ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins (original) (raw)
Abstract
The mechanisms responsible for the inverse relationship between plasma high-density lipoprotein (HDL) levels and atherosclerotic cardiovascular disease are poorly understood. The ATP-binding cassette transporter A1 (ABCA1) mediates efflux of cellular cholesterol to lipid-poor apolipoproteins but not to HDL particles that constitute the bulk of plasma HDL. We show that two ABC transporters of unknown function, ABCG1 and ABCG4, mediate isotopic and net mass efflux of cellular cholesterol to HDL. In transfected 293 cells, ABCG1 and ABCG4 stimulate cholesterol efflux to both smaller (HDL-3) and larger (HDL-2) subclasses but not to lipid-poor apoA-I. Treatment of macrophages with an liver X receptor activator results in up-regulation of ABCG1 and increases cholesterol efflux to HDL. RNA interference reduced the expression of ABCG1 in liver X receptor-activated macrophages and caused a parallel decrease in cholesterol efflux to HDL. These studies indicate that ABCG1 and ABCG4 promote cholesterol efflux from cells to HDL. ABCG1 is highly expressed in macrophages and probably mediates cholesterol efflux from macrophage foam cells to the major HDL fractions, providing a mechanism to explain the relationship between HDL levels and atherosclerosis risk.
A major theory to account for the inverse relationship between high-density lipoprotein (HDL) levels and cardiovascular risk is that HDL promotes the efflux of cholesterol from arterial wall macrophage foam cells and decrease atherosclerosis. This hypothesis appeared to be supported by the discovery that Tangier disease, a disorder characterized by very low HDL levels, macrophage foam cell accumulation, and increased atherosclerosis, is caused by mutations in the ATP-binding cassette transporter, ABCA1 (1-4). ABCA1 mediates efflux of cellular phospholipids and cholesterol to lipid-poor apolipoproteins, such as apoA-I and apoE (5, 6), initiating the formation of HDL. However, ABCA1 interacts poorly with HDL-2 and HDL-3 particles (5, 7) that constitute the bulk of the plasma HDL, and ABCA1 variants are not likely to account for a major part of the genetic variation in HDL levels in the general population (8). Thus, the activity of ABCA1 does not readily account for cholesterol efflux from foam cells to HDL, and the mechanism underlying the inverse relationship between HDL levels and atherosclerosis risk remains uncertain.
The oxysterol-activated transcription factors liver X receptor/retinoid X receptor (LXR/RXR) induce the expression of ABCA1, as well as a number of other molecules involved in cellular cholesterol efflux, transport, and excretion (9, 10). Treatment of macrophages with LXR activators increased net cholesterol efflux to HDL-2, suggesting the presence of unique LXR target genes mediating cholesterol efflux to HDL (11). Some ABCG family members are also LXR/RXR targets, such as ABCG5 and ABCG8, the defective genes in sitosterolemia (12-14). ABCG family members are half-transporters, largely of unknown function. These considerations led us to investigate the possibility that different members of the ABCG transporter family might be responsible for cellular cholesterol efflux to HDL.
Methods
Plasma Lipoprotein Preparations. HDL-2 (density 1.063-1.125 g/ml) and HDL-3 (density 1.125-1.210 g/ml) were isolated by preparative ultracentrifugation from normolipidemic human plasma and stored in PBS containing 1 mM EDTA. Low-density lipoprotein (LDL) was from Biomedical Technologies (Stoughton, MA). ABCA1-/- mice were kindly provided by O. Francone (Pfizer, Groton, CT), and macrophages isolated from the WT and knockout littermates were used for the experiments.
Plasmid Constructs and Cell Transfection. The plasmid constructs expressing mouse ABCG transporters were prepared by cloning mouse full-length cDNAs into pCMV-sport6 vector, and the cDNA sequence was confirmed by DNA sequencing. For transient transfection of human embryonic kidney (HEK)293 cells, cells in 12- or 24-well collagen-coated plates were transfected with various plasmid constructs with LipofectAMINE 2000 (Invitrogen) at 37°C overnight (≈20 h). To estimate transfection efficiency, a construct expressing GFP was routinely used in the experiment to visually monitor for transfection efficiency. The transfection efficiency of HEK293 cells was in the range of 60-80% of cells. Although transfection efficiency did vary from experiment to experiment, we found that the variation within the same experiment was small.
Cellular Lipid Efflux Assays. Generally, HEK293 cells were labeled by culturing for 24 h in 10% FBS/DMEM containing either 2 μCi/ml [3H]cholesterol for cholesterol efflux or 2 μCi/ml [3H]choline (1 Ci = 37 GBq) for phospholipid efflux. The next day, cells were washed with fresh media and then HDL, LDL, or cyclodextrin were added as acceptor and incubated for the indicated period before the media and cells were collected for analysis. Phospholipid and cholesterol efflux were expressed as the percentage of the radioactivity released from the cells into the medium relative to the total radioactivity in cells plus medium. For cholesterol mass efflux, the collected media were extracted with hexane:isopropanol (3:2 vol/vol) with β-sitosterol (5 μg per sample) added as the internal standard. The recovered lipid fractions were dried under nitrogen gas; 100 μl of chloroform was added, and the samples were subject to gas-liquid chromatographic analysis. For HDL cell association, cells were incubated with [125I]HDL (1.5 μg/ml) in 0.2% BSA/DMEM for 1 h at 37°C. After washing three times with fresh media, cells were lysed with 0.1% SDS and 0.1 M NaOH lysis buffer, and radioactivity was determined by γ-counter. To determine the free cholesterol mass in media after cholesterol efflux in the presence or absence of HDL, the lipid fraction was extracted from the media with hexane:isopropanol (3:2). After drying under nitrogen gas, the mass of free cholesterol dissolved in chloroform was determined by using gas chromatography.
Small Interfering (si)RNA-Mediated Macrophage RNA Interference (RNAi). cRNA oligonucleotides derived from the mouse ABCG1 and ABCG4 target sequences were obtained from Dharmacon (Lafayette, CO) and used to induce RNAi to suppress ABCG1 and ABCG4 expression in thioglycollate-elicited mouse peritoneal macrophages. Two target sequences were selected by using the program from Dharmacon: 5′-CGTGGATGAGGTTGAGACA-3′ and 5′-GGTGGACAACAACTTCACA-3′ for ABCG1; and 5′-GAAGGTGGAGAACCATATC-3′ and 5′-GCACTTGAACTACTGGTAT-3′ for ABCG4.
Where indicated, RNA oligonucleotides targeting both sequences were mixed and used to down-regulate ABCG1 or ABCG4 gene expression. The scrambled control RNA oligonucleotides also were obtained from Dharmacon. An independent set of siRNA targeting ABCG1 (5′-TCGTATCTTATCTGTAGAGAA-3′ or ABCG4 (5′-CCGGGTCAAGTCAAGTCTGAGAGATA-3′) was obtained from Qiagen and used where indicated. For cholesterol efflux assay, mouse peritoneal macrophages were plated in 24- or 48-well plates and cultured in 10% FBS and DMEM at 37°C for 24 h. Cell were then transfected with siRNA and LipofectAMINE 2000 at indicated concentration and labeled with isotopic cholesterol (2 μCi/ml [3H]cholesterol in 1% FBS) in the presence or absence of TO901317 (2 μM) for 48 h. Cells were washed twice and equilibrated for 30 min for the third wash, and then HDL or other acceptors were added for the indicated period. Levels of ABCG1 and ABCG4 mRNAs normalized against β-actin mRNA were determined by using TaqMan real-time quantititative RT-PCR. The primers and probes were from Applied Biosystems.
Results
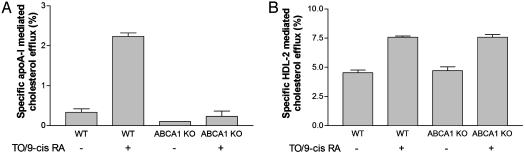
We previously showed that LXR activation in macrophages resulted in increased cholesterol efflux to HDL-2 (11). Because HDL-2 does not appear to interact with ABCA1 in transfected 293 cells (7), this finding suggested the possibility that LXR activation might induce an alternative pathway, leading to increased cholesterol efflux to HDL. To more directly test this possibility, macrophages from WT or ABCA1-/- mice were treated with LXR/RXR activators, and then cholesterol efflux to apolipoprotein (apo)A-I or HDL-2 was determined. LXR/RXR activation induced cholesterol efflux to both apoA-I (Fig. 1_A_) and to HDL-2 (Fig. 1_B_). Whereas deficiency of ABCA1 virtually abolished cholesterol efflux to apoA-I, there was no effect on cholesterol efflux to HDL, confirming an ABCA1-independent, LXR-induced efflux pathway to HDL. Our previous studies (11) ruled out a role of apoE or SR-BI in this process because LXR/RXR activation led to increased cholesterol efflux to HDL-2 in apoE-/- macrophages or macrophages treated with scavenger receptor (SR)-BI-neutralizing Ab. This finding led us to evaluate the possibility that unique ABC transporters that are LXR targets might mediate cholesterol efflux to HDL. Members of the ABCG family have been implicated in cholesterol transport (12), and several members of this family are known LXR targets (13, 15).
Fig. 1.
LXR/RXR activation increases macrophage cholesterol efflux to HDL independent of ABCA1. (A and B) Cholesterol efflux to apoA-I (15 μg/ml protein) (A) or HDL-2 (25 μg/ml protein) (B) was determined in mouse peritoneal macrophages isolated from WT or ABCA1-/- mice. The cells were labeled with [3H]cholesterol in cell culture media plus 10% FBS for 16 h and then treated with or without 5 μM TO901317 plus 5 μM 9-_cis_-retinoic acid for 16 h followed by cholesterol efflux for 4 h.
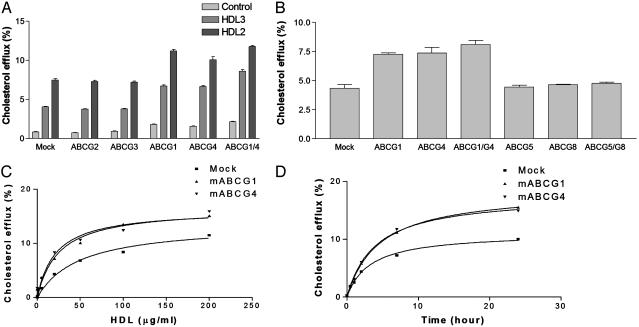
To examine the hypothesis that an ABCG transporter family member might be responsible for cholesterol efflux to HDL, we cloned all six members of the family that are expressed in mammalian cells and transiently expressed each cDNA in HEK293 cells labeled with isotopic cholesterol. Incubation of mock-transfected 293 cells with plasma HDL caused efflux of isotopic cholesterol (Fig. 2_A_), likely reflecting passive exchange of cholesterol between HDL and cells. Transient transfection with ABCG1 or ABCG4 resulted in stimulation of isotopic cholesterol efflux to both HDL-2 and HDL-3 (Fig. 2 A). HDL-specific cholesterol efflux (total minus control) was approximately doubled for HDL-3, whereas efflux to HDL-2 was increased by ≈50% (Fig. 2_B_). The combination of ABCG1 and ABCG4 resulted in a further small increase in cholesterol efflux. In contrast, other ABCG transporters that we examined, ABCG2, ABCG3 (Fig. 2 A), ABCG5, ABCG8, or ABCG5/ABCG8, co-expression (Fig. 2_B_) did not promote cholesterol efflux to HDL. Combination of ABCG1 or ABCG4 with any of the other ABCG transporters did not lead to a further increase in cholesterol efflux to HDL (data not shown). Time- and concentration-dependence experiments showed that efflux mediated by ABCG1 or ABCG4 continued to increase over 24 h and reached a maximum at ≈50 μg/ml HDL protein, similar to the concentration of HDL present in interstitial fluid (ref. 16 and Fig. 2 C and D).
Fig. 2.
Cells transfected with ABCG1 and ABCG4 cDNAs show increased cholesterol efflux to HDL. HEK293 cells were transiently transfected with plasmid constructs expressing ABCG transporters or control empty vector (mock), and cholesterol efflux was initiated by addition of HDL to media. (A) [3H]cholesterol efflux to HDL-2 or HDL-3 (25 μg/ml HDL protein) or media alone (control) for 4 h. (B) [3H]Cholesterol efflux to HDL-2 (25 μg/ml HDL protein) for 4 h. (C) [3H]Cholesterol efflux to HDL-2 at indicated concentrations for 4 h. (D)[3H]Cholesterol efflux to HDL-2 (25 μg/ml HDL protein) for the indicated period of time.
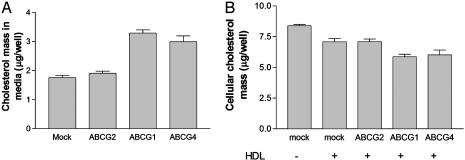
The efflux of isotopic cholesterol can result either from a net transfer process or from exchange of free cholesterol between the cell and HDL. Remarkably, gas-chromatographic measurement of cholesterol content in media indicated almost a doubling of HDL-free cholesterol mass after incubation with cells expressing ABCG1 or ABCG4, indicating a marked stimulation of net free cholesterol efflux (Fig. 3_A_). A similar increase in free cholesterol mass was observed both for total HDL (Fig. 3_A_) and for HDL-2 (data not shown). We also determined total cellular cholesterol mass in transfected 293 cells after HDL-mediated cholesterol efflux (Fig. 3_B_). HDL treatment slightly decreased cellular cholesterol mass (Fig. 3_B_) and ABCG1 or ABCG4 expression further reduced the cellular cholesterol content (Fig. 3_B_), reflecting the increased cholesterol efflux to HDL.
Fig. 3.
ABCG1 and ABCG4 expression increase cholesterol mass efflux to HDL and decrease cellular cholesterol content. (A) Free cholesterol mass in culture media determined in transfected 293 cells incubated with HDL-2 (25 μg/ml HDL protein) for 4 h. (B) Total cellular cholesterol was determined after 6 h incubation of transfected 293 cells with HDL-2 (25 μg/ml HDL protein).
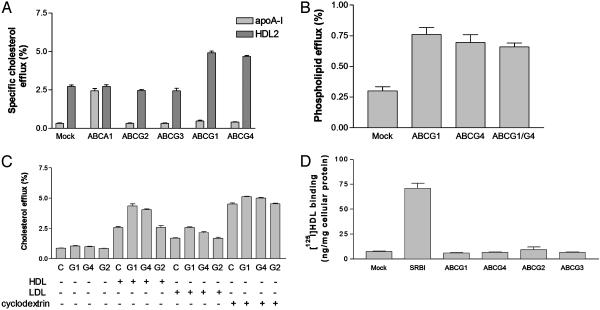
When incubated with lipid-poor apoA-I, cells transfected with ABCG1, ABCG4, or the other ABCG transporters did not stimulate cholesterol efflux to apoA-I (Fig. 4_A_). In marked contrast, ABCA1 stimulated efflux to apoA-I but not HDL-2, as reported (7). Cell transfection with ABCG1 or ABCG4 also resulted in a slight increase in efflux of phospholipid radioactivity to HDL (Fig. 4_B_). However, this transfection represented <1% of cellular phospholipid; by comparison, cells transfected with ABCA1 typically efflux several percentage of both cellular phospholipid and cholesterol to apoA-I (5, 7). Thus, ABCG1 and ABCG4 mediate prominent net cholesterol efflux to HDL but not to lipid-poor apoA-I.
Fig. 4.
ABCG1 and ABCG4 do not promote cholesterol efflux to apoA-I and do not bind HDL while promoting cholesterol efflux to HDL, LDL, and cyclodextrin. (A) HDL-2 (25 μg/ml HDL protein) and apoA-I (15 μg/ml protein) mediated cholesterol efflux during a 4-h incubation with 293 cells expressing ABCG transporters. (B) [3H]choline-containing phospholipid efflux to HDL-2 (25 μg/ml HDL protein) during a 4-h incubation with 293 cells expressing ABCG transporters. (C) Cholesterol efflux to HDL (25 μg/ml HDL protein), LDL (25 μg/ml LDL protein), or cyclodextrin (1 mM) during a 4-h incubation with 293 cells expressing ABCG transporters. (D) [125I]HDL binding to 293 cells expressing SR-BI or ABCG transporters.
We also determined the ability of ABCG1 and ABCG4 to stimulate cholesterol efflux to LDL and to an inert cholesterol acceptor, cyclodextrin (Fig. 4_C_). In cells transfected with ABCG1 or ABCG4, there was a small but significant stimulation of cholesterol efflux to LDL and to cyclodextrin, but this amount was less than observed with HDL. ABCA1 binds lipid-poor apolipoproteins, and this activity is closely correlated with its ability to mediate lipid efflux from cells (5, 17, 18). Similarly, SR-BI binds HDL and this result appears to be required for its selective uptake function (19). In contrast, cells transfected with ABCG1 or ABCG4 did not bind HDL above control levels (Fig. 4_D_).
The finding that ABCG1 and ABCG4 promote cholesterol efflux to HDL-2 but not to apoA-I could explain our earlier observations, suggesting an LXR-induced, ABCA1-independent pathway of cholesterol efflux in macrophages (Fig. 1 and ref. 11). ABCG1 and ABCG4 are expressed in macrophages and are induced by LXR/RXR activation with 22-OH cholesterol and 9-_cis_-retinoic acid (15, 20, 21). A specific LXR activator, TO901317 (2 μM) increased mRNA levels of ABCG1 and ABCG4 by 3- and 2-fold in mouse macrophages (data not shown), confirming the previous findings.
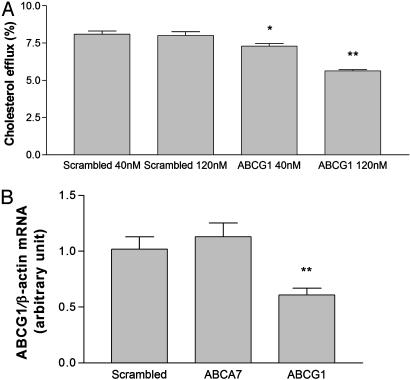
To determine if the induction of cholesterol efflux to HDL-2 is due to expression of ABCG1 and/or ABCG4, we used RNAi induced by synthetic siRNA in mouse peritoneal macrophages pretreated with the LXR activator TO901317 (2 μM). Knockdown experiments were conducted at two different concentrations of siRNA (40 and 120 nM). Suppression of ABCG1 resulted in a dose-dependent significant reduction in cholesterol efflux to HDL (Fig. 5_A_). At the higher dose, the suppression resulted in approximately a 30% reduction in isotopic efflux to HDL. By contrast, RNAi by using scrambled RNA or an irrelevant ABCA7 target sequence did not change cholesterol efflux. Measurements of mRNA levels by real-time RT-PCR indicated a specific suppression of ABCG1 mRNA, with ≈50% reduction at the higher level of RNAi (Fig. 5_B_). Initial results with siRNA against ABCG4 showed a reduced macrophage cholesterol efflux to HDL. However, ABCG1 mRNA was also reduced by the ABCG4 siRNAs (data not shown), probably due to homology in the target sequences (22). An independent set of siRNAs targeting different sequences in ABCG1 and ABCG4 confirmed a decreased cholesterol efflux to HDL (data not shown). Because the residual efflux of isotopic cholesterol likely includes a large component due to passive exchange processes, these data suggest that ABCG1, and possibly ABCG4, makes a major contribution to HDL-mediated cholesterol efflux in LXR-induced macrophages.
Fig. 5.
Suppression of ABCG1 expression by RNAi decreases macrophage cholesterol efflux to HDL. [3H]cholesterol efflux to HDL-2 was determined by using mouse peritoneal macrophages after transfection of the cells with synthetic siRNA against ABCG1 (A and B). Two different concentrations of siRNA were used, as indicated. As a control, scrambled siRNA or siRNA against ABCA7 were used. mRNA levels of ABCG1 (B) normalized against β-actin mRNA from macrophages treated with 120 nM siRNA were determined by TaqMan real-time RT-PCR. *, P < 0.05; **, P < 0.01.
Discussion
Our previous studies (11) suggested the existence of an LXR-induced, ABCA1-independent pathway of cholesterol efflux to HDL. This hypothesis was confirmed by comparing efflux in WT and ABCA1-/- macrophages (Fig. 1). By using cell transfection and RNAi, we now show that two known LXR targets of unknown function, ABCG1 and ABCG4, mediate cholesterol efflux to the major HDL fractions HDL-2 and HDL-3 but not to lipid-poor apoA-I. In contrast, ABCA1 mediates cholesterol efflux to apoA-I and interacts poorly with HDL-2 and HDL-3 (5, 7). The ability of ABCG1, and possibly ABCG4, to mediate cholesterol efflux to HDL could be important in the athero-protective effect of HDL because the bulk of plasma HDL consists of such mature HDL particles.
Although it has been speculated that ABCG1 could have a role in cellular cholesterol efflux and reverse cholesterol transport (23, 24), no definite function of either ABCG1 or ABCG4 has been previously assigned. ABCG1 was initially identified as a macrophage LXR target (15). Schmitz and coworkers (21) reported that an antisense oligodeoxynucleotide to ABCG1 reduced macrophage cholesterol efflux to HDL-3. However, this group subsequently stated that the same oligodeoxynucleotide also reduced expression of apoE and questioned whether ABCG1 was directly involved in lipid efflux (23). Hepatocyte overexpression of ABCG1 by adenovirus infection in mice resulted in a slight lowering of HDL and increased biliary cholesterol secretion (24, 25). Physiological relevance of these experiments is somewhat uncertain because hepatic expression of ABCG1 is probably predominantly in Kupffer cells (26), whereas adenovirus is expressed mainly in hepatocytes. Thus, the function of ABCG1 has remained enigmatic and its role in reverse cholesterol transport is considered uncertain (24).
The present study suggests a major role of ABCG1 in HDL-mediated cholesterol efflux in macrophages while the role of ABCG4 in macrophages is less certain. Although ABCG4 mRNA is detectable in this and other studies by using RT-PCR methodology (20), these semiquantitative measurements suggest a low level of ABCG4 expression in mouse macrophages even after LXR activation (data not shown). However, ABCG4 is highly expressed in brain (27), and HDL-like particles are present in cerebrospinal fluids (28, 29). Therefore, ABCG4 could promote cholesterol efflux to these HDL particles in brain. This finding is of particular interest in light of recent studies of the role of cholesterol metabolism in development of Alzheimer's disease, which suggest that promotion of cholesterol efflux in neuronal cells decreases amyloid β peptide formation and secretion (30).
In addition to ABCG1 and ABCG4, we showed previously that SR-BI also facilitates cholesterol efflux to HDL, but not to lipid-poor apoA-I (31). SR-BI promotes the bidirectional flux of cholesterol between cells and HDL and when HDL is phospholipid-rich and cholesterol-poor, net cholesterol efflux can result. However, unlike the findings with ABCG1 and ABCG4, SR-BI does not create a gradient of cholesterol concentration from cells to HDL. Studies with bone marrow transplantation show an increased atherosclerotic lesion area in apoE-/--recipient mice treated with SR-BI-/- apoE-/- donor cells compared with apoE-/--recipient mice receiving SR-BI+/+ apoE-/- donor cells (32). However, the SR-BI knockout macrophages display no difference in cholesterol efflux to HDL compared with WT macrophages (32), suggesting that macrophage SR-BI does not have a major role in cholesterol efflux to HDL in mice.
ABCG1 is most closely related to ABCG4, and both may be mammalian homologs of the Drosophila white gene. ABCG transporters are thought to function either as heterodimers, e.g., ABCG5/ABCG8 (33), or homodimers/homomultimers, e.g., ABCG2 (34). Because overexpression of either ABCG1 or ABCG4 resulted in cholesterol efflux to HDL, it appears they can function as homodimers. Also, inconsistent with function as heterodimers, the distribution of the two mRNAs in various tissues does not appear to be strongly correlated (27, 35), and they did not make functional partners with other ABCG family members in our cell expression experiments. However, several different transcripts of ABCG1 are present in macrophages (35, 36), raising the possibility of different functions and possibly heterodimerization with other half-transporters. Moreover, different functions in various cell types and tissues are also possible (26).
When comparing the ability of different acceptors to take up cholesterol, we found that in addition to efflux to HDL, ABCG1 and ABCG4 caused a slight but significant stimulation of cholesterol efflux to LDL and to an inert cholesterol acceptor, cyclodextrin (Fig. 2). Thus, ABCG1 and ABCG4 can promote cholesterol efflux to a variety of lipoprotein and nonlipoprotein acceptors, suggesting that these transporters may increase availability of cholesterol at the plasma membrane or at sites that are readily accessible to the plasma membrane. Although specific binding of HDL to ABCG1 and ABCG4 was not observed, HDL can bind cell membranes by nonspecific lipid-lipid interactions (37), perhaps acting to facilitate cholesterol efflux in ABCG1- or ABCG4-expressing cells (38). Rapid and slow components of cholesterol efflux to cyclodextrin have been described (39). The slow component of efflux is ATP dependent and may reflect cholesterol movement from the endocytic recycling compartment to the plasma membrane (40). A possible relationship of ABCG1 and ABCG4 to these efflux pathways warrants further investigation.
At equal protein concentrations, the level of cholesterol efflux to LDL was ≈55% of that observed for HDL (Fig. 3). Because HDL protein concentration in plasma normally exceeds that of LDL, this finding suggests that HDL will represent the major acceptor in normal plasma. However, in subjects with high LDL and low HDL levels, cholesterol efflux to LDL could predominate, giving rise to a futile cycle if LDL particles are subsequently ingested by macrophages.
Epidemiological studies indicate that both HDL-2 and HDL-3 are inversely related to atherosclerosis risk (41). Much of the difference in HDL levels among individuals reflects different levels of HDL-2, and HDL levels are in major part genetically determined by variation at the hepatic lipase and apoA-I/apoCIII/apoA-IV loci (8). Because ABCA1 does not directly interact with the main fraction of HDL (5), and does not likely account for a major part of the genetic variation in HDL levels in the general population (8, 42), ABCA1-HDL interactions or associations do not readily explain the protective effect of HDL. In contrast, our demonstration that ABCG1 promotes net cellular cholesterol efflux to HDL has the potential to provide a mechanistic understanding of the relationship of HDL to atherosclerosis risk. HDL infusions in humans are being carried out with reconstituted HDL particles (43), and HDL-raising therapies such as niacin (44) or cholesterol ester transfer protein inhibition (45) primarily block HDL catabolism and cause an increase in larger particles that are unlikely to interact with ABCA1. Cholesterol ester transfer protein inhibitors are in advanced human trials (46). Our findings suggest a mechanism to explain how these HDL-directed therapies could lead to cholesterol efflux from macrophage foam cells.
Acknowledgments
This work was supported by National Institutes of Health Grant HL 54591.
Abbreviations: HDL, high-density lipoprotein; LDL, low-density lipoprotein; ABC, ATP-binding cassette; LXR, liver X receptor; RXR, retinoid X receptor, siRNA, small interfering RNA; RNAi, RNA interference; HEK, human embryonic kidney; apo, apolipoprotein; SR-BI, scavenger receptor BI.
References
- 1.Orso, E., Broccardo, C., Kaminski, W. E., Bottcher, A., Liebisch, G., Drobnik, W., Gotz, A., Chambenoit, O., Diederich, W., Langmann, T., et al. (2000) Nat. Genet. 24**,** 192-196. [DOI] [PubMed] [Google Scholar]
- 2.Rust, S., Rosier, M., Funke, H., Real, J., Amoura, Z., Piette, J. C., Deleuze, J. F., Brewer, H. B., Duverger, N., Denefle, P., et al. (1999) Nat. Genet. 22**,** 352-355. [DOI] [PubMed] [Google Scholar]
- 3.Bodzioch, M., Orso, E., Klucken, J., Langmann, T., Bottcher, A., Diederich, W., Drobnik, W., Barlage, S., Buchler, C., Porsch-Ozcurumez, M., et al. (1999) Nat. Genet. 22**,** 347-351. [DOI] [PubMed] [Google Scholar]
- 4.Brooks-Wilson, A., Marcil, M., Clee, S. M., Zhang, L. H., Roomp, K., van Dam, M., Yu, L., Brewer, C., Collins, J. A., Molhuizen, H. O., et al. (1999) Nat. Genet. 22**,** 336-345. [DOI] [PubMed] [Google Scholar]
- 5.Wang, N., Silver, D. L., Costet, P. & Tall, A. R. (2000) J. Biol. Chem. 275**,** 33053-33058. [DOI] [PubMed] [Google Scholar]
- 6.Oram, J. F., Lawn, R. M., Garvin, M. R. & Wade, D. P. (2000) J. Biol. Chem. 275**,** 34508-34511. [DOI] [PubMed] [Google Scholar]
- 7.Wang, N., Silver, D. L., Thiele, C. & Tall, A. R. (2001) J. Biol. Chem. 276**,** 23742-23747. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J. C., Wang, Z., Grundy, S. M., Stoesz, M. R. & Guerra, R. (1994) J. Clin. Invest. 94**,** 2377-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costet, P., Luo, Y., Wang, N. & Tall, A. R. (2000) J. Biol. Chem. 275**,** 28240-28245. [DOI] [PubMed] [Google Scholar]
- 10.Repa, J. J., Turley, S. D., Lobaccaro, J. A., Medina, J., Li, L., Lustig, K., Shan, B., Heyman, R. A., Dietschy, J. M. & Mangelsdorf, D. J. (2000) Science 289**,** 1524-1529. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W., Sun, Y., Welch, C., Gorelik, A., Leventhal, A. R., Tabas, I. & Tall, A. R. (2001) J. Biol. Chem. 276**,** 43564-43569. [DOI] [PubMed] [Google Scholar]
- 12.Berge, K. E., Tian, H., Graf, G. A., Yu, L., Grishin, N. V., Schultz, J., Kwiterovich, P., Shan, B., Barnes, R. & Hobbs, H. H. (2000) Science 290**,** 1771-1775. [DOI] [PubMed] [Google Scholar]
- 13.Repa, J. J., Berge, K. E., Pomajzl, C., Richardson, J. A., Hobbs, H. & Mangelsdorf, D. J. (2002) J. Biol. Chem. 277**,** 18793-18800. [DOI] [PubMed] [Google Scholar]
- 14.Yu, L., York, J., von Bergmann, K., Lutjohann, D., Cohen, J. C. & Hobbs, H. H. (2003) J. Biol. Chem. 278**,** 15565-15570. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, M. A., Venkateswaran, A., Tarr, P. T., Xenarios, I., Kudoh, J., Shimizu, N. & Edwards, P. A. (2001) J. Biol. Chem. 276**,** 39438-39447. [DOI] [PubMed] [Google Scholar]
- 16.Reichl, D. (1990) Eur. Heart J. 11**,** Suppl. E, 230-236. [DOI] [PubMed] [Google Scholar]
- 17.Chroni, A., Liu, T., Fitzgerald, M. L., Freeman, M. W. & Zannis, V. I. (2004) Biochemistry 43**,** 2126-2139. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald, M. L., Morris, A. L., Chroni, A., Mendez, A. J., Zannis, V. I. & Freeman, M. W. (2004) J. Lipid Res. 45**,** 287-294. [DOI] [PubMed] [Google Scholar]
- 19.Krieger, M. (2001) J. Clin. Invest. 108**,** 793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel, T., Lorkowski, S., Lueken, A., Rust, S., Schluter, B., Berger, G., Cullen, P. & Assmann, G. (2001) Biochem. Biophys. Res. Commun. 288**,** 483-488. [DOI] [PubMed] [Google Scholar]
- 21.Klucken, J., Buchler, C., Orso, E., Kaminski, W. E., Porsch-Ozcurumez, M., Liebisch, G., Kapinsky, M., Diederich, W., Drobnik, W., Dean, M., et al. (2000) Proc. Natl. Acad. Sci. USA 97**,** 817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G. & Linsley, P. S. (2003) Nat. Biotechnol. 21**,** 635-637. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz, G., Langmann, T. & Heimerl, S. (2001) J. Lipid Res. 42**,** 1513-1520. [PubMed] [Google Scholar]
- 24.Brewer, H. B., Jr., & Santamarina-Fojo, S. (2003) Am. J. Cardiol. 91**,** 3E-11E. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T. (2003) Drug News Perspect. 16**,** 490-492. [DOI] [PubMed] [Google Scholar]
- 26.Hoekstra, M., Kruijt, J. K., Van Eck, M. & Van Berkel, T. J. (2003) J. Biol. Chem. 278**,** 25448-25453. [DOI] [PubMed] [Google Scholar]
- 27.Oldfield, S., Lowry, C., Ruddick, J. & Lightman, S. (2002) Biochim. Biophys. Acta 1591**,** 175-179. [DOI] [PubMed] [Google Scholar]
- 28.Borghini, I., Barja, F., Pometta, D. & James, R. W. (1995) Biochim. Biophys. Acta 1255**,** 192-200. [DOI] [PubMed] [Google Scholar]
- 29.Koch, S., Donarski, N., Goetze, K., Kreckel, M., Stuerenburg, H. J., Buhmann, C. & Beisiegel, U. (2001) J. Lipid Res. 42**,** 1143-1151. [PubMed] [Google Scholar]
- 30.Sun, Y., Yao, J., Kim, T. W. & Tall, A. R. (2003) J. Biol. Chem. 278**,** 27688-27694. [DOI] [PubMed] [Google Scholar]
- 31.Ji, Y., Jian, B., Wang, N., Sun, Y., Moya, M. L., Phillips, M. C., Rothblat, G. H., Swaney, J. B. & Tall, A. R. (1997) J. Biol. Chem. 272**,** 20982-20985. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, W., Yancey, P. G., Su, Y. R., Babaev, V. R., Zhang, Y., Fazio, S. & Linton, M. F. (2003) Circulation 108**,** 2258-2263. [DOI] [PubMed] [Google Scholar]
- 33.Graf, G. A., Yu, L., Li, W. P., Gerard, R., Tuma, P. L., Cohen, J. C. & Hobbs, H. H. (2003) J. Biol. Chem. 278**,** 48275-48282. [DOI] [PubMed] [Google Scholar]
- 34.Xu, J., Liu, Y., Yang, Y., Bates, S. & Zhang, J. T. (2004) J. Biol. Chem. 279**,** 19781-19789. [DOI] [PubMed] [Google Scholar]
- 35.Savary, S., Denizot, F., Luciani, M., Mattei, M. & Chimini, G. (1996) Mamm. Genome 7**,** 673-676. [DOI] [PubMed] [Google Scholar]
- 36.Croop, J. M., Tiller, G. E., Fletcher, J. A., Lux, M. L., Raab, E., Goldenson, D., Son, D., Arciniegas, S. & Wu, R. L. (1997) Gene 185**,** 77-85. [DOI] [PubMed] [Google Scholar]
- 37.Tabas, I. & Tall, A. R. (1984) J. Biol. Chem. 259**,** 13897-13905. [PubMed] [Google Scholar]
- 38.Yancey, P. G., Bortnick, A. E., Kellner-Weibel, G., de la Llera-Moya, M., Phillips, M. C. & Rothblat, G. H. (2003) Arterioscler. Thromb. Vasc. Biol. 23**,** 712-719. [DOI] [PubMed] [Google Scholar]
- 39.Yancey, P. G., Rodrigueza, W. V., Kilsdonk, E. P., Stoudt, G. W., Johnson, W. J., Phillips, M. C. & Rothblat, G. H. (1996) J. Biol. Chem. 271**,** 16026-16034. [DOI] [PubMed] [Google Scholar]
- 40.Hao, M., Lin, S. X., Karylowski, O. J., Wustner, D., McGraw, T. E. & Maxfield, F. R. (2002) J. Biol. Chem. 277**,** 609-617. [DOI] [PubMed] [Google Scholar]
- 41.Stampfer, M. J., Sacks, F. M., Salvini, S., Willett, W. C. & Hennekens, C. H. (1991) N. Engl. J. Med. 325**,** 373-381. [DOI] [PubMed] [Google Scholar]
- 42.Knoblauch, H., Bauerfeind, A., Toliat, M. R., Becker, C., Luganskaja, T., Gunther, U. P., Rohde, K., Schuster, H., Junghans, C., Luft, F. C., et al. (2004) Hum. Mol. Genet. 13**,** 993-1004. [DOI] [PubMed] [Google Scholar]
- 43.Nissen, S. E., Tsunoda, T., Tuzcu, E. M., Schoenhagen, P., Cooper, C. J., Yasin, M., Eaton, G. M., Lauer, M. A., Sheldon, W. S., Grines, C. L., et al. (2003) J. Am. Med. Assoc. 290**,** 2292-2300. [DOI] [PubMed] [Google Scholar]
- 44.Blum, C. B., Levy, R. I., Eisenberg, S., Hall, M., III, Goebel, R. H. & Berman, M. (1977) J. Clin. Invest. 60**,** 795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitlock, M. E., Swenson, T. L., Ramakrishnan, R., Leonard, M. T., Marcel, Y. L., Milne, R. W. & Tall, A. R. (1989) J. Clin. Invest. 84**,** 129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brousseau, M. E., Schaefer, E. J., Wolfe, M. L., Bloedon, L. T., Digenio, A. G., Clark, R. W., Mancuso, J. P. & Rader, D. J. (2004) N. Engl. J. Med. 350**,** 1505-1515. [DOI] [PubMed] [Google Scholar]