Two Acquired Immunodeficiency Syndrome-Associated Burkitt’s Lymphomas Produce Specific Anti-i IgM Cold Agglutinins Using Somatically Mutated VH4-21 Segments (original) (raw)
. Author manuscript; available in PMC: 2015 Dec 2.
Published in final edited form as: Blood. 1994 May 15;83(10):2952–2961.
Abstract
We analyzed the reactivity and the structure of the VH and VL segments of two IgM monoclonal antibodies (MoAbs) produced by spontaneously in vitro outgrowing cell lines, HBL-2 and HBL-3, established from two acquired immunodeficiency syndrome (AIDS) patients with Epstein-Barr virus (EBV)-negative Burkitt’s lymphoma (BL). These B-cell clones were representative of the respective neoplastic parental clones, as determined by immunophenotypic and molecular genetic analysis. The IgM MoAbs were highly specific for the i determinant on red blood cells (cold agglutinins), but bound none of the other eight self and nine foreign antigens (Ags) tested, including those most commonly recognized by natural antibodies or autoantibodies. Structural analysis showed that the IgM MoAb VH segment sequences were 93.5% and 84.2% identical with that of the germline VH4-21 gene, which encodes the vast majority of cold agglutinins that are specific for the i/I carbohydrate Ag and are produced under chronic lymphoproliferative conditions. The HBL-2 MoAb VH4-21 gene segment was juxtaposed with 20P3 and JH6 genes and paired with a Vλ1 segment, the sequence of which was 95.5% identical to that of the germline Humlv117 gene; the HBL-3 MoAb VH4-21 gene segment was juxtaposed with DXP′1 and JH5 genes and paired with a Vλ1 segment, the sequence of which was 86.7% identical to that of the germline Humlv1L1 gene. The high degree of conservation of the VH4-21 gene in the human population, the nature of the nucleotide differences in the expressed VH4-21 segments, and the presence of nucleotide substitutions in the HBL-2 and HBL-3 IgM MoAb JH and/or Jλ segments suggested that the MoAb V segments underwent a process of somatic hypermutation. This was formally shown in the HBL-3 MoAb VHsegment, by differentially targeted polymerase chain reaction amplification of the HBL-3 MoAb-producing cell genomic DNA. In addition, cloning and sequencing of the genomic DNA from fibroblasts of the same patient whose neoplastic B cells gave rise to the HBL-3 cell line yielded a germline copy of the VH4-21 gene. Thus, the expression of VH4-21 gene products may be involved in a self Ag-driven process of clonal B-cell expansion and selection associated with BL in these AIDS patients.
Autoimmune phenomena can occur in association with several human B-cell disorders, such as cold agglutinin disease,1,2 lymphoma,3–6 and the B-cell expansion and hypergammaglobulinemia occurring in human immunodeficiency virus (HIV)-infected patients.7–9 Although the precise role of different self antigens (Ags) in the B-cell clonal selection associated with the above pathologic conditions remains to be defined, circumstantial evidence for a role of self Ags in clonal expansion and selection in autoimmune humans and mice has been provided.10–16 The crucial role of Ags in inducing clonal expansion and selection in the normal B-cell repertoire is well documented.17–21 Recently, it has been suggested that Ag stimulation also plays a role in the B-cell expansion and selection preceding and/or associated with development of lymphomas of various histologic types.5,6,22–24
The assessment of a potential role for Ags in the clonal B-cell expansion and selection associated with lymphoma or leukemia entails, first of all, the definition of the specificity and of VH and VL segment structure of the tumor-derived antibody. The analysis of antibody specificity and VH and VL segment structure depends on the availability of a homogeneous in vitro growing and Ig-producing tumor cell population representative of the in vivo neoplastic clone. This is a critical requirement in view of the findings of oligoclonal or polyclonal, not necessarily neoplastic, B-cell populations accompanying the predominant neoplastic clone, as found particularly in bioptic specimens of Burkitt’s lymphoma (BL) emerging in patients with acquired immunodeficiency syndrome (AIDS).25,26 Possibly due to the improved treatment and longer survival rate, these patients display a 60-fold increased incidence, relative to that expected for the general population, of lymphomas, mainly of the Burkitt’s type.27
We analyzed the Ag reactivity and the structure of the VH and VL segments of the IgM monoclonal antibodies (MoAbs) produced by two spontaneously in vitro outgrowing cell lines established from 2 AIDS patients with Epstein-Barr virus (EBV)-negative BL.28 The absolute identity between the in vitro growing monoclonal cell lines and their respective in vivo neoplastic clones was established by immunophenotypic and molecular genetic analysis. The IgM MoAbs from both cell lines strongly bound to the i Ag on red blood cells (RBCs; cold agglutinins), but to none of the other self and foreign Ags tested. The structural correlate for such Ag-binding specificity was provided by segments encoded by VH4-21 and Vλ1 genes in somatically mutated configuration. Thus, a process of selection by the i self Ags may have played a role in the B-cell clonal expansion preceding and/or associated with the development of BL in these AIDS patients.
MATERIALS AND METHODS
Generation and characterization of the monoclonal AIDS BL cell lines
The HBL-2 and HBL-3 MoAb-producing cell lines were established using B lymphocytes spontaneously outgrowing from two tumors histologically and immunophenotypically classified as small noncleaved cell lymphoma (SNCCL).28 Both SNCCL arose in patients with AIDS. Immunophenotypic analysis was performed by fluorescence flow cytometry of isolated cells using a FACScan (Becton Dickinson Corp, Mountain View, CA) and a panel of labeled murine MoAbs, including those to CD19, HLA-DR, CD10, CD21, and CD5.28 The clonality of the cell lines and their absolute relatedness to the tumors were determined by Ig gene rearrangement analysis using a JH probe on _Hind_III, _Eco_RI, and _Bam_HI DNA digests.29 The c-myc translocations were detected by cytogenetic analysis.28 The status of the c-myc locus was analyzed by hybridization of_Eco_RI- and _Hind_III-digested DNA to the human c-myc probe MC413RC, representative of the third exon of the c-myc gene.30 The presence of the EBV genome was investigated using a probe specific for the EBV genomic termini (5.2-kb_Bam_HI-_Eco_RI fragment isolated from the fused _Bam_HI terminal fragment NJ-het).28 The presence of HIV sequences was investigated using the λ7A/2 probe on Hind_III and_Sac I DNA digests, and that of HTLV-I sequences was investigated using an HTLV-env probe on Bam_HI and_Pst I DNA digests.28
Analysis of the AIDS BL cell line-derived MoAb
The IgM MoAbs produced by the HBL-2 and HBL-3 cell lines were analyzed for their binding to polyclonal human IgG Fc fragment (Organon Teknika-Cappel, Malvern, PA); calf thymus DNA (Sigma Chemical Co, St Louis, MO); insulin (Sigma); human recombinant tumor necrosis factor-α(TNF-α), TNF-β, and interleukin-1_β_ (IL-1_β_; BASF Biotech Corp, Cambridge, MA); human thyroglobulin; HIV-1 and cytomegalovirus (CMV) and parvovirus B19 recombinant glycoproteins; lipopolysaccharide (LPS) and β_-galactosidase from_Escherichia coli (Sigma); phosphorylcholine chloride (Sigma); Pneumococcus polysaccharides, including types 1, 3, and 4; and tetanus toxoid (Massachusetts Public Health Biological Laboratories, Jamaica Plain, MA), using specific enzyme-linked immunosorbent assay (ELISA) involving plates coated with 1 to 5 _μ_g/mL of these Ags.31–34 The IgM MoAb specific i/I blood group cold agglutinin activity was tested by hemagglutination using papain-treated and untreated group 0+ umbilical cord or adult human erythrocytes in a 100 _μ_L reaction volume.35 Cold agglutinating titers were expressed as the smallest amount (nanograms) of IgM MoAb agglutinating 107 papain-treated human RBCs. Finally, the presence of the cross-reacting 9G4 idiotype on the IgM HBL-2 and HBL-3 MoAbs was analyzed using the appropriate IgG2a rat antibody.1,2,36
Cloning and sequencing of the MoAb VH and Vλ genes
Poly(A)+ RNA was isolated from the HBL-2 and HBL-3 MoAb-producing B-cell lines using the Micro Fast-Track mRNA isolation kit (Invitrogen Corp, San Diego, CA) and reverse-transcribed using Moloney murine leukemia virus (MMLV) reverse transcriptase (SuperScript RNase H-Reverse Transcriptase; GIBCO BRL Life Technologies, Gaithersburg, MD). cDNA (100 ng) was submitted to polymerase chain reaction (PCR) in a volume of 50_μ_L containing 200 _μ_mol/L of each dNTP, 2.5 U of Taq polymerase (Perkin Elmer Corp, Norwalk, CT), and 10 pmol of each primer. The following sense degenerated primers encompassing a portion of the leader region of different VHfamilies plus an Eco_RI site were used: ALT 1 (VHI family) [5′ GGGAATTCATGGACTGGACTGGACCTGGAGG(AG)TC(CT)TCT(GT)C 3′]; ALT2 (VHIII family) [5′ GGGAATTCATGGAG(CT)TTGGGCTGA(CG)CTGG(CG)TTT(CT)T 3′]; ALT4 (VHIV family) [5′ GGGAATTCATGAA(AG)CA(TC)CTGTGGTTCTT(CT)(AC)T(CT)CT(CG)C 3′]; and HI-3, an oligonucleotide primer encompassing a portion of the leader sequence of VHIII family, [5′ TTGGGCTGTGCTGGGTTTTCCT 3′]. The antisense primer consisted of the reverse complement [5′ CCGAATTCAGCCGAGGGGGAAAAGGGTTT 3′] of a 21 nucleotide 5′ C_μ sequence plus an_Eco_RI site. The oligonucleotide primers specific for the 5′ portions of the Vλ chains were as follows: VλI[5′ ATG(GA)CC(TG)GCT(CT)CCCTCTCCTCCT 3′], and VλII–VI (VλII, VλIII, VλIV, VλVI chains) [5′ ATG(AG)C(CT)TGGACCC(CT)(AT)CTC(CT)(TG)(TG)TT 3′]. The antisense Cλ chain primer consisted of the [5′ TTGGCTTGAAGCTCCTCAGAGGA 3′] oligonucleotide. For VHand Vλ gene amplification, 30 cycles of PCR were performed under denaturing, annealing, and extending conditions of 94°C (1 minute), 52°C (1 minute), and 72°C (2 minutes), respectively. PCR products were sized and isolated on low melting agarose gel and ligated into pCR 1000 plasmid vector (Invitrogen Corp). The ligation mixture was used to transform INV_α_F′ competent cells according to the manufacturer’s protocol. Recombinant clones were selected according to the length of the insert and sequenced by the dideoxy chain termination method using the Taq Track Sequencing Kit (Promega Corp, Madison, WI). Each VH or Vλ sequence was derived from the analysis of at least three independent clones. Differences in nucleotide sequences among different recombinant clones were observed in few cases (<0.001/base) and such variants were excluded from the sequence analysis. DNA sequences were analyzed using the software package of the Genetics Computer Group of the University of Wisconsin, version 7.1, and a Model 6000-410 VAX computer (Digital Equipment Corp, Marlboro, MA). DNA sequence identity searches were performed using the GenBank database and the FASTA method.37
Analysis of the putative germline IgVH segment that gave rise to the expressed HBL-3 VH gene
Genomic DNA was extracted from the monoclonal HBL-3 B cells and autologous fibroblasts obtained from the same bioptic sample used for the generation of the tumoral HBL-3 cell line. B-cell or fibroblast genomic DNA (100 ng) was supplemented with the appropriate sense and antisense oligonucleotide primers (10 pmol each). PCR amplification was performed in a 50_μ_L reaction volume using _Taq_polymerase under denaturing, annealing, and extending conditions of 94°C (1 minute), 60°C (1 minute), and 72°C (1 minute), respectively, for 30 cycles. The oligonucleotides used were as follows: (1) the sense VH4-21 FR1 primer, encompassing a portion (residues 10 to 27) of the FR1 sequence of germline VH4-21 gene38 and differing in two nucleotides from the corresponding sequence of the expressed HBL-3 VH gene [5′ CTACAGCAGTGGGGCGCA 3′]; (2) the sense HBL-3 leader primer, encompassing a portion of the leader sequence of the HBL-3 VH gene and differing in one nucleotide (C instead of G at position −31) from the corresponding area of the germline V58 gene,39 the member of the VHIV gene family displaying the highest degree of identity with the VH4-21 gene; and (3) the antisense VH4-21 FR3 primer, consisting of the reverse complement [5′ GTGTCCGCGGCGGTCACAGA 3′] of a FR3 sequence (residues 250 to 269) shared by the expressed HBL-3 VH gene and the germline VH4-21 gene. Part of the amplified DNA was fractionated through a 1.5% agarose gel, transferred to a nylon membrane (Hybond; Amersham Life Sciences, Arlington Heights, IL) and hybridized at 48°C with the HBL-3 complementarity-determining region 1 (CDR1) oligonucleotide probe labeled with [_γ_-32P] ATP (DuPont NEN Research Products, Boston, MA) by T4 polynucleotide kinase. The HBL-3 CDR1 oligonucleotide encompassed a FR1-CDR1 sequence [5′ GTTCCGTGATTACCTCTGGACA 3′] (residues 84 to 105) of the expressed HBL-3 VH gene, differing in seven bases from that of the corresponding area of the germline VH4-21 gene. After hybridization, the membrane was washed twice with 2× SSC/0.1% sodium dodecyl sulfate (SDS) at room temperature for 30 minutes and twice with 1× SSC/0.1% SDS at 54°C for 30 minutes. Autoradiography was performed using Kodak XAR-5 film (Eastman Kodak Co, Rochester, NY). Other amplified DNA was inserted into the pCR II plasmid vector (Invitrogen Corp) for cloning and sequencing.
RESULTS
Characterization of the IgM MoAb-producing HBL-2 and HBL-3 cell lines
The features of the HBL-2 and HBL-3 cell lines established from 2 AIDS patients with SNCCL and their identity with the respective primary tumoral tissues have been reported28and are summarized in Table 1. Both tumor samples and respective cell lines expressed surface Ig_μ_ and λ chains and the B-cell–restricted marker CD19. This was consistent with the B-cell origin of the cell lines and with the histologic diagnosis of SNCCL.40 In addition, the surface expression of CD10 but not CD21 was consistent with the cell line phenotype of sporadic BL.40 The monoclonality of the AIDS BL cell lines and their absolute relatedness to the respective tumors was formally established by the analysis of the Ig H chain gene rearrangements and that of the c-myc oncogene translocations. The Southern blotting analyses of _Eco_RI- and_Hind_III-digested DNA were consistent with two distinct patterns of c-myc activation. In the HBL-2 cells, the breakpoint was located 5′ (3 to 5 kb) to the c-_myc_first exon; in the HBL-3 cells, the breakpoint was within a ~4-kb region 3′ of the c-myc exon 3. Both HBL-2 and HBL-3 cells were negative for EBV, HIV, and HTLV-1 sequences and so were their respective original tumor cells.
Table 1.
Features of the AIDS BLs and Related Cold Agglutinin MoAbs
| Tumor | MoAb | VHSegment | Vλ1 Segment | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell Line | Source | Histology | EBV DNA | HIV DNA | HTLV-I DNA | c-myc | Ag Specificity | VH Gene | Nucleotide(amino acid)Identity* (%) | Nucleotide Differences in | D Gene† | JH Gene | Vλ1 Gene‡ | Nucleotide(amino acid)Identity* (%) | Nucleotide Differences in | Jλ Gene | ||||||||
| Chains | CDR | FR | CDR | FR | ||||||||||||||||||||
| H | L | R | S | R | S | R | S | R | S | |||||||||||||||
| HBL-2 | Pleural fluid | SNCC | No | No | No | t(8;14) | μ | λ | i 4§ | VH4-21[| | ](#TFN5) | 93.5 (88.6) | 5 (3.2)¶ | 1 | 7 (10.1)¶ | 6 | 20P3 | JH6 | Humlv117 | 95.5 (92.8) | 5 (2.7)¶ | 4 | 2 (6.9)¶ | 2 |
| HBL-3 | Liver mass | SNCC | No | No | No | t(8;22) | μ | λ | i 0.6§ | VH4-21 | 84.2 (80.4) | 11 (7.8)¶ | 5 | 8 (26.2)¶ | 22 | DXP′1 (inverted) | JH5 | Humlv1L1 | 86.7 (80.6) | 9 (8.0)¶ | 8 | 10 (20.8)¶ | 12 | Jλ1 |
The Ag-binding activity of the IgM MoAbs produced by the HBL-2 and HBL-3 cell lines
The HBL-2 and HBL-3 IgM MoAbs specifically bound to the i determinant on human erythrocytes, as shown by their strong agglutination of cord (smallest agglutinating doses: 4 and 0.6 ng/107 RBCs, respectively), but not adult (no agglutination by HBL-2 MoAb; HBL-3 MoAb smallest agglutinating dose, 38.8 ng/107 RBCs) papain-treated human RBCs. The specificity of the HBL-2 and HBL-3 IgM MoAbs was further strengthened by the MoAbs’ failure to bind to any of the eight self and nine foreign Ags tested, including IgG Fc fragment, human thyroglobulin, ssDNA, phosphorylcholine, insulin, human recombinant TNF-α, human recombinant TNF-β, human recombinant IL-1_β_, _β_-galactosidase and LPS from E coli, tetanus toxoid, HIV-1, recombinant glycoproteins of CMV and Parvovirus B19, and _Pneumococcus_polysaccharides (Table 1). Binding to each of these Ags by HBL-2 and HBL-3 MoAbs yielded an absorbance of less than 0.05 at 492 nm; negative and positive controls were always less than 0.05 and more than 1.00, respectively. The HBL-3 but not the HBL-2 MoAb expressed the cross-reacting idiotype defined by the anti-idiotypic 9G4 MoAb.1,2,36
The HBL-2 and HBL-3 MoAb VH segments
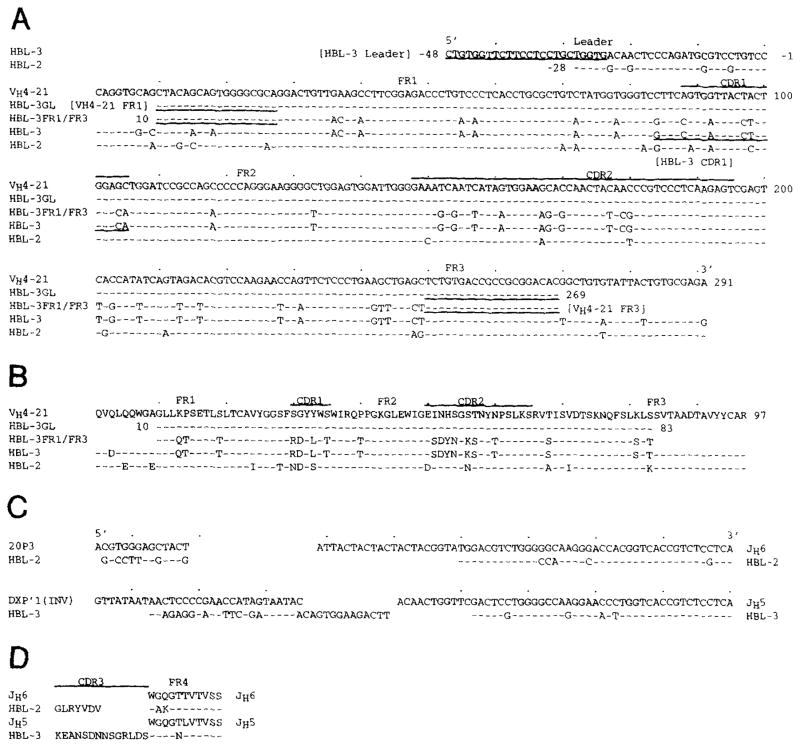
Figure 1 depicts the nucleotide (A) and deduced amino acid (B) sequences of the HBL-2 and HBL-3 IgM-MoAb VH genes and that of the closest reported germline VHgene. The differences between sequences are summarized in Table 1. The HBL-2 and HBL-3 MoAb VH gene sequences were 93.5% and 84.2% identical, respectively, to that of VH4-21 gene, a member of the VHIV gene family.38 Accurate inspection of the VH4-21–related sequences available in the GenBank showed that the HBL-2 VH gene sequence displayed an identical nucleotide difference at position 82, A instead of T, resulting in the same replacement of a Ser with a Thr, as found in the sequence of the VH4-21 allele HumigvH4c.41 Twelve of the 19 nucleotide differences displayed by the HBL-2 VH gene sequence resulted in putative amino acid replacements, yielding replacement to silent (R:S) mutation ratios of 5.0 in the CDR and 1.1 in the framework regions (FR). Nineteen of the 46 nucleotide differences displayed by the HBL-3 VH sequence resulted in putative amino acid replacements, yielding R:S mutation ratios of 2.2 in the CDR and 0.3 in the FR.
Fig. 1.
Top clusters: nucleotide (A) and deduced amino acid (B) sequences of the VH genes used by the HBL-2 and HBL-3 IgM MoAbs. The top sequence is given for comparison and represents the published germline VH gene (VH4-21) displaying the highest degree of identity to the expressed VH genes. The VH4-21 gene belongs to the VHIV family. Dashes indicate identity. Solid lines on the top of each cluster depict CDR. Underlined nucleotides define the sequences or the reverse complement sequences of the primers adopted for PCR amplification. HBL-3 GL and HBL-3 FR1/FR3 are the sequences amplified from fibroblast and HBL-3 cell genomic DNA, respectively, using the combination of the VH4-21 FR1 and the VH4-21 FR3 oligonucleotide primers (see Materials and Methods for details). Bottom clusters: nucleotide (C) and deduced amino acid (D) sequences of the D and JH genes used by the HBL-2 and HBL-3 IgM MoAbs. Germline D genes are given for comparison. Dashes indicate identity. DXP′1 (Inv) is the reverse complement of the germline DXP′1 sequence. The present sequences are available from EMBL/GenBank/DDBJ under accession numbers L29115 and L29116.
The HBL-2 and HBL-3 MoAb D and JH genes
The nucleotide and deduced amino acid sequences of the HBL-2 and HBL-3 IgM MoAb D and JH genes, and those of their closest germline D42–46 and JH genes42 are depicted in Fig 1. The HBL-2 MoAb D gene sequence displayed some identity with that of the expressed fetal 20P3 D gene47; the HBL-3 MoAb D gene displayed a stretch of similarity to that of the reverse complement of the germline DXP′1 gene, suggesting a possible inverted D gene joining origin (Fig 1C).48 Both expressed D genes were flanked by untemplated nucleotide additions. The entire length of the D segment ranged from 13 nucleotides in HBL-2 to 36 nucleotides in HBL-3. The HBL-2 and HBL-3 MoAbs used truncated and mutated forms of JH6 and JH5 genes, respectively (Fig 1C). The deduced amino acid sequences of the D-JH genes are depicted in Fig 1D, as segregated in CDR3 and FR4 stretches, according to Kabat et al.49The CDR3 sequences were highly divergent in length and composition. The HBL-2 and HBL-3 FR4 sequences were invariable in length and displayed two and one amino acid replacements, respectively.
The HBL-2 and HBL-3 MoAb Vλ and Jλ genes
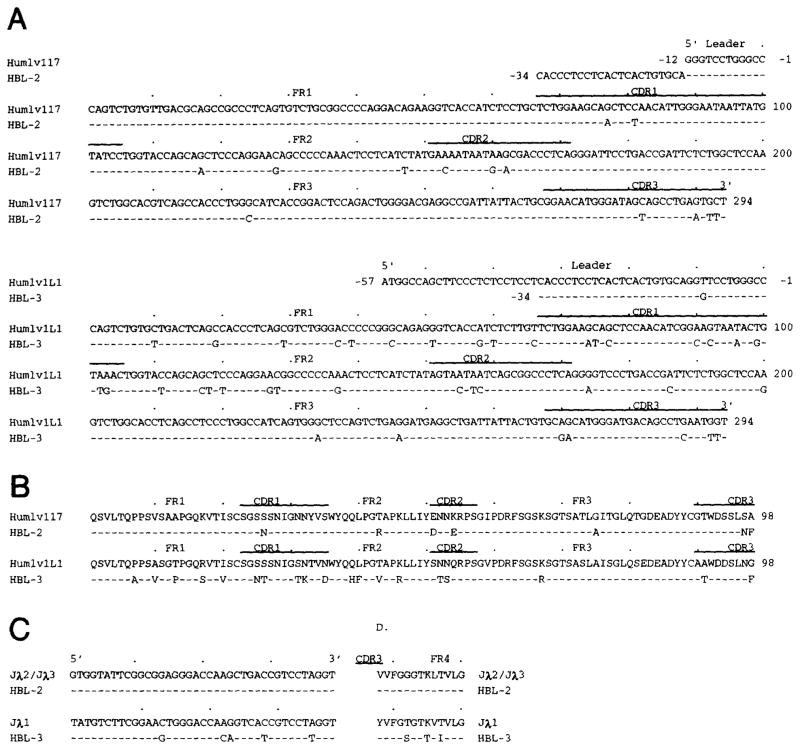
Figure 2 depicts the nucleotide (A) and deduced amino acid (B) sequences of the HBL-2 and HBL-3 MoAb Vλ genes and those of the closest reported germline Vλ genes. The differences between sequences are summarized in Table 1. HBL-2 and HBL-3 MoAbs used two members of the Vλ1 subgroup, the Humiglv117 and Humiglv1L1 genes, respectively.14,50 When compared with the germline gene, the HBL-2 Vλ1 gene sequence displayed nine and four nucleotide differences in the CDRs and FRs, respectively, yielding four and two amino acid replacements, and R:S mutation ratios of 1.2 and 1.0, respectively. When compared with the germline gene, the HBL-3 Vλ1 gene sequence displayed 39 nucleotide differences. These were scattered throughout the CDRs and FRs, yielding a total of 19 amino acid replacements and R:S mutation ratios of 1.1 and 0.8, respectively. Figure 2 depicts the nucleotide (C) and deduced amino acid (D) sequences of the MoAb Jλ segments and their respective germline Jλ templates. The HBL-2 MoAb used a Jλ2/Jλ3 segment in germline configuration; the HBL-3 MoAb used a Jλ1 segment with five nucleotide mutations resulting in three amino acid replacements.
Fig. 2.
Top clusters: nucleotide (A) and deduced amino acid (B) sequences of the Vλ genes used by the HBL-2 and HBL-3 IgM MoAbs. In each cluster, the top sequence is given for comparison and represents the published germline Vλ gene displaying the highest degree of identity with the expressed Vλ genes. Dashes indicate identity. Solid lines on the top of each cluster depict CDR. The Humlv117 and Humlv1L1 genes belong to the Vλ1 subgroup. Bottom clusters: nucleotide (C) and deduced amino acid (D) sequences of the Jλ segments used by the HBL-2 and HBL-3 MoAbs. Dashes indicate identity. The present sequences are available from EMBL/GenBank/DDBJ under accession numbers L29113 and L29114.
Somatic mutations in the HBL-3 MoAb VH segment
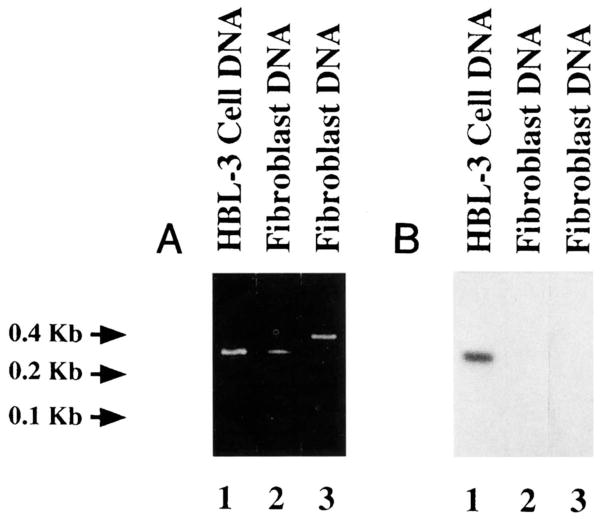
Because of conservation of the VH4-21 gene in humans, the high number of nucleotide differences displayed by the HBL-3 VH gene sequence when compared with that of the VH4-21 germline gene, and the detection of mutations in the HBL-3 MoAb JH5 and Jλ1 segments, we hypothesized that the HBL-3 MoAb VH segment consisted of a somatically mutated form of the VH4-21 gene. PCR amplifications were performed using ad hoc designed oligonucleotide primers and genomic DNA from the HBL-3 cell line or autologous fibroblasts. The sense VH4-21 FR1 primer, encompassing an FR1 sequence (residues 10 to 27) shared by the VH4-21 segment and the expressed HBL-3 VH gene, was used in conjunction with the antisense VH4-21 FR3 primer, encompassing an FR3 sequence (residues 250 to 269) shared by the germline VH4-21 and the expressed HBL-3 genes. The two combined primers amplified DNA from both fibroblasts and HBL-3 cells. The molecular size of the amplified product (~260 bp) was consistent with that of the sequence spanning residues 10 to 269 of the VH4-21 gene sequence (Fig 3A, lanes 1 and 2). The same antisense VH4-21 FR3 primer was also used to amplify fibroblast DNA, in conjunction with the sense HBL-3 leader primer, encompassing a stretch of the leader sequence of HBL-3 VH gene (residues −49 to −25) and differing in only one nucleotide from that of the corresponding area of the V58 gene, the VHIV family member displaying the highest degree of identity with the VH4-21 gene. The molecular size of the amplified product (~400 bp) was consistent with that of the sequence spanning residues −49 to 269 of the HBL-3 VH gene, including the untranslated intervening intron (Fig 3A, lane 3).
Fig. 3.
Evidence for somatic mutations in the HBL-3 VH, gene. (A) Ethidium bromide staining of amplified DNA after fractionation in agarose gel electrophoresis. Using the VH4-21 FR1 and VH4-21 FR3 oligonucleotide primers, amplification products of identical and appropriate size were obtained by priming genomic DNA from the HBL-3 cells (lane 1) and autologous fibroblasts that had been established in primary culture from the BL bioptic sample of the patient whose neoplastic B cells gave rise to the HBL-3 cell line (lane 2). Using the HBL-3 leader and VH4-21 FR3 oligonucleotide primers, a product of appropriate size was amplified by priming genomic DNA from fibroblasts (lane 3). (B) Southern blot hybridization of the PCR products shown in (A) with 32P-labeled oligonucleotide HBL-3 CDR1 probe encompassing an FR1-CDR1 sequence of the HBL-3 MoAb VH gene. A strong positive hybridization signal was detected only with DNA amplified from the HBL-3 MoAb-producing B-cell line (lane 1), but not with DNA amplified from autologous fibroblasts (lanes 2 and 3).
The three DNA amplification products were analyzed for their ability to hybridize with the [_γ_-32P]-labeled HBL-3 CDR1 oligonucleotide. This encompassed a stretch of the HBL-3 VH gene FR1-CDR1 sequence that displayed seven putative mutations when compared with the corresponding germline VH4-21 gene sequence. The [_γ_-32P]-labeled HBL-3 CDR1 oligonucleotide strongly hybridized with the ~260 bp DNA amplified from the HBL-3 cell line (Fig 3B, lane 1), but not with the ~260 or ~400 bp DNA amplified from the autologous fibroblasts (Fig 3B, lanes 2 and 3, respectively). To identify the autologous germline VH gene that putatively gave rise to the expressed HBL-3 VH gene, the product amplified from fibroblast DNA using the sense VH4-21 FR1 and the antisense VH4-21 FR3 primers was cloned and sequenced. The sequences of the eight independent clones were all identical to each other and to that of the VH4-21 germline gene throughout the overlapping area (residues 28 through 249) (Fig 1A and B; HBL-3GL sequence). DNA amplified from the HBL-3 cell line using the sense VH4-21 FR1 and the antisense VH4-21 FR3 primers was also cloned and sequenced. The sequences of three independent clones were identical to each other and to that of the expressed HBL-3 VH gene throughout the overlapping area (residues 28 through 249) (Fig 1A and B; HBL-3 FR1/FR3 sequence). These experiments proved that the expressed HBL-3 VH segment was somatically point-mutated, and suggested that VH4-21 is the germline gene that gave rise to it.
Ag-selection of the IgV genes expressed by the HBL-2 and HBL-3 BL cells
In absence of negative or positive selective pressure on a gene product, R and S mutations are scattered randomly throughout the protein sequence. If a DNA segment displays a number of R mutations lower than that expected by chance only, it is likely that pressure to maintain the germline-encoded protein structure was exerted. Conversely, if a DNA segment displays a number of R mutations higher than that expected by chance only, it is likely that a positive pressure to select R mutations was exerted. The numbers of expected R mutations in the HBL-2 and HBL-3 MoAb VH and Vλ segment CDRs and FRs were calculated using the formula n × R_f_ × CDR_f_ (or FR_f_), where n is the total number of observed mutations, R_f_ is the expected proportion of R mutations (0.75),51 and CDR_f_ is the relative size of the CDRs (or FRs) (0.23 and 0.77 for VH4-21 CDRs and FRs, respectively; 0.29 and 0.71 for Vλ1 CDRs and FRs, respectively). Both HBL-2 and HBL-3 MoAb V segments displayed higher and lower numbers of R mutations in the CDRs and FRs, respectively, than those theoretically expected (Table 1). Because of the primary role played by the VH4.21 segment in the binding to the i/I Ag, we calculated the probabilities that the excess and the scarcity of R mutations arose by chance in the HBL-2 and HBL-3 MoAb VH segment CDRs and FRs, respectively, using the binomial distribution model P = [n!/k!(_n_− k)!] _q_k(1 − q)n−k, where _q_is the probability an R mutation will locate to the CDRs (q_= 0.23 × 0.75) or FRs (q = 0.77 × 0.75), and k is the number of observed R mutations in the CDRs or FRs.52 The likelihood that the excess R mutations arose by chance in the VHsegment CDR were P = .11 in HBL-2 MoAb and_P = .06 in HBL-3 MoAb. The probability that the scarcity of R mutations in the VH segment FR resulted from chance were P = .03 in HBL-2 MoAb and _P_= .00000002 in HBL-3 MoAb. In their original report on the application of the binomial distribution model to the analysis of the R point-mutations in Ag-selected IgV segment CDRs, Shlomchik et al52 suggested that the observed number of FR R mutations should be doubled to account for the fact that some of these mutational events will never be observed because they are deleterious to the Ig structure. Although this correction was inferred from some experimental observations, it is approximate and may not be applicable as such to all Ig VH genes. When the analysis of the VH segment somatic mutation pattern was performed with the adjustment of doubling the number of observed FR R mutations, the probabilities that excess R mutations arose by chance in the VH segment CDR were P = .49 and P = .35 in HBL-2 MoAb and HBL-3 MoAb, respectively.
DISCUSSION
In the present studies, we established from bioptic specimens of 2 AIDS patients with BL two MoAb-producing cell lines representative of the respective tumors, and analyzed the Ag-binding activity and the V segment structure of these MoAbs. We found that both IgM MoAbs were cold agglutinins highly specific for the i blood group determinant, and both MoAbs bore Ag-combining sites consisting of point-mutated VH4-21 segments in conjunction with Vλl segments.
The exquisite specificity of the HBL-2 and HBL-3 IgM MoAb cold agglutinins for the i repetitive N-acetyllactosamine units was strengthened by the MoAb failure to bind any of the other eight self Ags and nine foreign Ags tested. The putative use of the VH4-21 gene segment by the HBL-2 and HBL-3 MoAbs is consistent with the use of the same segment by the majority of the reported cold agglutinins from patients with idiopathic cold agglutinin disease, FL, and Waldenstrom’s macroglobulinemia.1,2,53,56 A primary role of the VH4-21 segment in the binding to the i Ag is further supported by the divergence in composition and length of the H chain CDR3 sequences as well as by the heterogeneity of the VL, segments of the present two IgM and the 10 reported VH4-21+ cold agglutinins, which use a V_k_I segment three times, a V_k_II segment once, a V_k_IIIa segment once, a V_k_IIIb four times, and a Vλ1 segment once.1,54–56 Thus, the VH4-21 gene restriction in the cold agglutinin system may result from a selection process based on an inherent affinity of this VH gene product for the i/I carbohydrate structure.1,2,54,56 Nevertheless, the VH4-21 segment is not an absolute requirement for i/I-dependent RBC agglutination, because cold agglutinins using VH segments of the VHIII family have been reported.57 The restricted usage of the VH4-21 by anti-i/I cold agglutinins is intriguing and may be related to the overrepresentation of the VH4-21 –expressing clones in the normal B-cell repertoire, as determined by the VH4-21–related 9G4 idiotype studies in the circulating blood of adults, as well as cord blood and fetal tissues.4 In this regard, it is not known whether these circulating VH4-21 –expressing B cells also have anti-i/I specificity. If this were the case, one could speculate that the abundant representation of VH4-21 –expressing B cells in the periphery results from positive selection by i/I Ag, which are present not only on RBCs but also on lymphocytes.58 Expression of the i Ag on human erythrocytes is developmentally regulated. It is maximal in fetal and neonatal life and decreases in adult life, in which the expression of the I Ag prevails.59
An Ag-driven process of clonal selection may play a role in the emergence and/or expansion of certain neoplastic B lymphocytes. Consistent with their putative germinal center origin, FL B cells, which represent the neoplastic equivalents of the elements recruited in a secondary Ag-specific response, display somatic mutations that resemble in nature and distribution those characteristic of an affinity maturation process.22–24 A recent thorough documentation of the somatic mutation and clonal evolution of an FL expressing a VH4-21 gene, the antigenic specificity of which had not been determined, showed at least three amino acid mutations, the anti-i/I “characteristic” Gly to Asn mutation at position 31, a Val to Ile mutation at position 71, and a Ser to Thr mutation at position 83, which are shared by the HBL-2 and HBL-3 MoAb VH4-21 segments.24 A role for clonal selection by self Ag during the evolution of anti-“Pr2”–specific B-cell lymphoma has been documented in detail.5,6 The Pr2Ag is a sialoglycoprotein and provides, along with the multiple N-acetyllactosamine i/I Ag, the target for autoimmune phenomena that occur in association with several human clonal B-cell disorders.
The sequences of the DNA amplified from the HBL-3 MoAb-producing cell line and autologous fibroblast genomic DNA, using the HBL-3 leader, VH4-21 FR1, and VH4-21 FR3 primers, as well as the differential hybridization of the HBL-3 CDR1 oligonucleotide (encompassing an FR1-CDR1 sequence of the HBL-3 MoAb VH segment) with the above amplification products, formally proved the mutated status of the HBL-3 VH segment, and suggested that VH4-21 was the germline gene that gave rise to it. The somatically mutated status of the HBL-3 and, possibly, HBL-2 MoAbs was further strengthened by the high degree of conservation of the VH4-21 gene sequence in humans, and the extension of the point-mutations to the, in general, highly conserved, JH and/or Jλ segments. An Ag-selection of the point-mutations in the HBL-2 and HBL-3 MoAb VH segments was suggested by the differential R:S mutation ratios in the CDR and FR (HBL-2 MoAb, 5.0 and 1.1, respectively; HBL-3 MoAb, 2.2 and 0.3, respectively) and the accumulation in the CDR of the HBL-2 and/or HBL-3 VH segments of amino acid replacements that are shared by other anti-i/I cold agglutinins and might have increased the affinity of the VH4-21 segment for the i Ag, including the Gly to Asp mutation at position 31, which is shared by the FS-1, FS-2, FS-4, and KAU VH cold agglutinins,54,55 the Ser to Thr mutation at position 35, which is shared by FS-1, FS-4, and FS-6 cold agglutinins, and the His to Tyr at position 53, which is shared by the FS-3 cold agglutinin.55 However, a positive clonal selection of R mutations in the HBL-2 and HBL-3 MoAb VH segment CDRs was not further substantiated by the statistical analysis according to the binomial distribution model with the correction for FR R mutations, as proposed by Shlomchik et al.52 This finding may be consistent with a putatively inherent anti-i/I activity of the unmutated VH4-21 gene product, and, perhaps, a clonal selection against R mutations in the HBL-2 and HBL-3 MoAb VH segments CDRs, similar to that shown for other B-cell tumor anti-RBC autoantibodies.6 The substitution of the Val with an Ile in the HBL-3 MoAb VH segment FR1 Ala-Val-Tyr (residues 23 to 25) triplet, which provides the structural correlates for the anti-idiotypic 9G4 antibody binding, as recently shown by Potter et al,60 is possibly responsible for the lack of 9G4 reactivity of the HBL-2 MoAb.
In the present AIDS-associated BLs, it is unclear whether the initiation of the anti-i/I Ag autoantibody response constituted a crucial event in the neoplastic transformation. The putative anti-self Ag clonal expansion and selection may have preceded the genetic accident, ie, c_-myc_ proto-oncogene chromosomal translocation.61,62 Alternatively, in these BLs, the specific B-cell expansion and selection may have followed the chromosomal translocation, resembling the series of events that have been paradigmatically illustrated in relationship to _bcl-2 proto-oncogene chromosomal t(14;18) translocation by Zelenetz et al23in a FL for which, however, a specific Ag could not be identified. Knowledge of the sequential order of activating, proliferating, and transforming events, including c_-myc translocation and activation, Ag-dependent B-cell amplification, somatic hypermutation, and clonal selection, is crucial for a better understanding of the molecular pathogenesis of AIDS BL and, possibly, other BLs. These issues could be best addressed by the use of a tumor-specific Ig H chain CDR3 sequence oligonucleotide to identify tumor-related Ig VH-D-JHsequences in nonmalignant B-cell progenitors.
Acknowledgments
Supported by the US Public Health Service Grants No. AR-40908, CA-16087, and CFAR 5P30 AI-27742 (to P.C.), and CA-37895 (to R.D.-F.) and the Ministry of Health, I.S.S. (Italy) AIDS Project 8206-07 1993 (to P.R.). G.G. was supported by a fellowship for AIDS research from the Ministry of Health, I.S.S. (Italy). P.C. is a Kaplan Cancer Scholar. This is publication 22 from The Jeanette Greenspan Laboratory for Cancer Research.
We thank Dr M. Gorny for testing the HBL-2 and HBL-3 MoAbs for binding to HIV-1, CMV, and parvovirus.
References
- 1.Silberstein LE, Jefferies LC, Goldman J, Friedman D, Moore JS, Nowell PC, Roelcke D, Pruzanski W, Roudier J, Silverman GJ. Variable region gene analysis of pathologic human autoantibodies to the related i and I red blood cell antigens. Blood. 1991;78:2372. [PubMed] [Google Scholar]
- 2.Pascual V, Victor K, Lelsz D, Spellberg MB, Hamblin TJ, Thompson KM, Randen I, Natvig J, Capra JD, Stevenson FK. Nucleotide sequence analysis of the V regions of two IgM cold agglutinins. Evidence that the VH4-21 gene segment is responsible for the major cross-reactive idiotype. J Immunol. 1991;146:4385. [PubMed] [Google Scholar]
- 3.Miller DG. The association of immune disease and malignant lymphoma. Ann Intern Med. 1967;66:507. doi: 10.7326/0003-4819-66-3-507. [DOI] [PubMed] [Google Scholar]
- 4.Stevenson FK, Smith GJ, North J, Hamblin TJ, Glennie MJ. Identification of normal B-cell counterparts of neoplastic cells which secrete cold agglutinins of anti-I anti-i specificity. Br J Haematol. 1989;72:9. doi: 10.1111/j.1365-2141.1989.tb07643.x. [DOI] [PubMed] [Google Scholar]
- 5.Silberstein LE, Litwin S, Carmack CE. Relationship of variable region genes expressed by a human B cell lymphoma secreting pathologic anti-PR2 erythrocyte autoantibodies. Exp Med. 1989;169:1631. doi: 10.1084/jem.169.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman DF, Cho EA, Goldman J, Carmack CE, Besa EC, Hardy RR, Silberstein LE. The role of clonal selection in the pathogenesis of an autoreactive human B cell lymphoma. J Exp Med. 1991;174:525. doi: 10.1084/jem.174.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye BR. Rheumatologic manifestations of infection with human immunodeficiency virus (HIV) Ann Intern Med. 1989;111:158. doi: 10.7326/0003-4819-111-2-158. [DOI] [PubMed] [Google Scholar]
- 8.Freter CE. Acquired immunodeficiency syndrome-associated lymphomas. J Natl Cancer Inst Monogr. 1990;10:45. [PubMed] [Google Scholar]
- 9.Raphael BG, Knowles DM. Acquired immunodeficiency syndrome-associated non-Hodgkin’s lymphoma. Semin Oncol. 1990;17:361. [PubMed] [Google Scholar]
- 10.Shlomchik MJ, Marshak-Rothstein A, Wolfowicz CB, Rothstein TL, Weigert MG. The role of clonal selection and somatic mutation in autoimmunity. Nature. 1987;328:805. doi: 10.1038/328805a0. [DOI] [PubMed] [Google Scholar]
- 11.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, Weigert MG. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harindranath N, Goldfarb IS, Ikematsu H, Burastero SE, Wilder RL, Notkins AL, Casali P. Complete sequence of the genes encoding the VH and VL regions of low and high affinity monoclonal IgM and IgA1 rheumatoid factors produced by CD5+ B cells from a rheumatoid arthritis patient. Int Immunol. 1991;3:865. doi: 10.1093/intimm/3.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Randen I, Brown D, Thompson KM, Hughens-Jones N, Pascual V, Victor K, Capra JD, Forre O, Natvig JB. Clonally related IgM rheumatoid factors, undergo affinity maturation in the rheumatoid synovial tissue. J Immunol. 1992;148:3296. [PubMed] [Google Scholar]
- 14.Olee T, Liu EW, Huang D-F, Soto-Gil RW, Deftos M, Kozin F, Carson DA, Chen PP. Genetic analysis of self-associating immunoglobulin G rheumatoid factors from two rheumatoid synovia implicates an antigen driven response. J Exp Med. 1992;167:840. doi: 10.1084/jem.175.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Es JH, Aanstoot H, Gmelig-Meyling FHJ, Derksen RHWM, Logtenberg T. A human systemic lupus erythematosus-related anti-cardiolipin/single-strand DNA autoantibody is encoded by somatically mutated variant of the developmentally restricted 51P1 VH gene. J Immunol. 1992;149:2234. [PubMed] [Google Scholar]
- 16.Mantovani L, Wilder RL, Casali P. Human rheumatoid B-1a (CD5+ B) cells make somatically hypermutated high affinity IgM rheumatoid factors. J Immunol. 1993;151:473. [PMC free article] [PubMed] [Google Scholar]
- 17.McKean D, Huppi K, Bell M, Staud L, Gerhard W, Weigert MG. Generation of antibody diversity in the immune response of Balb/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1984;81:3180. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berek C, Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987;96:23. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 19.Manser T. Evolution of antibody structure during the immune response. The differentiative potential of a single B lymphocyte. J Exp Med. 1989;170:1211. doi: 10.1084/jem.170.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueki Y, Goldfarb IS, Harindranath N, Gore M, Koprowski H, Notkins AL, Casali P. Clonal analysis of a human antibody response. Quantitation of precursors of antibody-producing cells and generation and characterization of monoclonal IgM, IgG, and IgA to rabies virus. J Exp Med. 1990;171:19. doi: 10.1084/jem.171.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikematsu H, Harindranath N, Ueki Y, Notkins AL, Casali P. Clonal analysis of a human antibody response. II. Sequences of the VH genes of human monoclonal IgM, IgG and IgA to rabies virus reveal preferential utilization of the VHIII family members and somatic hypermutation. J Immunol. 1993;150:1325. [PMC free article] [PubMed] [Google Scholar]
- 22.Levy S, Mendel E, Kon S, Avnur Z, Levy R. Mutational hot spots in Ig V region genes of human follicular lymphomas. J Exp Med. 1988;168:475. doi: 10.1084/jem.168.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zelenetz AD, Chen TT, Levy R. Clonal expansion in follicular lymphoma occurs subsequent to antigenic selection. J Exp Med. 1992;176:1137. doi: 10.1084/jem.176.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahler DW, Levy R. Clonal evolution of a follicular lymphoma: Evidence for antigen selection. Proc Natl Acad Sci USA. 1992;89:6770. doi: 10.1073/pnas.89.15.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelicci P-G, Knowles DM, Arlin ZA, Wieczorek R, Luciw P, Dina D, Basilico C, Dalla-Favera R. Multiple monoclonal B cell expansions and c-myc oncogene rearrangements in acquired immune deficiency syndrome-related lymphoproliferative disorders. J Exp Med. 1986;164:2049. doi: 10.1084/jem.164.6.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaidano G, Dalla-Favera R. Biologic aspects of human immunodeficiency virus-related lymphoma. Curr Opin Oncol. 1992;4:900. doi: 10.1097/00001622-199210000-00013. [DOI] [PubMed] [Google Scholar]
- 27.Knowles DM, Chadburn A. Lymphadenopathy and the lymphoid neoplasms associated with the acquired immune deficiency syndrome (AIDS) In: Knowles DM, editor. Neoplastic Hematology. Baltimore, MD: Williams & Wilkins; 1992. p. 773. [Google Scholar]
- 28.Gaidano G, Parsa NZ, Tassi V, Della-Latta P, Chaganti RSK, Knowles DM, Dalla-Favera R. In vitro establishment of AIDS-related lymphoma cell lines: Phenotypic characterization, oncogene and tumor suppressor gene lesions, and heterogeneity in Epstein-Barr virus infection. Leukemia. 1993;7:1621. [PubMed] [Google Scholar]
- 29.Korsmeyer SJ, Hyeter PA, Ravetch JV, Poplack DG, Waldmann TA, Leder P. Developmental hierarchy of immunoglobulin gene rearrangement in human leukemic pre-B cells. Proc Natl Acad Sci USA. 1981;78:7096. doi: 10.1073/pnas.78.11.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc oncogene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorny MK, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:1624. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura M, Burastero SE, Notkins AL, Casali P. Human monoclonal rheumatoid factor-like antibodies from CD5 (Leu-1)+ B cells are polyreactive. J Immunol. 1988;140:4180. [PubMed] [Google Scholar]
- 33.Kasaian MT, Ikematsu H, Casali P. Identification and analysis of a novel CD5 – B lymphocyte subset producing natural antibodies. J Immunol. 1992;148:2690. [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura M, Burastero SE, Ueki Y, Larrick JW, Notkins AL, Casali P. Probing the normal and autoimmune B cell repertoire with Epstein-Barr virus. Frequency of B cells producing monoreactive high affinity autoantibodies in patients with Hashimoto’s disease and systemic lupus erythematosus. J Immunol. 1988;141:4165. [PubMed] [Google Scholar]
- 35.Silberstein LE, Jefferies LC, Goldman J, Spitalnik SL. Production of carbohydrate-specific human monoclonal antibodies in vitro. Methods Enzymol. 1989;179:299. doi: 10.1016/0076-6879(89)79131-x. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson FK, Wrightham M, Glennie MJ, Jones DB, Cattan AR, Feizi T, Spelleberger MB, Hamblin TJ, Stevenson GT. Antibodies to shared idiotypes as reagents for analysis and therapy for human B cell tumors. Blood. 1986;68:430. [PubMed] [Google Scholar]
- 37.Parson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 38.Sanz I, Kelly P, Williams C, Scholl S, Tucker P, Capra JD. The smaller human VH gene families display remarkably little polymorphism. EMBO J. 1989;8:3741. doi: 10.1002/j.1460-2075.1989.tb08550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KH, Matsuda F, Kinashi T, Kodaira M, Honjo T. A novel family of variable region genes of the human immunoglobulin heavy chain. J Mol Biol. 1987;195:761. doi: 10.1016/0022-2836(87)90482-7. [DOI] [PubMed] [Google Scholar]
- 40.Magrath IT. The pathogenesis of Burkitt’s Lymphoma. Adv Cancer Res. 1990;55:133. doi: 10.1016/s0065-230x(08)60470-4. [DOI] [PubMed] [Google Scholar]
- 41.van Es JH, Heutink M, Aanstoot H, Logtenberg T. Sequence analysis of members of the human Ig VH4 gene family derived from a single VH locus. Identification of novel germ-line members. J Immunol. 1992;149:492. [PubMed] [Google Scholar]
- 42.Ravetch JV, Siebenlist U, Korsmeyer SJ, Waldmann T, Leder P. Structure of the human immunoglobulin μ locus. Characterization of embryonic and rearranged J and D genes. Cell. 1981;27:583. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- 43.Siebenlist V, Ravetch JV, Korsmeyer SJ, Waldmann T, Leder P. Human immunoglobulin D segments encoded in tandem mutigene families. Nature. 1981;294:631. doi: 10.1038/294631a0. [DOI] [PubMed] [Google Scholar]
- 44.Ichihara Y, Matsuoka H, Kurosawa Y. Organization of human immunoglobulin heavy chain diversity gene loci. EMBO J. 1988;7:4141. doi: 10.1002/j.1460-2075.1988.tb03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buluwela L, Albertson DG, Sherrington P, Rabbitts PH, Spurr N, Rabbitts TH. The use of chromosomal translocations to study human immunoglobulin gene organization: Mapping DH segments within 35 kb of the Cμ gene and identification of a new DH locus. EMBO J. 1988;7:2003. doi: 10.1002/j.1460-2075.1988.tb03039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuda F, See KH, Nakai S, Sato T, Kodaira M, Zong SQ, Ohno H, Fukuhara S, Honjo T. Dispersed localization of D segments in the human immunoglobulin heavy-chain locus. EMBO J. 1988;7:1047. doi: 10.1002/j.1460-2075.1988.tb02912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder HW, Jr, Hillson JL, Perlmutter RM. Early restriction of the human antibody repertoire. Science. 1987;238:791. doi: 10.1126/science.3118465. [DOI] [PubMed] [Google Scholar]
- 48.Meek KD, Hasemann CA, Capra JD. Novel rearrangements at the immunoglobulin D locus. Inversion and fusion add to IgH somatic diversity. J Exp Med. 1989;170:39. doi: 10.1084/jem.170.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabat EA, Wu IT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. Washington, DC: US Department of Health and Human Services; 1991. [Google Scholar]
- 50.Siminovitch KA, Misener V, Kwong PC, Song Q-L, Chen PP. A natural antibody is encoded by germline heavy and lambda light chain variable region genes without somatic mutation. J Clin Invest. 1989;84:1675. doi: 10.1172/JCI114347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jukes TH, King JL. Evolutionary nucleotide replacements in DNA. Nature. 1979;281:605. doi: 10.1038/281605a0. [DOI] [PubMed] [Google Scholar]
- 52.Shlomchik MJ, Aucoin AH, Pisetsky DS, Weigert MG. Structure and function of anti-DNA autoantibodies derived from a single autoimmune mouse. Proc Natl Acad Sci USA. 1987;84:9150. doi: 10.1073/pnas.84.24.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverman GJ, Carson DA. Structural characterization of human monoclonal cold agglutinins: Evidence for a distinct primary sequence-defined VH4-21 idiotype. Eur J Immunol. 1990;20:351. doi: 10.1002/eji.1830200218. [DOI] [PubMed] [Google Scholar]
- 54.Leoni J, Ghiso J, Goni F, Frangione B. The primary structure of the Fab fragment of protein KAU, a monoclonal immunoglobulin M cold agglutinin. J Biol Chem. 1991;266:2836. [PubMed] [Google Scholar]
- 55.Pascual V, Victor K, Spellerberg M, Hamblin TJ, Stevenson FK, Capra JD. VH restriction among human cold agglutinins. The VH4-21 gene segment is required to encode anti-I and anti-i specificities. J Immunol. 1992;149:2337. [PubMed] [Google Scholar]
- 56.Pascual V, Capra JD. VH4-21, a human VH gene segment overrepresented in the autoimmune repertoire. Arthritis Rheum. 1992;35:11. doi: 10.1002/art.1780350103. [DOI] [PubMed] [Google Scholar]
- 57.Jeffreys LC, Carachidi CM, Goldman J, Silberstein LE. Naturally occurring anti-i/I cold agglutinins may be encoded by different VHIII genes as well as by VH4.21 gene segments. J Clin Invest. 1993;92:2821. doi: 10.1172/JCI116902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grillot-Courvalin C, Brouet J-C, Piller F, Rassenti LZ, Labaume S, Silverman GJ, Silberstein L, Kipps TJ. An anti-B cell autoantibody from Wiskott-Aldrich syndrome which recognizes i blood group specificity on normal human B cells. Eur J Immunol. 1992;22:1781. doi: 10.1002/eji.1830220717. [DOI] [PubMed] [Google Scholar]
- 59.Fukuda M, Fukuda MN, Papayannopoupou T, Hakomori SI. Membrane differentiation in human erythroid cells: Unique profiles of cell surface glycoproteins expressed in erythroblasts in vitro from three ontogenic stages. Proc Natl Acad Sci USA. 1980;77:3473. doi: 10.1073/pnas.77.6.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Potter KN, Li Y, Pascual V, Williams RC, Jr, Byres LC, Spellberg M, Stevenson FK, Capra JD. Molecular characterization of a cross-reactive idiotope on human immunoglobulins utilizing the VH4-21 gene segment. J Exp Med. 1993;178:1419. doi: 10.1084/jem.178.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pelicci P-G, Knowles DM, Magrath IT, Dalla-Favera R. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt’s lymphoma. Proc Natl Acad Sci USA. 1986;83:2984. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neri A, Barriga F, Knowles DM, Magrath IT, Dalla-Favera R. Different regions of the immunoglobulin heavy-chain locus are involved in chromosomal translocations in distinct pathogenetic forms of Burkitt’s lymphoma. Proc Natl Acad Sci USA. 1988;85:2748. doi: 10.1073/pnas.85.8.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]