Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females (original) (raw)
Abstract
Osteoarthritis (OA) is a leading cause of disability in Western society with multiple risk factors, including a complex genetic pattern. Identifying loci involved in the heredity of OA might lead to insights into the molecular pathogenesis of this common disorder. A previous genome scan mapped a primary hip OA susceptibility locus to chromosome 2q with a maximum multipoint logarithm of odds score of 1.6 in 378 affected sibling pair families. Here, microsatellite targeting of eight candidate genes in this region from 2q23-2q32 demonstrated significant associations with the tumor necrosis factor α-induced protein 6 gene in all probands and the integrin α_6_ and frizzled motif associated with bone development (FRZB) genes in female probands. However, genotyping showed lack of association for a nonsynonymous single-nucleotide polymorphism in tumor necrosis factor α-induced protein 6, whereas a single-nucleotide polymorphism in FRZB resulting in an Arg324Gly substitution at the carboxyl terminus was associated with hip OA in the female probands (P = 0.04). This association was confirmed in an independent cohort of female hip cases (n = 338; P = 0.04). In addition, a haplotype coding for substitutions of two highly conserved arginine residues (Arg200Trp and Arg324Gly) in FRZB was a strong risk factor for primary hip OA, with an odds ratio of 4.1 (P = 0.004). FRZB encodes secreted frizzled-related protein 3, which is a soluble antagonist of wingless (wnt) signaling. Variant secreted frizzled-related protein 3 with the Arg324Gly substitution had diminished ability to antagonize wnt signaling in vitro. Hence, functional polymorphisms within FRZB confer susceptibility for hip OA in females and implicate the wnt signaling pathway in the pathogenesis of this disease.
Osteoarthritis (OA; Online Mendelian Inheritance in Man no. 165720) is the most prevalent form of arthritis and a common cause of disability. Epidemiological studies have demonstrated a major genetic component to primary OA, with estimated heritability of 40% for the knee, 60% for the hip, and 65% for the hand (1). Several genome-wide linkage scans have been performed on cohorts of affected sibling pairs or affected relative pairs concordant for hip, knee, or hand OA (reviewed in refs. 2 and 3). These scans revealed suggestive linkages with logarithm of odds (LOD) scores >1.5 on 12 chromosomes, underscoring the complex nature of the transmittance of OA susceptibility. Five loci were detected in more than one scan on chromosomes 2p (4, 5), 2q (6, 7), 7p (4, 8), 11q (4, 9), and 16p (10, 11). The regions identified by multiple genome-wide scans warranted further investigation to further validate these loci.
Previously, we mapped the 2q locus more precisely by using 378 families that contained one or more sibling pairs concordant for hip OA (6). The linkage centered at microsatellite marker D2S2284 (2q31.1) and had a maximum multipoint LOD score (MLS) of 1.6. Here, eight candidate genes spanning the region from 2q23-2q32 were selected based on their known roles in skeletal development or homeostasis. A microsatellite analysis revealed association to three of these genes: tumor necrosis factor α_-induced protein 6_ (TNFAIP6), integrin α_6_ (ITGA6), and frizzled motif associated with bone development (FRZB). Nonsynonymous single-nucleotide polymorphisms (SNPs) for these three genes were identified by using public databases and tested for association to hip OA. Only a FRZB substitution was associated.
FRZB encodes for secreted frizzled-related protein 3 (sFRP3), a glycoprotein that antagonizes the signaling of wingless (wnt) ligands through the frizzled membrane-bound receptors (12). Wnt signaling regulates the accumulation of cytoplasmic β-catenin, which is sequestered by a multiprotein complex, including glycogen synthase kinase 3β (13, 14). The phosphorylation of β-catenin by glycogen synthase kinase 3β marks it for ubiquitination and degradation. Inhibition of this activity by wnt/frizzled signaling allows unbound β-catenin to accumulate in the cytoplasm, translocate into the nucleus, and activate T cell factor/lymphoid-enhancing factor-dependent transcription (15). In vitro transfection assays demonstrated that sFRP3 can inhibit β-catenin nuclear translocation and T cell factor/lymphoidenhacing factor-dependent transcriptional activation. Here we show that sFRP3 with the Arg324Gly substitution identified in the SNP analysis has diminished ability to antagonize wnt signaling. The canonical wnt pathway is critical in skeletal and joint patterning in embryogenesis (16-19) and recently has been implicated as a determining factor of mature adult bone mass (20-22). We suggest that the reduced ability of variant sFRP3 to antagonize wnt signaling may predispose to hip OA in females by altering the development or stability of cartilage or bone in weight-bearing joints.
Materials and Methods
Patients for Association Analysis. Association analysis was carried out on the probands (220 females, 158 males) from 378 hip-affected sibling pair families described in ref. 6. These 378 hip families are composed of 328 families containing an affected sibling pair (136 female pairs, 77 male pairs, and 115 female-male pairs), 38 families containing an affected sibling trio (10 female trios, 5 male trios, and 23 mixed-sex trios), 6 families containing an affected sibling quartet (all mixed sex), and 6 families containing an affected sibling pair plus an affected uncle, aunt, or cousin. A second cohort comprised 558 unrelated hip OA cases (338 female and 220 male). The probands and cases were ascertained through the Nuffield Orthopaedic Centre in Oxford and had undergone total hip replacement for primary OA. The primary status was supported by clinical, radiological, operative, and histological findings. The control individuals consisted of 760 members (399 females and 361 males) of the general population who had not undergone joint replacement surgery. All cases and controls were aged 55 or over and were of U.K. Caucasian origin. Ethical approval for the study was obtained from appropriate ethics committees, and informed consent was obtained from all subjects.
Microsatellite Discovery and Genotyping. Microsatellite markers were identified for each of the eight candidate genes by using a sequence-based predictive test (23). The markers were PCR amplified, and the alleles were sized by using genescan and genotyper software (Applied Biosystems). Specific primer sequences are listed in Table 6, which is published as supporting information on the PNAS web site.
SNP Genotyping. The FRZB SNPs in exon 4 [+6] and exon 6 [+109] were genotyped by PCR-restriction enzyme analysis. The PCR reactions had a final volume of 15 μl and contained standard reagents and 50 ng of genomic DNA. The primers and restriction enzymes used are listed in Table 7, which is published as supporting information on the PNAS web site. Digestion products were electrophoresed through 3% agarose and scored after ethidium bromide staining. The accuracy of the genotyping was confirmed by the direct sequencing of the two SNPs in a panel of 48 individuals. The TNFAIP6 SNP was genotyped as described in ref. 24.
Molecular Haplotyping. FRZB SNPs exon 4 [+6] and exon 6 [+109] are 3752 bp apart. For compound heterozygotes, we amplified the two SNPs in a single fragment by using the exon 4 [+6] forward primer and the exon 6 [+109] reverse primer listed above and Elongase polymerase (Invitrogen). The 3922-bp product was then subcloned into Escherichia coli by using the TOPO XL PCR cloning kit (Invitrogen). Transformed colonies were picked into 10 μl of water, 5 μl of which was used to genotype the exon 4 [+6] SNP, and 5 μl was used to genotype the exon 6 [+109] SNP, as described above. Ten clones were genotyped for each compound heterozygote.
Statistical Analysis. clump (www.mds.qmw.ac.uk/statgen/dcurtis/software.html) was used to test each microsatellite marker for association to OA (25). Significance was assessed for each marker by carrying out 1,000 simulations to generate tables with the same marginal totals as the original data. Empirical P values were obtained by counting the number of times the χ2 value of the real data was achieved by the simulated tables. clump calculates four statistics: T1-T4. T1 is a standard contingency table χ2; T2 is a χ2 calculated after collapsing together alleles with a small expected value (<0.05); T3 is a 2 × 2 χ2 in which each allele is compared in turn to the rest; T4 is a 2 × 2 χ2 in which alleles demonstrating a frequency difference between cases and controls are clumped together and compared to the remainder. SNP association and Hardy-Weinberg equilibrium for the distribution of genotypes was tested by χ2 analysis with Yates' correction. Odds ratios were calculated with 95% confidence intervals. Multipoint linkage analysis was performed by using aspex (http://aspex.sourceforge.net).
FRZB Plasmid Constructs. IMAGE clone 360887 was purchased from Invitrogen and sequenced. Based on reference sequence GenBank NM_001463.1, there was a polymorphism at position 526 representing a C→G base change. The cDNA was subcloned into pcDNA3 (Invitrogen), and the 526 polymorphism was reverted to the reference sequence (pcFRZB) by using the QuikChange XL site-directed mutagenesis kit (Stratagene). The C→G change at exon 6 [+109] was engineered by subcloning a PCR fragment between the internal _Hin_dIII site and an _Xba_I site following the stop codon (pcFRZB-Arg324Gly). The C→T polymorphism at exon 4 [+6] was introduced into pcFRZB and pcFRZB-Arg324Gly (creating pcFRZB-Arg200Trp and pcFRZB-Arg200Trp/Arg324Gly, respectively) by using the QuikChange XL kit. Further details are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Cell Transfections. The human embryonic kidney cell line HEK293 (American Type Culture Collection) was transfected by using SuperFect (Qiagen, Valencia, CA). For luciferase assays, typically cocktails of 0.25 μg of pTOPFLASH-Luc reporter gene vector, 0.025 μg of β-galactosidase expressing plasmid pACB-Z, 0.25 μg of β-catenin-expressing plasmid (kindly provided by Hans Clevers, University Medical Center, Utrecht, The Netherlands) or 0.25 μg of pUSE-Wnt1 (Santa Cruz Biotechnology), and 1 μg of expression plasmid or pcDNA3 were used. Control cultures were transfected with pTOPFLASH-Luc and pACB-Z to establish the basal levels for the luciferase and β-galactosidase assays. After 24 h, the luciferase and β-galactosidase activities were determined by using the Dual Light kit (Applied Biosystems) and a microtiter plate luminometer (MicroBeta TriLux, Gaithersburg, MD). The luciferase values were normalized for variations in transfection efficiency by using the β-galactosidase internal control and are expressed as fold stimulation of luciferase activity. All of the transfection results represent means of a minimum of three independent transfections.
Immunohistochemistry. Histologic sections were prepared and stained by the Rheumatic Disease Center Core Facility at the University of California at San Diego. Cartilage samples were obtained from patients with clinical and radiographic OA at the time of joint replacement surgery. The studies were approved by the University of California at San Diego Human Subjects Research Protection Program. The samples were embedded in TissueTek OCT compound (Miles), frozen, sectioned at 6 μm, and fixed in cold acetone for 10 min. After blocking with 1% BSA, 5% horse serum, and 5% human serum, polyclonal goat sera (Santa Cruz Biotechnology) or anti-recombinant human sFRP-3 antibody (R & D Systems) was applied for 1 h at room temperature. The sections were washed and incubated with biotinylated horse anti-goat IgG (Vector Laboratories). After endogenous peroxidase was depleted by quenching with 0.3% H2O2 for 20 min, the sections were incubated with ABC peroxidase reagent (Vector Laboratories) for 30 min. The slides were then washed with PBS, developed with diaminobenzidine substrate, and counterstained with hematoxylin.
Immunoblotting. Whole-cell lysates or separate cytosol and nuclear fractions were prepared, and 30 μg of protein was separated by electrophoresis. The proteins were transferred to a polyvinylidene difluoride membrane. After being blocked, the blots were incubated with anti-sFRP3 antibody (R & D Systems), or anti-β-catenin. Horseradish peroxidase-conjugated anti-IgG (Santa Cruz Biotechnology) was used as the secondary antibody. The membranes were developed by using a chemiluminescence system (ECL detection reagent, Amersham Pharmacia) and scanned. The membranes were stripped with Re-Blot Western blot recycling kit (Chemicon) and reprobed by using an actin monoclonal antibody (Sigma). Further details are provided in Supporting Materials and Methods.
Results
Candidate Genes and Microsatellite Association Analysis. The linkage to chromosome 2q centered at microsatellite marker D2S2284 (2q31.1, 171.7 megabases) with a MLS of 1.6 (6). Because the low LOD score implied that the susceptibility gene could be positioned at some distance from the point of maximum linkage (8), we searched a broad region from 2q23 to 2q32 for candidate genes that had been reported to be expressed in joint tissue. Eight candidates were identified and chosen for further investigation: TNFAIP6, activin A receptor, fibroblast activation protein α, ITGA6, activating transcription factor 2, integrin α_4, FRZB_, and integrin α_V_. The 378 probands from the previous linkage study and 760 age-matched control individuals were genotyped for microsatellite markers that targeted these eight genes. Association was tested by using the clump program, which has been designed to overcome the problems of sparse contingency tables as found in a χ2 analysis of multiallelic markers, such as microsatellites (Table 1). Only the TNFAIP6 marker demonstrated association in the unstratified analysis, with all four T statistics being significant at P < 0.05. Stratification by sex revealed significant evidence for association to the ITGA6 and FRZB microsatellites in the female probands. The integrin α_V_ microsatellite provided some evidence for association in the male probands but was only significant for the T3 statistic.
Table 1. Association analysis of candidate gene microsatellites in the OA probands.
| CLUMP P values | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unstratified | Female probands | Male probands | ||||||||||
| Gene | T1 | T2 | T3 | T4 | T1 | T2 | T3 | T4 | T1 | T2 | T3 | T4 |
| TNFAIP6 | 0.006 | 0.009 | 0.02 | 0.003 | 0.06 | 0.06 | 0.02 | 0.06 | 0.1 | 0.2 | 0.2 | 0.05 |
| ACVR1 | 0.8 | 0.6 | 0.5 | 0.5 | 0.5 | 0.4 | 0.7 | 0.3 | 0.2 | 0.3 | 0.4 | 0.3 |
| FAP | 0.8 | 0.9 | 0.8 | 0.7 | 0.4 | 0.4 | 0.3 | 0.4 | 1.0 | 0.8 | 0.8 | 1.0 |
| ITGA6 | 0.4 | 0.6 | 0.3 | 0.4 | 0.005 | 0.03 | 0.02 | 0.006 | 0.8 | 0.8 | 0.5 | 0.8 |
| ATF2 | 0.9 | 0.6 | 0.9 | 0.7 | 0.6 | 0.8 | 0.6 | 0.6 | 0.8 | 0.4 | 0.3 | 0.5 |
| ITGA4 | 0.2 | 1.0 | 0.9 | 0.1 | 0.8 | 0.9 | 0.8 | 0.8 | 0.1 | 0.9 | 0.8 | 0.1 |
| FRZB | 0.7 | 0.7 | 0.5 | 0.6 | 0.02 | 0.01 | 0.09 | 0.007 | 0.9 | 0.8 | 0.8 | 0.9 |
| ITGAV | 0.5 | 0.3 | 0.2 | 0.4 | 0.7 | 0.7 | 0.6 | 0.8 | 0.2 | 0.1 | 0.02 | 0.07 |
SNP Association Analysis. By using the ensembl (www.ensembl.org) and University of California, Santa Cruz (http://genome.ucsc.edu), genome browsers, we searched TNFAIP6, ITGA6, and FRZB for nonsynonymous SNPs. One was identified in TNFAIP6, two in ITGA6, and two in FRZB (Table 2). The genotyping of 48 unrelated individuals revealed that the two ITGA6 SNPs were nonpolymorphic. We genotyped the TNFAIP6 SNP and the two FRZB SNPs in the 378 probands from our hip OA families and in the 760 age-matched controls. All three SNPs were in Hardy-Weinberg equilibrium in the controls (P > 0.05). There was no significant difference in allele frequencies between the probands and the controls (Table 3).
Table 2. Nonsynonymous SNPs identified in TNFAIP6, ITGA6, and FRZB with the ENSEMBL and University of California, Santa Cruz, genome browsers.
| dbSNP reference† | Change | |||
|---|---|---|---|---|
| Gene | Location* | Nucleotide | Amino acid substitution | |
| TNFAIP6 | Exon 4 [+37] | rs1046668 | A→G | Gln 144Arg |
| ITGA6 | Exon 18 [+52] | rs2737085 | G→T | Asp805Tyr |
| ITGA6 | Exon 23 [+10] | rs10209072 | G→A | Val969Met |
| FRZB | Exon 4 [+6] | rs288326 | C→T | Arg200Trp |
| FRZB | Exon 6 [+109] | rs7775 | C→G | Arg324Gly |
Table 3. Association analysis of the TNFAIP6 and FRZB SNPs in the 378 probands.
| Controls | Probands | |||||
|---|---|---|---|---|---|---|
| Gene | SNP | Strata | Allele frequency* | Strata | Allele frequency* | P value |
| TNFAIP6 | Exon 4 [+37] | All | 0.12 (187/1,323) | All | 0.11 (78/664) | 0.2 |
| Female | 0.11 (89/703) | Female | 0.09 (38/396) | 0.2 | ||
| Male | 0.14 (98/620) | Male | 0.13 (40/268) | 0.9 | ||
| FRZB | Exon 4 [+6] | All | 0.13 (189/1,321) | All | 0.13 (101/651) | 0.6 |
| Female | 0.11 (91/703) | Female | 0.15 (64/374) | 0.1 | ||
| Male | 0.14 (98/618) | Male | 0.12 (37/277) | 0.5 | ||
| FRZB | Exon 6 [+109] | All | 0.07 (111/1,399) | All | 0.09 (69/679) | 0.2 |
| Female | 0.07 (56/734) | Female | 0.11 (47/391) | 0.04 | ||
| Male | 0.08 (55/665) | Male | 0.07 (22/288) | 0.9 |
Stratification by sex revealed no significant difference for the _TNFAIP_6 or the FRZB exon 4 SNPs in either sex. There was also no significant difference in the male stratum for the FRZB exon 6 SNP. However, the frequency of the G allele of this SNP was significantly elevated in the female OA probands compared with the female controls (0.11 versus 0.07, respectively; P = 0.04), similar to the microsatellite data for FRZB. To exclude a type I error we genotyped the two FRZB SNPs in a separate cohort of 558 unrelated hip OA cases (Table 4). Confirming the results with the probands, the G allele of the exon 6 SNP had a significantly increased frequency only in the female cases (0.10 versus 0.07 in the female controls, P = 0.04). The frequency of this allele in the combined female probands and female cases (hereafter referred to as the affected females) was 0.10 (P = 0.02; Table 4). The odds ratio for the G allele was 1.5 (95% confidence interval = 1.1-2.1). There was not a gene-dose effect, with 1.6% of the affected females being homozygous for the G allele compared with 1.3% of the female controls (P = 0.9). Direct sequencing of the coding region of 35 unrelated individuals revealed that FRZB contained no other nonsynonymous SNPs. The only other common (rare allele frequency > 0.05) variant identified was a G→A transition in the 3′ UTR that was in complete linkage disequilibrium (_r_2 = 1.0) with the exon 6 SNP.
Table 4. Association analysis of the FRZB SNPs in the cases and in the cases and probands combined.
| Controls | Cases | Cases and probands combined | ||||||
|---|---|---|---|---|---|---|---|---|
| FRZB SNP | Strata | Allele frequency* | Strata | Allele frequency* | P value | Strata | Allele frequency* | P value |
| Exon 4 [+6] | All | 0.13 (189/1,321) | All | 0.13 (145/959) | 0.7 | All | 0.13 (246/1,610) | 0.6 |
| Female | 0.11 (91/703) | Female | 0.13 (86/580) | 0.4 | Female | 0.14 (150/954) | 0.2 | |
| Male | 0.14 (98/618) | Male | 0.13 (59/379) | 1.0 | Male | 0.13 (96/656) | 0.7 | |
| Exon 6 [+109] | All | 0.07 (111/1,399) | All | 0.08 (89/1,007) | 0.5 | All | 0.09 (158/1,686) | 0.2 |
| Female | 0.07 (56/734) | Female | 0.10 (68/598) | 0.04 | Female | 0.10 (115/989) | 0.02 | |
| Male | 0.08 (55/665) | Male | 0.05 (21/409) | 0.09 | Male | 0.06 (43/697) | 0.2 |
FRZB Haplotype Analysis. The FRZB exon 4 and exon 6 SNPs were in linkage equilibrium (_r_2 = 0.01, _D_′ = 0.1). By examining compound genotypes, we determined the percentage of affected females and female controls, who contained a T allele at the exon 4 SNP together with a G allele at the exon 6 SNP. The figures were 6.2% (34 of 549 affected females) and 1.8% (7 of 394 female controls), respectively (P = 0.003; odds ratio = 3.6; 95% confidence interval = 1.6-8.3) (Table 8, which is published as supporting information on the PNAS web site). We subsequently determined the frequency of the four haplotypes for the two exonic SNPs in the affected females and in the female controls (Table 5). Haplotypes in the compound heterozygotes were determined by molecular haplotyping. There was a significant difference in frequency (P = 0.007). The T-G haplotype, which contains the minor allele of both SNPs, showed the greatest proportionate increase: 28 affected females possessed the T-G haplotype (2.6%) compared with five female controls (0.6%). This haplotype, which will result in both of the conserved arginines being substituted in the same sFRP3 molecule, was a particular risk factor with an odds ratio of 4.1 (95% confidence interval = 1.6-10.7).
Table 5. Frequency of the four possible haplotypes for the exon 4 [+6] and the exon 6 [+109] FRZB SNPs in combined female probands and cases (affected females) and in the female controls.
| Haplotype exon 4 [+6] -exon 6 [+109] | Frequency (n) | T allele of exon 4 SNP and G allele of exon 6 SNP | |
|---|---|---|---|
| Affected females | Female controls | ||
| C-C | 0.79 (863) | 0.82 (649) | No |
| C-G | 0.08 (87) | 0.06 (50) | No |
| T-C | 0.11 (120) | 0.11 (84) | No |
| T-G | 0.026 (28) | 0.006 (5) | Yes |
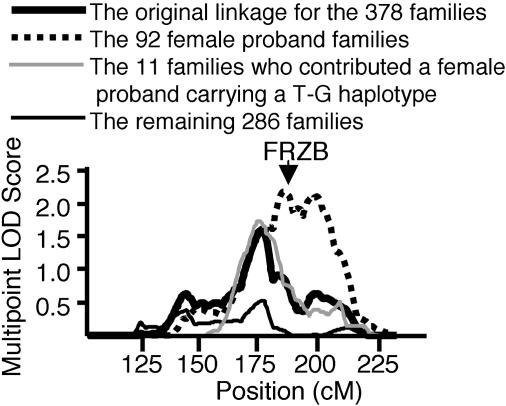
Effect of the FRZB SNPs on the Original Linkage. We next assessed the effect that the exon 4 and exon 6 SNPs had on our original multipoint linkage. Of our 378 families, 92 had contributed a female proband who carried either a T allele of exon 4 [+6] (47 families), a G allele of exon 6 [+109] (31 families), or a copy of both alleles (14 families). The MLS for these 92 families was 2.2, an increase on the MLS of 1.6 that we had obtained for all 378 families (Fig. 1). Of our 378 families, 11 had contributed a female proband who carried the T-G haplotype. The MLS was 1.7 for these 11 families. The 286 families from the 378 who had not contributed a female proband carrying a minor allele at either SNP had an MLS of only 0.5. These data imply that the T allele of the exon 4 SNP and the G allele of the exon 6 SNP account for our original 2q linkage. To reinforce this conclusion, we needed to determine whether the increased LOD scores observed for each of our subsets of 92 and 11 families was a result of our selection criteria rather than random sampling. For each subset, 1,000 permutations of the multipoint linkage analysis were performed on 92 (or 11) families, randomly selected from the main dataset of 378 families. After each permutation, the linkage interval was screened for a MLS ≥ 2.2 (for our subset of 92 families) or a MLS ≥ 1.7 (for our subset of 11 families). Thus, an empirical P value was obtained for each subset. The empirical P value for the 92-family subset was 0.05; the empirical P value for the 11-family subset was 0.06. These results suggest that the stratified linkage data should be considered as corroborative but not as definitive evidence for a role of the FRZB SNPs in the 2q OA susceptibility.
Fig. 1.
The effect of FRZB SNPs on the original multipoint linkage plot. The thick black line is the original multipoint linkage plot of chromosome 2q for the 378 hip OA families (6). The dashed line is the plot for the 92 families from the 378 who contributed a female proband who carried either a T allele of the exon 4 [+6] SNP (47 families), a G allele of the exon 6 [+109] SNP (31 families), or a copy of both alleles (14 families). Also shown is the plot for the 11 families from the 378 who contributed a female proband carrying a T-G haplotype (gray line). The thin black line is the linkage plot for the remaining 286 families. The position of FRZB is marked. The genetic position on the x axis was determined by using the deCODE Genetics (Reykjavik, Iceland) sex-averaged map (35). cM, centimorgans.
sFRP3 Expression in OA Cartilage and Functional Effects of the Conserved Arginines. sFRP3 is expressed by human chondrocytes during skeletal development (26, 27), but it had not been analyzed in adults. However, immunoreactive sFRP3 was easily detectable in OA articular cartilage by immunohistochemistry (Fig. 2). The Arg200Trp substitution is within the netrin-like domain of sFRP3, whereas the Arg324Gly substitution lies penultimate to the carboxyl-terminus. The ability of wild-type protein and of the Arg200Trp- and Arg324Gly-substituted proteins to antagonize wnt signaling was compared after transient transfection of HEK293 cells (Fig. 3). The cells were cotransfected with either (i) a wnt1 (Fig. 3_B_) or a β-catenin expression vector (Fig. 3_C_) to induce signaling, (ii) the β-catenin-dependent reporter gene TOPflash, (iii) a β-galactosidase expressing plasmid to control for transfection efficiency, or (iv) a FRZB wild-type gene or genes containing the Arg200Trp or Arg324Gly substitutions cloned into pcDNA3. Whereas the wild-type FRZB vector efficiently inhibited TOPflash activity, the Arg324Gly substitution and the Arg200Trp/Arg324Gly double substitution had diminished activity, despite adequate protein expression (Fig. 3). Whether the Arg200Trp mutation had an effect in addition to the Arg324Gly variation could not be determined with certainty. Similarly, HEK293 cells transiently transfected with the plasmid containing the Arg324Gly substitution required higher levels of expressing plasmid to modestly decrease detectable free cytosolic and nuclear levels of β-catenin (Fig. 3_D_). These data highlight the important role that the Arg-324 residue has in the wnt antagonistic activity of sFRP3.
Fig. 2.
sFRP3 expression in human chondrocytes. Human cartilage from an OA patient undergoing total knee replacement was harvested intraoperatively, embedded in OCT compound, frozen, and sectioned. The sections were immunostained with polyclonal goat serum (A) or goat anti-human sFRP3 (B) and are shown at ×400 original magnification. The arrows indicate some of the chondrocytes expressing sFRP3.
Fig. 3.
sFRP3 with the Arg324Gly substitution is less efficient at antagonizing wnt signaling. (A) HEK293 cells were transfected with 1.0 μg, 0.5 μg, 0.25 μg, or 0.125 μg of wild-type (wt) pcFRZB, pcFRZB-Arg200Trp, pcFRZB-Arg324Gly, or pcFRZB Arg200Trp/Arg324Gly or were left untransfected (Control, C). After 24 h, the cells were lysed and 30 μg of protein was separated by SDS/PAGE under reducing conditions. The protein was transferred to a polyvinylidine difloride membrane and then serially probed with anti-sFRP3 and β-actin. (B) HEK293 cells were cotransfected in triplicate with 0.25 μg of pTOPFLASH-Luc, 0.025 μg of pACB-Z, 0.25 μg of pUSE-Wnt1, and 1 μg of either pcFRZB, pcFRZB-Arg200Trp, pcFRZB-Arg324Gly, pcFRZB-Arg200Trp/Arg324Gly, or vector DNA. (C) Similar transfections were performed with 0.25 μg of a β-catenin-expressing plasmid instead of the Wnt1-expressing plasmid. Control cultures were transfected with pTOPFLASH-Luc and pACB-Z to establish basal enzyme activity levels. Additional control cultures were transfected with pTOPFLASH-Luc, which confirmed specificity of the assay (data not shown). After 24 h, the cells were lysed and the normalized luciferase activities were determined. The fold induction values are the ratios of the normalized luciferase activities in cells transfected with both expression and reporter plasmids compared with activities in cells receiving the respective reporter plasmids alone. Shown are the mean ± SEM of triplicates. (D) Triplicate cultures of HEK293 cells were transfected with 1 μg, 0.3 μg, or 0.1 μg of pcFRZB or pcFRZB-Arg324Gly. Control cultures were transfected with empty vector. After 48 h, the cells were lysed and the cytosolic and nuclear fractions were separated by SDS/PAGE, transferred to a polyvinylidine difloride membrane, and then probed with antibodies for β-catenin and β-actin.
Discussion
Primary OA is a common late-onset arthritis with a complex mode of transmittance, including joint-site and sex-specific heterogeneity. Because of disease heterogeneity and the variable role of environmental factors, it is unlikely that any single genetic effect would by itself be associated with the majority of affected patients. Thus, the aim of our genetic studies in OA was to reveal previously unsuspected biochemical pathways that could explain the pathogenesis of this perplexing illness and lend opportunities for prevention, early diagnosis, and treatment. The data reported here imply that the wnt signaling pathway could play a role in the etiology of OA. We identified common functional variants within the gene for the wnt regulator sFRP3 that were associated with hip OA in females. Sex differences in OA susceptibility have been reported in several epidemiologic studies (reviewed in ref. 2). Indeed, the five OA loci that we previously mapped all demonstrate a female restriction (3), which is consistent with a large twin study demonstrating a greater female heritability for OA (28),
The influence of the FRZB SNPs on our original 2q linkage was from our female hip probands, and there was little or no residual linkage left once the FRZB SNPs were taken into account. However, there may be other contributing loci that are clustered in this region. Of the eight candidate genes from within the 2q linkage interval, three candidates (_TNFAIP_6, _ITGA_6, and FRZB) provided evidence for association by microsatellite analysis. Although we did not identify a nonsynonymous polymorphism in _TNFAIP_6 and _ITGA_6 associated with OA, we have not excluded the possibility that other, perhaps uncommon, variants exist that may encode for OA susceptibility.
The haplotype coding for substitutions of two highly conserved arginine residues (Arg200Trp and Arg324Gly) in FRZB was a strong risk factor for primary hip OA. Transfection studies indicated that substitutions of these positively charged residues diminished the ability of the sFRP3 protein to disrupt wnt signaling. Although the amino-terminal, cysteine-rich domain of sFRP3 interacts with wnt proteins (29), the carboxyl terminus has heparan-binding activity (12, 30). Some truncated forms of SFRPs that retained wnt binding were not able to antagonize wnt signaling (29, 31), suggesting that both domains are necessary for optimal function. The variant sFRP3 with the Arg324Gly substitution may have reduced interaction with acidic components of the cartilage extracellular matrix, such as the heparan sulfate proteoglycans (32, 33). This interaction may be required for the efficient localization and functional activity of sFRP3 in the joints. Inhibition of wnt signaling, mediated by sFRP3 may be required to retain chondrocytes in their immature prehypertrophic state and maintain the integrity of the cartilage-bone junction (26, 34).
FRZB is expressed by developing and mature chondrocytes (27). During long-bone development, the chondrocytes at the epiphyseal tip produce the articular cartilage. The expression of sFRP3 Arg324Gly may result in a structurally occult hip dysplasia, which may manifest as hip OA later in life after exposure to mechanical forces. In general, candidate genes for a late-onset disease may include those genes that have a fundamental role during development. The complex wnt pathway includes multiple proteins encoded by >30 genes that are variously expressed during tissue patterning and development. Patients with the functional sFRP3 substitutions that we have described here lived into old age before developing symptoms. Thus, it is likely that regulatory networks in the wnt signaling cascade can compensate for a deficiency in any one component.
The Arg200Trp-Arg324Gly haplotype was carried by 2.6% of all female hip OA patients. The high incidence of hip OA in aging females and the relative prevalence of this haplotype should make prospective analyses of OA disease susceptibility in this defined population feasible and could facilitate interactive studies directed at the underlying defect.
Supplementary Material
Supportinhg Information
Acknowledgments
We thank Professor Andrew Carr and Ms. Kim Clipsham for organizing the collection of patient and family samples used in this study; Drs. H. Hoffman, D. Holderbaum, and R. Moscovitz for assistance and review of the manuscript; Dr. G. Firestein and D. Boyle for providing cartilage samples; K. Pekny for technical assistance; and N. Noon for secretarial support. The plasmid sequencing was performed by the Cancer Center Sequencing Core Facility (University of California at San Diego), and the immunohistochemistry sections were prepared and stained by the Rheumatic Disease Center Core Facility. This work was supported in part by the Arthritis Research Campaign, the Help the Aged Research into Ageing program (J.L.), the National Institutes of Health (D.A.C. and M.C.), and the University of California BioSTAR Program (M.C.).
Abbreviations: OA, osteoarthritis; sFRP3, secreted frizzled-related protein 3; wnt, wingless; TNFAIP6, tumor necrosis factor α-induced protein 6; ITGA6, integrin α_6_; FRZB, frizzled motif associated with bone development; LOD, logarithm of odds; MLS, multipoint logarithm of odds score; SNP, single-nucleotide polymorphism.
References
- 1.Spector, T. D. & MacGregor, A. J. (2004) Osteoarthritis Cartilage 12**,** Suppl., S39-S44. [DOI] [PubMed] [Google Scholar]
- 2.Loughlin, J. (2002) Rheum. Dis. Clin. N. Am. 28**,** 95-109. [DOI] [PubMed] [Google Scholar]
- 3.Loughlin, J. (2003) Curr. Opin. Pharmacol. 3**,** 295-299. [DOI] [PubMed] [Google Scholar]
- 4.Demissie, S., Cupples, L. A., Myers, R., Aliabadi, P., Levy, D. & Felson, D. T. (2002) Arthritis Rheum. 46**,** 946-952. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson, S. E., Jonsson, H., Ingvarsson, T., Manolescu, I., Jonsson, H. H., Olafsdottir, G., Palsdottir, E., Stefansdottir, G., Sveinbjornsdottir, G., Frigge, M. L., et al. (2003) Am. J. Hum. Genet. 72**,** 1448-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loughlin, J., Dowling, B., Mustafa, Z., Southam, L. & Chapman, K. (2002) Rheumatology (Oxford) 41**,** 955-956. [DOI] [PubMed] [Google Scholar]
- 7.Slagboom, P. E., Heijmans, B. T., Beekman, M., Westendorp, R. G. & Meulenbelt, I. (2000) Ann. N.Y. Acad. Sci. 908**,** 50-63. [DOI] [PubMed] [Google Scholar]
- 8.Roberts, S. B., MacLean, C. J., Neale, M. C., Eaves, L. J. & Kendler, K. S. (1999) Am. J. Hum. Genet. 65**,** 876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, K., Mustafa, Z., Irven, C., Carr, A. J., Clipsham, K., Smith, A., Chitnavis, J., Sinsheimer, J. S., Bloomfield, V. A., McCartney, M., et al. (1999) Am. J. Hum. Genet. 65**,** 167-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loughlin, J., Mustafa, Z., Irven, C., Smith, A., Carr, A. J., Sykes, B. & Chapman, K. (1999) Am. J. Hum. Genet. 65**,** 1795-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingvarsson, T., Stefansson, S. E., Gulcher, J. R., Jonsson, H. H., Jonsson, H., Frigge, M. L., Palsdottir, E., Olafsdottir, G., Jonsdottir, T., Walters, G. B., et al. (2001) Arthritis Rheum. 44**,** 2548-2555. [DOI] [PubMed] [Google Scholar]
- 12.Jones, S. E. & Jomary, C. (2002) BioEssays 24**,** 811-820. [DOI] [PubMed] [Google Scholar]
- 13.Rubinfeld, B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S. & Polakis, P. (1996) Science 272**,** 1023-1026. [DOI] [PubMed] [Google Scholar]
- 14.Yost, C., Torres, M., Miller, J. R., Huang, E., Kimelman, D. & Moon, R. T. (1996) Genes Dev. 10**,** 1443-1454. [DOI] [PubMed] [Google Scholar]
- 15.Behrens, J., von Kries, J. P., Kuhl, M., Bruhn, L., Wedlich, D., Grosschedl, R. & Birchmeier, W. (1996) Nature 382**,** 638-642. [DOI] [PubMed] [Google Scholar]
- 16.Capdevila, J. & Izpisua Belmonte, J. C. (2001) Annu. Rev. Cell Dev. Biol. 17**,** 87-132. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann, C. & Tabin, C. J. (2001) Cell 104**,** 341-351. [DOI] [PubMed] [Google Scholar]
- 18.Russell, J., Gennissen, A. & Nusse, R. (1992) Development (Cambridge) 115**,** 475-485. [DOI] [PubMed] [Google Scholar]
- 19.Zakany, J. & Duboule, D. (1993) Nature 362**,** 546-549. [DOI] [PubMed] [Google Scholar]
- 20.Gong, Y., Slee, R. B., Fukai, N., Rawadi, G., Roman-Roman, S., Reginato, A. M., Wang, H., Cundy, T., Glorieux, F. H., Lev, D., et al. (2001) Cell 107**,** 513-523. [DOI] [PubMed] [Google Scholar]
- 21.Little, R. D., Carulli, J. P., Del Mastro, R. G., Dupuis, J., Osborne, M., Folz, C., Manning, S. P., Swain, P. M., Zhao, S. C., Eustace, B., et al. (2002) Am. J. Hum. Genet. 70**,** 11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuguchi, T., Furuta, I., Watanabe, Y., Tsukamoto, K., Tomita, H., Tsujihata, M., Ohta, T., Kishino, T., Matsumoto, N., Minakami, H., et al. (2004) J. Hum. Genet. 49**,** 80-86. [DOI] [PubMed] [Google Scholar]
- 23.Denoeud, F., Vergnaud, G. & Benson, G. (2003) Genome Res. 13**,** 856-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nentwich, H. A., Mustafa, Z., Rugg, M. S., Marsden, B. D., Cordell, M. R., Mahoney, D. J., Jenkins, S. C., Dowling, B., Fries, E., Milner, C. M., et al. (2002) J. Biol. Chem. 277**,** 15354-15362. [DOI] [PubMed] [Google Scholar]
- 25.Sham, P. C. & Curtis, D. (1995) Ann. Hum. Genet. 59**,** 97-105. [DOI] [PubMed] [Google Scholar]
- 26.Enomoto-Iwamoto, M., Kitagaki, J., Koyama, E., Tamamura, Y., Wu, C., Kanatani, N., Koike, T., Okada, H., Komori, T., Yoneda, T., et al. (2002) Dev. Biol. 251**,** 142-156. [DOI] [PubMed] [Google Scholar]
- 27.Hoang, B., Moos, M., Jr., Vukicevic, S. & Luyten, F. P. (1996) J. Biol. Chem. 271**,** 26131-26137. [DOI] [PubMed] [Google Scholar]
- 28.Kaprio, J., Kujala, U. M., Peltonen, L. & Koskenvuo, M. (1996) BMJ 313**,** 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, K., Wang, S., Julius, M. A., Kitajewski, J., Moos, M., Jr., & Luyten, F. P. (1997) Proc. Natl. Acad. Sci. USA 94**,** 11196-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong, J. M., Uren, A., Rubin, J. S. & Speicher, D. W. (2002) J. Biol. Chem. 277**,** 5134-5144. [DOI] [PubMed] [Google Scholar]
- 31.Uren, A., Reichsman, F., Anest, V., Taylor, W. G., Muraiso, K., Bottaro, D. P., Cumberledge, S. & Rubin, J. S. (2000) J. Biol. Chem. 275**,** 4374-4382. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, N. B. & Domowicz, M. (2002) Glycobiology 12**,** 57R-68R. [DOI] [PubMed] [Google Scholar]
- 33.Perrimon, N. & Bernfield, M. (2000) Nature 404**,** 725-728. [DOI] [PubMed] [Google Scholar]
- 34.Pacifici, M., Koyama, E., Iwamoto, M. & Gentili, C. (2000) Connect. Tissue Res. 41**,** 175-184. [DOI] [PubMed] [Google Scholar]
- 35.Kong, A., Gudbjartsson, D. F., Sainz, J., Jonsdottir, G. M., Gudjonsson, S. A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. (2002) Nat. Genet. 31**,** 241-247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supportinhg Information