miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation (original) (raw)
. Author manuscript; available in PMC: 2015 Nov 24.
Summary
Chronic inflammation is a contributing factor to most life shortening human diseases. However, the molecular and cellular mechanisms that sustain chronic inflammatory responses remain poorly understood making it difficult to treat this deleterious condition. Using a mouse model of age-dependent inflammation that results from a deficiency in miR-146a, we demonstrate that miR-155 contributed to the progressive inflammatory disease that emerged as _Mir146a_−/− mice grew older. Upon analyzing lymphocytes from inflamed versus healthy middle-aged mice we found elevated numbers of T follicular helper (Tfh) cells, germinal center (GC) B cells and autoantibodies, all occurring in a miR-155-dependent manner. Further, _Cd4-cre Mir155_fl/fl mice were generated and demonstrated that miR-155 functions in T cells, in addition to its established role in B cells, to promote humoral immunity in a variety of contexts. Taken together, our study discovers that miR-146a and miR-155 counter-regulate Tfh cell development that drives aberrant GC reactions during chronic inflammation.
Introduction
Chronic, low-grade inflammation is driven by sustained production of inflammatory factors such as cytokines, reactive oxygen species and autoantibodies, and is associated with 7 of the top 10 causes of mortality in the United States such as heart disease, neurodegeneration, metabolic disorders, autoimmunity and cancer (Akbaraly et al., 2013; Howcroft et al., 2013; Michaud et al., 2013). This deleterious condition, which typically lacks clinical symptoms, can cause damage to important organ systems over time, and serve as a primer for many age-related disorders. Despite our growing knowledge of immune regulation during acute inflammatory responses, our understanding of molecular and cellular mechanisms governing chronic, low-grade inflammation remains tenuous. This deficiency has prevented the development of targeted therapies to reduce chronic inflammation and the plethora of disorders associated with one’s inflammatory status during the aging process. By the year 2050 it is projected that those 60 years and older will make up 22% of the world’s population, compared to only 10% in the year 2000 (Dorshkind et al., 2009). Thus, finding therapeutic solutions to chronic inflammatory conditions has become more important than ever before.
It is now appreciated that most of the human genome is transcribed into noncoding RNAs that carry out diverse cellular functions including differentiation, survival and proliferation (Esteller, 2011). Among noncoding RNAs, microRNAs (miRNAs) regulate gene expression post-transcriptionally and have been shown to modulate a wide range of biological systems (Mendell and Olson, 2012). Further, several miRNAs have been shown to control inflammation in young mice subjected to infection by pathogens or during antigen-induced autoimmunity (Baumjohann et al., 2013; Kang et al., 2013; O'Connell et al., 2010b; Oertli et al., 2011; Rodriguez et al., 2007). Despite their emerging connection to acute inflammation, little is known about the functions of miRNAs during chronic inflammation and diseases associated with aging. Recently, the anti-inflammatory miR-146a has emerged as a molecular safeguard against age-dependent inflammatory disease (Boldin et al., 2011; Zhao et al., 2011; Zhao et al., 2013). Mice deficient in miR-146a have increased serum concentrations of interleukin-6 (IL-6) and autoantibodies, and display splenomegaly, myeloproliferation and inflammatory damage to several tissues as they reach middle age. When _Mir146a_−/− mice grow even older, they succumb to different types of cancers and hematopoietic neoplasms that reduce their lifespans compared to wild type (Wt) controls. These findings clearly demonstrate that specific miRNAs have evolved to regulate chronic, low-grade inflammation, and establish _Mir146a_−/− mice as an excellent model with which to study this clinically relevant condition. While miR-146a functions to prevent chronic inflammation, we hypothesized that other miRNAs act to promote this deleterious process.
miR-155 has emerged as a multi-faceted regulator of immunity that impacts different types of inflammatory responses in young mice (Hu et al., 2013; Huffaker et al., 2012; O'Connell et al., 2010b; Rodriguez et al., 2007; Thai et al., 2007). Further, previous studies find that constitutive overexpression of miR-155 in the hematopoietic compartment causes a chronic inflammatory disease (O'Connell et al., 2008) or leukemia (Costinean et al., 2006), shortening the animal’s lifespan. In the present study, we investigated the role of endogenous miR-155 during chronic, low-grade inflammation that develops in _Mir146a_−/− mice.
Results
miR-155-dependent accumulation of activated T cells in _Mir146a_−/− mice
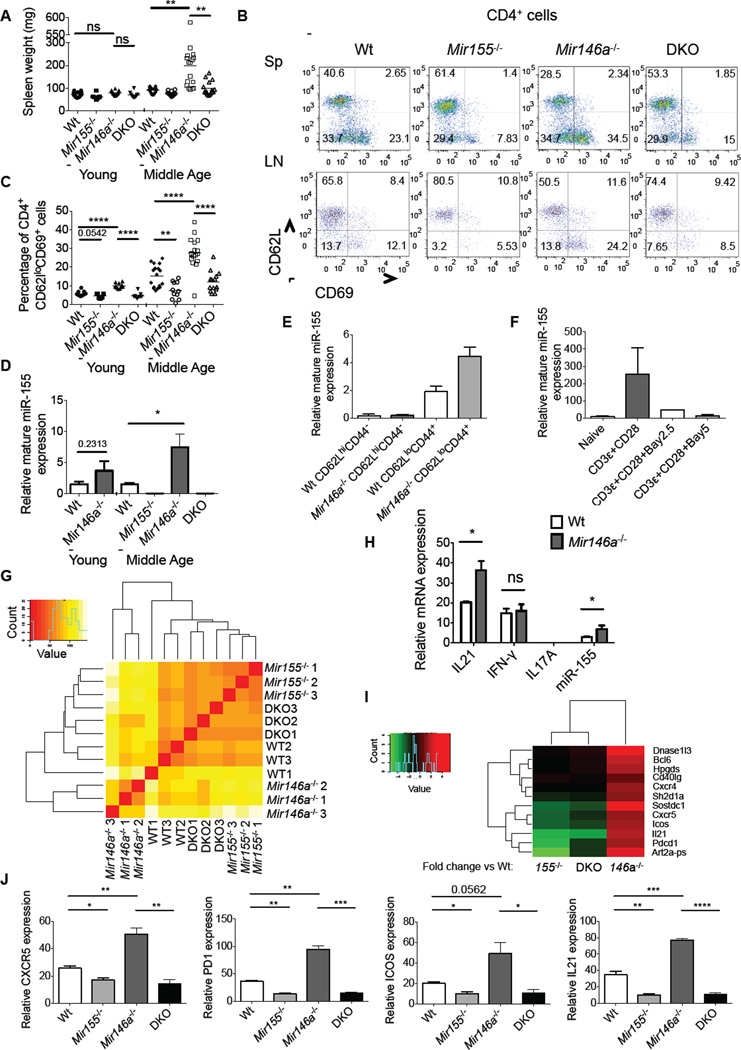
To determine if endogenous miR-155 plays a role in promoting age-dependent disease in _Mir146a_−/− mice, we aged _Mir155_−/− _Mir146a_−/− (DKO) and control mice for 7–10 months (middle-age). As previously reported (Boldin et al., 2011; Zhao et al., 2011; Zhao et al., 2013), middle-aged but not young _Mir146a_−/− mice had enlarged spleens (Figures 1A). Elevated amounts of activated T cells (CD4+CD69+CD62L−) were also evident in middle-aged _Mir146a_−/− mice, both in the spleen and lymph nodes, and this activated T cell phenotype did begin to emerge in young mice (Figures 1B, 1C and S1). In contrast, middle-aged _Mir155_−/− _Mir146a_−/− mice had spleen weights and activated CD4+ T cell levels that were similar to middle-aged Wt mice, indicating that miR-155 promotes these phenotypes in _Mir146a_−/− animals (Figures 1A–1C and S1).
Figure 1. miR-155 is required for expansion of CD4+ T cells with a Tfh cell gene expression profile in middle-aged _Mir146a_−/− mice.
(A) Spleen weights from 2 (young) and 10 (middle-age) month-old age- and sex-matched Wt, miR155−/−, _Mir146a_−/− and _Mir155−/− Mir146a_−/− (DKO) mice. (B) Representative FACS analysis of activated (CD69+CD62L) CD4+ T cells in the spleens and lymph nodes of 10-months old mice. (C) Spleen data from multiple young and middle-aged mice in (B). (D) miR-155 expression in young and middle-aged CD4+ T cells of the indicated genotypes (n=3). (E) CD4+CD3+CD62lloCD44+ (activated) and CD4+CD3+CD62lhiCD44 (naïve) cells were sorted from 10-month old mouse spleens and miR-155 expression was assayed. (F) miR-155 expression in activated CD4+ T cells cultured with or without an NF-κB inhibitor (Bay 11-7082) at the indicated concentration (2.5 or 5 µM) for 24 hours. (G) Total RNA was extracted from splenic CD4+ T cells taken from 10-month old mice of the indicated genotypes and subjected to an RNA-Seq analysis (n=3). Gene clustering of RNA-Seq data is shown. The magnitude of the gene expression differences between any two groups is indicated by color, and the scale is shown. (H) Relative mRNA expression of IL21, IFN-γ, IL17A and miR-155 in CD4+ T cells from middle-aged Wt and _Mir146a_−/− mice. (I) Heat map indicating the fold change in Tfh cell gene expression between _Mir146a_−/−, _Mir155_−/− or _Mir155_−/− _Mir146a_−/− compared to Wt cells. The color key is shown. (J) QPCR analyses of Tfh-related genes in aged CD4+ T cells. The values have been normalized to L32. * denotes a p value of <0.05 using a Student’s t test. Dashed line indicates that young and middle-aged mice were harvested on different days, and each dot represents an individual mouse. See also Figure S1.
The _Mir146a_−/− mouse phenotype is largely dependent upon lymphocytes (Zhao et al., 2013), and consistent with previous work (Yang et al., 2012), we found that an increase in activated CD4+ T cells precedes other disease manifestations in this model (Figures 1C and S1). Using bone marrow reconstitution experiments we also determined that this phenotype was primarily hematopoietic in nature (Figure S1). Based upon these findings, we next assessed miR-155 expression in CD4+ T cells. Results showed that miR-155 expression trended towards being higher in CD4+ T cells from young _Mir146a_−/− mice, compared to Wt controls, and reached even higher expression in CD4+ T cells taken from middle-aged _Mir146a_−/− mice (Figure 1D). This correlated with an increased proportion of activated T cells in the bulk CD4+ T cell populations from _Mir146a_−/− versus Wt mice. Upon sorting, we confirmed that activated CD4+ T cells expressed higher miR-155 than naïve T cells, and this expression was enhanced further in activated T cells lacking miR-146a (Figure 1E). It has been previously demonstrated that activated _Mir146a_−/− CD4+ T cells have elevated NF-κB activation, a pathway that we found to promote miR-155 expression in activated CD4+ T cells (Figure 1F). In contrast to T cells, enhanced expression of miR-155 was not observed in B220+ B cells from Mir146a −/− mice (Figure S1). Based on these results, we focused our subsequent analysis on the CD4+ T lymphocyte compartment to better understand the role of miR-155 during chronic inflammation.
Spontaneous T follicular helper cells, germinal center B cells and autoantibodies accumulate in _Mir146a_−/− mice
We examined gene expression patterns in CD4+ T cells from 10-month old Wt, _Mir155_−/−, _Mir146a_−/− and _Mir155_−/− _Mir146a_−/− mice by RNA-Seq. Gene expression profiles in _Mir146a_−/− CD4+ T cells were distinct from the other three genotypes according to a cluster analysis while _Mir155_−/− _Mir 146a_−/− profiles clustered closer to Mir155−/− than Wt or _Mir146a_−/− profiles (Figure 1G). IL-21 expression was significantly higher in _Mir146a_−/− compared to Wt middle-age CD4+ T cells, while there was little difference in interferon-γ (IFN-γ) mRNA amounts and undetectable expression of the IL-17A message in both genotypes (Figure 1H). IL-21 is produced by T follicular helper (Tfh) cells, and its elevated expression in _Mir146a_−/− T cells prompted us to examine additional Tfh genes. We observed increased expression of B cell lymphoma 6 protein (Bcl6), chemokine (C-X-C motif) receptor 5 (Cxcr5), programmed cell death 1 (Pd1), and inducible T cell co-stimulator (Icos) (among others) in _Mir146a_−/− CD4+ T cells, and decreased or unchanged expression of these genes in _Mir155_−/− and _Mir155_−/− _Mir146a_−/− T cells, compared to Wt controls (Figure 1I). This was confirmed by quantitative rtPCR (QPCR) (Figure 1J). These data suggested that _Mir146a_−/− CD4+ T cells from middle-aged mice are enriched in Tfh cells, and that this occurs through a miR-155-dependent mechanism.
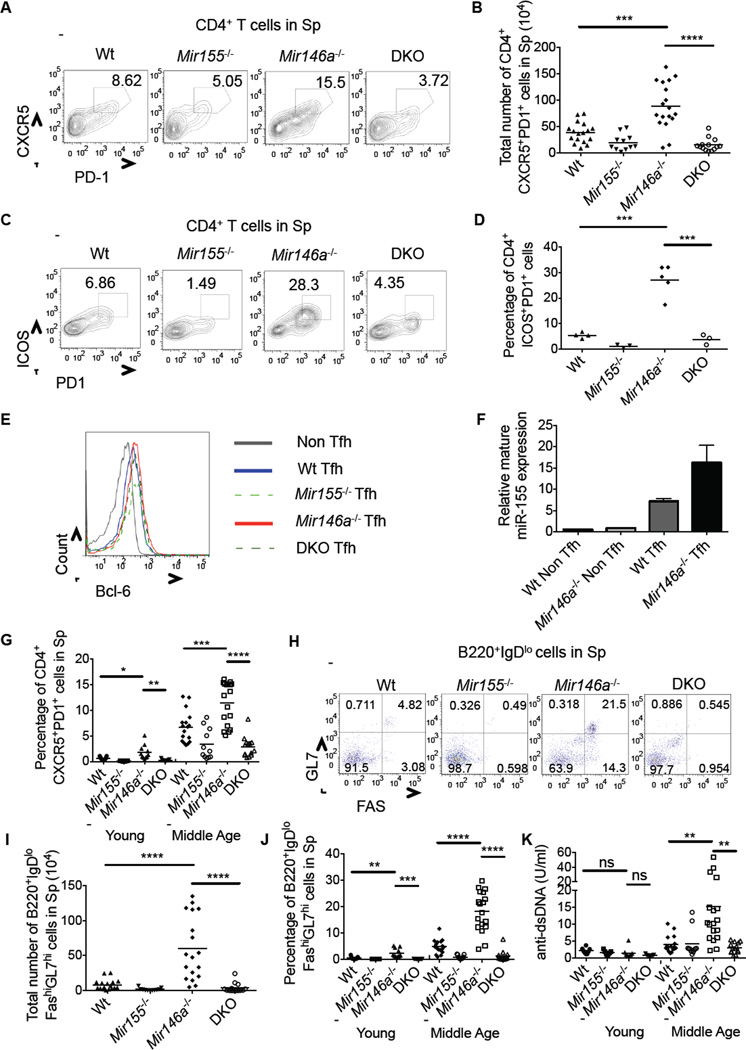
Using flow cytometry, we next detected increases in CD44+CD4+CXCR5+PD1+ Tfh cell numbers in the spleens and LNs of middle-aged _Mir146a_−/− mice compared to controls (Figures 2A, 2B and S2). Further, these cells also expressed ICOS and BCL6 consistent with their Tfh cell identity (Figures 2C–2E). miR-155 was expressed at higher amounts in Wt Tfh compared to non-Tfh cells, and further enhanced in _Mir146a_−/− Tfh cells (Figure 2F). This Tfh cell phenotype began to emerge in young _Mir146a_−/− mice, suggesting that it may be an early step in disease progression. We also observed an overall increase in Tfh cells in the CD4+ T cell compartment as a function of age in all groups (Figure 2G).
Figure 2. miR-155-dependent accumulation of Tfh cells, GC B cells and autoantibodies in _Mir146a_−/− mice.
(A) Flow cytometry plots showing CD4+CXCR5+PD1+ Tfh cells in the spleens of middle-aged mice. (B) Average total number of Tfh cells in the spleens of middle-aged mice 7–10 months old. (C) Flow cytometry plots showing CD4+ICOS+PD1+ Tfh cells in the spleens of middle-aged mice. (D) Average percentage of CD4+ICOS+PD1+ Tfh cells in the spleens of middle-aged mice. (E) Bcl6 expression in Tfh and non-Tfh cells in the spleens of middle-aged mice. (F) CD4+CXCR5+PD1+ Tfh or CD4+CXCR5−PD1− non-Tfh cells were sorted from the spleens of middle-aged mice and miR-155 expression was tested. (G) Average percentage of Tfh cells in the spleens of young and middle-aged mice. (H) Flow cytometry plots showing B220+IgDloGL7hiFAShi GC B cells in the spleens of middle-aged mice. (I) Average total number of GC B cells in the spleens of middle-aged mice. (J) Average percentage of GC B cells in the spleens of young and middle-aged mice. (K) ELISA of anti-dsDNA autoantibodies in the serum of young and middle-aged mice. Error bars represent +/− SEM. * denotes a p value of <0.05 using a Student’s T test. Dashed line indicates that young and middle-aged mice were harvested on different days, and each dot represents an individual mouse. Data represent at least two independent experiments. See also Figure S2.
Tfh cells promote antibody class-switching and production by germinal center (GC) B cells (Johnston et al., 2009; Nurieva et al., 2008), and play established roles in driving autoimmunity when their development becomes dysregulated (Linterman et al., 2009). Consistent with this, increases in B220+IgDloFashiGL7hi GC B cells were observed in young and middle-age _Mir146a_−/− mice compared to controls, and elevations in GC B cells were also observed as a function of age in all groups (Figure 2H–J). Heightened serum concentrations of anti-dsDNA autoantibodies were detected in middle-aged mice, but unlike Tfh and GC B cells, not young _Mir146a_−/− mice (Figure 2K). This suggests that in some cases long-term GC reactions may be necessary for production of autoantibodies, which are associated with inflammation in a variety of autoimmune settings. Of relevance, this entire phenotype was dependent upon miR-155, as _Mir155_−/−_Mir146a_−/− mice resembled Wt or _Mir155_−/− animals in both age groups (Figures 2A–D, 2G–K and S2). These findings indicate that miR-155 is required for spontaneous Tfh cell development and aberrant humoral responses in _Mir146a_−/− mice.
miR-155 promotion of GC formation in _Mir146a_−/− mice precedes the onset of systemic tissue inflammation
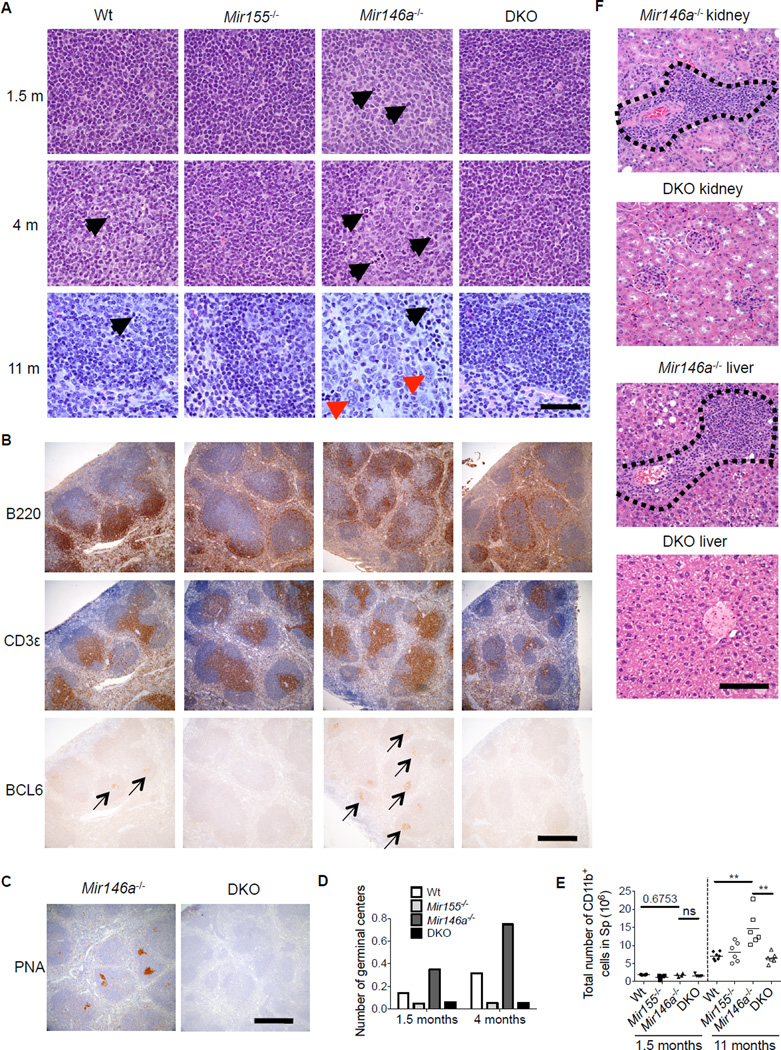
To further assess the miR-155-dependent inflammatory disease that develops in _Mir146a_−/− mice, H&E stained splenic sections from Wt, _Mir155_−/−, _Mir146a_−/− and _Mir155_−/− _Mir146a_−/− mice at 1.5, 4 and 11 months of age were prepared (Figures 3A and S3). Immmunohistochemistry (IHC) was also performed on splenic sections from 1.5 month old mice to detect B (B220) and T (CD3) cells, and serial sections were also stained for BCL6 (Figures 3B and S3). Peanut agglutinin (PNA) staining was also performed (Figure 3C). Histological analysis demonstrated the highest number GCs in _Mir146a_−/− spleens, reduced amounts in Wt mice and the lowest numbers in _Mir155_−/− and _Mir155_−/− _Mir146a_−/− mice at 1.5 and 4 months of age (Figures 3A–3D). The number of germinal centers increased with age in Wt and _Mir146a_−/− mice, but remained low in _Mir155_−/− and _Mir155_−/− _Mir146a_−/− mice. Consistent with this, there were increased numbers of BCL6+ and PNA+ cells in lymphoid follicles in _Mir146a_−/− spleens compared to spleens from the other genotypes (Figure 3B, 3C and S3). All of these observations are concordant with our flow cytometry data, and further demonstrate that the aberrant GC phenotype in _Mir146a_−/− mice begins in young mice and becomes more severe with age.
Figure 3. Histological assessment of miR-155-dependent, spontaneous GC formation and chronic inflammatory disease in _Mir146a_−/− mice.
(A) Representative H&E stained sections of Wt, _Mir155_−/−_Mir146a_−/− and _Mir155_−/−_Mir146a_−/− (DKO) male mouse spleens from the indicate ages. Black arrows represent examples of germinal center elements including large centroblasts, mitotic figures and apoptotic bodies. Red arrowheads represent myeloid cells. Scale bar: 40 microns. B. Spleen sections from 1.5 month-old mice were also subjected to IHC to detect T cells (CD3), B cells (B220) and BCL6+ cells. Arrows indicate regions with BCL6 positive cells. Scale bar: 400 microns. (C) Representative PNA staining of spleens from _Mir146a_−/− and _Mir155_−/−_Mir146a_−/− mice at 1.5 months of age. Scale bar: 100 microns. (D) The number of GCs per splenic lymphoid follicle was determined at 1.5 and 4 months of age. (E) CD11b+ myeloid cell numbers were measured using flow cytometry. (F) Representative H&E staining of kidneys and livers _from Mir146a_−/− an_d Mir155−/− Mir146a_−/− mice at 12 months of age. Scale bar: 100 microns. Inflammatory infiltrate is located within the dashed boarder. * denotes a p value of <0.05 using a Student’s T test. See also Figure 3S.
By 11 months of age, _Mir146a_−/− spleens had disrupted lymphoid follicles that were replaced by increased myeloid cells and other hematopoietic elements, and this was generally not observed in the other age-matched genotypes (Figures 3A, 3E and S3). Further, inflammatory infiltrates into the liver and kidneys of _Mir146a_−/− mice were found by 12 months of age (Figure 3F), indicating a multi-organ inflammatory condition was emerging. In contrast, these features were not observed in the _Mir155_−/− _Mir146a_−/− mice analyzed suggesting that miR-155 promotes chronic inflammation in multiple tissues when miR-146a is absent.
miR-155 regulates Tfh cell development through a T cell intrinsic mechanism following immunization
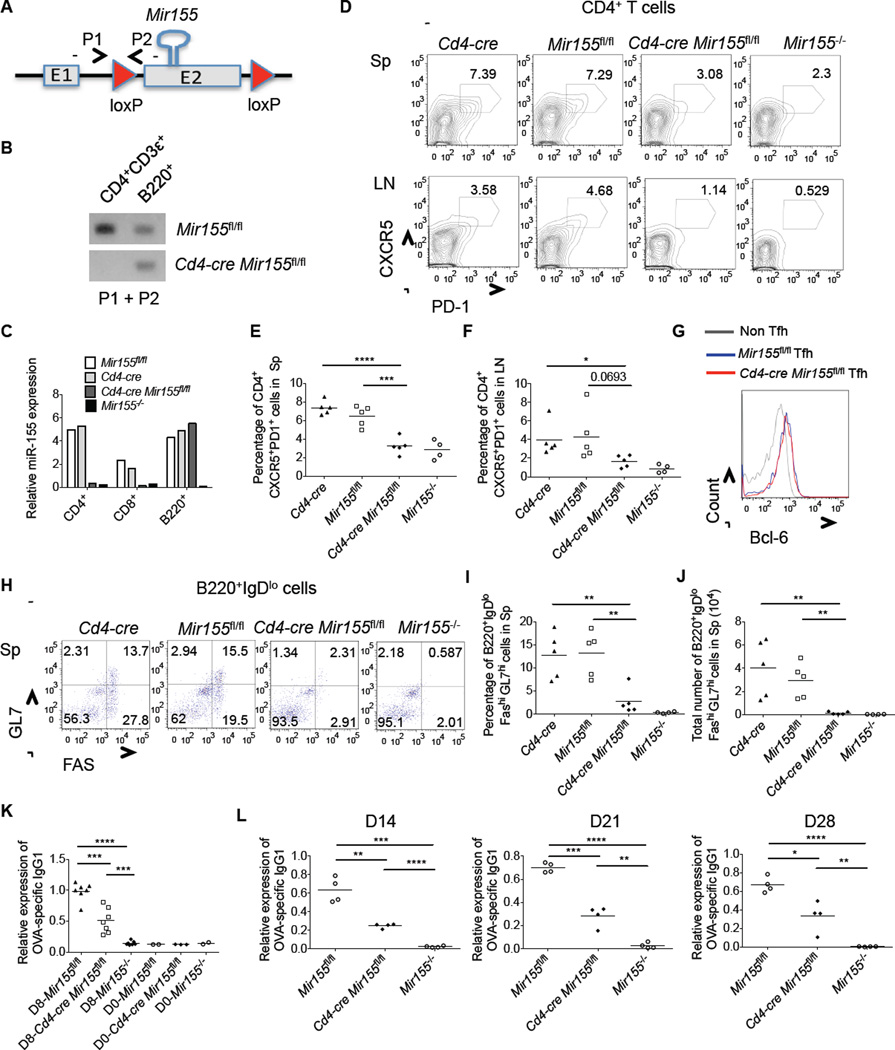
Tfh cell formation involves both cell-intrinsic and -extrinsic mechanisms (Tangye et al., 2013). To determine if miR-155 plays a cell-autonomous role in Tfh cell development, we created T cell-specific miR-155 deficient mice (_Cd4-cre Mir155_fl/fl) where the miR-155 gene is floxed and Cre is driven by the CD4 locus (Figures 4A, 4B, Figure S4 and Table S1). In addition to confirming specific deletion of miR-155 in T cells (Figure 4C and S4), we also determined that _Cd4-cre Mir155_fl/fl mice and controls had similar percentages of CD4+, CD8+ and B220+ cells in the spleens under steady-state conditions (Figure S4). Next, we immunized _Cd4-cre Mir155_fl/fl mice and relevant controls with Ovalbumin (Ova) in complete freund’s adjuvant (CFA). After 8 days we measured the development of Tfh and GC B cells, and found that miR-155 expression was specifically required by T cells for proper development of these cell lineages in the spleen and lymph nodes (Figures 4D–4J and S5). Further, reduced anti-Ova IgG1 serum antibodies were detected in _Cd4-cre Mir155_fl/fl mice at multiple time points post-immunization (Figures 4K, 4L and S5), albeit amounts were not as low as in whole body _Mir155_−/− mice because miR-155 also functions in B cells (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2013). Despite substantial reductions in GC phenotype B cells in the spleens of _Cd4-cre Mir155_fl/fl mice, only partial decreases in antigen specific antibodies were observed, perhaps reflecting contributions or compensation by extra follicular B cells. Together, our data demonstrate that miR-155 has a T cell-intrinsic role during Tfh cell development and the GC response.
Figure 4. T cell-intrinsic role for miR-155 during Tfh cell development and T-dependent humoral responses.
(A) Schematic diagram showing the loxP sites flanking the miR-155 hairpin sequence in miR-155fl/fl mice, and location of genotyping primers. (B) Genomic DNAs were isolated from flow cytometry sorted CD4+CD3ε+ T and B220+ cells from the indicated mouse spleens and the primers that flank the 5’ loxP site were used for PCR. A representative agarose gel is shown to demonstrate T cell specific deletion of miR-155. (C) CD4+ T, CD8+ T and B220+ B cells were isolated from the indicated mouse spleens by flow cytometry sorting and expression of mature miR-155 by each cell type was assayed by QPCR. (D-L) The indicated mice were immunized with Ova in CFA. Following 8 days, spleens and LNs were isolated and subjected to staining with the indicated antibodies and analyzed by flow cytometry. (D) Representative FACS plots showing CD4+CXCR5+PD1+ Tfh cells in the spleens (upper panel) and LNs (lower panel). (E and F) Average percentage of Tfh cells in spleens (E) and LNs (F). (G) Bcl6 expression in Tfh and non-Tfh cells in the spleens of the indicated mice. (H) Flow cytometry plots showing GC B cells in the spleens. (I and J) Average percentage (I) or total number (J) of B220+IgDloFashiGL7hi GC B cells in the spleens. (K and L) Relative serum amounts of Ova-specific IgG1 antibodies in OVA immunized mice from the indicated time points and genotypes (D=Day). Error bars represent +/− SEM. * denotes a p value of <0.05 using a Student’s t test. Data represent at least three independent experiments. See also Figures 4S and 5S.
miR-155 is required for early Tfh cell differentiation by antigen-specific, naïve CD4+ T cells
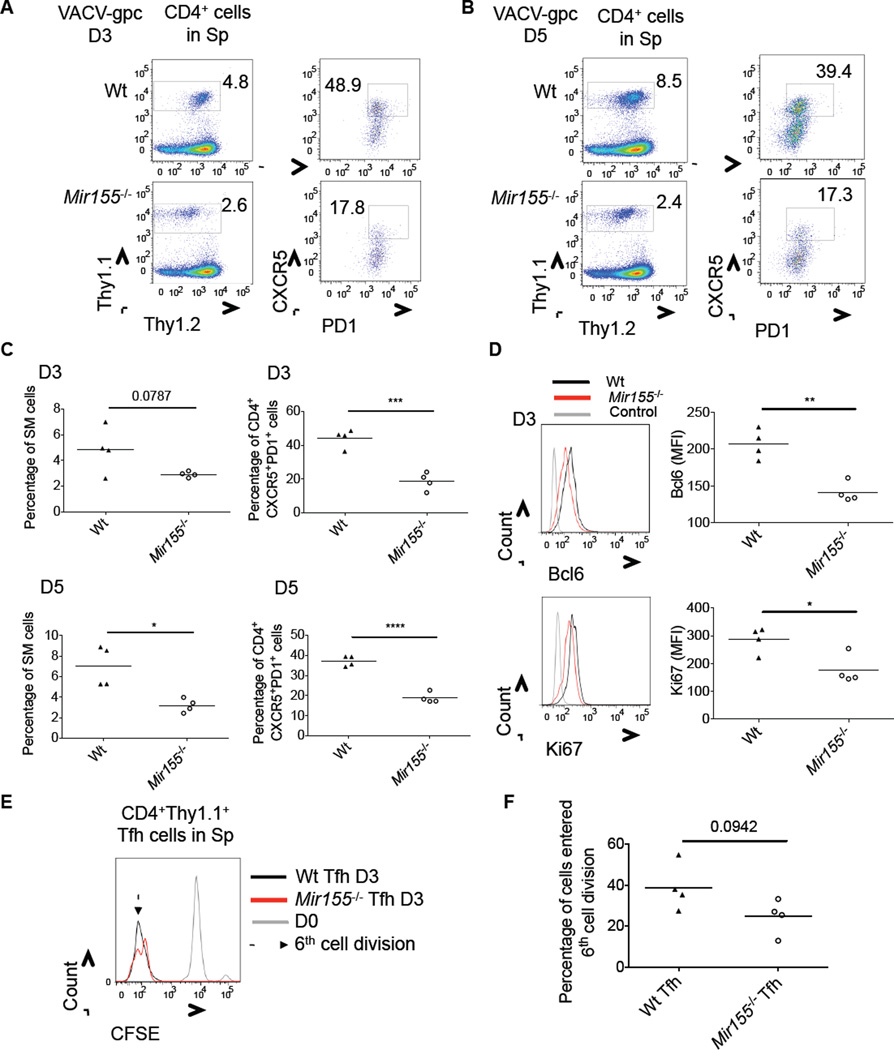
To gain further insight into the role of miR-155 during Tfh cell development, we crossed _Mir155_−/− mice with Smarta (SM) TCR Tg mice. Equal numbers of naïve splenic Wt or _Mir155_−/− SM cells were injected into Wt B6 recipients and mice were infected with Vaccinia virus expressing the lymphocytic choriomeningitis virus (LCMV) glycoprotein (VACV-gpc) recognized by SM T cells. Following 3 or 5 days of infection, _Mir155_−/− SM cells were defective in their production of CXCR5+PD1+ Tfh cells compared to Wt controls at both time points (Figures 5A–C). We also observed defective expression of BCL6 by _Mir155_−/− SM cells at the 3-day time point (Figure 5D, top). _Mir155_−/− SM cells demonstrated modest yet significant reductions in the proliferation marker Ki67 (Figure 5D, bottom), suggesting that reduced proliferation could be contributing to their diminished numbers compared to Wt SM cells. Further, _Mir155_−/− CXCR5+PD1+ SM cells also displayed subtle defects in proliferation as assayed by loss of CFSE, as fewer cells had entered the 6th cell division by 3 days post-infection when compared to Wt controls (Figures 5E and 5F). No difference in SM cell death was observed between the genotypes according to 7-AAD staining, and homeostatic amounts of SM cells were not reduced by a loss in miR-155 in the absence of infection (Figure S5). These data demonstrate that naïve, antigen-specific CD4+ T cells require miR-155 for robust Tfh cell development at early stages during the response, and that miR-155 appears to influence both Tfh cell differentiation and expansion.
Figure 5. Robust Tfh cell differentiation by naïve antigen specific CD4+ T cells in response to VACV-gpc infection requires miR-155.
8×104 (D3) or 2×104 (D5) Wt or _Mir155_−/− SM cells were transferred into B6 Wt mice which were infected with VACV-gpc the following day. Mice were analyzed at D3 or D5 post-infection. (A-B) Representative flow cytometry plots showing the percentage of Wt and _Mir155_−/−CD4+Thy1.1+ SM cells (left panel) and percentage of CXCR5+PD1+ Tfh cells within this population (right panel), in the spleens at the indicated time point. (C) Average percentage of SM T cells, or Tfh cells within this population, in the spleens at the indicated time points post-infection. (D) Representative histogram of Bcl6 or Ki67 staining of Wt or _Mir155_−/− SM cells at the 3-day time point. The average MFI of each stain is shown to the right. (E) Representative histogram showing CFSE expression by Wt or _Mir155_−/− SM Tfh cells at 3 days post infection. (F) The average percentage of Tfh cells having entered the 6th cell division. * denotes a p value of <0.05 using a Student’s t test. Data represent at least two independent experiments. See also Figure 5S.
Identification of miR-155 targets in Tfh cells
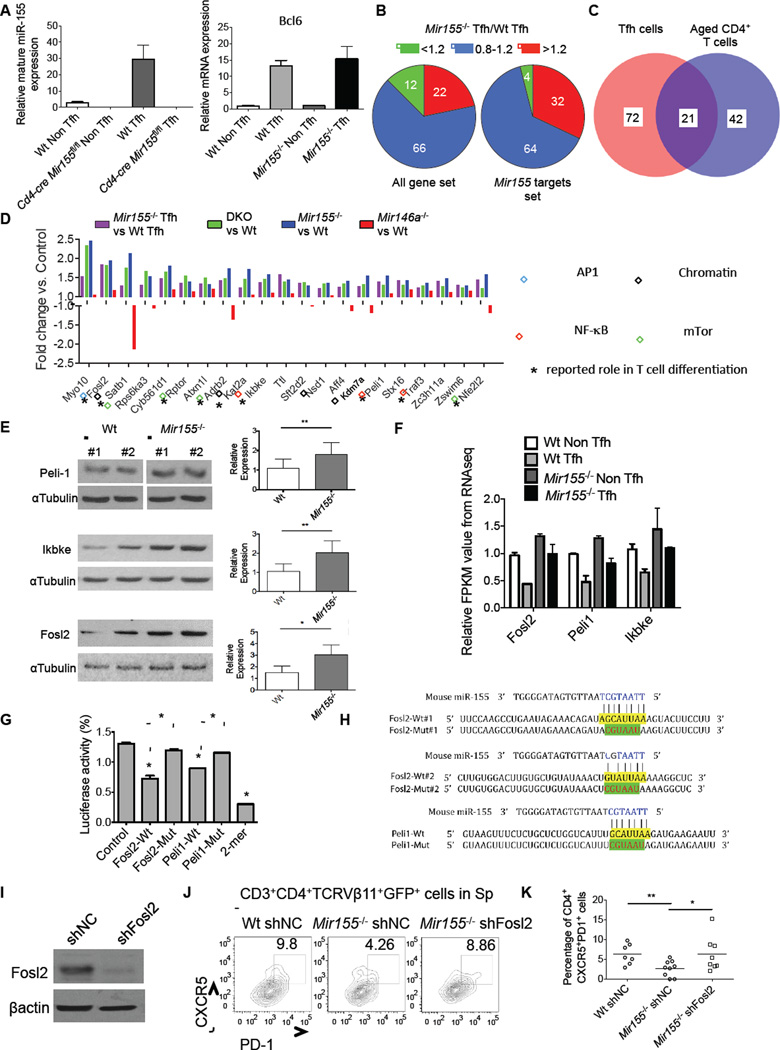
To identify miR-155 target genes involved in Tfh cell development in the context of chronic inflammation we utilized an integrated approach (Figure S6). RNA was extracted from sorted Tfh cells isolated 8 days after immunizing Wt and _Mir155_−/− mice with Ova and expression of Bcl6 was confirmed in sorted Tfh compared to non Tfh cells by QPCR (Figure 6A). The RNA was next subjected to RNA-Seq to profile gene expression in Wt and _Mir155_−/− Tfh cells, and cluster analysis revealed that the two genotypes had disparate profiles (Figure S6). However, expression of several Tfh-related genes was not significantly changed between Wt and _Mir155_−/− Tfh cells on a per cell basis, suggesting that miR-155 regulates the quantity and not quality of Tfh cells (Figure S6). Among miR-155 target mRNAs predicted bioinformatically by Targetscan and determined experimentally using Ago HITS-CLIP (Loeb et al., 2012), we observed a bias towards higher expression of these genes in _Mir155_−/− versus Wt Tfh cells, consistent with miR-155 target genes being de-repressed (Figures 6B and Table S2).
Figure 6. Identification of Tfh cell-relevant miR-155 target genes in CD4+ T cells from middle-aged mice.
(A) QPCR analyses of mature miR-155 and BCL6 mRNA expression in sorted Tfh and non-Tfh cells from the indicated mouse spleens following immunization. (B) Pie charts indicating the percentage of genes that are upregulated (>1.2) or downregulated (<0.8) in _Mir155_−/− vs. Wt Tfh cells based on all genes in the dataset or specifically miR-155 targets genes. (C) Venn diagram showing the number of common miR-155 targets in purified Tfh cells and CD4+ T cells from middle-aged mice. (D) Genes targeted by miR-155 according to the overlap in (C). Bars represent relative fold change of gene expression in the mutant cells compared to Wt controls based on RNA-Seq data. Boxes indicate different pathways in which these genes function, and * denotes that a role for the gene has been described in T cell differentiation. (E) Representative Western blot analysis of Peli1, Ikbke, Fosl2 and αTubulin expression in activated Wt and _Mir155_−/− CD4+ T cells (left). Quantification of Peli1 (n=9), Ikbke (n=6) and Fosl2 (n=4) expression in T cells from multiple mice normalized to αTubulin (right). (F) Relative expression of Fosl2, Peli1 and Ikbke in sorted CD4+ non Tfh and Tfh cells from OVA immunized Wt and _Mir155_−/− mice based on RNA-Seq. (G) Luciferase reporter assays demonstrating direct repression of the 3’ UTRs by miR-155. For Fosl2, the mutant 3’ UTR has both sites disrupted. (H) Schematic layout of the 3’ UTR sequence of mouse Fosl2 and Peli1 and the seed region (yellow) where miR-155 is predicted to bind. Mutant sequence is shown in green. (I) Representative Western blot analysis of Fosl2 expression in activated Wt CD4+ T cells after silencing with a shFosl2 expressing retroviral vector. (J) Representative flow cytometry plots showing the percentage of CXCR5+PD1+ Tfh cells among CD3+CD4+Vβ11+GFP+ cells in the spleens following 7 days of immunization with MOG35–55. (K) Average percentage of Tfh cells among CD3+CD4+Vβ11+GFP+ cells from (I). Error bars represent +/− SEM. * denotes a p value of <0.05 using a Student’s t test. See also Figure 6S.
To identify candidate miR-155 target genes involved in Tfh cell formation during chronic inflammation we determined which miR-155 targets were 1) elevated in sorted _Mir155_−/− Tfh cells from immunized mice (Table S2), 2) elevated in middle-aged _Mir155_−/− and _Mir155_−/− _Mir146a_−/− CD4+ T cells (Table S3), and 3) unchanged or repressed in aged _Mir146a_−/− CD4+ T cells. Using this stringent approach, we identified 21 candidate miR-155 targets putatively involved in the development of Tfh cells in _Mir146a_−/− mice (Figures 6C and 6D). We confirmed some of these by QPCR (Figure S6), and further validated higher expression of Peli1, Ikbke and Fosl2 at the protein level in _Mir155_−/−CD4+ T cells compared to Wt controls (Figure 6E). We also found that expression of Peli1, Fosl2 and Ikbke was lower in Wt Tfh compared to non Tfh cells taken from immunized mice, while their expression values in _Mir155_−/− Tfh cells were similar to Wt non Tfh cells and well above amounts in Wt Tfh cells (Figure 6F). To confirm direct targeting of Peli1 and Fosl2, both genes that regulate T cell differentiation, we cloned their 3’ UTRs downstream from luciferase. Luciferase assays confirmed that miR-155 directly repressed protein expression through its binding sites in these 3’ UTRs, as repression was observed with Wt 3’ UTRs but not when miR-155 binding sites were mutated (Figures 6G and 6H).
Pathway analyses indicated that a subset of these targets regulate the NF-kB (Ikbke and Peli1) and AP-1 (Fosl2) pathways, both involved in Tfh cell development and autoimmunity (Chang et al., 2011; Clark et al., 2011) (Figure 6D). Another subset of genes repressed by miR-155 has been shown to regulate the mTOR pathway (including Rptor, Rps6ka3, Adrb2, Ikbke and Nfe2l2), and there were multiple target genes involved in chromatin modifications (Satb1, Kat2a, Kdm7a and Nsd1) (Figure 6D). Many of these targets have been shown to influence T helper cell development (Figure 6D and S6).
shRNA silencing of Fosl2 in adoptively transferred _Mir155_−/− 2D2 TCR-transgenic CD4+ T cells, which are TCRVβ11+, resulted in improved Tfh cell development following immunization with MOG35–55 (Figure 6I–6K). Silencing of Peli1 also trended towards rescuing the phenotype, supporting our view that multiple miR-155 targets are likely involved in this process (Figure S6). Taken together, our findings suggest that miR-155 facilitates Tfh cell accumulation during chronic, low-grade inflammation through a complex mechanism involving multiple target genes and signaling pathways that instruct early Tfh cell formation.
T cell-specific expression of miR-155 drives spontaneous Tfh and GC B cell development in _Mir146a_−/− mice
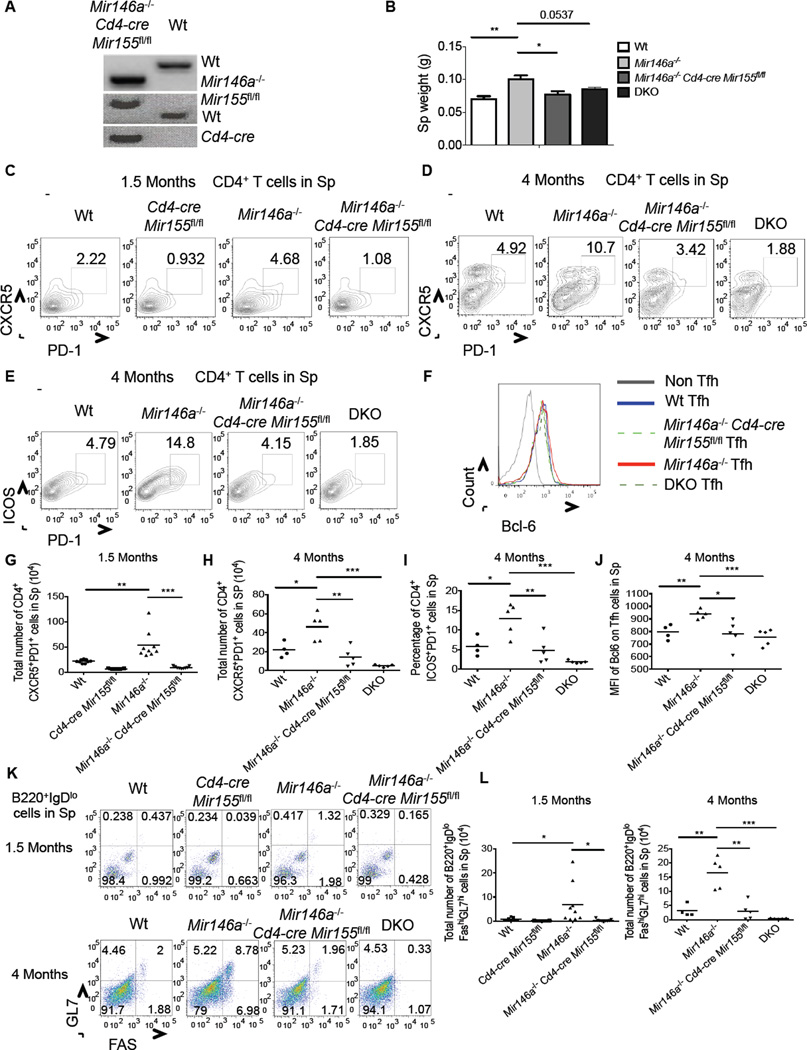
To determine the role of T cell-intrinsic miR-155 during the early stages of chronic, low-grade inflammation, when expanded Tfh cell populations are first observed, we crossed _Mir146a_−/− mice with _Cd4-cre Mir155_fl/fl animals (Figure 7A). By 4 months of age we began to observe mild splenomegaly in _Mir146a_−/− mice, which was not seen in _Mir146a_−/− Cd4-cre Mir155fl/fl mice (Figure 7B). Upon further analyzing 1.5 and 4 months old _Mir146a_−/− Cd4-cre Mir155 fl/fl mice we detected reduced numbers of Tfh cells (Figures 7C–7J and S7) and GC B cells (Figures 7K, 7L and S7) compared to the numbers found in age-matched _Mir146a_−/− mice. Further, at the 4 month time point we also noticed that early signs of the myeloproliferative disease that emerged as _Mir146a_−/− mice grew older were observed in _Mir146a_−/− but not _Mir155_−/− _Mir146a_−/− or _Mir146a_−/−_Cd4-cre Mir155_fl/fl mice (Figure S7). These included elevated CD11b+ and decreased Ter119+ cells in the bone marrow. Overall, these findings demonstrate that miR-155 plays a T cell intrinsic role in promoting spontaneous germinal center reactions in _Mir146a_−/− mice.
Figure 7. T cell-intrinsic miR-155 is required for Tfh cell expansion in _Mir146a_−/− mice.
(A) Genotyping gel confirming the creation of _Mir146a_−/− CD4-Cre: miR-155fl/fl mice. (B) Spleen weights from 4 month old mice of the indicated genotypes. (C) Flow cytomtery plots showing CD3+CD4+CXCR5+PD1+ Tfh cells in the spleens of 1.5-month or (D) 4-month old mice. (E) Representative flow cytomtery plots of CD3+CD4+ICOS+PD1+ Tfh cells from 4-month old spleens (F) Histogram showing Bcl6 expression by Tfh and non-Tfh cells in the spleens of the indicated genotypes at 4 months of age. (G-J) Average values for multiple mice from (C-F). (K) Flow cytometry plots showing B220+IgDloFashiGL7hi GC B cells in the spleens of the indicated mice at the 1.5 and 4 month time points. (L) Average total number of GC B cells in the spleens from (K). Error bars represent +/− SEM. * denotes a p value of <0.05 using a Student’s t test. Data represent two independent experiments. See also Figure 7S.
Discussion
The humoral response gradually loses its potency against novel, exogenous antigens and begins to initiate responses against self-tissues as a function of age (Dorshkind et al., 2009; Linterman, 2014). Our study indicates that cells with a Tfh cell phenotype may be involved in this age-dependent conversion, as both their numbers and downstream effects (e.g. increases in GC B cells) increase before the onset of autoantibody production in _Mir146a_−/− mice. This concept is also in accordance with clinical studies that report increases in the Tfh cell growth factors IL-6 (Akbaraly et al., 2013) and IL-21 (Agrawal et al., 2012), memory phenotype T cells (Moro-Garcia et al., 2013), autoantibodies (Nagele et al., 2013) and autoimmune disease (Yung and Julius, 2008) in some older versus younger individuals. During future studies, it will be important to assess both the quality and quantity of Tfh cell populations as a function of age in human tissues, and to determine if different features of chronic, low-grade inflammation in human populations involves aberrations to this cellular population.
Mechanistically, our results identify opposing roles for miRNAs in controlling progressive, spontaneous Tfh cell expansion, where miR-146a restricts and miR-155 promotes this program in mice. This suggests that factors controlling the ratio of miR-155:miR-146a can influence this process. Both of these miRNAs are transcriptionally induced by inflammatory stimuli or T cell receptor (TCR) engagement (Haasch et al., 2002; Yang et al., 2012). Further, genetic variants in the miR-155 gene locus has been linked to autoimmune disease (Paraboschi et al., 2011), while specific polymorphisms in the passenger strand of miR-146a lead to reduced production of mature miR-146a (Jazdzewski et al., 2008). These observations indicate that both genetic and environmental factors are involved in controlling the concentrations of these miRNAs and thus their abilities to regulate humoral responses during the aging process. Furthermore, while we found a variety of age-related inflammatory phenotypes in _Mir146a_−/− mice to involve miR-155 through the use of whole body _Mir155_−/− _Mir146a_−/−mice, and focused on miR-155’s T cell-intrinsic role in promoting GC reactions in this setting, future investigation is needed to determine if miR-155 functions in either Tfh or non-Tfh cell types to promote other aspects of the disease that emerge in this model. It is also plausible that other miR-146a–dependent phenotypes are independent of miR-155.
In addition to its well-established function in B cells during Ig class-switching and affinity hyper-mutation (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007), our data identify a previously unappreciated role for miR-155 in the CD4+ T cells as they provide help to B cells during the germinal center reaction. In particular, we describe a decreased capacity by _Mir155_−/− CD4+ T cells to develop into the Tfh cell lineage following immunization, viral infection or during age-related inflammatory disease. Because we observe decreased Tfh cell numbers, while our expression analysis indicates that effector function may be intact on a per cell basis, it is possible that miR-155 is involved in Tfh cell differentiation and expansion as opposed to their functions once mature. Our findings also indicate that multiple miRNAs are involved in regulating Tfh cell biology, as recent studies have described roles for the miRNAs 17~92 family (Baumjohann et al., 2013; Kang et al., 2013) and miR-10a (Takahashi et al., 2012) during Tfh cell formation.
We identified 21 direct miR-155 targets in Tfh cells that regulate critical signaling pathways including NF-κB, AP-1 and mTor, in addition to several genes that regulate chromatin modifications. Consistent with many previous studies (Hu et al., 2013; Huffaker et al., 2012; Loeb et al., 2012), our results continue to support a model whereby miR-155 regulates T cell biology through a multi-target mechanism that enables development of different T effector cell subsets in distinct contexts. However, it remains unclear if miR-155 targets unique sets of genes to regulate the distinct effector T cell lineages that it has been linked to, including regulatory T (Treg) cells (Lu et al., 2009), Th17 cells (Kurowska-Stolarska et al., 2011; O'Connell et al., 2010b), Th1 cells (Oertli et al., 2011), Th2 cells (Malmhall et al., 2013), and now Tfh cells, or if there is a core “targetome” that is commonly required to license the formation of these subtypes. This will be an important area of future research that will require target identification in multiple T cell types in parallel using the same technology.
Our data also provide evidence that Fosl2, and to some extent Peli1, are functionally relevant miR-155 targets. Fosl2 is a repressor of CD4+ T cell plasticity (Ciofani et al., 2012) that binds to Jun proteins and is thought to compete with Batf for DNA binding sites. Batf-containing AP-1 complexes bind cooperatively with IRF4 to defined DNA elements called AP-1-IRF composite elements (AICEs) (Glasmacher et al., 2012), and both of these factors are necessary for Tfh cell development (Betz et al., 2010; Bollig et al., 2012). However, Fosl2 containing complexes are unable to recruit IRF4 once bound to these sites (Glasmacher et al., 2012), pointing to a mechanistic basis for its repressive role during T cell differentiation. Thus, because Fosl2 is at higher amounts in _Mir155_−/− CD4+ T cells, our data suggest that it is able to hinder Tfh cell development by interfering with normal Batf and IRF4 functions. In the case of Peli1, it has been shown to inhibit NF-kB activation, which is also involved in the induction of Tfh cell-associated genes (Chang et al., 2011; Chen et al., 2010). Future work will investigate these connections further, and determine their relevance in other Th cell lineages, including Th17 and Treg cells where miR-155 and Fosl2 have been shown to have opposing function.
Finally, our observations have many translational implications with relevance to human disease. First, the relative expression of miR-155 and miR-146a in middle-aged individuals might have predictive, diagnostic or prognostic value in the context of chronic, low-grade inflammation. Next, therapeutic targeting of miR-155 or miR-146a with antisense oligonucleotides in patients with chronic, low-grade inflammation could be an effective strategy to reduce certain disorders that stem from aberrant humoral responses. Conversely, from a vaccine development standpoint, these findings indicate that manipulation of miR-155 in both T and B cells might have a synergistic effect on the production of high affinity, class switched antibodies that can be induced via immunization to target tumor cells or pathogenic microbes.
Experimental Procedures
Mice
All experiments were approved by the University of Utah Institutional Animal Care and Use Committee (IACUC). _Mir155_−/− _Mir146a_−/− (DKO) mice were generated as previously described (Huffaker et al., 2012). _Mir155_fl/fl mice were generated at Taconic and crossed with Cd4-cre mice to create Cd4-cre Mir155 fl/fl mice. Additionally, Mir146a −/− mice were crossed with _Cd4-cre Mir155_fl/fl mice to generate _Mir146a_−/− Cd4-cre Mir155fl/fl mice, and Wt SMARTA TCR Tg+ mice were crossed with Mir155−/− mice to generate _Mir155_−/− SMARTA TCR Tg+. _Mir155_−/− 2d2 TCR Tg+ mice were described previously (Hu et al., 2013).
Flow Cytometry
Fluorophor-conjugated antibodies against the indicated surface markers were used to stain RBC-depleted splenocytes, LN cells, BM cells and peripheral blood cells. For intracellular staining, cells were first surface stained for lineage markers, fixed, permeabilized and stained with antibodies against Bcl6, IL-17A, IFNγ, or Ki67 (Biolegend). Stained cells were analyzed using a BD LSR Fortessa flow cytometer, and further data analysis was carried out using FlowJo software.
Bone marrow reconstitution
RBC-depleted bone marrow cells from Wt mice expressing the congenic marker CD45.1 were mixed with bone marrow from Wt, miR155−/−, _Mir146a_−/−, or _Mir155−/−Mir146a_−/− mice expressing CD45.2 in equal proportions and injected into lethally irradiated (1050 Rads) Wt mice expressing CD45.1, similar to (O'Connell et al., 2010a).
Adoptive transfer of SM T cells and VACV-gpc infection
Untouched naive (Thy1.1+CD44lo) CD4+ T cells were isolated from Wt or Mir155−/− SMARTA mice (SM T cells) using magnetic beads, per the manufacturer’s instructions (Stem cell technology), and injected intravenously into C57BL/6 mice. The next day mice were infected with recombinant Vaccinia virus that expresses the LCMV glycoprotein (referred to as VACV-gpc). For CFSE labeling, T cells were incubated with 0.625 µM CFSE (final concentration) for 10 minutes and washed with FBS before adoptive transfer. 7-AAD staining was used to determine viability.
Adoptive transfer of shRNA expressing 2d2 TCR Tg CD4+ T cells and MOG35–55 Immunization
Untouched naive CD44lo CD4+ T cells were injected intravenously into B6 mice following spin infection with control or shRNA retroviral vectors, as described (Hu et al., 2013). The sequences targeting Fosl2 and Peli1 are shown in Table S4. The next day the mice were immunized with MOG35–55 (0.5 mg/ml) emulsified in complete Freund's adjuvant (CFA) at the base of the tail (200 µl each mouse), as described (Hu et al., 2013).
ELISAs
Titers of autoantibodies against dsDNA in the serum of aged mice were measured using a commercial ELISA test (BioVendor) according to the manufacturer’s protocol. Serum from Ova-immunized mice (0.5 mg/ml emulsified in complete Freund's adjuvant) was also collected, and Ova antigen-specific IgG and IgG1 antibodies were measured by ELISA, as described (O'Connell et al., 2010b).
QPCR
Sybrgreen-based quantitative real-time PCR (QPCR) was conducted to assay relative mRNA amounts using the Light Cycler 480 PCR system (Roche) and gene-specific primers (Table S4). For mature miR-155 and miR-146a expression analyses, gene-specific primers were purchased from Exiqon. 5S or L32 were used to normalize.
RNA Sequencing
For both experiments, total RNA was isolated using the miRNeasy kit (Qiagen). Stranded RNA sequencing (following RiboZero treatment and library preparation) was conducted using Illumina HiSeq 2000 Sequencing and carried out by the University of Utah core facility (https://bioserver.hci.utah.edu/microarrayweb/ordering.html). The analysis approach is described in our supplemental methods. All RNA Seq data has been deposited into the NCBI GEO database under the accession number GSE58373.
Immunoblotting
Cell extracts were subjected to gel electrophoresis and transferred onto a nitrocellulose membrane followed by antibody staining and detection of Peli1, Ikbke, Fosl2, βActin or αTubulin, as described (Hu et al., 2013).
Luciferase Assay
The 3’ UTR regions of mouse Fosl2 and Peli1 that contain the miR-155 binding sites, or mutant versions, were synthesized by GeneArt technology (Life Technologies) and cloned into pMiR reporter plasmid. Experiments were performed using 293T cells, as described (Hu et al., 2013).
Histological analyses
Tissue preparation and H&E staining were performed as described previously (O'Connell et al., 2008). IHC was performed with antibodies against B220, CD3, and BCl6 or PNA.
Statistical Analysis
Statistical significance was determined by performing an unpaired t test using Graphpad Prism. All quantitative data are reported as mean ± SEM or mean. Significance is denoted as *** P ≤0.001, ** P ≤0.01, * P ≤0.05, and ns P>0.05.
Supplementary Material
1
2
Acknowledgements
We would like to thank the University of Utah Gene Expression and Bioinformatics core facilities for help with RNA-Seq and data analysis. We also thank the University of Utah Flow Cytometry core facility for assistance with cell sorting. This work was supported by the NIH New Innovator Award DP2GM111099-01 (RMO), the NHLBI Pathway to Independence Award R00HL102228-05 (RMO), an American Cancer Society Research Grant (RMO), the Edward Mallinckrodt Jr. Foundation (JLR), Pew Scholars Program (JLR), NSF CAREER award IOS-1253278 (JLR), Packard Fellowship in Science and Engineering (JLR), NIAID K22 AI95375 (JLR), NIAID AI107090 (JLR), the NIH training grant 5T32DK007115-39 (DAK), R03NS070141 (GAG and TM) and R01CA166450-02 (DSR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
R.H. and R.M.O. designed the study. R.H. carried out all experimental work with assistance from D.A.K., T.B.H., M.C.R, J.L., MA. and E.B. and guidance from R.M.O., M.A.W. and J.L.R. W.S., T.M. and G.A.G. contributed to the miR-155fl/fl mice. D.S.R. performed histology analyses. R.H. and R.M.O. wrote the manuscript with contributions from all authors.
Competing financial interests
The authors declare no competing financial interests.
References
- Agrawal A, Su H, Chen J, Osann K, Agrawal S, Gupta S. Increased IL-21 secretion by aged CD4+T cells is associated with prolonged STAT-4 activation and CMV seropositivity. Aging (Albany NY) 2012;4:648–659. doi: 10.18632/aging.100490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbaraly TN, Hamer M, Ferrie JE, Lowe G, Batty GD, Hagger-Johnson G, Singh-Manoux A, Shipley MJ, Kavimaki M. Chronic inflammation as a determinant of future aging phenotypes. CMAJ : Canadian Medical Association journal = journal de l'Association medicale canadienne. 2013 doi: 10.1503/cmaj.122072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. The microRNA cluster miR-17 approximately 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840–848. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, Taparowsky EJ. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig N, Brustle A, Kellner K, Ackermann W, Abass E, Raifer H, Camara B, Brendel C, Giel G, Bothur E, et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci U S A. 2012;109:8664–8669. doi: 10.1073/pnas.1205834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Jin W, Chang JH, Xiao Y, Brittain GC, Yu J, Zhou X, Wang YH, Cheng X, Li P, et al. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat Immunol. 2011;12:1002–1009. doi: 10.1038/ni.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hardy K, Bunting K, Daley S, Ma L, Shannon MF. Regulation of the IL-21 gene by the NF-kappaB transcription factor c-Rel. J Immunol. 2010;185:2350–2359. doi: 10.4049/jimmunol.1000317. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM, Aleksiyadis K, Martin A, McNamee K, Tharmalingam T, Williams RO, Memet S, Cope AP. Inhibitor of kappa B epsilon (IkappaBepsilon) is a non-redundant regulator of c-Rel-dependent gene expression in murine T and B cells. PLoS One. 2011;6:e24504. doi: 10.1371/journal.pone.0024504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshkind K, Montecino-Rodriguez E, Signer RA. The ageing immune system: is it ever too old to become young again? Nat Rev Immunol. 2009;9:57–62. doi: 10.1038/nri2471. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cellular immunology. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- Howcroft TK, Campisi J, Louis GB, Smith MT, Wise B, Wyss-Coray T, Augustine AD, McElhaney JE, Kohanski R, Sierra F. The role of inflammation in age-related disease. Aging (Albany NY) 2013;5:84–93. doi: 10.18632/aging.100531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, Round JL, O’Connell RM. MicroRNA-155 Confers Encephalogenic Potential to Th17 Cells by Promoting Effector Gene Expression. J Immunol. 2013;190:5972–5980. doi: 10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O’Connell RM. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2:1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Liu WH, Lu P, Jin HY, Lim HW, Shepherd J, Fremgen D, Verdin E, Oldstone MB, Qi H, et al. MicroRNAs of the miR-17 approximately 92 family are critical regulators of T(FH) differentiation. Nat Immunol. 2013;14:849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA. How T follicular helper cells and the germinal centre response change with age. Immunol Cell Biol. 2014;92:72–79. doi: 10.1038/icb.2013.77. [DOI] [PubMed] [Google Scholar]
- Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Molecular cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmhall C, Alawieh S, Lu Y, Sjostrand M, Bossios A, Eldh M, Radinger M. MicroRNA-155 is essential for T2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2013 doi: 10.1016/j.jaci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Balardy L, Moulis G, Gaudin C, Peyrot C, Vellas B, Cesari M, Nourhashemi F. Proinflammatory Cytokines, Aging, and Age-Related Diseases. Journal of the American Medical Directors Association. 2013 doi: 10.1016/j.jamda.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Moro-Garcia MA, Alonso-Arias R, Lopez-Larrea C. When Aging Reaches CD4+ T-Cells: Phenotypic and Functional Changes. Front Immunol. 2013;4:107. doi: 10.3389/fimmu.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele EP, Han M, Acharya NK, DeMarshall C, Kosciuk MC, Nagele RG. Natural IgG autoantibodies are abundant and ubiquitous in human sera, and their number is influenced by age, gender, and disease. PLoS One. 2013;8:e60726. doi: 10.1371/journal.pone.0060726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010a;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010b;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Muller A. MicroRNA-155 Is Essential for the T Cell-Mediated Control of Helicobacter pylori Infection and for the Induction of Chronic Gastritis and Colitis. J Immunol. 2011 doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- Paraboschi EM, Solda G, Gemmati D, Orioli E, Zeri G, Benedetti MD, Salviati A, Barizzone N, Leone M, Duga S, Asselta R. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci. 2011;12:8695–8712. doi: 10.3390/ijms12128695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O’Shea JJ. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye SG, Ma CS, Brink R, Deenick EK. The good, the bad and the ugly - TFH cells in human health and disease. Nat Rev Immunol. 2013;13:412–426. doi: 10.1038/nri3447. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012 doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung RL, Julius A. Epigenetics, aging, and autoimmunity. Autoimmunity. 2008;41:329–335. doi: 10.1080/08916930802024889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL, Rao DS, Boldin MP, Taganov KD, O’Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a–deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL, Rao DS, O’Connell RM, Garcia-Flores Y, Baltimore D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. eLife. 2013;2:e00537. doi: 10.7554/eLife.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2