (_K_d = 0.4 nM). In contrast, binding analysis with product DNA indicated the R181A mutant enzyme bound with ~3-fold weaker affinity than the wild-type enzyme (Fig. 2b).
To further probe the role of R181, we conducted kinetic measurements of AP-site incision using the same DNA substrate as in the binding studies. In these experiments the reaction mixture contained an excess of substrate DNA. The biphasic time courses of product formation demonstrated that catalysis during the first enzymatic turnover was more rapid than the subsequent steady-state phase for both R181A and wild-type enzymes indicating that a step after chemistry limited enzyme cycling (i.e., product release). For wild-type enzyme, the observed rate constant of the burst phase was 36 s−1 followed by an apparent linear rate (_v_ss) of 9 nM/s. With this analysis, the burst amplitude represents the apparent active enzyme concentration (11 nM) so that the steady-state rate (_k_ss = _v_ss/APE1active) was 0.8 s−1. For the mutant enzyme, the observed rate constant of the burst phase was 14.2 s−1 followed by an apparent linear rate (_v_ss) of 20 nM/s corresponding to a steady-state rate of 1.9 s−1. Since the observed burst and steady-state rates for the mutant enzyme are not well separated (i.e., _k_burst/_k_ss) < 10), they are used to calculate _k_incision and _k_off,product for the mutant enzyme; 11.9 and 2.3 s−1, respectively (Online Methods). This represents a 3-fold decrease in the incision rate and a 3-fold increase in the product dissociation rate constant for the mutant enzyme (Fig. 2c and Supplementary Table 1). Together, these binding and kinetic results indicate that R181 facilitates product DNA binding.
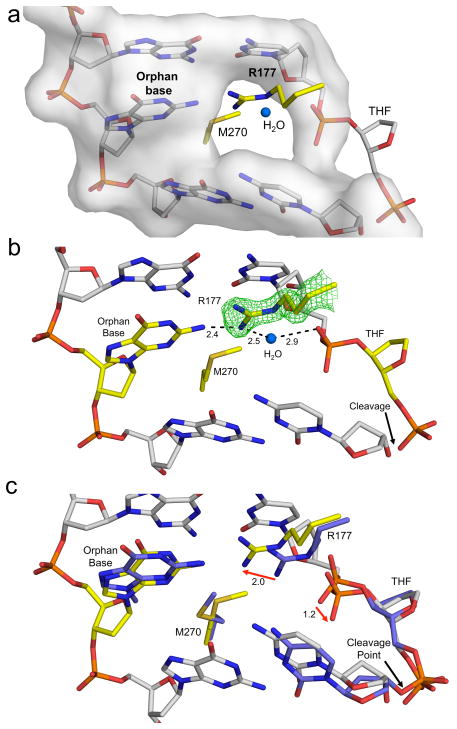
An essential aspect of the reaction mechanism is the flipping of the AP-site into the active site, leaving an orphan base in the opposing strand (Fig. 1a). R177 acts as a surrogate base by intercalating into the major groove to form a base stacking interaction (Fig. 3a). Previously reported structures show R177 contacting only the backbone non-bridging oxygen of the AP-site in both the product and substrate complexes11. Those putative interactions do not adequately account for the differential loss in substrate and product binding reported for the R177A mutant (7- and 500-fold decrease, respectively)27,28. Instead, product specific stabilization by R177 can be rationalized from our product structure. This structure indicates that R177 comes within hydrogen bonding distance of the orphan base and a water molecule that can form a hydrogen bond to the backbone of the cleaved AP-site (Fig. 3b). These contacts are specific to product DNA based on an overlay of the substrate and product complexes. The overlay indicates that R177 swings 2.0 Å toward the orphan base with the AP-site backbone moving 1.2 Å away from R177 following cleavage (Fig. 3c). Thus, R177 enhances product binding while only moderately impacting substrate binding.
Figure 3. R177 intercalates into the major groove anchoring the orphan base.
(a) The DNA in our product complex excluding the THF is shown in a transparent grey surface representation. R177 and M270 are shown in yellow stick format intercalating into the major and minor groove, respectively. A structural water is shown as a blue sphere within this cavity. (b) The same orientation as panel A with the orphan base and THF shown in yellow. An omit map contoured at 3σ is shown for R177 and the distances (Å) are indicated. (c) Overlay of the previous substrate (PDB ID 1DE8) and our product complex is shown with key residues in purple or yellow, respectively. The DNA in grey corresponds to our product complex. Shifts following product formation are shown with red arrows with the distances (Å) indicated.
High-resolution APE1 substrate complexes
The high enzyme concentrations and long incubations necessary for crystallographic studies render enzyme substrate complexes difficult to trap. The robust incision rate of APE1 and minor reagent Mg2+ contamination adds to this difficulty. To overcome these challenges and obtain a substrate complex structure, we utilized a modified DNA with 2′-O-methyl phosphorothioate backbone modification 5′ to the AP-site. This modification contains a sulfur substitution for a non-bridging oxygen that reduces incision16,29. The resulting substrate complex diffracted to 1.65 Å, which is substantially higher than a previously reported 3.0 Å substrate complex (Table 1). A difficulty in using the phosphorothioate substrate is the existence of two isomers, _S_p and _R_p. In the crystal structure, we observed both isomers in the active site with equal occupancy (Supplementary Fig. 2). However, the _R_p isomer is shifted away from the active site by 2.1 Å (Supplementary Fig. 2b). Only the _S_p isomer is in the proper orientation to coordinate key active site residues: N174, Y171, H309, and the nucleophilic water. Therefore, the subsequent description focuses on the structure with the _S_p isomer.
The identity of the nucleophile during APE1 mediated cleavage is uncertain. Here we have captured the requisite structural snapshots to identify key catalytic groups involved in APE1 backbone incision. Figure 4 shows a detailed view of the active site with substrate DNA containing the _S_p isomer of O-methyl phosphorothioate. The AP-site backbone oxygen (O5′) is coordinated to N174, while Y171 and H309 coordinate the non-bridging oxygen and sulfur, respectively (Fig. 4b). These contacts stabilize the active position of the AP-site and are observed in both the product and substrate complexes. The high resolution of our structure identifies a water molecule in position to act as the nucleophile (Fig. 4b). This well ordered water is coordinated by the non-bridging oxygen and sulfur atoms of the backbone phosphate positioning the water 2.8 Å from the phosphorous atom (Fig. 4c). N212 and D210 oxygen atoms come within hydrogen bonding distance of the nucleophilic water molecule. In this orientation, D210 (OD1) is both posed to activate the water for nucleophilic attack and comes within hydrogen bonding distance of N68. Additionally, D210 (OD2) is within hydrogen bonding distance of N212 (backbone nitrogen). This hydrogen-bonding network likely alters the p_K_a of D210 to facilitate attack of the nucleophilic water (Fig. 4c).
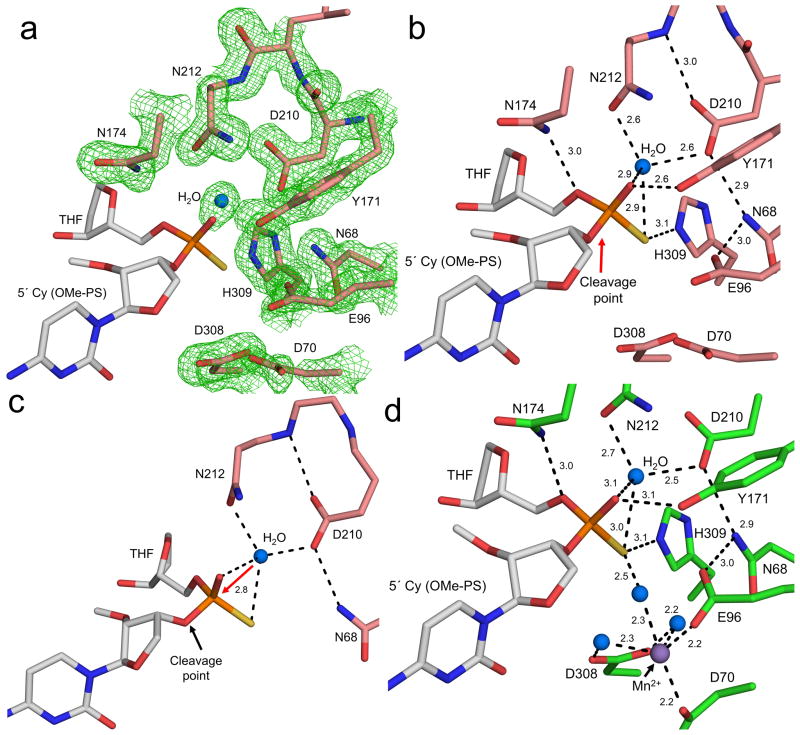
Figure 4. High resolution APE1:DNA substrate complex.
The APE1 active site with (a) and without (b) density is shown with key residues, cleavage point, and distances (Å) indicated. The protein side chains are shown in salmon and DNA in grey. The nucleophilic water is shown as a blue sphere. The omit map (green) is contoured at 3σ. (c) A focused view of the nucleophilic water with the in-line attack shown with a red arrow. The distance (Å) between the nucleophilic water and phosphate group is indicated. Key contacts promoting the activation of the water molecule are highlighted with dashes. (d) The complete APE1 active site following a soak in MnCl2 is shown with key residues and distances indicated. The protein side chains are shown in green and DNA in grey. The Mn2+ and water ions are shown in purple and blue, respectively.
The substrate complex lacks a clearly identifiable metal ion within the active site even with MgCl2 present during crystallization. This is consistent with previous structural snapshots indicating high disorder for Mg2+ within the active site26. It has previously been shown that APE1 is a metal dependent enzyme, but the identity of the metal binding site(s) and the number of metal ions required for catalysis remains controversial13,18,26. To provide insight into the role of metal ions during catalysis, we briefly soaked the O-methyl-phosphorothioate substrate crystals in a cryo-solution containing MnCl2 in an equimolar concentration to MgCl2 (Table 1). Figure 4d shows the 1.8 Å substrate complex with both the nucleophilic water and Mn2+ ion bound within the active site. The presence of Mn2+ was verified by its anomalous signal (Supplementary Fig. 3a). This Mn2+ coordinates D308, E96, D70, and three water molecules (Fig. 4d). One of these coordinating water molecules is within hydrogen bonding distance to the non-bridging sulfur. This positions Mn2+ only 2.7 Å from the location of the metal in the product structure (Supplementary Fig. 3c). Overlaying the metal bound and metal free substrate complexes indicates that E96 and D70 shift 1.2 and 1.9 Å upon metal binding, respectively (Supplementary Fig. 3b). The nucleophilic water remains in the same position in both substrate complexes and is located 7 Å from Mn2+. The position of this metal binding pocket near the site of cleavage likely facilitates catalysis and stabilizes reaction intermediates after bond breakage (see Discussion).
APE1 processing an AP-site with a 5′ mismatch
The presence of a mismatch 5′ to the AP-site lesion has been shown to dramatically reduce catalytic activity of APE129,30. A biologically relevant mismatch that APE1 may encounter during BER is a T:G mispair arising from deamination of the epigenetic marker 5-methylcytosine in CpG dinucleotide repeats where the guanine is damaged (e.g., 8-oxoguanine) and undergoing repair31–34. Previous kinetic studies had shown that a 5′ T:G mismatch reduces AP-site incision by ~5400-fold35. Currently, the molecular basis for this reduction remains unclear. To probe this catalytic defect, we characterized a catalytically dead double mutant enzyme (E96Q D210N) in combination with a natural DNA substrate containing a T:G mismatch 5′ to the AP-site lesion36. The resulting crystal diffracted to 1.8 Å with APE1 bound to substrate DNA in a similar global conformation as observed with correctly base paired DNA and wild-type APE1 (Table 1). Overlaying the wild-type substrate complex with the double mutant provides insight into how this enzyme binds DNA with similar affinity, but is unable to incise an AP-site. The Q96, N68, and N210 residues form a catalytically dead triad by undergoing moderate rotameric shifts to hydrogen bond with one another (Supplementary Fig. 4). This prevents E96Q from coordinating the metal because NE2 is pointed toward the metal binding pocket and OE1 is coordinating ND2 of N68. A similar phenomenon occurs with the N210 substitution. N210 (ND2) coordinates OD1 of N68, resulting in ND2 coordinating the nucleophilic water molecule. The nucleophilic water molecule is in the same location as observed with the phosphorothioate substrate complex, but is unable to be activated by N210 (Supplementary Fig. 4b). This combination prevents APE1 from cleaving DNA, while validating the substrate contacts observed in the phosphorothioate complex are not artifacts of the modified DNA.
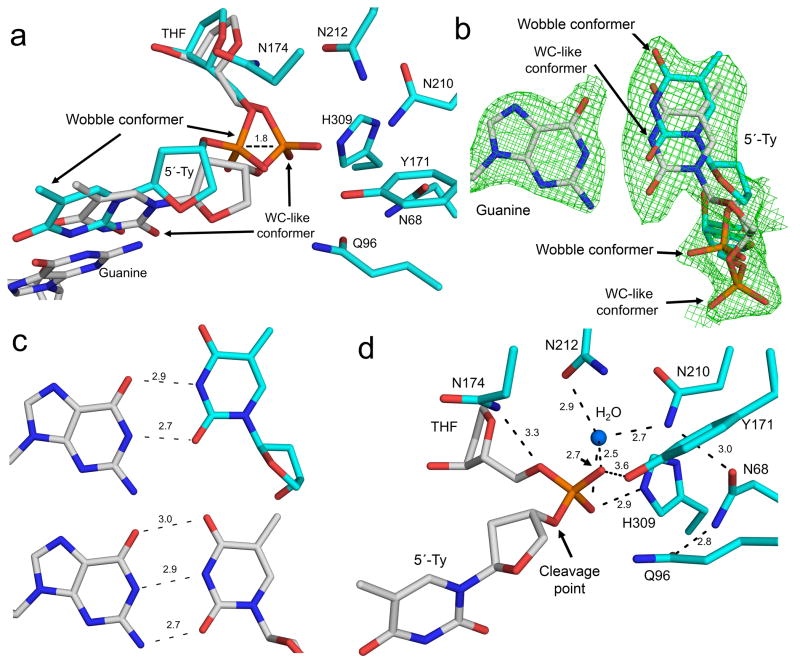
A close-up view of the active site shows the T:G mispair being accommodated in the substrate complex by forming either a wobble or Watson-Crick (WC)-like mismatch within a single structural snapshot (Fig. 5a, b, c). This conformer is likely not active because the backbone phosphate is shifted 1.8 Å out of the active site (Fig. 5a). In contrast, the WC-like mismatch is formed through either an ionic or tautomeric interaction between the bases (Fig. 5c). The WC-like mismatch represents the active conformer because the phosphate backbone remains within the active site (Fig. 5a). In this active conformer, the nucleophilic water is 2.5 Å from the phosphate and only 2.7 and 2.5 Å from its non-bridging oxygens (Fig. 5d). In addition, key active site contacts with Y171 and N174 are not within a stable hydrogen bonding distance of the phosphate backbone (Fig. 5d). The loss of these contacts and alternate conformers of the T:G mispair reduce the cleavage activity of APE1.
Figure 5. APE1 E96Q/D210N mismatch substrate complex.
(a) Close-up of the mutant APE1 active site is shown with key amino acid residues indicated. The 5′-thymidine (relative to THF) mispaired with guanine adopts a wobble conformer (cyan) and Watson-Crick (WC, grey) like mismatch conformer, as indicated. The backbone shift in the wobble conformer is indicated by dashes. (b) Focused view of the wobble and WC-like conformer is shown in cyan and grey, respectively. Omit map is contoured at 3σ for the T:G mispair. The phosphate backbone location of each conformer is indicated. (c) The top panel is the T:G wobble base pairing interaction and the bottom panel correspond to the WC-like mismatch conformer. The distances (Å) between base-pairing groups are indicted. (d) Same view as panel A with the inactive wobble base pair removed. The distances (Å) to key active site residues and the nucleophilic water are shown.
To capture the product state, we utilized wild-type APE1 to form the product complex containing a 5′-T:G mispair (Table 1). Figure 6a shows density corresponding only to the product complex following cleavage. The key contacts to the product phosphate with N174, Y171, D210, N212, and H309 are maintained in the product complex (Fig. 6b). The phosphate backbone is also coordinating a single Mg2+. E96 and three water molecules that are coordinated by D70 and D308 coordinate this Mg2+. In contrast to the substrate complex, the T:G mispair has adopted only a wobble base upon cleavage of the phosphate backbone (Fig. 6c). This wobble base pair conformation results in a shift of 3.0 Å of O3′ compared to the product complex without a mismatch (Fig. 6d) thereby removing O3′ from the active site Mg2+ coordination sphere in the product mismatch complex (Fig. 6b). The shift to a wobble base pairing indicates the strain within the active site is not relieved until backbone incision.
Figure 6. APE1 T:G mismatch product complex.
The APE1 active site following cleavage with a T:G mismatch 5′ to the THF is shown with (a) and without (b) density. Water and magnesium ions are shown as blue and red spheres with key residues, site of cleavage, and distances (Å) indicated. (c) Omit map density (3σ) for the wobble T:G base pairing is shown with distances (Å). (d) Overlay of the APE1 product complex with a 5′ matched and mismatched base pair in yellow and magenta, respectively. The base pairing interaction for the T:G wobble base pair is shown as dashes and the shift arising from the mismatch is highlighted with a red arrow with the corresponding distance (Å). The Mg2+ and key residues for each structure are indicated.
DISCUSSION
APE1 has been implicated in a number of critical biological pathways such as BER, RNA cleavage, 3′-end processing, redox regulation, cell proliferation, and nucleotide incision repair37–42. Consistent with a critical biological role, mouse model studies indicate that APE1 gene deletion is embryonic lethal43. Even subtle APE1 point mutations or alterations in expression levels have been associated with the development of cancer, aging, cardiovascular and neurological diseases44,45. Some aggressive cancers show increased expression of APE1; which is presumed to protect metabolically active cancer cells from the high level of reactive oxygen species during enhanced oxidative phosphorylation46. This protective effect has resulted in APE1 emerging as a therapeutic drug target to reduce enzymatic activity during chemotherapy41.
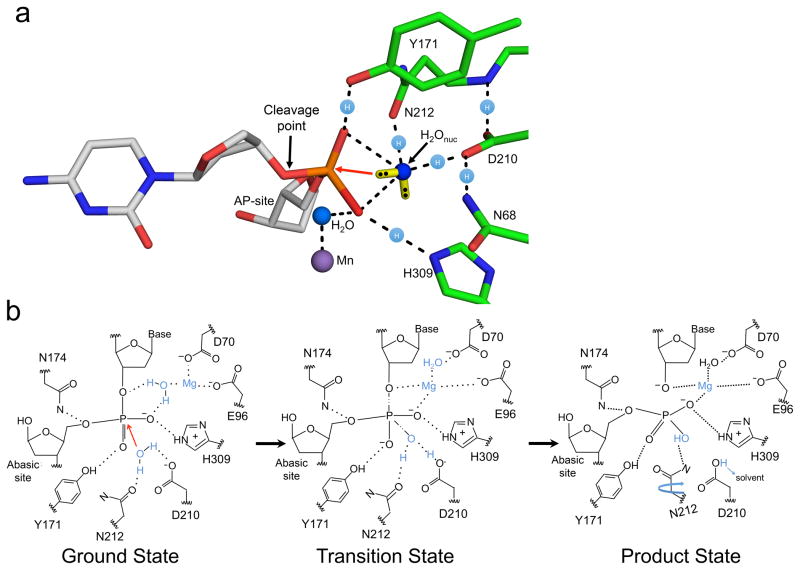
The lack of APE1 active site structural detail has resulted in mechanistic ambiguity11,14,15. The pre-chemistry structures reported here indicate that a water molecule is in position for an in-line nucleophilic attack in all three substrate complexes, even with mutant APE1 or modified DNA. This location is similar to that of an ordered water molecule reported in an apoenzyme APE1 structure26; thus, this water binding site exists prior to DNA binding. Development of the transition state appears to involve the enzyme molding the DNA substrate into the active site to position the AP-site near this ordered active site water (Fig. 7a). This water is in close contact to both D210 and N212 so that it can share protons with these hydrogen bond acceptors. The lone electron pairs of the water are illustrated (yellow bars in Fig 7a) with an orientation avoiding a clash with the non-bridging phosphate oxygens. Instead, these electron pairs are in position to conduct an in-line nucleophilic attack on the phosphate (red arrow in Fig. 7a). This attack is facilitated by charge neutralization afforded by two hydrogen bonds to the phosphate non-bridging oxygens from Y171 and H309. Importantly, this water molecule is in position to directly donate a proton to D210, but not H309 as proposed previously14,24. N212 also plays a key role in coordinating the nucleophilic water, consistent with the dramatic loss of activity with the N212A mutant47. The position of this water is similar to that observed with other nucleases such as TDP248, endonuclease IV (Nfo)18, and the Neisseria meningitides APE149. As with APE1, these nucleases utilize side-chain contacts to facilitate cleavage of the DNA backbone by hydrogen bonding to a water molecule.
Figure 7. APE1 mechanism during strand cleavage.
(a) The organization of the active site prior to the attack by the nucleophilic water is shown with a red arrow indicating the orientation of attack. The cleavage point, key side chains, and AP-site are indicated. The Mn2+ (purple) and water (blue) molecules are shown as spheres. The location of the free electron pairs for the nucleophilic water (H2Onuc) are shown as a yellow bar with black dots for the electrons. These locations represent the distributed cloud determined by Guassian57. Locations of the proton atoms are shown as light blue spheres with an (H) on the dashed lines. (b) Based on our high-resolution structural snapshots we propose a refined APE1 mechanism. Key observations are highlighted in blue. The ground and product states are based on our structural snapshots. The transition state model is inferred from the two structures.
It had previously been postulated that APE1 utilizes a single divalent metal based on an incised product complex and apoenzyme crystal structures that showed a single metal ion coordinated by E96 (site A)11,26,50. A second metal binding site (site B) was identified using lead as a co-factor, but the role of this site was unclear12. Our metal bound substrate complex includes all the necessary atoms to support catalysis and is consistent with a single metal ion mechanism bound near site A. This binding site is similar to that observed with the apoenzyme crystal structure (Supplementary Fig. 5a). An overlay of this structure with the lead bound apoenzyme APE1 indicates that site B is the nucleophilic water site, explaining why lead inhibits catalysis by displacing the nucleophilic water (Supplementary Fig. 5b). It is also not surprising that lead would occupy this water-binding site due to their similar coordination spheres. Computational studies have proposed a moving metal hypothesis that involves metal binding at both sites13,17. The structures reported make this unlikely since there is a lack of space in the ground state structure to accommodate both a nucleophilic water molecule and a metal in site B. Our results indicate a single metal at site A that repositions itself to coordinate the leaving group oxygens of the scissile bond upon formation of the transition state (Supplementary Fig. 3c and Fig. 7b). In this scenario, the metal mediates the developing negative charge in the active site and promotes the formation of product following nucleophilic attack.
Together, our structural snapshots are consistent with the mechanistic features outlined in Figure 7b. In the ground state, the AP-site is flipped into the active site, positioning the phosphate backbone 5′ to the AP-site for cleavage. A single metal is bound to D70, D96 and a water molecule that contacts the non-bridging oxygen of the phosphate. The nucleophilic water is positioned for an in-line attack on the phosphorus atom through short hydrogen bonding interactions with N212 and D210. These short hydrogen bonds combined with the proximity (2.8 Å) of the water oxygen to the phosphorus suggest a formal oxyanion may not be required for nucleophilic attack (Fig. 7b). The resulting pentacovalent transition state intermediate is stabilized by Mg2+, Y171, H309, D210, and N212. The Mg2+ shifts to coordinate a phosphate non-bridging oxygen and the leaving group oxygen that becomes O3′. During or immediately after cleavage, the proton from the nucleophilic water molecule is transferred to D210 and subsequently released to solvent. The final product state results in a 5′-sugar phosphate and 3′-hydroxyl that are coordinated by Mg2+, N174, D70, E96, H309, Y171, and N212. N212 rotates to provide a hydrogen bond donor and stabilize the product state. The interactions observed here appear to be fundamental to catalysis, since alterations that eliminate each of these interactions strongly diminish catalytic activity.
The epigenetic control of gene expression occurs in tracks of CpG dinucleotides through the methylation of cytosine to form 5-methylcytosine51. In mammalian promoters containing CpG tracks, 70–80% of the cytosines undergo methylation32. These modified 5-methylcytosine residues will often undergo spontaneous deamination to thymine to form a T:G mispair within CpG islands33. In addition the neighboring guanine in CpG tracks is susceptible to oxidative DNA damage such as 8-oxo-7,8-dihydroguanine (8-oxoG)52. These hot spots for DNA lesions will result in tandem modifications of CpG islands that alter the epigenetic status of cells. The repair of 8-oxoG in CpG islands is through DNA glycosylase-initiated BER where the oxidized guanine is removed and APE1 processes the resulting AP site. In this situation, APE1 may encounter a 5′ flanking T:G mispair that has arisen from 5-methylcytosine deamination, and this drastically reduces the APE1 catalytic rate35. With the reference structures described here, we can now assess the catalytic repercussions of DNA abnormalities encountered by APE1 in the context of an adjacent T:G mismatch.
Base pairing interactions in DNA can occur through wobble base pairing, rare tautomers, or anionic forms of the nucleobases. These alternative base pairing interactions are believed to contribute to spontaneous mutations and translation errors, but have been difficult to characterize structurally. Previous structural work has observed alternative base pairing interactions within duplex DNA and DNA polymerase active sites53–55. The structures captured here indicate that the mispair shifts between the wobble and a WC-like geometry conformer within the APE1 active site (Fig. 5). Noteworthy, only the WC-like mismatch is aligned for catalysis by contacting key active site residues (Fig. 5d). This implies the wobble conformer results in a non-productive complex that cannot undergo cleavage, consistent with the reduction in the overall catalytic rate. In contrast, the product complex relieves the strain imposed by the WC-like mismatch by shifting solely to the wobble conformer (Fig. 6). Taking the substrate and product structures into account implies the enzyme and DNA are undergoing a “tug of war”: APE1 is trying to sculpt the AP-site into the active site by favoring a WC-like mismatch, while the DNA favors the wobble conformer. This tug of war between protein and DNA highlights the strain APE1 places on the DNA to precisely position the phosphate backbone with the nucleophilic water and metal ion prior to cleavage.
The observation that a 5′-flanking mismatch reduces APE1 activity due to decreased strain provides an important mechanistic detail. Previous structures have utilized ideal substrates and products to characterize APE1 binding to DNA. In these structures, the WC base pairing interaction was maintained and the AP-site was kinked and forced into the active site. By using the T:G mismatch we observe multiple conformers of substrate that indicate the system was undergoing strain while inserting the phosphate backbone into the active site. This is consistent with the rigid platform APE1 provides upon binding the DNA and the need to precisely position the DNA into the active site; this is in contrast to enzymes that undergo conformational changes to accommodate DNA. This mechanism optimizes APE1 for correctly base paired DNA flanking the AP site and DNA abnormalities reduce the rate of incision. Since APE1 must deal with AP sites in a large number of DNA contexts, it may have evolved a rapid incision rate to provide a kinetic buffer to overcome situations where it must incise an AP site in a less than ideal situation.
Given the high number of AP sites in the genome, it is essential that they are rapidly processed to maintain genomic integrity. The rapid APE1 incision rate is facilitated by utilizing water as a nucleophile and a preformed metal binding site to stabilize the developing charge during the course of the reaction. However, incision generates a cytotoxic DNA break that could lead to chromosomal rearrangements. To prevent this, APE1 has evolved a slow product release step that enables channeling to other DNA repair enzymes56.
ONLINE METHODS
DNA Sequences
To generate the 21-mer for crystallization the following DNA sequences were used (IDT): non-damaged strand, 5′-GGA-TCC-GTC-GGG-CGC-ATC-AGC-3′; modified matched strand 5′-GCT-GAT-GCG-CXC-GAC-GGA-TCC-3′ where the underlined X represents THF; modified mismatched strand 5′-CTGATGCGTXCGACGGATCC-3′; 2′-O-methyl phosphorothioate-containing strand 5′-GCT-GAT-GCG-2′OMe(C(ps))-XCG-ACG-GAT-CC-3′. The kinetic and binding studies utilized a 19-mer hairpin DNA to prevent end binding, as described previously58. The substrate was formed using a 46-mer 5′-CTG-GAG-CTTG-CT-CCA-GCG-CXC-GGT-CGA-T*C-GTAA-GAT-CGA-CCG-TGC-G-3′ purchased from Midland Certified Reagent Company with a 5′-phosphate and fluorescein label (indicated by *). The underlined residues form the hairpin loop. This 46-mer oligonucleotide containing 5′-phosphate was annealed by slowly cooling down in Tris-EDTA buffer and ligated to generate a circular DNA dumbbell substrate with a THF. The ligation reaction was performed in a reaction mixture (100 μl) containing 1000 pmol oligonucleotide, buffer X1 for T4 DNA ligase (New England Biolabs Inc.), and 100 mM ATP. The reaction was initiated by addition of 4000 U of T4 DNA ligase (New England Biolabs Inc.), and the incubation was at 20 °C overnight, 1 h at 30 °C and overnight at 4 °C. The enzyme was inactivated by incubating for 5 min at 95 °C. The nicked product DNA for the anisotropy binding studies was generated using a similar hairpin 46-mer to form the 5′-nick product substrate without the ligation step. The oligonucleotides were separated from other DNA species by electrophoresis on an 18% polyacrylamide gel containing 8 M urea in TBE buffer. Purified DNA dumbbell substrate was annealed in the buffer containing 50 mM Tris and 50 mM KCl and the concentration was determined by absorbance at 260 nm.
Protein expression and purification
Human wild-type APE1 was expressed from the pxc53 clone described previously59. The truncated APE1 clone was generated from pxc53 through removal of the N-terminal 43 amino acids by PCR. All mutagenesis was carried out in either the full length or truncated clones. APE1 was expressed in E. coli BL21(DE3) and induced as described59. After induction the cells were lysed at 4 °C by sonication in 50 mM Hepes, pH 7.4, 100 mM NaCl, and protease inhibitors (1 mM AEBSF, 1 mM benzamidine, 1ug/ml pepstatin A, 1ug/ml leupeptin). The lysate was pelleted at 27,000 × g for 1 h. The resulting supernatant was passed over a HiTrap Heparin HP (GE Health Sciences) equilibrated in lysate buffer. APE1 was eluted from the heparin column using a linear gradient of NaCl up to 1 M. APE1 eluting at high salt was buffer exchanged into 50 mM NaCl and loaded onto a Mono S HR 10/10 column. The purified protein was eluted using a linear gradient to 1 M NaCl. Purified APE1 was subsequently loaded onto a HiPrep 26/60 Sephacryl S-200 HR (GE Health Sciences). The resulting pure fractions were pooled and the concentration was determined by nano-drop (Thermo-Scientific).
Crystallization and structure determination
APE1:DNA complex crystals with a central THF were grown by annealing 1 mM of two 21-mer oligonucleotides in a 1:1 ratio using a PCR thermocycler by heating 10 min at 90 °C and cooling to 4 °C (1 °C/min). The annealed DNA was mixed with truncated APE1 to achieve a final concentration of 0.5 mM DNA and 10 mg/ml APE1. The product DNA was generated by allowing APE1 to cleave the DNA (MgCl2 in annealing buffer) in solution for 10 min on the bench prior to setting up the crystallization drops. APE1:DNA complexes were crystallized by vapor diffusion. The reservoir solution for crystal formation was 10% PEG20000, 100 mM sodium citrate, pH 5.0, and 200 mM MgCl2. Crystals grew within a week at 20 °C. APE1:DNA complex crystals were transferred to a cryosolution containing the mother liquor with 25% ethylene glycol. The MnCl2 soak was performed in the same manner with the addition of 200 mM MnCl2 for 20 s. All crystals were frozen at 100K prior to data collection at the home source. Data collection was done on a SATURN92 or SATURN94 CCD detector system mounted on a MiraMax-007HF rotating anode generator at a wavelength of 1.54 Å. This allows for anomalous data detection after phasing by molecular replacement with high redundancy. Data were processed and scaled using the HKL2000 software package60. Initial models were determined using molecular replacement with two APE1 molecules (PDB ID 1DEW)11 and the DNA was built using Coot. Refinement was carried out using PHENIX and model building using Coot61. The metal-ligand coordination restraints were generated by ReadySet (PHENIX) and not utilized until the final rounds of refinement. The figures were prepared in PyMol and all density maps were generated after performing simulated annealing (http://www.pymol.org). Ramachandran analysis determined 100% of non-glycine residues lie in allowed regions and at least 96% in favored regions.
Kinetic characterization
A rapid quench-flow system was employed for activity measurements. The fluorescein-containing dumbbell DNA substrate containing THF (F) was used to measure APE1 incision activity (37 °C). The reaction buffer was 50 mM Hepes, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, and 0.1 mg/ml bovine serum albumin. The final concentrations (after mixing) were 100 nM DNA substrate and 30 nM APE1. At time intervals, aliquots were quenched by mixing with 100 mM NaOH. An equal volume of DNA gel loading buffer (10 M urea, 100 mM EDTA, 0.01% bromophenol blue, and 0.01% xylene cyanol) was added to the quenched reaction mixture. After incubation at 95 °C for 5 min, the reaction products were separated by 15% denaturing polyacrylamide gel. A Typhoon PhosphorImager was used for gel scanning and imaging, and the data were analyzed with ImageQuant software. The biphasic time courses were fit to Product = A{1-e−k_obs_t}+vsst where A represents the amplitude of the rising exponential and _k_obs the first-order rate constant. The steady-state rate constant (_k_ss) is the steady-state velocity (_v_ss)/A where A represents the fraction of actively bound enzyme. When the observed burst and steady-state rate constants are not well-separated (i.e., _k_burst/_k_ss) < 10), these values do not represent intrinsic rate constants. The intrinsic rate constants can be calculated from _k_burst = (_k_incision + _k_off,product) and _k_ss = (_k_incision * _k_off,product)/(_k_incision + _k_off,product).
Anisotropy binding studies
Fluorescence anisotropy measurements were used to quantify binding of wild-type APE1 and a R181A mutant to a DNA dumbbell as described above. Fluorescence anisotropy measurements were carried out on Horiba FluoroLog Fluorimeter at 20 °C in a buffer consisting of 50 mM Tris/HCl, pH 7.0, 25 mM NaCl, 10 mM EDTA and 0.1 mM DTT. The excitation and emission wavelengths were 485 and 520 nm respectively, each with a 14 nm slit width. For all DNA titrations, fluorescein-labelled DNA dumbbell concentration ranged from 0.5 nM to 10 nM, adjusted depending on initial _K_d value determinations to be below the _K_d. The lower limit (0.5 nM) is based on instrument configuration limitations, with insufficient signal to noise observed when the DNA probe was below this level. Fluorescence anisotropy changes were normalized to the number of binding sites and fit to either a one-site binding model or to a two-site binding model using the program Origin 862. A minimum of three independent titrations was averaged to determine _K_d values.
Supplementary Material
1
2
Acknowledgments
We thank the Collaborative Crystallography group at NIEHS for help with data collection and analysis. We thank L. Pedersen and L. Perera for valuable discussions. This research was supported in part by the Intramural Research Program of the US National Institutes of Health, National Institute of Environmental Health Sciences (project numbers Z01-ES050158 (S.H.W) and Z01-ES050161 (S.H.W)). A part of this research was performed at Oak Ridge National Laboratory’s Spallation Neutron Source, sponsored by the United States Department of Energy, Office of Basic Energy Sciences (M.J.C.). N.S.D is supported in part by Eli Lilly and Co. and the United States Department of State, as part of the United States-Russia Collaboration in the Biomedical Sciences US National Institutes of Health Visiting Fellows Program.
Footnotes
Accession codes. Coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 5DFF, 5DFI, 5DFJ, 5DFH, and 5DG0.
Competing financial interests
The authors declare no competing financial interests.
Author contributions
B.D.F designed the project. B.D.F. carried out crystallography. N.S.D. did the kinetic analyses. M.J.C. did the binding studies. B.D.F., W.A.B., and S.H.W. prepared the manuscript. All authors discussed the results and commented on the manuscript.
References
- 1.Kingma PS, Corbett AH, Burcham PC, Marnett LJ, Osheroff N. Abasic sites stimulate double-stranded DNA cleavage mediated by topoisomerase II: DNA lesions as endogenous topoisomerase II poisons. J Biol Chem. 1995;270:21441–21444. doi: 10.1074/jbc.270.37.21441. [DOI] [PubMed] [Google Scholar]
- 2.Cuniasse P, Fazakerley GV, Guschlbauer W, Kaplan BE, Sowers LC. The abasic site as a challenge to DNA polymerase: a nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Biol Chem. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- 3.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 4.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 5.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radical Bio Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 6.Dianov GL, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YJ, Wilson DM., III Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, et al. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase β. J Biol Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 11.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 12.Beernink PT, et al. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, ape1: implications for the catalytic mechanism. J Mol Biol. 2001;307:1023–1034. doi: 10.1006/jmbi.2001.4529. [DOI] [PubMed] [Google Scholar]
- 13.Oezguen N, et al. A “moving metal mechanism” for substrate cleavage by the DNA repair endonuclease APE-1. Proteins: Struct, Funct, Bioinf. 2007;68:313–323. doi: 10.1002/prot.21397. [DOI] [PubMed] [Google Scholar]
- 14.Gorman MA, et al. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mundle ST, et al. Novel role of tyrosine in catalysis by human AP endonuclease 1. DNA Repair. 2004;3:1447–1455. doi: 10.1016/j.dnarep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Mundle ST, Delaney JC, Essigmann JM, Strauss PR. Enzymatic mechanism of human apurinic/apyrimidinic endonuclease against a THF AP site model substrate. Biochemistry. 2009;48:19–26. doi: 10.1021/bi8016137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oezguen N, et al. MD simulation and experimental evidence for Mg2+ binding at the B site in human AP endonuclease 1. Bioinformation. 2011;7:184–198. doi: 10.6026/97320630007184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutakawa SE, et al. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J Biol Chem. 2013;288:8445–8455. doi: 10.1074/jbc.M112.422774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipton AS, et al. Characterization of Mg2+ binding to the DNA repair protein apurinic/apyrimidic endonuclease 1 via solid-state 25Mg NMR spectroscopy. J Am Chem Soc. 2008;130:9332–9341. doi: 10.1021/ja0776881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–68. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, Zhao Y, Yamagata Y, Hua Y-j, Yang W. Watching DNA polymerase η make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molina R, et al. Visualizing phosphodiester-bond hydrolysis by an endonuclease. Nature structural & molecular biology. 2015;22:65–72. doi: 10.1038/nsmb.2932. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen LH, Barsky D, Erzberger JP, Wilson DM., III Mapping the protein-DNA interface and the metal-binding site of the major human apurinic/apyrimidinic endonuclease1. J Mol Biol. 2000;298:447–459. doi: 10.1006/jmbi.2000.3653. [DOI] [PubMed] [Google Scholar]
- 24.Erzberger JP, Wilson DM., III The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J Mol Biol. 1999;290:447–457. doi: 10.1006/jmbi.1999.2888. [DOI] [PubMed] [Google Scholar]
- 25.Maher RL, Bloom LB. Pre-steady-state kinetic characterization of the AP endonuclease activity of human AP endonuclease 1. J Biol Chem. 2007;282:30577–30585. doi: 10.1074/jbc.M704341200. [DOI] [PubMed] [Google Scholar]
- 26.He H, Chen Q, Georgiadis MM. High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I. Biochemistry. 2014;53:6520–6529. doi: 10.1021/bi500676p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi T, Schein CH, Oezguen N, Feng Y, Braun W. Effects of backbone contacts 3′ to the abasic site on the cleavage and the product binding by human apurinic/apyrimidinic endonuclease (APE1) Biochemistry. 2003;43:684–689. doi: 10.1021/bi0346190. [DOI] [PubMed] [Google Scholar]
- 28.Peddi SR, Chattopadhyay R, Naidu CV, Izumi T. The human apurinic/apyrimidinic endonuclease-1 suppresses activation of poly(adp-ribose) polymerase-1 induced by DNA single strand breaks. Toxicology. 2006;224:44–55. doi: 10.1016/j.tox.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 29.Wilson DM, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 30.Schermerhorn KM, Delaney S. Transient-state kinetics of apurinic/apyrimidinic (AP) endonuclease 1 acting on an authentic AP site and commonly used substrate analogs: the effect of diverse metal ions and base mismatches. Biochemistry. 2013;52:7669–7677. doi: 10.1021/bi401218r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanyushin BF, Tkacheva SG, Belozersky AN. Rare bases in animal DNA. Nature. 1970;225:948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- 32.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen JC, Rideout WM, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sassa A, Beard WA, Prasad R, Wilson SH. DNA sequence context effects on the glycosylase activity of human 8-oxoguanine DNA glycosylase. J Biol Chem. 2012;287:36702–36710. doi: 10.1074/jbc.M112.397786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassa A, Ağlayan M, Dyrkheeva NS, Beard WA, Wilson SH. Base excision repair of tandem modifications in a methylated CpG dinucleotide. J Biol Chem. 2014;289:13996–14008. doi: 10.1074/jbc.M114.557769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNeill DR, Wilson DM., 3rd A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 37.Chohan M, Mackedenski S, Li W, Lee CH. Human apurinic/apyrimidinic endonuclease 1 (APE1) has 3′ RNA phosphatase and 3′ exoribonuclease activities. J Mol Biol. 2014;427:298–311. doi: 10.1016/j.jmb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Barnes T, et al. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gros L, Ishchenko AA, Ide H, Elder RH, Saparbaev MK. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thakur S, et al. APE1/Ref-1 as an emerging therapeutic target for various human diseases: phytochemical modulation of its functions. Exp Mol Med. 2014;46:e106. doi: 10.1038/emm.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ Mol Mutagen. 2015;56:1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meira LB, et al. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- 45.Illuzzi JL, et al. Functional assessment of population and tumor-associated APE1 protein variants. PLoS ONE. 2013;8:e65922. doi: 10.1371/journal.pone.0065922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Kanazhevskaya LY, Koval VV, Lomzov AA, Fedorova OS. The role of Asn-212 in the catalytic mechanism of human endonuclease APE1: stopped-flow kinetic study of incision activity on a natural AP site and a tetrahydrofuran analogue. DNA Repair. 2014;21:43–54. doi: 10.1016/j.dnarep.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Schellenberg MJ, et al. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol. 2012;19:1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu D, et al. Structural basis for the recognition and cleavage of abasic DNA in Neisseria meningitidis. Proc Natl Acad Sci USA. 2012;109:16852–16857. doi: 10.1073/pnas.1206563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manvilla BA, Pozharski E, Toth EA, Drohat AC. Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg2+ cofactor. Acta Crystallogr D Biol. 2013;69:2555–2562. doi: 10.1107/S0907444913027042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delgado S, Gómez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalam MA, Basu AK. Mutagenesis of 8-oxoguanine adjacent to an abasic aite in simian kidney cells: tandem mutations and enhancement of G→T transversions. Chem Res Toxicol. 2005;18:1187–1192. doi: 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- 53.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-Hashimi HM. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015;519:315–320. doi: 10.1038/nature14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bebenek K, Pedersen LC, Kunkel TA. Replication infidelity via a mismatch with Watson–Crick geometry. Proc Natl Acad Sci USA. 2011;108:1862–1867. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci USA. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 57.Chesnut DB. An electron localization function study of the lone pair. J Phys Chem A. 2000;104:11644–11650. [Google Scholar]
- 58.Eustermann S, et al. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J Mol Biol. 2011;407:149–170. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strauss PR, Beard WA, Patterson RA, Wilson SH. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J Biol Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 60.Otwinowski Z, Minor W. Processsing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lefurgy ST, Leyh TS. Analytical expressions for the homotropic binding of ligand to protein dimers and trimers. Anal Biochem. 2012;421:433–438. doi: 10.1016/j.ab.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2