Single-cell transcriptome analysis reveals coordinated ectopic gene expression patterns in medullary thymic epithelial cells (original) (raw)
. Author manuscript; available in PMC: 2016 Mar 1.
Published in final edited form as: Nat Immunol. 2015 Aug 3;16(9):933–941. doi: 10.1038/ni.3246
Abstract
Expression of tissue-restricted self-antigens (TRAs) in medullary thymic epithelial cells (mTECs) is essential for self-tolerance induction and prevents autoimmunity, with each TRA being expressed in only a few mTECs. How this process is regulated in single mTECs and coordinated at the population level, such that the varied single-cell patterns add up to faithfully represent TRAs, is poorly understood. Here we used single-cell RNA-sequencing and provide evidence for numerous recurring TRA co-expression patterns, each present in only a subset of mTECs. Co-expressed genes clustered in the genome and showed enhanced chromatin accessibility. Our findings characterize TRA expression in mTECs as a coordinated process, which might involve local re-modeling of chromatin and thus ensures a comprehensive representation of the immunological self.
Self-non-self-discrimination, including self-tolerance, is a hallmark of the adaptive immune system, and in case this subtle distinction fails, various autoimmune diseases have been shown to develop1, 2. Self-tolerance of T cells, as imposed in the thymus (i.e., central tolerance), relies on the exhaustive scanning of self-antigens by maturing T cells3. Distinct types of thymic antigen presenting cells (APCs) display a broad range of self-antigens in a partly redundant and partly complementing fashion4. Among the various thymic APCs, medullary thymic epithelial cells (mTECs) stand out due to their unique ability to ectopically express a wide range of tissue-restricted antigens (TRAs)5, 6. In mTECs, TRAs, whose expression outside of the thymus is tightly controlled in time and space, become accessible to developing T cells when they are still most responsive to tolerance imprinting. Self-tolerance induction operates via two modes, either via elimination of self-reactive T cells or by cell fate diversion towards the regulatory T cell lineage3, 4, 7, 8, 9. Typically, each TRA protein is only expressed in 1-3% of mTECs, and thus, TRA expression follows a mosaic pattern. Therefore, self-antigen availability is a potential limiting factor during self-tolerance induction4, 10, 11, 12.
Many aspects of the complex molecular regulation of thymic TRA expression are poorly understood; the transcriptional regulator Aire, which is responsible for expression of a large part of ectopically expressed TRAs in the thymus, represents a notable exception1, 13, 14, 15. Aire targets inactive chromatin either directly by binding the repressive chromatin mark H3K4me0 with its PHD1 finger domain16, 17, or indirectly through its binding partners such as the ATF7ip-MBD1 complex18 or the Cdh4 protein19. These proteins are thought to recruit Aire to methylated CpG dinucleotides at repressed promoters and polycomb-silenced chromatin, respectively. Upon recruitment to silent chromatin, Aire is believed to promote ectopic expression of TRA-encoding genes by releasing stalled polymerase II from their promoters20. These studies imply that Aire preferentially targets inactive chromatin, potentially using multiple mechanisms. However, it remains unclear which underlying rules govern patterning of thymic TRA expression at the single-cell level, such that the composite of mTECs reliably covers the combined transcriptomes of peripheral tissues. It is also unclear whether each mTEC samples a random set of TRAs or whether there are constraints on the set of TRAs that individual mTECs express. Likewise, it remains elusive how thymic TRA expression is coordinated at the intra- and inter-cellular levels in time and space, and how stable these patterns are throughout the lifetime of an individual mTEC.
Previous studies have addressed some of these questions by applying bulk transcriptome analysis, single-cell multiplex PCR and single-cell RNA-sequencing (scRNA-seq)10, 12, 19, 21. These studies indicated that single mTECs express TRA genes of diverse functional categories, thus arguing against the notion that thymic TRA expression mimics tissue-specific gene expression patterns at the single-cell level. However, while multiple studies using single-cell approaches did not discern TRA co-expression patterns in single mouse mTECs10, 19, 21, a recent study on human mTECs provided evidence for TRA co-regulation within single cells12. Identifying the molecular mechanisms that regulate thymic TRA expression in single cells is key to understanding how self-antigen diversity, a prerequisite of self-tolerance, is generated in the mTEC compartment.
Hence, we applied scRNA-seq to mouse mTECs and studied single-cell expression profiles of 203 mature (MHCIIhi) mTECs, as well as 3 mature mTEC subsets that were selected for the expression of particular TRAs. We focused our study on mature mTECs, as they represent the mTEC subset mainly responsible for inducing self-tolerance in developing T cells by expressing the largest diversity of TRA-encoding genes. At the same time they are fully competent antigen presenting cells (APCs) expressing high levels of surface MHCII and CD80. Using this genome-wide approach, we found that the mature mTEC population at large is composed of numerous distinct TRA gene co-expression clusters. Each co-expression cluster comprises only a fraction of all genes, and individual clusters are expressed only in a small subset of mTECs. Our findings characterize thymic TRA expression as a highly regulated process, which ensures representation of the full diversity of self-antigens in the mTEC compartment by assembling a population composite of recurrent and complementing co-expression clusters that are present in individual cells.
Results
Comprehensive coverage of the immunological self by mature mTECs
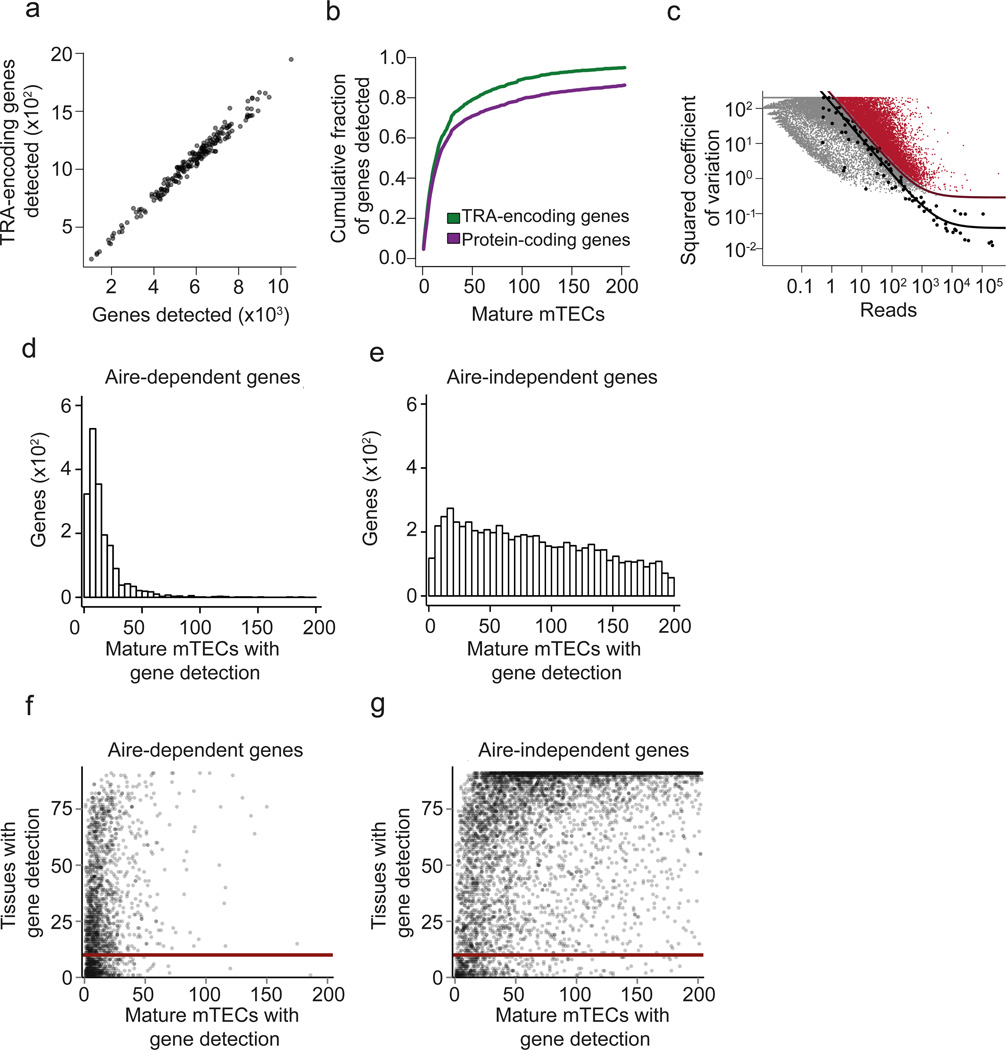
To investigate the extent of heterogeneity and patterning of thymic TRA expression in single mTECs, we performed scRNA-seq on mature MHCIIhi mouse mTECs (hereafter referred to as mature mTECs). Single mature mTECs (PI− CD45− Ly51− EpCAM+ MHCIIhi) were sorted from pooled thymic tissue of 5-20 female C57BL/6 mice (4-6 weeks old), and 211 single-cell cDNA libraries were generated using a modified version of the Smart-seq2 method22, 23. After data quality control, 203 cells (96%) were retained for further analysis (Supplementary Code I, Section 1). For each mTEC, we counted the number of protein coding genes and the number of TRA-encoding genes (i.e. a subset of protein coding genes) whose expression was detected by scRNA-seq. We found that the number of detected TRA-encoding genes within single cells was proportional to the total number of detected genes (19 ± 3.6% of detected genes were classified as TRAs) (Fig. 1a and Supplementary Fig. 1). We did not observe evidence for cell-to-cell variation in the proportion of expressed TRA-encoding genes, as the variation in the number of detected TRAs per mTEC can be explained by varying sequencing coverage (Fig. 1a). Moreover, 95% of the previously reported 3,976 TRA-encoding genes12 were cumulatively detected in the 203 analyzed mature mTECs (Fig. 1b). In addition, the scRNA-seq assay cumulatively detected the expression of 86% of all annotated protein-coding genes in the 203 analyzed mature mTECs (19,619 out of 22,740 genes; Ensembl release 75) (Fig. 1b), indicating that close to 90% of the protein-coding genome was sampled across a few hundred mature mTECs. These data document a comprehensive representation of the immunological self in mature mTECs at the population level, as recently suggested19, 24.
Figure 1.
Mature mTECs are heterogeneous at the single-cell level but express a comprehensive set of TRAs as a population. (a) Scatterplot of scRNA-seq assay showing the number of detected TRA genes versus the number of total genes detected in single mature mTECs (n=203) isolated from pooled thymic tissue of 4-6 weeks old C57BL/6 wild type mice. Semitransparent coloring has been used to ameliorate over-plotting. (b) Cumulative fraction of detected TRA-encoding genes (green line) and all protein coding genes (purple line) with increasing number of mTEC transcriptomes (n=203). (c) Identification of 9,689 significantly highly variable genes across single mature mTECs (n=203) using a published method.25. Genes with a biological squared coefficient of variation (SCV) of more than 0.25 at 10% FDR were classified as highly variable (colored in red). Black points represent ERCC RNA spike-ins, the solid black line shows the model fit for the technical noise, and the purple line depicts the threshold of 0.25 biological SCV (i.e. 50% CV). (d) Histogram showing the number of Aire-dependent genes (y-axis) as a function of the number of mature mTECs (n=203) for which the gene was detected (x-axis). (e) Same plot as in Fig. 1d, but showing the data for Aire-independent genes. (f) Scatterplot of the number of tissues in which individual genes are detected in the FANTOM dataset26 (y-axis) plotted as a function of the number of mature mTECs (n=203) in which the gene was detected. Each data point represents one Aire-dependent gene. The solid red line shows the value of 10 on the y-axis (i.e. threshold on the number of tissues described in the main text). (g) Same plot as in Fig. 1f, but showing the data for Aire-independent genes.
Next, we identified genes whose expression was highly variable across the 203 single mTECs using a published method25. This analysis revealed a high degree of gene expression heterogeneity across mTECs, with 9,689 genes having a biological coefficient of variation (CV) larger than 50% (i.e. a squared coefficient of variation (SCV) larger than 0.25) at 10% FDR (Fig. 1c). When compared to all protein coding genes, this set of highly variable genes was enriched for TRA-encoding genes (odds ratio=2.2, _p_-value < 2.2×10−16, Fisher’s Exact Test). More specifically, 26% of the highly variable genes were TRA-encoding, while only 14% of the genes not detected as highly variable were TRA-encoding (Supplementary Fig. 2). Thus, mature mTECs represent a cell type that is highly heterogeneous at the level of individual cells, and yet collectively seem to reliably express most of the genome.
TRA-encoding genes are generally expressed mosaically
Next, we investigated the Aire dependence of TRA expression in single mature mTECs. For this analysis, we integrated our single-cell gene expression data with the transcriptome atlas of 91 cell types (88 primary cell types and 3 cell lines) acquired by the FANTOM consortium26 and a list of Aire-regulated genes19. We found that Aire-dependent genes were expressed in a smaller fraction of mTECs compared to Aire-independent genes (Fig. 1d,e). Moreover, we found that genes with tissue-restricted expression patterns in the periphery of the body were expressed at low frequencies in single mTECs, irrespective of Aire regulation (Fig. 1f,g). When considering a set of 912 genes that were detected in at most 10 out of the 91 cell types from the FANTOM data set, 522 genes were Aire-dependent and 390 were Aire-independent. Out of the 522 Aire-dependent genes, 94% (492) were detected in less than 15% of our single mature mTECs (Fig. 1f). In a similar manner, out of the 390 Aire-independent genes, 68% (265) were detected in less than 15% of mTECs (Fig. 1g). These results indicate that genes whose expression tends to be restricted to fewer cell types in the periphery of the body are generally expressed at low frequencies in mature mTECs, with a more pronounced effect for Aire-dependent genes.
TRA expression patterns in single mature mTECs are non-random
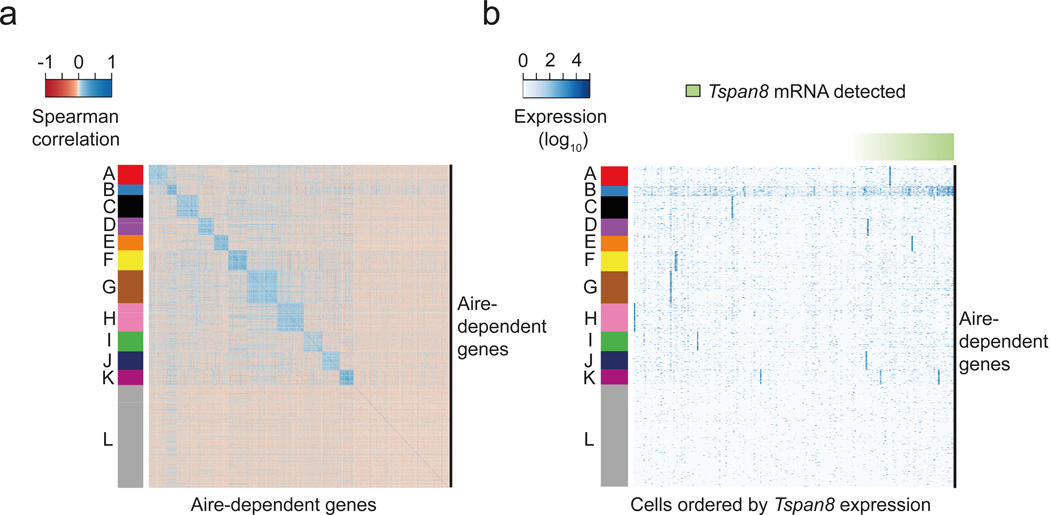
Next, we addressed whether TRA expression in single mTECs occurs randomly, i.e. without noticeable gene co-expression patterns10, 19, 21 or instead is governed by rules of gene co-regulation12. Because the cell cycle was a potential confounding factor, due to many genes being co-regulated in a cell-cycle dependent manner, we first regressed out cell cycle variation from the 203 mature mTEC single-cell transcriptomes using the scLVM method27. Next, we used k-medoids clustering to group highly variable Aire-dependent genes based on their level of expression across cells, and assessed the statistical stability of the clustering by resampling (Supplementary Code I, Section 6)28. We identified 11 stable gene clusters (A-K) that showed patterns of co-expression, and one cluster (cluster L) that grouped together genes for which the data provided no evidence for co-expression (Fig. 2a). Most of these co-expression patterns showed high expression only in a small fraction of mature mTECs (Fig. 2b). This is consistent with the previous identification of three distinct co-expression groups at low cell frequencies in human mTECs12. We observed a notable exception for co-expression cluster B, which was present in a larger fraction of cells (Fig. 2a,b). These results suggest that co-expression patterns exist in single mTECs and that the regulation of TRA-encoding genes follows discernible patterns in individual mature mTECs.
Figure 2.
The mature mTEC population consists of numerous low-frequency TRA co-expressed gene sets. (a) Heatmap depicting the pair-wise Spearman correlation matrix of expression profiles of 2,174 highly variable Aire-dependent genes (identified in Fig. 1c) across mature mTECs (n=203). The colors of the vertical bar depict 12 co-expressed gene sets identified by k-medoids clustering. (b) Heatmap representation of the gene expression levels of highly variable Aire-dependent genes across individual mature mTECs (n=203). The row ordering is the same as in Fig 2a. Columns represent individual mature mTECs ordered by the expression levels of Tspan8 (green horizontal bar). Cluster B (colored in blue) represents the set of genes co-expressed with Tspan8. Cluster L (colored in grey) contains genes for which no evidence for co-expression was found.
TRA co-expression occurs irrespective of Aire dependence
To further evaluate the concept of co-expression patterns in single mTECs, we chose an independent in silico analytical approach to test for co-expression of TRA-encoding genes within mature mTECs (n=203). To this end, we selected an Aire-dependent TRA, Tspan8 (Tetraspanin-8), which belonged to gene cluster B (identified in Fig. 2a). We detected Tspan8 expression in 66 out of the 203 mature mTECs (~33%). Next, we tested each of the 9,689 highly variable genes (identified in Fig. 1c) for whether they were more highly expressed in the 66 cells for which we detected Tspan8 mRNA than in the remaining 137 mTECs that were Tspan8-negative. Because both Aire-dependent and -independent genes are concomitantly up-regulated upon differentiation into mature mTECs, both gene sets were considered for testing. Using this approach, we identified 595 genes as co-expressed with Tspan8 at a FDR of 10%, further referred to as the “Tspan8 co-expressed gene set” (Supplementary Table 1). The Tspan8 co-expressed gene set consisted of 129 Aire-dependent genes and 466 Aire-independent genes (Supplementary Table 1). Consistent with the k-medoids clustering analysis (Fig. 2a), the 129 Aire-dependent genes showed a high overlap with the genes from cluster B as compared to the other gene clusters (p-value < 2.2×10−16, odds-ratio=22, Fisher’s Exact Test) (Supplementary Fig. 3).
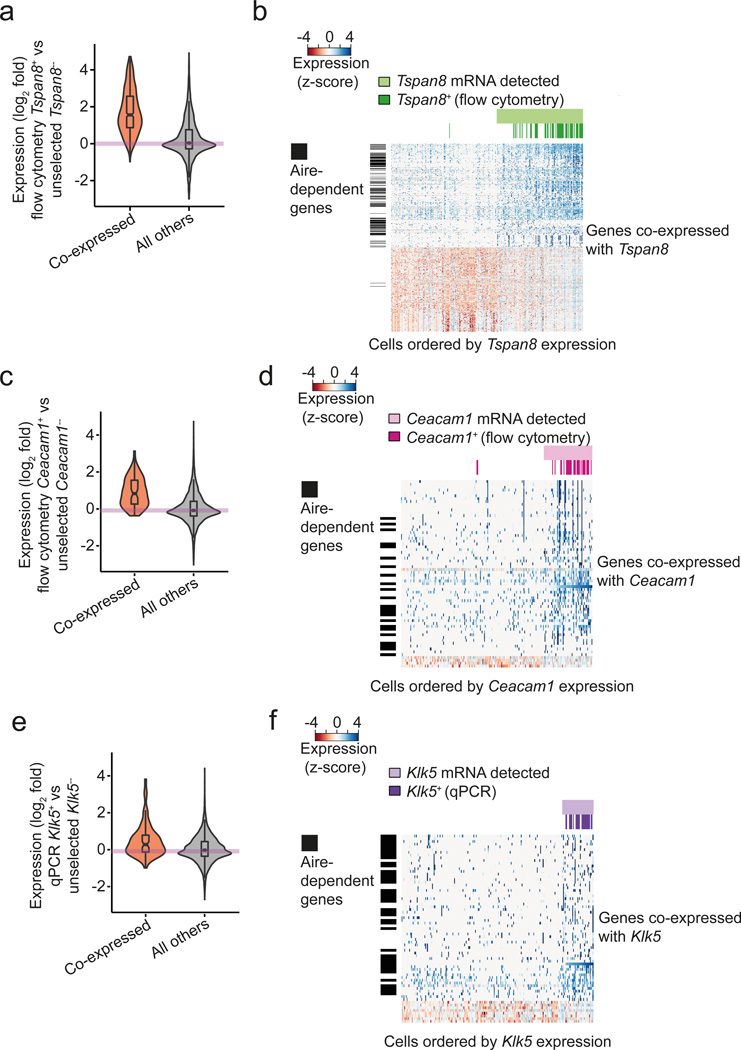
We then independently validated the Tspan8 co-expressed gene set by using flow cytometry to sort single mTECs expressing Tspan8 on the cell surface, using a setup published recently for human mTECs12. We sequenced single-cell cDNA libraries from 48 Tspan8+ mature mTECs (PI− CD45− CDR1− EpCAM+ MHCIIhi Tspan8+). We found that the patterns of co-expression for both Aire-dependent and -independent genes were highly concordant between these 48 sorted Tspan8+ mTECs and the 66 unselected mature mTECs for which the expression of Tspan8 mRNA was detected ad hoc (Fig. 3a,b). Specifically, 96% of the genes belonging to the Tspan8 co-expressed gene set were also up-regulated in the 48 sorted Tspan8+ cells (Fig. 3a and Supplementary Fig. 4, p-value < 2.2×10−16, t-test).
Figure 3.
Co-expressed gene sets are validated by independent experimental approaches. (a) Distribution of gene expression fold-changes (logarithm base 2) between the 48 FACS-selected Tspan8+ mature mTECs and the 137 unselected mature mTECs for which Tspan8 mRNA was not detected by scRNA-seq. The orange violin represents the co-expressed gene set (Supplementary Table 1) and the gray violin represents the data for all other genes. The distribution of fold changes is different between the two gene sets (p < 2.2×10−16). (b) Heatmap representation of expression levels of genes in the Tspan8 co-expressed gene set across the unselected mTECs (n=203) and the pre-selected Tspan8+ mTECs (n=48). Columns represent individual cells (ordered by increasing Tspan8 transcript levels as measured by scRNA-seq) and the rows represent genes co-expressed with Tspan8 (Supplementary Table 1). Cells for which Tspan8 expression was detected by scRNA-seq are labeled in light green, and the preselected Tspan8+ mTECs are colored in dark green. Vertical black bars label Aire-dependent genes. (c) Analogous results as shown in Fig. 3a for the Ceacam1 co-expressed gene set (unselected Ceacam1− mTECs n=172; preselected Ceacam1+ mTECs (n=30). The distribution of fold changes is different between the two gene sets (p = 9.8×10−11). (d) Analogous results as shown in Fig. 3b for the Ceacam1 co-expressed gene set (unselected mTECs (n=203); preselected Ceacam1+ mTECs (n=30). (e) Analogous results as shown in Fig. 3a for the Klk5 co-expressed gene set (unselected Klk5− mTECs n=190; preselected Klk5+ mTECs n=24). The distribution of fold changes is different between the two gene sets (p = 8.2×10−5). (f) Analogous results as shown in Fig. 3b for the Klk5 co-expressed gene set (unselected mTECs (n=203); preselected Klk5+ mTECs (n=24)).
To further corroborate co-expression in mature mTECs for both Aire-dependent and Aire-independent genes, we repeated the strategy followed for Tspan8 for two additional TRAs. First, we selected the cell adhesion protein Ceacam1, an Aire-independent TRA that was detected as co-expressed with Tspan8 (Supplementary Table 1). As for Tspan8, we screened the 203 mature mTECs for the presence of Ceacam1 transcripts and detected the expression of Ceacam1 in 15% of the mature mTECs (31 out of the 203 cells). We found 65 genes (23 Aire-dependent and 42 Aire-independent ones) to be co-expressed with Ceacam1 at a FDR of 10%, further referred to as the “Ceacam1 co-expressed gene set” (Supplementary Table 1). Next, we validated the Ceacam1 co-expressed gene set by sequencing 30 single mTECs that were selected by flow cytometry for the surface expression of Ceacam1 (PI− CD45− CDR1− EpCAM+ MHCIIhi Ceacam1+)(Fig. 3c,d). Out of the 65 genes belonging to the Ceacam1 co-expressed gene set, 92% showed consistent up-regulation in the FACS-selected Ceacam1+ mTECs when compared to the unselected Ceacam1− mTECs (Fig. 3c,d and Supplementary Fig. 4, p-value = 9.8×10−11, t-test).
Both Tspan8 and Ceacam1 are relatively frequently expressed across the mature mTEC population (33% and 15% respectively). Thus, we also tested a TRA-encoding gene that was expressed at a more representative frequency, the TRA gene Klk5, which was assigned to the cluster D in the k-medoids clustering. As for Tspan8 and Ceacam1, we defined the Klk5 co-expressed gene set based on 13 out of the 203 mature mTECs (6.4%) for which we detected the presence of Klk5 transcripts (Supplementary Table 1). The Klk5 co-expressed gene set consisted of 68 genes, i.e. 39 Aire-dependent and 29 Aire-independent genes (Supplementary Table 1). Consistent with the k-medoids clustering, these 39 Aire-dependent genes were significantly enriched among the genes from cluster D as compared to the rest of the clusters (Supplementary Fig. 5, odds-ratio=4.7, p-value = 8.2×10−5 Fisher’s exact test).
We validated the Klk5 co-expressed gene set experimentally by screening 562 mature mTEC cDNA libraries that had been confirmed to be positive for the housekeeping gene Ubc by qPCR. 28 out of the 562 mTECs (5.0%) were also positive for Klk5 as determined by qPCR (data not shown). Next, we sequenced the transcriptomes of 24 of the Klk5+ mTECs (see Methods). In agreement with the 13 unselected mature mTECs for which we detected the expression of Klk5 transcripts, 71% of the genes from this defined Klk5 co-expressed gene set (Supplementary Table 1) showed a consistent up-regulation in the qPCR-selected Klk5+ mature mTECs (Fig. 3e,f and Supplementary Fig. 4, p-value = 8.2×10−5, t-test). Interestingly, this concordance was particularly pronounced for the genes neighboring Klk5 in the genome (discussed below).
In addition, while we found that the three co-expressed gene sets were enriched for TRA-encoding genes (Tspan8 p-value < 2.2×10−16, Ceacam1 p-value = 7×10−15 and Klk5 p-value = 1.3×10−4, Fisher’s exact test), they were not restricted to genes classified as TRAs (according to the TRA definition used in this study). Taken together, we identified patterns of co-expression by ad hoc transcriptome analysis of 203 single unselected mature mTECs and by transcriptome sequencing of subsets of mature mTECs pre-selected based on the surface expression of three TRA-encoding genes of varying population frequency, Tspan8, Ceacam1 and Klk5.
Potential genealogies within mTEC co-expression groups
We found a statistically significant overlap of the genes from the Ceacam1 and Tspan8 co-expression groups (p-value < 2.2×10−16 odds-ratio=23.5, Fisher’s Exact Test). Specifically, 39 genes belonging to the Ceacam1 co-expressed gene set (i.e. 60%) were also detected as being co-expressed with Tspan8 (Supplementary Table 1). Despite this high overlap, we also identified 27 genes (40% of the Ceacam1 co-expressed gene set) detected as being co-expressed only with Ceacam1 (Supplementary Table 1); and 557 (93% of the Tspan8 co-expressed gene set) detected as being co-expressed only with Tspan8 (Supplementary Table 1). A model in which single mTECs would sequentially shift through distinct co-expression groups throughout their lifespan has been previously suggested12, implying the existence of overlapping co-expression patterns in mTECs during their transition between distinct groups.
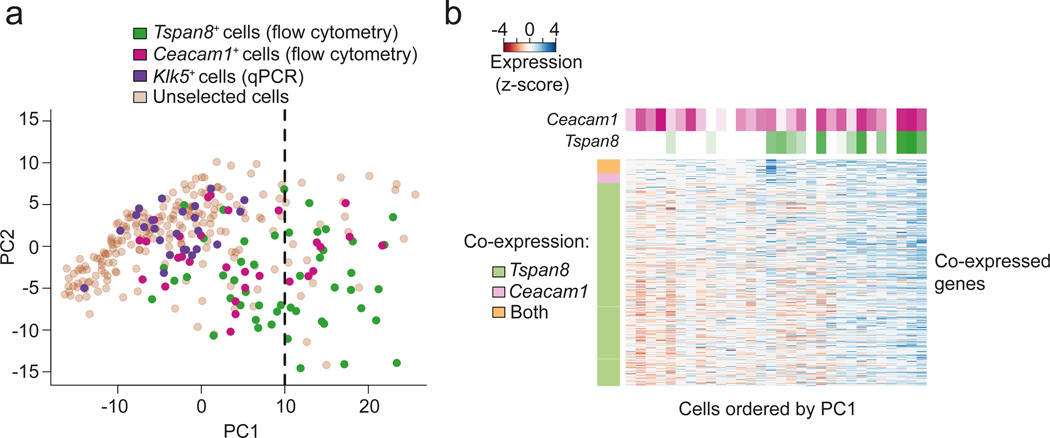
To explore this hypothesis, we visualized the interrelationships between the expression profiles of all single cells (n=305; i.e. 203 unselected, 48 FACS-selected Tspan8+, 30 FACS-selected Ceacam1+, and 24 qPCR-selected Klk5+ mature mTECs) by Principal Component Analysis (PCA). PCA was carried out on the expression data of all co-expressed genes in the Ceacam1 and Tspan8 co-expressed gene sets (i.e. the union of the two co-expressed gene sets; Fig. 4a). The dominant axis of gene expression variation, principal component 1 (PC1), separated the FACS-selected Tspan8+ (n=48) and Ceacam1+ (n=30) cells from the rest of the cells, with the Tspan8+ cells being further separated than the Ceacam1+ cells (Fig. 4a). 52% of the FACS-selected Tspan8+ mature mTECs had a PC1 projection (position along the x-axis) higher than 10, compared to 27% of the FACS-selected Ceacam1+ cells (Fig. 4a). Only 10% of the unselected mTECs and none of the qPCR-selected Klk5+ cells had a PC1 projection higher than 10 (Fig. 4a). These results suggest that a single gene expression program underlies most of the observed cell-to-cell variability of the selected genes, and that the Tspan8+ mTECs show a more pronounced adoption of this program compared to the Ceacam1+ mTECs.
Figure 4.
Tspan8 and Ceacam1 co-expressed gene sets overlap and corresponding mTECs are organized along a gradient of Tspan8 expression. (a) Principal Component Analysis (PCA) of all sequenced mature mTECs (n=305, i.e. 203 unselected mTECs, 48 Tspan8+, 30 Ceacam1+, 24 Klk5+ mTECs). Expression levels of genes in the union of the Tspan8 and Ceacam1 co-expressed gene sets were used for the PCA. Tspan8+ cells are colored in green, Ceacam1+ cells in magenta, Klk5+ cells in purple, and the unselected cells in brown. The dashed line indicates the value of 10 along the PC1 projection (i.e. the threshold used in the main text). (b) Heatmap representation of genes detected as being co-expressed with Tspan8 and Ceacam1 in the mature preselected Ceacam1+ mTECs (n=30). Rows correspond to genes of the Tspan8 and the Ceacam1 co-expressed gene sets, and the vertical color bar on the left indicates whether a gene was detected as co-expressed with only one or both of the surface TRA markers. Columns correspond to individual mature preselected Ceacam1+ mTECs and are ordered according to the first principal component from Fig. 4a. The horizontal color bars at the top indicate the mRNA expression levels of Tspan8 and Ceacam1 in individual mTECs.
To further expand this finding, we quantified the expression of Tspan8 mRNA (from scRNA-Seq) within the Tspan8+ and the Ceacam1+ mTECs. We found that Tspan8 mRNA expression correlated with the mean expression of all genes from the union of the Tspan8 and Ceacam1 co-expressed gene sets (Spearman correlation=0.62; Supplementary Code I, Section 7.4). The correlation was still present when considering exclusively the Ceacam1+ mTECs (Spearman correlation = 0.35, Fig. 4b). Thus, the amount of Tspan8 mRNA in Ceacam1+ mTECs was concomitant with increased expression of the co-expressed genes and increasing similarity to Tspan8+ mTECs. These data are consistent with the hypothesis of transitioning of individual mTECs from one co-expression group to another12.
Co-expressed genes cluster in the genome
One possible mechanism for the generation of non-random co-expression patterns could be local chromatin configurations that would allow the ectopic expression of neighboring genes, irrespective of their regulation in peripheral tissues6. Ectopic expression of gene clusters has been reported in human and mouse mTECs10, 12, 15, 29, 30. However, because inference of clustered gene expression from heterogeneous cell populations would be misleading due to averaging of different gene expression patterns from individual cells, only transcriptome-wide single-cell analysis can adequately address this point. Thus, for each of the 11 co-expression clusters, we calculated the median genomic distance between each gene to its nearest co-regulated gene neighbor within the same cluster. For each of the 11 clusters, we constructed a null model that allowed us to estimate the expected median genomic distance between genes given the size of the respective cluster (Methods and Supplementary Code I, Section 8). Based on these null models, the genes from 8 out of the 11 gene clusters were located in significant genomic proximity (FDR of 10%) (Supplementary Fig. 6). In order to visualize these effects, we plotted the localization of each of the 11 gene clusters resulting from the k-medoids clustering in a karyogram representation (Supplementary Fig. 7). Despite being dispersed across the genome, numerous genes from the same gene co-expression cluster were densely clustered in specific loci (exemplified by co-expression Cluster D; Fig 5a,b). Some of these loci comprised gene families encompassing structurally and functionally related genes. For example, 4 genes in cluster D belonging to the ‘BPI fold-containing family B’ (Bactericidal permeability-increasing protein-like 1) were located consecutively in the genome on chromosome 2 (Supplementary Fig. 8a), while two genes (Gstm2 and Gstm7) from the Glutathione S-transferase Mu gene family were close neighbors in the genome on chromosome 3 (Supplementary Fig. 8b). Importantly, we also identified clusters of neighboring genes that were co-expressed, but had no obvious functional relationship (Supplementary Fig. 8c).
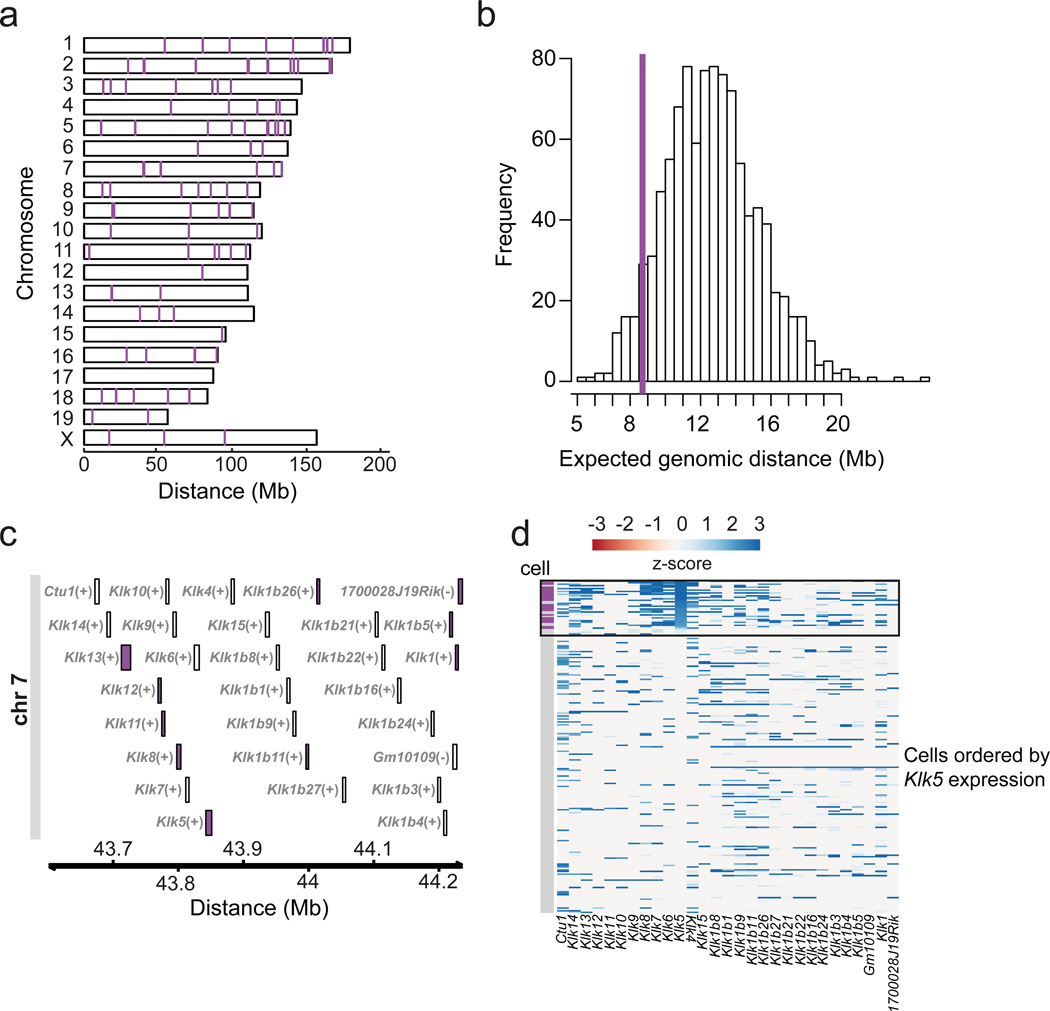
Figure 5.
Co-expressed genes cluster in the genome. (a) Karyogram depicting genomic localization of the genes from co-expressed gene set D (Fig.2 a, b). (b) Distribution of the expected median genomic distance between two genes in the genome (based on 1,000 permutations selecting random sets of genes of the same size as co-expressed gene set D). The purple line depicts the median distance observed for the 115 genes belonging to the co-expressed gene set D, which deviates from the null model (FDR = 10%). (c) Genomic region on chromosome 7 hosting the Kallikrein related-peptidase (Klk) protein family. The purple color indicates the genes assigned to cluster D by the k-medoids clustering (Fig. 2a). (d) Heatmap of gene expression profiles for the Klk gene family locus across single unselected mature mTECs (n=203) and qPCR-selected mature Klk5+ mTECs (n=24). Individual mTECs (y-axis) are ordered by decreasing Klk5 expression levels (from top to bottom). The order of genes (x-axis) corresponds to the genomic position of the genes (as in Fig. 4c). The black box highlights mTECs for which Klk5 transcripts are detected by scRNA-seq. Rows marked in purple (vertical bar) correspond to mature Klk5+ mTECs preselected by qPCR (n=24).
The Kallikrein protease gene cluster (Fig. 5c) provides a prominent example for a structurally and functionally related gene family. The locus consists of 27 genes that are located nearby on chromosome 7 (Fig. 5c). Nine of these genes, including Klk5, were assigned to cluster D (Fig. 5c). Moreover, we explored the gene expression patterns of the Klk genomic locus in both our unselected mature mTECs (n=203) and in the qPCR-selected Klk5+ mature mTECs (n=24). We found that Klk5 expression is a proxy for the expression of the neighboring Klk genes (Fig. 5d and Supplementary Fig. 9). These results show that TRA expression in mTECs involves co-expressed groups of genes located in close neighborhood in the genome.
Promoters of co-expressed TRAs map to accessible chromatin
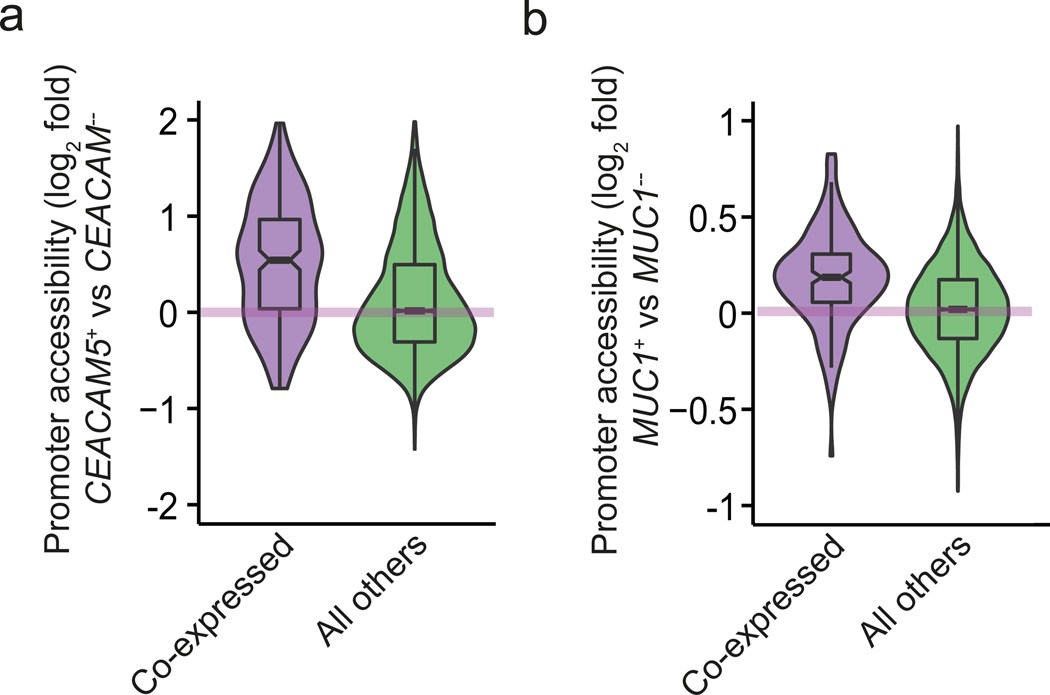
In order to directly assess the chromatin state for co-expressed genes, we assayed genome-wide DNA accessibility using the ATAC-seq method31, which is based on the preference of the TN5 transposase to integrate into un-compacted chromatin and thus allows a direct measurement of chromatin accessibility. To obtain a sufficient number of surface TRA-specific mTECs required for this assay, we used human thymic tissue and sorted for two previously published human co-expressed gene sets, namely the CEACAM5 and MUC1 gene sets12. The ATAC-seq experiments were performed in biological triplicates using mTECs from the respective surface TRA-positive and TRA-negative mTEC fractions. When accounting for all protein-coding genes, there was no difference in chromatin accessibility between the TRA-positive and -negative mTECs (Fig. 6a,b). However, we observed that gene loci that were co-expressed with the respective TRA-positive subsets (either CEACAM5 or MUC1) were significantly more accessible in the TRA-positive as compared to the TRA-negative mTECs (t-test, p-value=1.2×10−15 for CEACAM5 and p-value=1.1×10−14 for MUC1; Fig. 6a,b). Thus, gene co-expression in distinct mTEC subsets goes along with enhanced chromatin accessibility at the promoter regions of the respective gene loci.
Figure 6.
Promoters of co-regulated genes show increased chromatin accessibility. Violin plots showing moderated logarithmic fold changes (MLFC; base 2) in chromatin accessibility between human surface TRA-positive and respective surface TRA-negative mTEC subsets assayed by bulk ATAC-seq (n=3). MLFCs were calculated using the DESeq2 method44. (a) Violin representation of data from the human CEACAM5 co-expressed gene set (288 genes). The MLFC between CEACAM5-positive and CEACAM5-negative mTECs are depicted on the y-axis. The promoters (x-axis) are stratified into genes that have been shown previously12 to be co-expressed with CEACAM5 (purple violin) and the rest of the protein coding genes (green violin). Chromatin accessibility is higher for co-expressed genes (p = 1.2×10−15, t-test). (b) Violin representation as in Fig. 6a using data for the human MUC1 co-expressed gene set (219 genes), yielding analogous results (p = 1.1×10−14, t-test).
Discussion
TRA expression in mTECs is essential for self-tolerance induction. Yet, its molecular regulation remains poorly understood. One open question relates to the regulation of TRA expression in single mTECs, i.e. to what extent the process is random or follows rules. Here, we applied scRNA-seq22, 23, 32, 33, 34, 35, 36 and provide evidence for numerous recurring co-expression patterns in mature mTECs. These patterns generally occur at low cell frequencies. Co-expressed genes cluster in the genome, and their promoters display enhanced chromatin accessibility. Co-expressed gene sets form mosaic patterns that faithfully add up at the population level to present a comprehensive set of TRAs.
Mosaic gene expression patterns have been reported previously in the thymus10, 11, 12, and they allow for a high diversity of antigens to be presented at the population level, while limiting the number of TRA genes expressed in individual mTECs. As mTECs have a limited capacity for antigen presentation, restricting the number of ectopically expressed genes per cell appears crucial to ensure epitope presentation at sufficient density to transmit a tolerogenic signal to maturing T cells.
It has been proposed that mosaic expression patterns arise by random induction of TRA genes in single mTECs10, 19, 21; this model has been challenged by the discovery that subsets of human mTECs that were FACS-selected for the expression of particular TRAs displayed differential gene expression patterns12. However, these preselected mTEC subsets analyzed previously represented only a narrow subset of the mTEC population, because they were constrained by the availability of antibodies suitable for flow cytometry. The data provided herein substantially advance these findings, because the single-cell approach used here addressed the question of co-expression in a genome-wide unbiased way (i.e. no pre-selection required). The current depth of analysis allowed us to identify 11 novel co-expression patterns within the mature mTEC population. As the number of sequenced mTECs was limited (n=203), we expect this number to be an underestimate.
Nevertheless, even this relatively small number of mTECs covered 95% of the reported TRA-encoding genes. Given the size of the murine mTEC compartment of ~105 cells10, this finding implies that the complete TRA repertoire would be covered multiple times within the thymic medulla, even when allowing for a generous error margin in our calculations. Hence, T cells would only have to scan sub-domains of this compartment for efficient self-tolerance induction.
Moreover, by zooming in on the identified co-expression groups, we observed a positive correlation between Tspan8 transcript levels and increased expression of genes co-expressed with Tspan8 in both Ceacam1+ and Tspan8+ cells. This finding would be in line with a transitioning of individual cells between different co-expression groups, a concept recently proposed in a model12 that postulates that individual mTECs transit between different TRA co-expression patterns and thus might express a sizeable portion of the TRA repertoire during their lifetime. Such a mechanism could further reduce the minimal number of mTECs any single T cell would need to interact with to encounter the full TRA repertoire, because a given mTEC could express different TRAs when re-encountering the same T cell during its sojourn in the medulla37.
We could assign 71% of TRAs to a co-expression group based on 203 single mature mTECs. The remaining TRAs either escaped co-expression detection due to limited sample size, or represent some features of random sampling. In addition, the extent to which mono- versus bi-allelic expression, slipping promoter usage resulting in truncated mRNA isoforms and variable splicing patterns play a role is unclear6, 21, 38, 39. These latter features might extend the diversity of thymic self-antigen presentation; at the same time, they could represent pitfalls of thymic TRA expression that potentially undermine the process of tolerance-induction and may lead to auto-immunity38, 39.
Our single-cell data show that co-expressed genes tend to cluster in the genome. In conjunction with our ATAC-seq experiments, this suggests a potential mechanism for the generation of intra- and inter-chromosomal co-expression patterns. Such a mechanism would rely on local chromatin remodeling that allows neighboring genes to be co-expressed in a coordinated fashion in single mTECs, irrespective of their distinct tissue-specific regulation in the periphery. Although the definition of TRAs is operational and highly dependent on the thresholds employed, our observation that co-expressed gene sets also contain non-TRA genes could imply that TRA expression also promotes the expression of other genes adjacent to TRA genes. However, co-expressed gene sets were enriched for TRA genes, suggesting that the mechanism underlying co-expression patterns in mTECs is predominantly targeting genes whose expression in the periphery of the body is restricted to a small number of tissues.
Chromatin re-modeling can affect nearby genes on the same chromosome but also genes nearby in the 3-dimensional architecture of the nucleus. A correlation between gene co-expression and co-localization in transcription factories has been described for lineage-specific gene regulation40, and this might also be the case for thymic TRA expression12. The fact that co-expressed gene clusters can contain genes of unrelated biological function further supports our proposition that genomic positions influences thymic TRA expression.
Epigenetic signatures specifying such “accessible” chromatin stretches in mTECs have not yet been investigated genome-wide. However, a study focusing on the casein gene locus in murine mTECs showed ectopic gene expression of the casein beta gene to correlate with marks of active transcription41. Thus, it will be interesting to identify the molecular pathways that target co-expressed gene clusters; and moreover, to define the transcriptional regulators that promote transcription. In this context, spatially localized activation of gene expression by epigenetic remodeling, as proposed here for TRA expression in mTECs, has been reported for embryonic stem cells42 and cancer cells43.
Why mTEC-mediated tolerance induction, which presumably evolved in early vertebrates, employs coordinated co-expression patterns in single cells remains an intriguing question. If cells were to coordinate their expression programs with each other (e.g. to avoid expressing the same genes and thus ensure maximal coverage), then co-expression groups could provide an economic means to implement such coordination, compared to a fully independent, cell-autonomous choice of every single gene.
Online Methods
Mice
C57BL/6 mice were used in this study for the isolation of mTECs. All breeding and cohort maintenance were performed in the central animal laboratory of the German Cancer Research Center (DKFZ) under approved conditions in accordance with the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes and the German Legislation.
Isolation of mouse medullary thymic epithelial cells
Mouse mTECs were isolated and purified as described previously45 pooling cells from 5-20 mice per experiment. The pre-enriched stromal cell fraction, sorted for unselected mature mTECs (n = 211 cells) was stained using the following antibodies: anti-CD45-PerCP (clone 30-F11, BD Pharmingen), anti-EpCAM-Alexa647 (G8.8, described by Farr et al.46), anti-CDR1-Pacific Blue (CDR1 hybridoma, described by Rouse et al.47) or anti-Ly51-FITC (clone 6C3, BD Biosciences), and anti-MHCII-PE (clone 16-10A1, BD Biosciences).
For the surface TRA-selected mTECs Tspan8 (n = 48) and Ceacam1 (n = 30), additional antibodies were added to the antibody mix namely: anti-I-A(b)-FITC (clone AF6-120.1, BD Pharmingen), anti-Tspan8-PE (clone 657909, R&D Systems) and anti-CD66a-PE (i.e., anti-Ceacam1, clone CC1, eBioscience). Dead cells were excluded using propidium iodide (PI) in a final concentration of 0.2 µg/ml. Cells were sorted on BD FACSAria™ III cell sorter (BD Biosciences) using the single-cell sorting mode as previously described10. Single mature mTEC used in all the experiments represent cells from pooled thymic tissue.
Single-cell RNA-seq
Single-cell sequencing libraries were prepared as reported previously22, 23 with the following modifications: 1 µl of a 1:1,000,000 dilution of the ERCC spike-in mix (Life Technologies) in RNase-free water was included in a total volume of 5 µl lysis buffer. During analysis, sequencing reads mapping to ERCC spike-ins were used to estimate technical noise levels and call significantly highly variable genes using the method of Brennecke et al.25. We used 19 cycles of initial PCR amplification and used a ratio of 0.6:1 (instead of a 1:1 ratio) of Ampure XP beads (Beckman Coulter) for the first PCR purification to minimize primer dimer carryover. After the first PCR amplification, cDNA libraries were screened via qPCR (we used a 1:10 dilution of purified cDNA libraries for qPCR reactions) for expression of a mouse housekeeping gene (Ubc), and library size distribution was checked on the Bioanalyzer instrument (Agilent) as reported previously22, 23. Only cDNA libraries that passed both quality controls were processed further. We used 100 pg of cDNA for the tagmentation reaction and applied 12 cycles for the final enrichment PCR. The last purification step was performed using a 0.8:1 ratio of Ampure SPRIselect beads (Beckman Coulter). We multiplexed 24 samples per Illumina HiSeq 2500 lane and used 105 bp paired-end sequencing. A HiSeq sequencing lane typically yielded between ~150 and ~200 million reads.
ATAC-Seq
Human thymic tissue was obtained from children (biological triplicates) in the course of corrective cardiac surgery at the Department of Cardiac Surgery, Medical School of the University of Heidelberg, Germany. Studies on human samples were approved by the Institutional Review Board of the University of Heidelberg (367/2002), and informed consent was obtained from all patients. Human mTEC subsets (surface TRA-positive and -depleted cells that were MHCIIhigh) were isolated and FACS sorted as described previously12. ATAC-seq experiments were performed as reported previously31 with the following modifications: 5,000 – 50,000 pooled cells (depending on mTEC subset frequencies) were sorted in FACS buffer (PBS containing 5% FCS) and used for ATAC-seq experiments. We used 50% of each purified tagmentation reaction for the enrichment PCR (without 5 cycle pre-amplification). Enrichment PCRs were monitored individually using a StepOnePlus Real-Time PCR System (Life Technologies) and the amplification reaction was stopped as soon as amplification approached saturation. After the enrichment PCR and subsequent PCR product purification, we performed a gel extraction (QIA MinElute Gel Extraction Kit, QIAGEN) to remove primer dimers. Final multiplexed sequencing libraries were quantified using qPCR and sequenced on a HiSeq 2500 machine (Illumina). 105 bp paired-end sequencing was used and samples yielded between 16,867,055 and 40,820,441 sequenced fragments.
Klk5 co-expressed gene set validation by qPCR
Single-cell cDNA libraries of mature mTECs were prepared as described above. Libraries were purified after 19 cycles of PCR amplification using a ratio of 0.6:1 of Ampure XP beads (Beckman Coulter). 1:10 dilutions (in nuclease-free water) of the cDNA libraries were used for subsequent qPCR pre-screening. Primers were designed using the NCBI Primer-BLAST tool. Single-cell cDNA libraries that were positive for both the Klk5 gene and the housekeeping gene Ubc were processed further for Illumina sequencing. Since we used the 24-sample Illumina dual indexing kit, only 24 out of the 28 Klk5 positive cells (instead of the 28 identified) were subjected to Illumina sequencing.
Bioinformatics
For the single-cell data, we mapped the sequenced read fragments using GSNAP version 2014-07-04 to the Mouse reference genome (ENSEMBL release 75). Only uniquely mapped sequenced fragments were considered for further analysis. For each single-cell transcriptome, we tabulated the number of sequenced fragments overlapping with each gene using HTSeq, and normalized for sequencing depth using the method of Anders et al.48. In order to account for technical variation, we used the method by Brennecke et al.25 to identify genes whose biological coefficients of variation were larger than 50%, and we used this subset for further analysis. We used the method by Buettner et al.27 to regress out the variation on the data explained by the cell cycle. We identified groups of co-regulated genes using the partitioning around medoids (pam) method of the R package “cluster” and assessed their stability using the R package “clue”. In order to identify genes as being co-expressed with TRA genes, we tested for association using the Wilcoxon test. Multiple testing corrections were done using the Benjamini-Hochberg method. The ATAC-seq data were mapped to the Human reference genome (ENSEMBL release 75) using GSNAP 2014-07-04.
Code availability
In Supplementary Code I we provide a comprehensive and reproducible workflow containing the documented R code used for the analysis of all the data, including the generation of all reported figures and summary statistics.
Supplementary Material
C ode
Figures
Table
Acknowledgements
The authors thank K. Hexel and S. Schmitt (FACS facility, DKFZ) for single-cell sorts, and S. Egle (DKFZ) for technical help. C. Sebening and T. Loukanov (University of Heidelberg) for providing human thymic tissue. The Genomics Core Facility (EMBL) for initial sequencing and M. Miranda and E. Hopmans (Stanford University) for support during subsequent sequencing at the Stanford Genome Technology Center. J. Buenrostro (Stanford University) and C. Chabbert (EMBL) for discussions regarding ATAC-seq experiments and data, respectively. C. Michel (DKFZ) and S. Anders (EMBL) for advice and comments on the manuscript. Wu Wei and Michael Sikora (Stanford University) for help with data transfer. The Central Animal Facility (German Cancer Research Center) for animal care taking. W.H. and A.R. acknowledge funding from the European Union’s 7th Framework Programme (Health) via Project Radiant. B.K., S.P. and K.R. acknowledge funding from The Helmholtz Center PhD Program Fellowship (K.R.), the SFB /DFG 938 (S.P.) and the European Research Council (Grant ERC-2012-AdG to B.K.). P.B., M.N. and L.S.M. acknowledge funding from the National Institutes of Health (NIH P01 HG000205 and NIH R01 GM068717).
Footnotes
Database accession numbers
The sequencing data were deposited to ArrayExpress under the accession identifiers E-MTAB-3346 and E-MTAB-3624.
Author Contributions
L.M.S., B.K., and W.H. supervised the project; L.M.S, B.K., P.B. and S.P. conceived the project; P.B., S.P., and K.R. designed experiments; P.B. performed single-cell sequencing experiments, Klk5 single-cell qPCR validation experiments, and ATAC-seq experiments; S.P. helped with the ATAC-seq experiments. S.P. and K.R. performed experimental mTEC preparations and single and bulk mTEC FACS; A.R. and W.H. designed analysis strategy and analyzed the data; A.R. prepared the figures; P.B., A.R., S.P., K.R., W.H., B.K. and L.M.S. interpreted the data and wrote the manuscript. M.N. and R.K. provided technical assistance.
Competing Financial Interest
The authors declare no competing financial interest.
References
- 1.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298(5597):1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 2.DeVoss JJ, Anderson MS. Lessons on immune tolerance from the monogenic disease APS1. Current opinion in genetics & development. 2007;17(3):193–200. doi: 10.1016/j.gde.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nature reviews Immunology. 2005;5(10):772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 4.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nature reviews Immunology. 2014;14(6):377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nature immunology. 2001;2(11):1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 6.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 7.Perry JS, Lio CW, Kau AL, Nutsch K, Yang Z, Gordon JI, et al. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 2014;41(3):414–426. doi: 10.1016/j.immuni.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015 doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 2013;339(6124):1219–1224. doi: 10.1126/science.1233913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci U S A. 2008;105(2):657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cloosen S, Arnold J, Thio M, Bos GM, Kyewski B, Germeraad WT. Expression of tumor-associated differentiation antigens, MUC1 glycoforms and CEA, in human thymic epithelial cells: implications for self-tolerance and tumor therapy. Cancer research. 2007;67(8):3919–3926. doi: 10.1158/0008-5472.CAN-06-2112. [DOI] [PubMed] [Google Scholar]
- 12.Pinto S, Michel C, Schmidt-Glenewinkel H, Harder N, Rohr K, Wild S, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A. 2013;110(37):E3497–E3505. doi: 10.1073/pnas.1308311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathis D, Benoist C. Aire. Annual review of immunology. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 14.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140(1):123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. The Journal of experimental medicine. 2005;202(1):33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A. 2008;105(41):15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO reports. 2008;9(4):370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterfield M, Khan IS, Cortez JT, Fan U, Metzger T, Greer A, et al. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nature immunology. 2014;15(3):258–265. doi: 10.1038/ni.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sansom SN, Shikama N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al. Population and single cell genomics reveal the Aire-dependency, relief from Polycomb silencing and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014 doi: 10.1101/gr.171645.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. 2012;109(2):535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci U S A. 2008;105(41):15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013 doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 23.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 24.St-Pierre C, Brochu S, Vanegas JR, Dumont-Lagace M, Lemieux S, Perreault C. Transcriptome sequencing of neonatal thymic epithelial cells. Scientific reports. 2013;3:1860. doi: 10.1038/srep01860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013 doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 26.Consortium F, the RP, Clst. Forrest AR, Kawaji H, Rehli M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015 doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nature cell biology. 2014;16(1):27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. The Journal of experimental medicine. 2004;199(2):155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnnidis JB, Venanzi ES, Taxman DJ, Ting JP, Benoist CO, Mathis DJ. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci U S A. 2005;102(20):7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012 doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5(3):516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 35.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, et al. Highly multiplexed and strand-specific single-cell RNA 5' end sequencing. Nat Protoc. 2012;7(5):813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 37.Le Borgne M, Ladi E, Dzhagalov I, Herzmark P, Liao YF, Chakraborty AK, et al. The impact of negative selection on thymocyte migration in the medulla. Nature immunology. 2009;10(8):823–830. doi: 10.1038/ni.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto S, Sommermeyer D, Michel C, Wilde S, Schendel D, Uckert W, et al. Misinitiation of intrathymic MART-1 transcription and biased TCR usage explain the high frequency of MART-1-specific T cells. European journal of immunology. 2014;44(9):2811–2821. doi: 10.1002/eji.201444499. [DOI] [PubMed] [Google Scholar]
- 39.Klein L, Klugmann M, Nave KA, Tuohy VK, Kyewski B. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nature medicine. 2000;6(1):56–61. doi: 10.1038/71540. [DOI] [PubMed] [Google Scholar]
- 40.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42(1):53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tykocinski LO, Sinemus A, Rezavandy E, Weiland Y, Baddeley D, Cremer C, et al. Epigenetic regulation of promiscuous gene expression in thymic medullary epithelial cells. Proc Natl Acad Sci U S A. 2010;107(45):19426–19431. doi: 10.1073/pnas.1009265107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 43.Bert SA, Robinson MD, Strbenac D, Statham AL, Song JZ, Hulf T, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer cell. 2013;23(1):9–22. doi: 10.1016/j.ccr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rattay K, Claude J, Rezavandy E, Matt S, Hofmann TG, Kyewski B, et al. Homeodomain-interacting protein kinase 2, a novel autoimmune regulator interaction partner, modulates promiscuous gene expression in medullary thymic epithelial cells. Journal of immunology. 2015;194(3):921–928. doi: 10.4049/jimmunol.1402694. [DOI] [PubMed] [Google Scholar]
- 46.Farr A, Nelson A, Truex J, Hosier S. Epithelial heterogeneity in the murine thymus: a cell surface glycoprotein expressed by subcapsular and medullary epithelium. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1991;39(5):645–653. doi: 10.1177/39.5.2016514. [DOI] [PubMed] [Google Scholar]
- 47.Rouse RV, Bolin LM, Bender JR, Kyewski BA. Monoclonal antibodies reactive with subsets of mouse and human thymic epithelial cells. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1988;36(12):1511–1517. doi: 10.1177/36.12.2461413. [DOI] [PubMed] [Google Scholar]
- 48.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C ode
Figures
Table