Genetic Correction of Human Induced Pluripotent Stem Cells from Patients with Spinal Muscular Atrophy (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 22.
Published in final edited form as: Sci Transl Med. 2012 Dec 19;4(165):165ra162. doi: 10.1126/scitranslmed.3004108
Abstract
Spinal muscular atrophy (SMA) is among the most common genetic neurological diseases that cause infant mortality. Induced pluripotent stem cells (iPSCs) generated from skin fibroblasts from SMA patients and genetically corrected have been proposed to be useful for autologous cell therapy. We generated iPSCs from SMA patients (SMA-iPSCs) using nonviral, nonintegrating episomal vectors and used a targeted gene correction approach based on single-stranded oligonucleotides to convert the survival motor neuron 2 (SMN2) gene into an SMN1-like gene. Corrected iPSC lines contained no exogenous sequences. Motor neurons formed by differentiation of uncorrected SMA-iPSCs reproduced disease-specific features. These features were ameliorated in motor neurons derived from genetically corrected SMA-iPSCs. The different gene splicing profile in SMA-iPSC motor neurons was rescued after genetic correction. The transplantation of corrected motor neurons derived from SMA-iPSCs into an SMA mouse model extended the life span of the animals and improved the disease phenotype. These results suggest that generating genetically corrected SMA-iPSCs and differentiating them into motor neurons may provide a source of motor neurons for therapeutic transplantation for SMA.
Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive genetic disorder caused by a genetic defect in the survival motor neuron 1 (SMN1) gene resulting from deletions or other mutations (1, 2). SMA arises when there are no functional copies of SMN1, and the affected individual therefore must fully rely on the protein produced from SMN2, a gene paralogous to SMN1 (1, 2). A decrease in full-length SMN protein results in the selective degeneration of spinal cord motor neurons (3). Patients with SMA typically show generalized muscle weakness and paralysis that can progress very rapidly to early childhood death (4). There is no cure.
SMN2 shows a high sequence homology to SMN1, and the only critical difference is the C-to-T base change 6 base pairs (bp) inside exon 7. This alteration, but not other variations in the SMN genes, affects the splicing of SMN2 (5). This splicing change yields 10% of the full-length protein and high concentrations of an unstable, truncated protein lacking exon 7 (SMNΔ7) (5). The disease severity inversely correlates with SMN2 copy number (6). Worms, flies, and mice lack SMN2, which can be introduced only by transgenic modifications (7–10). Several therapeutic strategies have targeted increased exon 7 inclusion in SMN2 transcripts, and a human cell–based assay is a critical tool (10).
The differentiation of viral vector–generated induced pluripotent stem cells (iPSCs) from SMA patients into motor neurons has recently been demonstrated (11). Ex vivo correction of the SMN1 mutation in SMA-iPSCs might allow the generation of disease-free motor neurons for cell therapy. The vectors used, however, can produce insertional mutations that interfere with normal cell function, and transgene expression can modify differentiation into specific lineages (12) or even lead to tumorigenesis (13), limiting their usefulness.
Here, we have generated iPSCs from SMA patients who are free from vectors and transgenic sequences and have differentiated them into motor neurons. Motor neurons generated from SMA-iPSCs presented specific disease-related features indicative of selective motor neuron degeneration. We designed oligodeoxynucleotides to SMN2 that induced a permanent genetic modification of a single nucleotide in exon 7, thus modifying the coding region of SMN2 to a more SMN1-like sequence. This resulted in the inclusion of exon 7 and the production of a greater amount of full-length SMN2. We then compared the phenotypes of motor neurons derived from corrected and untreated SMA-iPSCs in vitro using gene expression and splicing genome-wide analysis, and in vivo after transplantation into the spinal cords of transgenic SMA mice.
Results
Nonviral vector generation and characterization of SMA-iPSCs
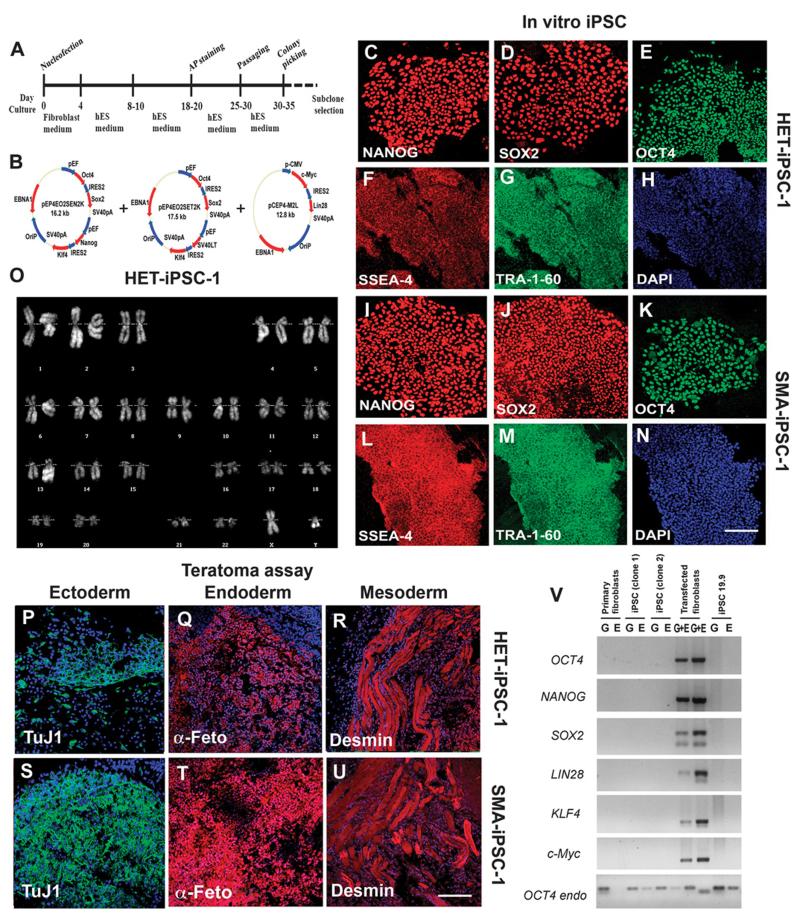
We generated iPSC lines from two type I SMA patients (SMA-iPSC-1 and SMA-iPSC-2) using a nonviral vector method based on nucleofection of adult fibroblasts with constructs encoding OCT4, SOX2, NANOG, LIN28, c-Myc, and KLF4 (14) (Fig. 1, A and B, and figs. S1 and S2). These plasmids are progressively lost from cells, leading to the generation of iPSCs free of vector and exogenous sequences. As a control, we generated iPSCs from the father of patient 1 (heterozygous, HET-iPSC-1), and we used an already generated wild-type cell line (iPSC 19.9). We isolated at least three clonal iPSC lines for each individual free from reprogramming vectors (Fig. 1V). All iPSC lines displayed embryonic stem (ES) cell morphology (Fig. 1, C to N) and expressed the pluripotency markers NANOG, SOX2, OCT4, SSEA-4, and TRA-1-60 (Fig. 1, F to H and L to N). All iPSC lines also maintained euploid karyotypes (Fig. 1O), could differentiate into all three germ layers in vitro (fig. S3), and could form teratomas in vivo (Fig. 1, P to U). DNA fingerprinting confirmed their origin from parental fibroblasts (table S1). These data and genome-wide gene expression analysis confirmed the reprogramming of fibroblasts to a pluripotent state (fig. S4).
Fig. 1.
Reprogramming human fibroblasts of an SMA patient and his father without genomic vector integration, and selection of iPSC clones. (A) Schematic representation of the nonviral reprogramming protocol for adult human fibroblasts. (B) Episomal vector maps. (C to N) Immunocytochemical characterization of iPSC clones derived from an SMA patient (SMA-iPSCs) and his heterozygous father (HET-iPSCs). These iPSCs express pluripotency transcription factors including NANOG (red), SOX2 (red), and OCT4 (green) (C to E and I to K), as well as stem cell surface markers (SSEA-4, red) and TRA-1-60 (green). Blue, 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain. (F) to (H) and (L) to (N) are representative images. (O) These cells (HET-iPSCs) were karyotypically normal. (P to U) SMA-iPSC clones and HET-iPSCs form teratomas in vivo that contain the three germ layers as shown by representative images from one SMA-iPSC and one HET-iPSC clone: ectoderm (TuJ1, green), mesoderm (desmin, red), and endoderm (α-fetoprotein, red). (V) PCR analysis of episomal DNA in iPSC clones. Genomic (G) and episomal (E) DNAs from nontransfected and vector-transfected adult human fibroblasts were used as negative (–) and positive (+) controls, respectively. No plasmid integration into iPSCs was observed. Scale bars, 100 μm (C to N); 150 μm (L to N) and (P to U).
SMN gene analysis and genetic conversion of SMN2 to SMN1 using oligodeoxynucleotides
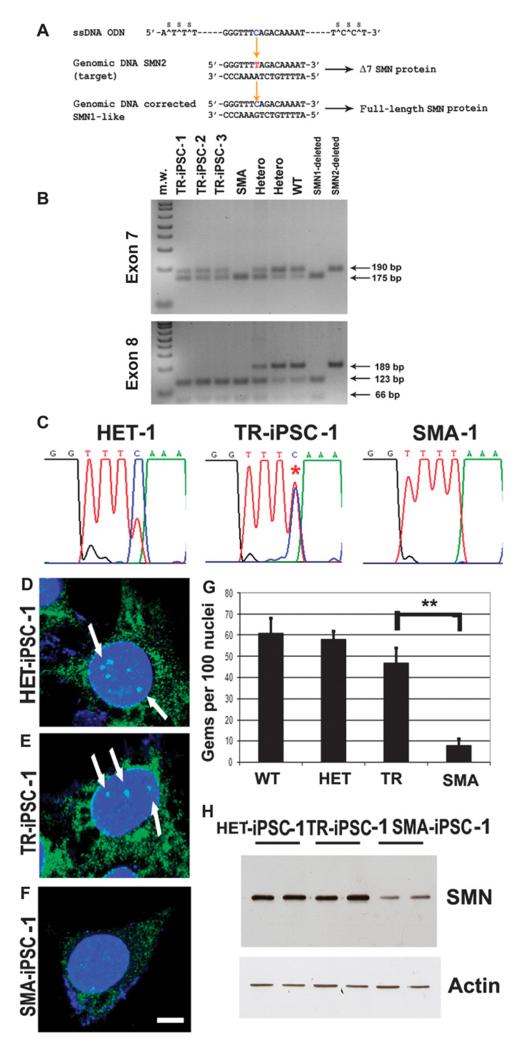
Previous work has described the possibility of genome editing at the single base level by introducing single-stranded DNA oligonucleotides into the cells (15). Taking advantage of this strategy, we used SMN2 sequence-specific oligodeoxynucleotides to direct the exchange of a T to C at position +6 of exon 7, thus modifying the coding region of SMN2 to a more SMN1-like sequence and therefore promoting the inclusion of exon 7. We transfected iPSCs with oligodeoxynucleotides 75 bases in length, designed to hybridize to the transcribed DNA strand of the SMN2 gene (15) (Fig. 2A). To demonstrate that the correction was permanent and heritable and to obtain a stable corrected cell population, we prepared clonal cell cultures of transfected cells. We screened the clones for genetic correction by detection of SMN protein nuclear foci (also known as Gemini bodies or gems) and polymerase chain reaction (PCR). We analyzed 47 subclones of the transfected SMA-iPSC-1 line, and we isolated three corrected clones that showed 50% SMN correction. We also performed the same gene editing in the second set of SMA-iPSC lines: 59, 61, and 65 subclones of SMA-iPSC-2 lines were analyzed, resulting in 2, 3, and 2 positive subclones, respectively. Therefore, the overall conversion efficiency (calculated as number of positive clones from the clones analyzed) was 10 of 232 (4%). The corrected genotype was stable during subsequent culturing for 12 passages, indicating that a permanent and heritable change had occurred.
Fig. 2.
Genetic correction by modification of the SMN2 gene using oligonucleotides in SMA-iPSCs. (A) Targeted SMN2_sequence and correcting oligonucleotides. The blue base in the oligonucleotides is the base that directs the targeting to the red base in the SMN2 sequence, which will render exon 7 in SMN2 like exon 7 in SMN1, resulting in its inclusion during splicing. (B and C) Restriction digest and cycle sequencing confirm base conversion in SMN2 in SMA-iPSCs. (B) The PCR products were subjected to restriction enzyme treatment: Dra1 for exon 7 (upper panel) and Dde1 for exon 8 (lower panel) (the enzymes Dra I and Dde I cleave the PCR products from SMN2 exons 7 and 8, respectively). In the corrected SMA-iPSCs, the presence of an uncut band corresponds to corrected exon 7, and the complete digested band (cut) corresponds to exon 8. This pattern corresponds to the modification of the_SMN2 gene into the SMN1_-like gene and rules out the possibility of a contamination with wild-type (WT) or heterozygous DNA. PCR electrophoresis showed the predicted band sizes: 175 bp for SMN1 and 190 bp for_SMN2 for exon 7 (upper panel). The three SMA-iPSC clones showed the presence of both bands, supporting the exon 7 base conversion to that of SMN1. (C) The results of the sequencing reaction confirm the conversion. The asterisk on a double peak indicates that both T and C have been detected, implying that this sample is heterozygous for SMN1 and SMN2. (D to G) The number of gems detected by SMN immunocytochemistry was higher in the HET-iPSCs (HET), WT iPSCs, and oligonucleotide-treated SMA-iPSCs (TR) when compared to untreated SMA-iPSCs (SMA) (**P ≤ 0.01). Mean ± SD, n = 5 independent experiments per condition [one-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test]. (H) Western blot analysis showed that corrected SMA-iPSCs had higher SMN protein concentrations than untreated SMA-iPSCs.
Using direct sequencing and allele-specific restriction fragment length polymorphism analysis, we evaluated the SMN genotype of the iPSC-corrected clones (Fig. 2, B and C, and fig. S5). The untreated iPSCs of SMA patient 1 and iPSCs treated with scrambled oligonucleotides carried a homozygous deletion of the SMN1 gene. In contrast, a 50% conversion of the SMN2 genotype to SMN1 was detected in corrected oligonucleotide-treated clones. The PCR products were subjected to restriction enzyme treatment: Dra1 for exon 7 and Dde1 for exon 8 (the enzymes Dra I and Dde I cleave the PCR products from SMN2 exons 7 and 8, respectively). These data demonstrated in the corrected iPSCs the presence of an uncut band corresponding to corrected exon 7, and the complete cut of exon 8 (Fig. 2B). This pattern corresponds to the conversion of SMN2 into SMN1 and rules out the possibility of a contamination with wild-type or heterozygous DNA. The father’s iPSCs presented a heterozygous genotype. The pluripotency features of the SMA-iPSCs were maintained after genetic correction.
The genetic correction resulted in a significantly increased number of detectable gems in oligodeoxynucleotide-treated cells, similar to that of the heterozygous father, whereas untreated SMA-iPSCs showed a lack of gems (SMA versus TR, P < 0.00001) (Fig. 2, D to G). We then evaluated SMN expression by Western blot and confirmed an increase in SMN protein in treated compared to untreated SMA-iPSCs. SMN protein expression in the treated SMA-iPSCs was similar to that in HET-iPSCs generated from the father of the patient (Fig. 2H). Together, these data demonstrated that oligonucleotides induced a direct and stable genetic correction in diseased human SMA-iPSCs.
Generation of spinal motor neurons from untreated and corrected SMA-iPSCs
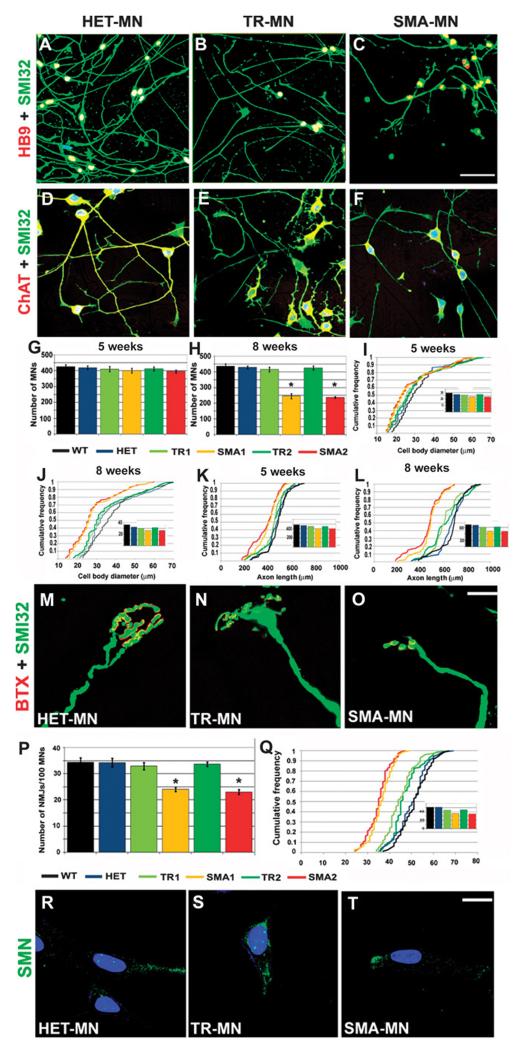
We next evaluated whether our nonviral vector–generated SMA-iPSCs could be lineage-committed toward a motor neuron fate to determine the effect of SMN1 deficiency on motor neurons and investigate whether genetic correction could rescue the SMA phenotype. Generation of spinal motor neurons from all iPSCs lines was achieved using a multistage differentiation protocol previously developed for human ES cells using retinoic acid and sonic hedgehog (SHH) (fig. S6) (16). After 4 to 5 weeks under differentiation conditions, cells that expressed the motor neuron–specific transcription factors HB9, ISLET1, and OLIG2 (spinal cord progenitor markers) and pan-neuronal markers such as TuJ1, Neurofilament, and MAP2 were generated. Most of these HB9/ISLET1-positive neurons expressed choline acetyltransferase (ChAT) and were positive for the motor neuron marker SMI32, demonstrating a motor neuron phenotype (fig. S7). The in vitro differentiation protocol yielded a mixed cell population that included nonmotor neuron cells. Given the limited availability of surface markers to isolate motor neurons and purify them further, we applied a physical strategy based on gradient centrifugation. After selection with this method, immunocytochemistry analysis showed that 78.2 ± 8.8% of cells were ChAT+SMI32+ derived from wild-type iPSCs, 76.7 ± 8.3% from HET-iPSCs, 74.1 ± 6.7% from untreated SMA-iPSCs, and 77.1 ± 7.2% from treated SMA-iPSCs. Quantification of astrocytic cells was performed. Less than 1% of cells differentiated from iPSCs expressed the astrocyte marker glial fibrillary acidic protein. Because there is a reduction in survival of SMA-iPSC–derived motor neurons in long-term culture, we increased the cell culture time (Fig. 3, A to F, and fig. S8). At 8 weeks, we found a reduced number of motor neurons in the untreated SMA-iPSC cultures relative to wild-type iPSCs and HET-iPSCs (P < 0.00001) (Fig. 3, G and H), and these motor neurons were reduced in size (P < 0.001) (Fig. 3, I and J). Notably, the cell number and size were increased in motor neurons derived from oligonucleotide-treated SMA-iPSCs compared to those derived from untreated SMA-iPSCs (treated motor neuron number, P < 0.00001; size, P = 0.002) (Fig. 3, G to J).
Fig. 3.
Differentiation of iPSCs from SMA patients into motor neurons. (A to F) After differentiation of iPSCs (HET-iPSCs, corrected and untreated SMA-iPSCs, TR, and SMA, respectively), the following motor neuron markers were expressed: SMI32- (green) and HB9-positive (red) (A to C) or ChAT (red) (yellow represents the merging of red and green colors). (D to F) Double-positive motor neurons were observed (merged, yellow). Nuclei are labeled with DAPI (blue). (G to L) Morphometric analysis of motor neurons. (G and H) Quantification of motor neurons at 5 (G) and 8 weeks (H) after differentiation from iPSCs, showing a reduced number of untreated SMA-iPSC–derived motor neurons compared to corrected SMA-iPSC–derived motor neurons (one-way ANOVA followed by a Tukey’s post hoc test) (*P < 0.001, 8 weeks). Values represent means ± SEM from five independent experiments performed in triplicate. (I to L) At 8 weeks, untreated SMA-iPSC motor neurons showed smaller cell diameters (J) and shorter axon lengths (L) than did motor neurons from the other three groups TR, WT, and HET (all, P < 0.001; size: SMA versus TR, P = 0.002). The difference was more evident at 8 weeks than at 5 weeks (I to K). Insets represent the respective medians. Five independent experiments performed in triplicate. (M to O) Neuromuscular junctions formed when motor neurons were cocultured with myotubes. The acetylcholine receptors were labeled with α-bungarotoxin (red) and axons with SMI32 (green). These are representative images. (P and Q) We evaluated the number (P) and size (Q) of neuromuscular junctions and found a reduced size and number of neuromuscular junctions in untreated SMA-iPSC motor neurons (SMA; patients 1 and 2) compared to corrected SMA-iPSC or HET-iPSC motor neurons (*P < 0.001, one-way ANOVA followed by a Tukey’s post hoc test). (P) Mean ± SEM, n = 5 independent experiments in triplicate. (Q) Inset represents the respective medians. (R to T) Gems are present in HET-MN (R) and TR-MN (S) but were absent in SMA-MN (T). Scale bars, 100 μm (A to C); 80 μm (D to F); 20 μm (M to O); 50 μm (R to T).
We then analyzed other characteristic elements of motor neurons that are likely to be critical in SMA pathogenesis: axonal elongation and neuromuscular junction formation. At 8 weeks, mean axon length was significantly shorter in SMA-iPSC–derived motor neurons with respect to wild-type and HET-iPSC–derived motor neurons (P < 0.001). However, motor neurons derived from corrected SMA-iPSCs had longer axons than those derived from untreated SMA-iPSCs (P < 0.001) (Fig. 3, K and L). Indeed, growth cones of SMA-iPSC–derived motor neurons were smaller compared to those derived from wild-type iPSCs and HET-iPSCs. However, after oligonucleotide correction of SMA-iPSCs, we observed an increase in size of the motor neurons derived from them (P < 0.001, treated versus untreated SMA motor neurons). We analyzed the difference in the ability of wild-type and SMA-iPSC–derived motor neurons to form neuromuscular junctions when cocultured with myotubes, evaluating the number and size of neuromuscular junctions formed (Fig. 3, M to O). Myotubes were stained with α-bungarotoxin to identify the acetylcholine receptor. SMA-iPSC–derived motor neurons generated fewer and smaller endplates compared to HET-iPSC–derived motor neurons. Furthermore, the mean neuromuscular junction size increased in motor neurons derived from treated versus untreated SMA-iPSCs (P < 0.001) (Fig. 3, P and Q).
Indeed, the quantification of nuclear gems demonstrated that gem number was increased in motor neurons derived from treated versus untreated SMA-iPSCs (Fig. 3, R and T). Thus, motor neurons can be obtained from nonviral vector–generated SMA-iPSCs, and cellular damage to the defective SMN1 gene can be rescued by SMN2 gene correction using oligonucleotides.
SMA-iPSC–derived motor neuron gene expression and splicing analysis
RNA and splicing abnormalities have recently been reported in SMA mice (17, 18), leading to the hypothesis that they could be primary mechanisms of selective motor neuron death. However, it remains unclear whether these alterations are present in motor neurons derived from iPSCs generated from SMA patient cells and if they can be rescued by gene correction. Thus, we performed a gene expression and exon array analysis of RNA from SMA motor neurons (SMA-iPSC-MN-1) in comparison to treated cells (TR-iPSC-MN-1). We compared different clones from SMA patient 1 and his father (HET-iPSC-MN-1). Detailed results are presented in the Supplementary Materials and Methods, tables S2 to S8, and figs. S9 and S10.
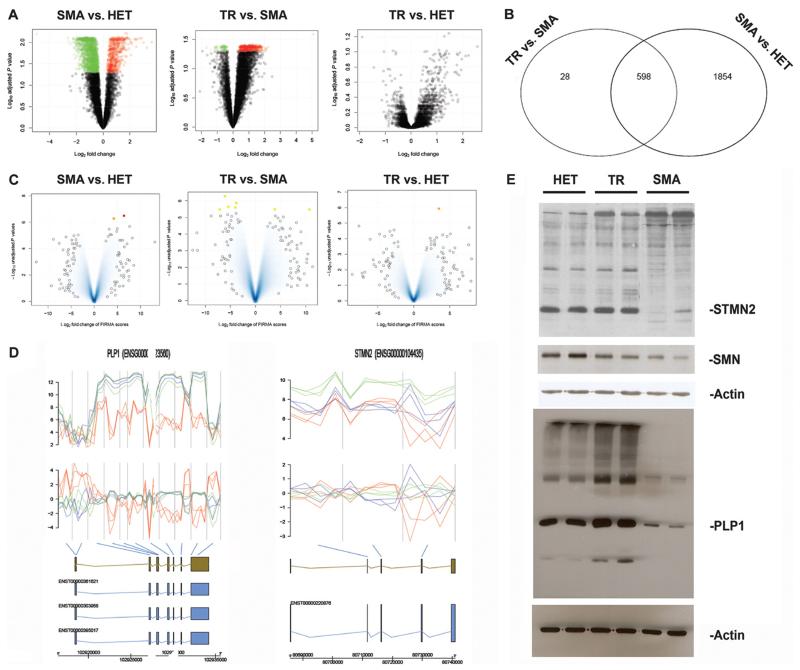
Regarding gene expression, a total of 2452 differentially expressed transcripts were identified in the comparison between SMA-iPSC–derived motor neurons and HET-iPSC–derived motor neurons as illustrated in Fig. 4, A and B (volcano plot), and table S2. Among the top down-regulated genes in SMA-iPSC motor neurons, we found genes encoding proteins involved in the cytoskeleton, neuronal development, and differentiation and synaptic generation. The expression of voltage-gated channels differed in SMA-iPSC–derived motor neurons compared to HET-iPSC control motor neurons (table S6). The comparison of treated versus untreated SMA-iPSC motor neurons revealed 626 differentially expressed transcripts (Fig. 4, A and B, and table S3). Remarkably, the genes that were most up-regulated in treated cells include some that were down-regulated in the comparison between SMA-iPSC motor neurons and HET-iPSC motor neurons, suggesting rescue with a shift toward the HET gene expression profile. Indeed, in the comparison between the HET-iPSC motor neurons and treated SMA-iPSC motor neurons, the volcano plot representation showed no differentially expressed transcripts (Fig. 4A and table S4). Overall, these analyses confirmed that correction with oligonucleotides shifted the expression profile toward HET. We then analyzed the mRNA splicing profiles of motor neurons derived from treated and untreated SMA-iPSCs and compared them to those of motor neurons derived from control HET-iPSCs. The results are illustrated in Fig. 4C and fig. S10, and a list of genes is provided in table S7.
Fig. 4.
Global gene expression and splicing analysis of iPSC-derived motor neurons. (A) Volcano plot for the class comparisons of gene expression. In the upper region of the picture are the most deregulated genes (green, down-regulated; red, up-regulated). (B) Venn diagram comparing the common differentially expressed genes. (C) Result for the three classes from the splicing analysis. The most significant data are marked in red, orange, and yellow and listed in the Supplementary Materials. (D) Splicing analysis of some of the most significantly deregulated genes. Figures illustrate the genes found to be differentially spliced, indicating from top to bottom the expression intensities and FIRMA scores for the individual probes in the probe sets [untreated (red) and corrected (blue) SMA-iPSC motor neurons; green, HET-iPSC motor neurons]. The gene, different transcripts, and exons are illustrated, with the blue bars indicating the association of the probe sets with the individual exons. The most significant probe set based on the FIRMA score is highlighted. (E) Western blot analysis of the deregulated proteins in motor neurons from untreated iPSCs (SMA), corrected iPSCs (TR), and HET-iPSCs. There is down-regulation of SMN in motor neurons from untreated iPSCs (SMA) compared to those from corrected iPSCs (TR) and HET-iPSCs. The most common_STMN2_ isoform as well as PLP1 appear to be down-regulated in the motor neurons from untreated iPSCs (SMA) compared to those from corrected iPSCs (TR) and HET-iPSCs. In the same Western blot, there are also some other differentially expressed bands between the two samples.
With the FIRMA method (19), the comparison between HET-iPSC motor neurons and treated SMA-iPSC motor neurons identified two differentially spliced genes: BCLAF1 and DOCK5 (Fig. 4D). BCLAF1 encodes a transcriptional repressor that interacts with several members of the BCL2 family of proteins. DOCK5 belongs to the family of guanine nucleotide exchange factors with a function in intracellular signaling networks. Expression of these two genes differed between motor neurons derived from treated and untreated SMA-iPSCs, indicating that oligonucleotide treatment can shift splicing toward the HET-iPSC–derived motor neuron profile.
In the expression intensities analysis (20), a total of 3345 differentially expressed probe sets were identified for the HET-iPSC and treated SMA-iPSC motor neuron comparison and 6 for the treated and untreated SMA-iPSC motor neuron comparison (fig. S10). The most prominent differentially spliced probe sets for the treated versus untreated SMA-iPSC motor neuron comparison included the following genes: BCLAF1, DOCK5, TPM3, COBL, OGFR, MSL3, and WDR7. The list of genes with the highest degree of differential splicing is summarized in table S7. It has been reported that decreased concentrations of SMN go along with a decreased assembly of the minor spliceosome (21). We tested whether some differentially spliced genes (table S7) were processed by the minor spliceosome. Two of these genes were shown to have U12 splicing introns (table S8). Overall, the genes with the greatest degree of differential splicing were members of functional classes mostly considered responsible for SMA pathogenesis, including axon guidance, motor neuron differentiation, DNA linkage, and signal transduction. We used Western blot analysis to demonstrate the expression of the most differentially expressed genes and its variation after oligonucleotide treatment (Fig. 4E). The STMN2 and PLP1 proteins presented different isoform patterns in motor neurons derived from SMA-iPSCs compared with those derived from control HET-iPSCs (Fig. 4E). In motor neurons from treated SMA-iPSCs, the level of expression of genes encoding these proteins was similar to that in motor neurons from control HET-iPSCs (Fig. 4E). Furthermore, Western blot analysis for the SMN protein revealed the expected down-regulation in the SMA-iPSC–derived motor neurons compared to that derived from HET-iPSCs, whereas in the motor neurons derived from SMA-iPSCs treated with oligonucleotides, the SMN protein concentration was restored (Fig. 4E).
Engraftment of motor neurons derived from SMA-iPSCs in the SMA spinal cord
Our next goal was to determine whether motor neurons derived from treated SMA-iPSCs survive and engraft appropriately within the spinal cord of SMA transgenic mice. We also wanted to determine the effect of disease environment on engrafted motor neurons and vice versa and whether transplantation can ameliorate the disease phenotype in SMA transgenic mice. Motor neurons derived from the three iPSC populations (heterozygous SMA, and treated and untreated SMA-iPSCs) were used for transplantation into the spinal cords of 1-day-old SMA mice (10,000 cells/1 μl × two horns, total: 20,000 for cervical C4 to C5; and total: 20,000 for lumbar L1 to L2 spinal cords). A group of mice was treated with vehicle alone. To track the fate of transplanted motor neurons, donor motor neurons were genetically modified with a lentiviral vector encoding a green fluorescent protein (GFP) reporter.
We evaluated whether motor neurons derived from human iPSCs survived and maintained an appropriate phenotype in the spinal cord of SMA mice. In all transplanted animals, we identified human transplant–derived GFP-labeled motor neurons within the ventral horn gray matter of the spinal cords of recipient mice (Fig. 5A and fig. S11). To define the neuronal phenotype of GFP+ motor neurons, we assessed neuronal markers in treated spinal cord sections. GFP+ motor neurons coexpressed a number of neuronal-specific pan-neuronal antigens, displayed a motor neuronal phenotype, and coexpressed HB9 and ChAT, indicative of cholinergic identity (Fig. 5A and fig. S11).
Fig. 5.
Transplanted motor neurons ameliorate the disease phenotype of SMA mice. (A) Engraftment of human donor iPSC-derived motor neurons in spinal cord of SMA mice. Representative images of GFP-tagged transplanted motor neurons, located in the anterior horns of the spinal cord, coexpressed motor neuron– specific proteins like ChAT. GFP (green), ChAT (red), merged (yellow); nuclei were stained with DAPI (blue). Scale bars, 75 μm (first to second rows); 50 μm (third row). The engraftment analysis was performed at the disease end stage; n = 24 per group. (B) Quantification of engrafted motor neurons and donor axons in the anterior roots; n = 24 per group. (C) Representative image of neuromuscular junctions formed with paravertebral muscles by axons (green) of donor transplanted treated SMA-iPSC–derived motor neurons. Cholinergic receptors are labeled with bungarotoxin (BTX, red). Merge, yellow. Scale bar, 20 μm. (D) Human motor neuron transplantation ameliorated muscle atrophy in SMA mice. Histograms of myofiber diameters: Mean tibialis anterior muscle cross-sectional area was reduced in vehicle-treated SMA mice compared with WT littermates (P < 0.00001) and was augmented after transplantation of motor neurons from different sources (P < 0.00001). Data represent mean values ± SD. Mean total myofiber number was reduced in vehicle-treated SMA mice compared with WT littermates (P < 0.00001) and increased after transplantation of motor neurons from different sources (P< 0.00001). n = 6 per group at postnatal day 13 (P13). (E) Gross appearance of an SMA mouse transplanted with motor neurons derived from corrected SMA-iPSCs (MNTR), a vehicle-treated SMA mouse (SMA), and a WT mouse showing that the transplanted SMA mice were larger than vehicle-treated SMA mice (P13). Survival was extended for mice transplanted with motor neurons compared with vehicle-treated and fibroblast-transplanted SMA mice (P < 0.00001) as shown by Kaplan-Meier survival curves. Transplanted SMA mice survived for HET-MN (median, 21 days), SMA-MN (19 days), oligodeoxynucleotide-corrected SMA-MN (TR-MN, 21 days), or human primary fibroblasts (13 days) or vehicle-treated mice (SMA, 14 days). Transplanted SMA mice presented increased weight compared with vehicle-treated SMA mice (P13; P < 0.00001), as shown by weight curves. All plots are shown as means of weight with error bars representing SD. The grip time was statistically different between the transplanted and vehicle-treated SMA mice (P < 0.00001) at 13 days. Error bars represent SD. At P13, the number of crossings of transplanted mice was increased with respect to vehicle-treated mice in the open-field test (P < 0.00001, ANOVA followed by a Tukey’s post hoc test for multiple comparisons).
Quantification analysis (Fig. 5B) demonstrated that the transplanted motor neurons from the untreated SMA-iPSC group showed reduced engraftment compared to those from the heterozygous SMA-iPSC group and that genetic correction rescued this defect (P < 0.00001). To evaluate whether iPSC-derived motor neurons could extend their axons outside the spinal cord in SMA mice, we analyzed the ventral roots. We observed the presence of GFP donor axons in the anterior roots (about 1%), suggesting that they can extend their axons toward the periphery (Fig. 5B). Further, we demonstrated that donor motor neurons can form a few new neuromuscular junctions (<10 neuromuscular junctions per condition) with skeletal muscles (Fig. 5C).
Improvement of muscle connections after motor neuron transplantation in SMA mice
We then performed histological analysis of muscle and spinal cord tissues at postnatal day 13 (n = 6 per group) and determined the number and size of transplanted motor neurons (fig. S12). The vehicle-treated SMA mice showed a reduction in motor neuron number and size compared with wild-type animals (P < 0.00001), whereas the number of motor neurons was significantly increased in transplanted (all three iPSC groups) versus vehicle-treated mice (P < 0.00001) (fig. S12). The stereological quantification, in the case of SMA mice receiving HET-iPSC–derived motor neurons, showed that donor neurons represented 27.5% of total motor neurons and that endogenous motor neurons were also increased by 6.8% compared to vehicle-treated SMA mice. The endogenous motor neurons of mice transplanted with HET-iPSC–derived motor neurons exhibited an increase in size versus vehicle-treated animals (P < 0.00001) (fig. S12). The same change was observed in animals transplanted with motor neurons from treated and untreated SMA-iPSCs versus vehicle control (P < 0.00001). Thus, the transplantation of human iPSC-derived motor neurons induced a neuroprotective effect relative to vehicle in SMA mice. However, the degree of this improvement was in proportion to the rate of engraftment and thus was higher for motor neurons derived from HET-iPSCs and treated SMA-iPSCs compared to those derived from untreated SMA-iPSCs.
Indeed, motor neuron transplantation resulted in a statistically significant increase in total muscle area (P < 0.00001) and mean myofiber diameter (P < 0.00001; Fig. 5D and fig. S13). These muscle area changes were observed also in the case of mice transplanted with motor neurons from treated and untreated SMA-iPSCs (P < 0.00001 versus vehicle-treated).
Amelioration of the disease phenotype and life extension after transplantation of iPSC-derived motor neurons in SMA mice
We determined the functional efficacy of motor neuron transplantation. After transplant, the physical appearance of the SMA mice was improved in comparison to that of vehicle-treated SMA mice (Fig. 5E, upper left panel, and movie S1). All mice receiving motor neuron transplants showed an increase in body weight that was statistically different at 13 days of age relative to vehicle-treated SMA mice (HET-iPSC motor neurons versus vehicle, untreated SMA-iPSC motor neurons versus vehicle, and treated SMA-iPSC motor neurons versus vehicle: P < 0.00001; Fig. 5E, bottom left panel).
To investigate whether transplanted motor neurons can rescue neuromuscular functions in SMA animals, we tested transplanted and control mice with the grip and open-field assays (22, 23). At 13 days of age, all three groups of transplanted SMA mice presented stable performance on the grip test for a few seconds, whereas nontransplanted SMA mice could not perform the task (HET-iPSC motor neuron transplant versus vehicle, untreated SMA-iPSC motor neuron transplant versus vehicle, and untreated SMA-iPSC motor neuron transplant versus vehicle: P < 0.00001) (Fig. 5E, bottom center panel). In addition, all motor neuron–transplanted SMA mice showed some locomotor activity and maintained some exploratory behavior compared to vehicle-treated SMA mice at 13 days of age (HET-iPSC motor neuron transplant versus vehicle, untreated SMA-iPSC motor neuron transplant versus vehicle, and untreated SMA-iPSC motor neuron transplant versus vehicle: P < 0.00001) (Fig. 5E, bottom right panel, and movie S1).
Mice that received motor neurons had improved survival rates when compared to their vehicle-treated counterparts. Figure 5E (upper right panel) illustrates the Kaplan-Meier survival curves. The median life spans of SMA mice receiving motor neuron transplants were 21 (HET-iPSC-MN), 19 (SMA-iPSC-MN), and 21 (treated SMA-iPSC-MN) days, whereas for vehicle-treated SMA mice, it was 14 days (n = 24 per group) (HET-iPSC, and treated and untreated SMA-iPSC motor neuron transplant versus vehicle, P < 0.00001). The maximum survival period observed was 24 days for mice receiving HET-iPSC motor neuron transplants and 16 days for vehicle-treated SMA mice. Survival was increased by 7 days for mice receiving HET-iPSC and treated SMA-iPSC motor neuron transplants, representing a gain of 50% over the lifetime. Compared to the HET-iPSC motor neuron–transplanted group, the untreated SMA-iPSC motor neuron–transplanted group showed a slightly reduced survival (P = 0.00027), whereas the treated SMA-iPSC motor neuron– transplanted group showed increased survival compared to the untreated group (P = 0.0018). SMA mice transplanted with human primary fibroblasts presented a survival trend similar to vehicle-treated SMA mice (median 13 days), demonstrating that the effect of motor neuron transplantation was specific (HET-iPSC motor neurons versus fibroblasts, P < 0.00001). Thus, motor neuron transplantation provided a survival benefit in an SMA mouse model.
Production of neuroprotective factors by transplanted human motor neurons
To analyze possible mechanisms for the observed improvement in SMA mice after transplantation, we evaluated whether human motor neurons derived from SMA-iPSCs, treated SMA-iPSCs, or HET-iPSCs by themselves can produce factors with a neuroprotective role on mouse motor neurons (Fig. 6, A to D, and fig. S14). We found that all of the human motor neurons release growth factors including neurotrophin 3 (NT3), NT4, vascular endothelial growth factor (VEGF), and nerve growth factor (NGF) (Fig. 6). They expressed moderate concentrations of ciliary neurotrophic factor (CNTF) and low concentrations of brain-derived neurotrophic factor (BDNF) and glial-derived neurotrophic factor (GDNF) (Fig. 6).
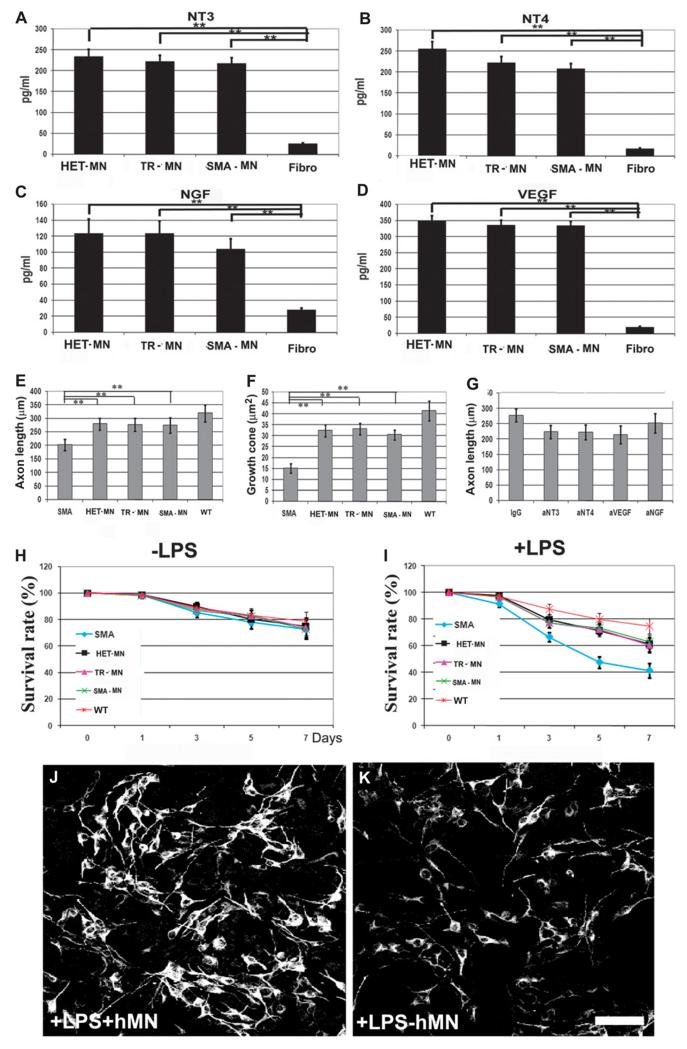
Fig. 6.
Human iPSC-derived motor neurons produce neuroprotective factors. (A to D) Amounts of (A) NT3, (B) NT4, (C) NGF, and (D) VEGF secreted in vitro by motor neurons and evaluated by ELISA. **P < 0.00001, two-tailed Student’s t test. Mean ± SD, n = 12 independent experiments for each cytokine. (E and F) Average length of axons (E) and growth cone area of motor neurons (F) from SMA mice, in coculture with human motor neurons. Motor neurons from SMA mice cocultured with human motor neurons exhibited an increase in axon length and size of growth cone with respect to SMA mouse motor neurons not cocultured with human motor neurons (**P < 0.00001, two-tailed Student’s t test; mean ± SD, n = 4 independent experiments for each condition). (G) The average axonal length of cocultured SMA mouse motor neurons was reduced after cytokine neutralization of cytokine production by human motor neurons (P < 0.05, two-tailed Student’s t test; mean ± SD, n = 4 independent experiments for each condition). (H and I) Quantification of the number of SMI32-positive SMA murine motor neurons in the presence of human motor neurons and microglial-conditioned media with (I) and without (H) lipopolysaccharide (LPS). The number of SMA mouse motor neurons in cultures with microglia + LPS was reduced but was increased in coculture with human motor neurons (mean ± SD, n = 4 independent experiments for each condition). (J and K) Representative SMI32 staining of SMA murine motor neurons (black and white were used to show the morphometric characteristics, that is, number and axonal length of cells) exposed to microglial-conditioned medium with LPS, with (J) or without (K) human motor neuron coculture. Scale bar, 50 μm.
To analyze the effect of donor human motor neurons on SMA pathogenesis, we used a coculture system with a bottom layer of primary motor neurons from SMA mice and a top layer of human motor neurons. As previously described, SMA mouse primary motor neurons alone presented a significant reduction in axon length relative to wild-type mouse primary motor neurons (P < 0.00001) (Fig. 3I). When SMA mouse primary motor neurons were cocultured in the presence of human motor neurons, axon length increased (Fig. 6E). Indeed, growth cones of SMN-deficient motor neurons from SMA mice were significantly smaller than those of HET-iPSC–derived motor neurons (P < 0.00001). Notably, we observed increased growth cone size for SMA mouse motor neurons after coculture with human motor neurons (P < 0.00001) (Fig. 6F). We hypothesized that up-regulation of neurotrophin expression may contribute to axon length enhancement. To test this hypothesis, we neutralized relevant cytokines individually or in combination by adding neutralizing antibodies to the motor neuron culture media. This treatment significantly reduced axon length for SMA mouse motor neurons (P < 0.05) (Fig. 6G). To study the effects of human motor neurons on neuronal survival in a neurotoxic environment, primary SMA mouse motor neurons were exposed to microglial-conditioned media or LPS-activated microglial-conditioned media in the presence of human motor neurons (Fig. 6, H and I). The findings demonstrated that human motor neurons secrete active neuroprotective factors that promote SMA mouse motor neurons in an inflammatory/toxic environment and enhance axonal elongation in vitro (Fig. 6, J and K).
The expression of neurotrophins by human motor neurons transplanted into the spinal cords of SMA mice, as evaluated by enzyme-linked immunosorbent assay (ELISA), demonstrated increased concentrations of neurotrophins when compared to vehicle-treated or fibroblast-transplanted SMA mouse controls (P < 0.00001) (fig. S14). To provide a mechanistic demonstration of the therapeutic role of neurotrophin expression by human motor neurons, we transplanted human motor neurons in which the relevant neurotrophin genes (NT3, NT4, and VEGF) were knocked down using short hairpin RNAs (shRNAs). These silenced cells are designated here as NT-kd-MNs. The ELISA analysis revealed a ≈70% reduction in neurotrophins compared with motor neurons transduced with nonsense control shRNAs (fig. S15, A and C). We then transplanted the NT-kd-MNs into the spinal cords of SMA mice. NT-kd-MNs extended survival less in comparison with HET-iPSC motor neuron transplants (P = 0.00002) but more than in vehicle-treated mice (P = 0.00094) (fig. S15D). Finally, we investigated expression of neurotrophins in transplanted NT-kd-MNs in SMA mouse spinal cord by ELISA. Under this condition, neutrophin expression was significantly reduced in NT-kd-MN–transplanted mice relative to scrambled-treated HET-iPSC motor neuron–transplanted mice (P < 0.00001) (fig. S15, E and G). Overall, these data suggest that motor neuron transplantation might exert a neuroprotective and axonogenic therapeutic effect by producing growth factors.
Discussion
SMA is a devastating neuromuscular disease without any effective treatment. Stem cells and iPSCs in particular represent a useful in vitro model as well as a cell source for potential therapeutic transplantation. To translate reprogramming technology from basic science toward clinical application, efficient derivation of human patient iPSCs that are free of foreign or chemical elements is crucial.
Here, we derived viral vector– and transgene-free human iPSCs from SMA patients using oriP/EBNA1-based vectors (12). Ebert et al. (11) previously described generating SMA-iPSCs and motor neurons using viral methods. Our work represents an advance because we achieved generation and characterization of SMA patient–specific motor neurons without genetic modification, thus increasing the possibility of applying these cells in basic research and transplantation regimens. Exploiting the full therapeutic potential of iPSCs will require precise and safe strategies for genetic modification in the case of the use of autologous cells from patients. In addition, four independent groups have recently described a complete rescue of the disease phenotype in SMA mice after intravenous injection of SMN-encoding adeno-associated virus (24–27), suggesting that cell and gene therapy are possibly complementary therapeutic strategies.
We used oligonucleotides to induce a permanent nonviral genetic correction of SMA-iPSCs such that SMN2 was converted into SMN1 by modifying a single base in exon 7. We demonstrate a genetic modification of human iPSCs from SMA patients with this technique. This strategy is specific, overcomes the limits of repeated treatment, does not introduce exogenous genes, and can be applied for other human genetic diseases. Furthermore, the SMN protein defect seems to affect motor neurons selectively. The use of SMA-iPSC–derived motor neurons described here can contribute to knowledge about the pathogenic mechanisms of SMA. We note that although we achieved a high purity of motor neurons from iPSCs, other cell populations were present at modest levels. In any case, our model can reproduce some aspect of SMA disease and potentially may be useful for in vitro drug screening.
We observed specific disease effects of the SMN defect on SMA motor neurons including a reduction in motor neuron survival and in their size, in line with that observed previously (11), although to a lesser degree. The centrifugation protocol used here might have selected for a stronger motor neuron population, resulting in overall improved results relative to those previously described. We also detected a reduction in axonal growth and neuromuscular junction formation in the motor neurons derived from SMA-iPSCs relative to control HET-iPSCs. Our data support the idea that more than one manifestation of motor neuron dysfunction plays a role in SMA, including a reduction in motor neuron survival and size, as well as in axonal growth and neuromuscular junction formation. Notably, we demonstrated that gene correction by converting SMN2 into SMN1 using oligonucleotides rescued neuropathological features of SMA in motor neurons and correlated with SMN expression.
We performed a detailed evaluation of transcriptional changes in the SMA-iPSC motor neurons in comparison to HET-iPSC and treated SMA-iPSC motor neurons. Our analysis identified several pathways that might shed light on SMA pathogenesis and its selectivity for motor neurons, and disease initiation and progression. We detected alterations in a subset of genes involved in RNA metabolism, motor neuron development, and axonal guidance. In particular, the expression of voltage-gated channels was altered in SMA-iPSC–derived motor neurons. A relatively restricted number of genes showed different splicing profiles in SMA-iPSC– versus HET-iPSC–derived motor neurons. Zhang et al. (17) reported widespread splicing abnormalities in SMA mouse tissues, including the spinal cord. Bäumer et al. (18) indicated that these events are a late occurrence in SMA and are therefore not likely to contribute to early disease pathogenesis. Our results do not support the hypothesis that widespread splicing abnormalities are present in motor neurons. Rather, they suggest the possibility that SMN deficiency critically affects splicing of one or a few transcripts and that the few splicing changes observed in our motor neuron models can contribute to SMA pathogenesis. Indeed, the genes with the higher degree of differential splicing belonged to classes of functions that are considered to have primary involvement in SMA pathogenesis, such as axon guidance, motor neuron differentiation, RNA metabolism, and signal transduction. The SMN defect can alter the minor spliceosome and, consequently, the genes that have U12 introns. Only two genes, DOCK5 and LPHN2, belong to this family. DOCK5 is involved in axon guidance and was a top differentially spliced gene identified in this work, warranting further analysis to define its role in SMA pathogenesis. Finally, our experiments demonstrated that most genes differentially expressed or spliced in motor neurons from SMA-iPSCs versus HET-iPSCs are shifted toward the HET pattern after gene correction, further supporting the efficacy of this gene repair strategy.
STMN2, a gene of particular interest from our analysis, is down-regulated and differentially spliced in SMA-iPSC versus HET-iPSC motor neurons as demonstrated both at the RNA and at the protein levels. Furthermore, the expression pattern of STMN2 was shifted toward the HET profile after oligonucleotide correction. The involvement of STMN2 in cytoskeleton metabolism and neuronal growth cone development and differentiation may be important in the context of SMA pathogenesis.
Another interesting gene was PLP1, which was down-regulated in SMA-iPSC motor neurons, and this down-regulation was rescued after oligonucleotide correction. PLP1 is expressed in nonmyelinating neuronal cell types and in motor neurons, where it plays a role in vesicular metabolism. These aspects may be important in the context of SMA pathogenesis and deserve further investigation.
Our in vivo experiments showed that iPSC-derived motor neurons can survive and integrate into the spinal cord of SMA mice and ameliorate the SMA type I phenotype. Donor-derived motor neurons correctly engrafted in the anterior spinal cords of transplanted mice and maintained a specific motor neuron phenotype. However, the SMA-iPSC motor neurons presented a slightly reduced survival and dimension as well as a decreased propensity for axonal elongation. These results suggest that SMA-iPSC motor neuron transplantation can produce an “in vivo model” of disease. The pathological features of SMA-iPSC motor neurons were rescued by genetic modification with oligonucleotides, opening the path to the possibility of autologous cell therapy. Transplanted SMA mice had a longer life span (about 50%) compared to vehicle-treated mice, an effect that was more pronounced with HET-iPSC and treated SMA-iPSC motor neurons than with untreated SMA-iPSC motor neurons. However, even if the difference in the therapeutic effect of wild-type motor neurons versus HET-iPSC and untreated SMA-iPSC motor neurons is small in our experimental settings, over the long-term, the reduced survival of these motor neurons would be problematic. We believe that ex vivo genetic correction of the SMA-iPSCs before transplantation would be important to obtain the maximum therapeutic benefit from this approach.
Indeed, the positive effects of transplanted motor neurons appeared to be specific because they were not observed after transplantation of fibroblasts. The increased survival time of SMA mice we observed after motor neuron cell transplantation, although limited with respect to other results like gene therapy, is noteworthy for two reasons: (i) The full potential of the motor neuron transplantation effect cannot be fully appreciated considering the limited time this model allowed for the formation of new neuromuscular junctions, and (ii) gene therapy, based on the experimental data available, seems to be most effective when applied in the very first stage of SMA (24–27). This suggests that a “window of opportunity” exists during the early stages of SMA in which gene therapy can have therapeutic benefits, at least in this experimental setting. The possibility of motor neuron replacement after the death of endogenous motor neurons is the ideal goal that has to be pursued to treat already symptomatic SMA patients, although such a goal is still a long way in the future.
Motor neuron transplantation also ameliorated defects in neuromuscular function compared to vehicle-treated SMA mice. This effect correlated with the number of donor-derived motor neurons, as well as with the change in endogenous motor neurons that were increased in both size and number, suggesting a neuroprotective effect of the transplanted motor neurons. We previously demonstrated that transplantation of murine neural stem cells, an undifferentiated population relative to the human postmitotic motor neurons used here, has a beneficial effect on motor neuron disease phenotypes in SMA mice (22, 23). In our past experiments (22, 23), mouse neural stem cells committed to a motor neuronal fate improved survival of SMA mice more than transplanting other cell types (undifferentiated cells, astrocytes, and fibroblasts), supporting the notion that transplantation of cholinergic precursors or motor neurons may be advantageous in the context of SMA spinal cord pathology. Regarding the possibility of transplanting undifferentiated iPSCs, this strategy is associated with a high risk of tumor development. Several studies report that undifferentiated iPSCs could also form teratomas in both immunocompromised and immunocompetent recipient animals (28, 29). Thus, the elimination of undifferentiated cells must be pursued. In our study, we wanted to investigate potential mechanisms that could account for the observed amelioration of the SMA phenotype after motor neuron transplantation. The formation of the few neuromuscular junctions likely contributed little to the phenotypic improvement. We demonstrated that human motor neurons derived from iPSC lines by themselves can produce several neurotrophic factors with a putative neuroprotective role through paracrine action on other motor neurons, as shown in the coculture experiments. Furthermore, we demonstrated that SMA spinal cords transplanted with human motor neurons showed increased concentrations of VEGF, NGF, NT3, and NT4.
After preventing production of neurotrophins by shRNAs, the therapeutic effect of motor neuron transplantation was reduced. These data suggest that motor neuron transplantation exerts a neuroprotective and axonogenic therapeutic effect by producing growth factors that might have therapeutic value beyond motor neuron replacement. Our experiments underscore the fact that motor neurons ameliorate some of the effects of SMA pathogenesis by providing neurotrophic support. The benefits of this cell population, however, will not be fully exploited until motor neurons can be stimulated to form functional neuromuscular junctions with the appropriate muscle targets in an efficient way. Future studies are needed to examine experimental strategies, like combining cell and pharmacological approaches to promote axonal elongation, as we have previously described (30), with the aim of achieving fully functional motor neurons. Our study has demonstrated transplantation of motor neurons derived from iPSCs generated from human SMA patients and their potential for ameliorating the SMA phenotype. Our findings may contribute to the development of cell therapies for SMA and other motor neuron diseases in the future.
Materials and Methods
See also the Supplementary Materials and Methods.
Reprogramming of human somatic cells
Reprogramming of human skin fibroblasts with oriP/EBNA1-based episomal vectors was carried out by nucleofection of the combinations of episomal plasmids (NHDF kit VPD-1001 with U-20 program, Amaxa). We used vectors that have been previously described (1). The complementary DNAs (cDNAs) for the open reading frames of the human genes OCT4, SOX2, NANOG, LIN28, c-Myc, and KLF4 were derived through direct PCR of human stem cell cDNA. We used two plasmid combinations (19 and 6) (14). After transfection, fibroblasts (1.0 × 106 cells per nucleofection) were plated onto 3 × 10–cm dishes covered with Matrigel (BD Biosciences) in fibroblast culture medium, which was changed every other day.
At 4 days from the transfection, we replaced the fibroblast culture medium with human ES cell culture medium (mTeSR, Stemcell Technologies Inc.) for 8 to 10 days. At day 18 after transfection, it was possible to identify the first colonies with an iPSC-like morphology. Between days 18 and 20 after transfection, we stained one of the 3 × 10–cm dishes of reprogramming culture with alkaline phosphatase (Millipore) to identify the eventual presence of human iPSC colonies. Between days 25 and 30, we passed the other two 10-cm dishes to fresh 10-cm Matrigel-covered dishes (1 ml each plate) at a ratio of 1:3. We then picked the iPSC colonies that were morphologically more similar to ES cells to further analyze and expand them. Efficiency of fibroblast reprogramming was about three to six colonies per 106 fibroblast cells, in line with that described previously (14).
Oligodeoxynucleotide transfection
A total of 500,000 cells were plated into 60-mm dishes and incubated overnight. The following day, the cells were transfected with oligodeoxynucleotide 75-T, targeted to the transcribed (T) strand of DNA. The oligodeoxynucleotides were previously designed so that the center nucleotide directed the base change (15). Different doses of oligodeoxynucleotide (3, 6, 10, or 20 μg/5 × 105 cells) were mixed with jetPRIME (Polyplus Transfection Inc.), and the complex was added directly to the medium. The oligodeoxynucleotides were allowed to incorporate overnight, after which the medium was replaced. The cells were collected at different time points for DNA, RNA, and protein analysis (24, 48, and 72 hours and 7 days). Transfection efficiencies were optimized with an oligodeoxynucleotide bearing a fluorescent tag (Cy3) as a tracer at different concentrations. A scrambled oligodeoxynucleotide sequence was used as control in all experiments. The oligodeoxynucleotides were produced by Sigma-Genosys. For generation of a clonal cell culture, at 24 hours from oligodeoxynucleotide transfection (10 μg) as described above, the cells were harvested and reseeded at clonal density (1 cell per well in a 96-well plate), and clones were grown. We screened the clones for the presence of genetic correction by PCR (see below).
iPSC differentiation in motor neurons
We generated spinal motor neurons using a multistage differentiation protocol developed for human ES cells (16). For the production of motor neurons from SMA-iPSCs, TR-iPSCs (patients 1 and 2), and HET-iPSCs, cells were plated with neuronal medium composed of Dulbecco’s modified Eagle’s medium/F12 (Gibco, Invitrogen), supplemented with MEM nonessential amino acids solution, N2, and heparin (2 μg/ml; Sigma-Aldrich). After 10 days, we added retinoic acid (0.1 μM; Sigma-Aldrich) for neural caudalization. At day 17, we collected the posteriorized neuroectodermal cells. These clusters were then suspended for a week in the same medium with retinoic acid (0.1 μM) and SHH (100 to 200 ng/ml; R&D Systems Inc.). On day 24, we added other growth factors like BDNF, GDNF, and insulin-like growth factor-1 (10 ng/ml; PeproTech).
Microarray analysis of iPSC expression
Microarray expression analysis of iPSCs (SMA, HET), iPSC 19.9 from the J. Thomson Lab (14), and parental fibroblasts was carried out with human genome U133 Plus 2.0 GeneChip arrays (Affymetrix). This analysis was performed by DNA Vision Laboratory. Total probe sets were 54,675, corresponding to 51,337 accession numbers as reported in the hgu133plus2 annotation package. The GeneChip IVT Express Kit and the GeneChip Hybridization wash and stain kit were used with 100 ng of total RNA that was mixed with 2 μl of 1:500,000 diluted spikes. Synthesis of the first strand and the second strand of cDNA was performed according to the kit instructions at 42°C, transcribed, and amplified into complementary RNA (cRNA). The cRNA was quantified by NanoDrop. Then, fragmentation of 12.5 μg of cRNA was performed and hybridized onto the GeneChip according to the Affymetrix protocol. An Affymetrix RUO platform was used, and data were processed with the Affymetrix GCOS program. Quality control was performed based on Affymetrix quality control metrics. All chips were normalized by the quantile method and background-corrected with robust multiarray analysis (RMA) (14). After normalization, the probe-level data were then summarized with median polish to obtain the probe set level measurement. We then collapsed 54,675 probe sets to 51,337 transcripts by keeping the average log intensity values for probe sets that represent a common accession number. Hierarchical cluster analyses were performed with 1 – PCC (Pearson correla on coefficient) as the distance parameter. The maximum distance between cluster members was used as the basis for merging lower-level clusters (complete linkage) into higher-level clusters. To assess the certainty of the existence of a cluster, we applied multiscale bootstrap resampling (10,000 bootstraps) to the hierarchical clustering of the 12 samples and calculated P values of the hypotheses as well as bootstrap probabilities for each cluster (14). Microarray data are available in the Gene Expression Omnibus (GEO) database (GSE27206).
Genome-wide and splicing analysis in iPSC-derived motor neurons
We then performed exon microarrays to determine whether there were any changes in expression and splicing in SMA cells compared to treated cells and unaffected cells. To do so, total RNA from isolated motor neurons, three SMA-iPSC–derived motor neuron samples (different subclones), three unaffected cells, and three SMA-treated cells (different subclones), each prepared and processed separately, were analyzed with the Affymetrix human exon 1.0 ST microarray. This analysis was performed by DNA Vision Laboratory according to the Affymetrix-recommended protocols. Affymetrix GeneChip Whole Transcript Sense Target labeling array and GeneChip Hybridization wash and stain kit were used: Ribosomal RNA was first reduced with the RiboMinus human/mouse kit (Invitrogen) starting from 1 μg of total RNA. Subsequently, purified RNA was used as a template in the synthesis of double-stranded cDNA according to instructions provided with the Affymetrix protocol. Double-stranded cDNA was in vitro–transcribed, producing cRNA. Finally, cRNA was newly retrotranscribed (second cycle), and single-stranded cDNA obtained through this passage was purified, quantified by NanoDrop, biotin-labeled, and fragmented according to the manufacturers’ protocols. The hybridization and scan protocols (Affymetrix RUO platform), suitable for Human Exon 1.0 ST microarrays, were provided by Affymetrix. RAW data were acquired with Affymetrix GCOS software. The groups of interest were compared with moderated t tests (20). Before adjustment for multiple comparisons (31), we filtered out the less-variable transcripts based on the interquartile range: A total of 8940 (3145 up-regulated and 5795 down-regulated) transcripts were kept. Differentially expressed transcripts were defined as having an adjusted P value of <0.05.
The results are illustrated with volcano plots in which the most significant transcripts are located toward the top of the figure and the largest fold changes are at the sides. The statistically differentially expressed genes (with an adjusted P value of <0.05, that is, a –log10 P value of >1.3) are shown in red (up-regulated) or green (down-regulated), respectively. We then analyzed the group of 8940 selected genes with a different statistical procedure. We calculated the ratio between all SMA samples and controls, obtaining nine values from each probe, from which we evaluated the mean value among values included in the tails of the distribution and with a homogeneous log2 ratio (meaning log2 ratio of the nine ratios to be positive in at least four cases for the up-regulated genes and negative in at least four cases for the down-regulated genes, to have a regular trend of the average and excluding the casual extreme values).
The detection of alternative splicing was performed to identify isoforms with the robust multichip analysis (FIRMA) method (19). This method is based on the RMA normalization (32, 33). The final goal of the FIRMA method is the gene expression valuation of each sample through the setting of a linear additive model with the following formula: log2(PMik) = ci + pk + εik, where ci is the chip effect for the chip i; pk is the probe effect for probe k; log2(PMik) is log2 of the exact match intensity for the probe k on the chip i corrected and normalized with the background; and εik is the remains for probe k on chip i. A sum value for the εik was evaluated for each probe set and used as scored. The differences in these scores are indicative of splicing phenomena. We have applied a moderated t test (20) to the FIRMA scores using the groups of interest. The results of each comparison are illustrated using two plots. The histograms illustrate the distribution of the adjusted P values. The individual adjusted P values are shown on the bottom of the figure as rungs, and the vertical dashed line represents the 0.05 threshold. The volcano plot is generated with unadjusted P values. Microarray data are available in the GEO database (GSE27205).
Animal models
The triple-mutant SMA mouse harbors two transgenic alleles and a targeted mutant. The Tg(SMN2*Δ7)4299Ahmb allele is an SMA cDNA lacking exon 7, whereas the Tg(SMN2)89Ahmb allele is the entire human SMN2 gene. Heterozygous Smn knockout mice with SMN2 transgenes were bred to obtain homozygous mice for the knockout Smn alleles (SMA mice, SMN2+/+;SmnΔ7+/+;mSmn−/−) [line 4299; FVB.Cg-Tg(SMN2*Δ7)4299Ahmb Tg(SMN2)89Ahmb Smn1tm1Msd]. The mice were genotyped with a PCR assay on genomic DNA from tail biopsies, as previously described (9).
All transgenic animals were obtained from the Jackson Laboratory. The in vivo experiments were approved by the University of Milan and Ministry of Health review boards, in agreement with U.S. National Institutes of Health guidelines (34).
Cell transplantation in SMA mice
Before cell transplantation, motor neurons were cultured and collected by gradient centrifugation as described and washed in phosphate-buffered saline (PBS). Human fibroblasts were used as control and were harvested with trypsin and then washed in PBS. To obtain GFP-positive cells, we used a lentiviral-based TurboGFP-encoding vector according to the manufacturer’s instructions (Mission TurboGFP Control Vector, Sigma-Aldrich). Cell culture medium was changed the day after transfection and the following 2 to 3 days after transduction, washing out any possible contaminating plasmid. We also transfected the control fibroblasts before transplantation. Under these conditions, we did not observe any motor neuron labeling. This experiment served as control for leakage of the plasmid into the host motor neurons.
iPSC-purified motor neurons were used for transplantation into the spinal cords of 1-day-old SMA mice (10,000 cells/1 μl × two horns, total: 20,000 for cervical C4 to C5; and total: 20,000 for lumbar L1 to L2 spinal cords) by direct injection as described (11, 19). There were four experimental groups of mice: Group 1 (HET motor neuron) received transplanted HET-iPSC–derived motor neurons; group 2 (SMA motor neuron) received SMA-iPSC–derived motor neurons; group 3 (TR motor neuron) received genetically corrected SMA motor neurons; and group 4 received vehicle (saline solution injection as mock transplantation, as the control group). Group 5 received human fibroblasts as an additional control group. Siblings were assigned equally in the groups. Each group had the same number of males and females. The immunosuppressor FK506 was administered intraperitoneally at 1.0 mg/kg to all animals in all groups for the entire length of the experiment.
These mice were monitored up to the end stage for neuromuscular function, survival, and histology of the human transplanted cell phenotype. Another series of mice (treated and vehicle-treated SMA mice and wild type) were analyzed for histological quantification (for each group per point: n = 6; 13 days).
Assessment of survival and phenotype
Transplanted and vehicle-treated SMA mice were assessed daily after transplantation for signs of disease. Observers were blinded to the treatments. The mice were sacrificed at the clinical endpoint, at which they showed problems in feeding, a clear downward progression (mice with 30% weight reduction and severe paralysis), and breathing difficulties, as described previously (22). Body weight was monitored daily. SMA mice were evaluated for grip strength as previously reported (22, 23). The investigators performing the functional analysis were blind to the treatment group. Wild-type mice as well as treated and vehicle-treated SMA mice were evaluated to see how long they could sustain their weight holding onto a metal bar suspended in midair (between 10 and 15 days of age). The data were analyzed by ANOVA followed by a Tukey’s post hoc test for multiple comparisons. Ambulatory function was investigated in an open-field test (23, 34). The apparatus was a wooden box (28 × 28 × 5 cm). The floor was partitioned into 16 equal squares of 7 × 7 cm. The mice were located in the center of the open field and observed individually. Each mouse was allowed to walk freely for 5 min, and the number of grids crossed was recorded.
Coculture of primary motor neurons of the SMA transgenic mice and human motor neurons
We obtained primary motor neuron cultures from embryonic day 12.5 SMA and wild-type mice as previously reported (22). The culture medium was a motor neuron medium supplemented with a cocktail of trophic factors as described above. We carried out a coculture assay to separate the primary motor neurons from the motor neurons with a microporous membrane, as previously described (22, 23).
Enzyme-linked immunosorbent assay
NT3, NT4, VEGF, NGF, CNTF, BDNF, GDNF, and TGF-α (transforming growth factor–α) secretion by MNs was determined with ELISAs, following the manufacturer’s instructions (R&D Systems). Medium was added (1.5 × 105 cells, 1.5 ml) and, 24 hours later, was removed for the ELISA test (12 independent experiments for each cytokine).
The same cytokines were investigated by ELISA in the lumbar spinal cord (n = 6 for each condition) after transplantation. The data were analyzed with Student’s t tests.
Supplementary Material
Supplementary Materials
Supplementary Movie
Acknowledgments
We thank M. Sironi for the revision of the manuscript and the Associazione Amici del Centro Dino Ferrari for their support. Funding: The financial support of research grants to S.C. and G.P.C. is gratefully acknowledged: FSMA and SMA Europe Grant, Telethon grant GGP09107, Telethon grant GGP10062; FIRB RBFR08RV86, Ministry of Health: GR-2009-1483560; Ass. Girotondo Onlus Grant; and SMArathon Onlus Grant.
Footnotes
Competing interests: The authors declare that they have no competing interests.
References and Notes
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Paslier D, Frézal J, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Munsat TL, Davies KE. International SMA consortium meeting. (26-28 June 1992, Bonn, Germany) Neuromuscul. Disord. 1992;2:423–428. doi: 10.1016/s0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Schrank B, Götz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossoll W, Prior TW, Morris GE, Burghes AH. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn−/− mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Stavarachi M, Apostol P, Toma M, Cimponeriu D, Gavrila L. Spinal muscular atrophy disease: A literature review for therapeutic strategies. J. Med. Life. 2010;3:3–9. [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr., Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMatteo D, Callahan S, Kmiec EB. Genetic conversion of an SMN2 gene to SMN1: A novel approach to the treatment of spinal muscular atrophy. Exp. Cell Res. 2008;314:878–886. doi: 10.1016/j.yexcr.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Hu BY, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat. Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdom E, Simpson KM, Robinson MD, Conboy JG, Lapuk AV, Speed TP. FIRMA: A method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonné R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum. Mol. Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D, Saladino F, Bordoni A, Fortunato F, Del Bo R, Papadimitriou D, Locatelli F, Menozzi G, Strazzer S, Bresolin N, Comi GP. Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J. Clin. Invest. 2008;118:3316–3330. doi: 10.1172/JCI35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D, Simone C, Falcone M, Papadimitriou D, Locatelli F, Mezzina N, Gianni F, Bresolin N, Comi GP. Embryonic stem cell-derived neural stem cells improve spinal muscular atrophy phenotype in mice. Brain. 2010;133:465–481. doi: 10.1093/brain/awp318. [DOI] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AH, Kaspar BK. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O’Riordan CR, Klinger KW, Shihabuddin LS, Cheng SH. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valori CF, Ning K, Wyles M, Mead RJ, Grierson AJ, Shaw PJ, Azzouz M. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- Dominguez E, Marais T, Chatauret N, Benkhelifa-Ziyyat S, Duque S, Ravassard P, Carcenac R, Astord S, Pereira de Moura A, Voit T, Barkats M. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Chen M, Yang B, Zhang F, Cao K. Intramyocardial transplantation of undifferentiated rat induced pluripotent stem cells causes tumorigenesis in the heart. PLoS One. 2011;6:e19012. doi: 10.1371/journal.pone.0019012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Kawai H, Tian F, Ohta Y, Abe K. Tumorigenic development of induced pluripotent stem cells in ischemic mouse brain. Cell Transplant. 2011;20:883–891. doi: 10.3727/096368910X539092. [DOI] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Del Bo R, Papadimitriou D, Locatelli F, Mezzina N, Gianni F, Bresolin N, Comi GP. Motoneuron transplantation rescues the phenotype of SMARD1 (spinal muscular atrophy with respiratory distress type 1) J. Neurosci. 2009;29:11761–11771. doi: 10.1523/JNEUROSCI.2734-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Corti S, Locatelli F, Papadimitriou D, Del Bo R, Nizzardo M, Nardini M, Donadoni C, Salani S, Fortunato F, Strazzer S, Bresolin N, Comi GP. Neural stem cells LewisX+CXCR4+ modify disease progression in an amyotrophic lateral sclerosis model. Brain. 2007;130:1289–1305. doi: 10.1093/brain/awm043. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- van der Steege G, Grootscholten PM, van der Vlies P, Draaijers TG, Osinga J, Cobben JM, Scheffer H, Buys CH. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995;345:985–986. [PubMed] [Google Scholar]
- Andreassi C, Angelozzi C, Tiziano FD, Vitali T, De Vincenzi E, Boninsegna A, Villanova M, Bertini E, Pini A, Neri G, Brahe C. Phenylbutyrate increases SMN expression in vitro: Relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- Gingras M, Gagnon V, Minotti S, Durham HD, Berthod F. Optimized protocols for isolation of primary motor neurons, astrocytes and microglia from embryonic mouse spinal cord. J. Neurosci. Methods. 2007;163:111–118. doi: 10.1016/j.jneumeth.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Wada T, Honda M, Minami I, Tooi N, Amagai Y, Nakatsuji N, Aiba K. Highly efficient differentiation and enrichment of spinal motor neurons derived from human and monkey embryonic stem cells. PLoS One. 2009;4:e6722. doi: 10.1371/journal.pone.0006722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Zhao W, Beers DR, Yen AA, Xie W, Henkel JS, Appel SH. Mutant SOD1G93A microglia are more neurotoxic relative to wild-type microglia. J. Neurochem. 2007;102:2008–2019. doi: 10.1111/j.1471-4159.2007.04677.x. [DOI] [PubMed] [Google Scholar]
- Curmi PA, Gavet O, Charbaut E, Ozon S, Lachkar-Colmerauer S, Manceau V, Siavoshian S, Maucuer A, Sobel A. Stathmin and its phosphoprotein family: General properties, biochemical and functional interaction with tubulin. Cell Struct. Funct. 1999;24:345–357. doi: 10.1247/csf.24.345. [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hübel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EC, Bongarzone ER, Campagnoni CW, Kampf K, Campagnoni AT. Soma-restricted products of the myelin proteolipid gene are expressed primarily in neurons in the developing mouse nervous system. Dev. Neurosci. 2003;25:96–104. doi: 10.1159/000072259. [DOI] [PubMed] [Google Scholar]
- Schmitt-John T, Drepper C, Mussmann A, Hahn P, Kuhlmann M, Thiel C, Hafner M, Lengeling A, Heimann P, Jones JM, Meisler MH, Jockusch H. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat. Genet. 2005;37:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- Swarup V, Julien JP. ALS pathogenesis: Recent insights from genetics and mouse models. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35:363–369. doi: 10.1016/j.pnpbp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Bürglen L, Seroz T, Miniou P, Lefebvre S, Burlet P, Munnich A, Pequignot EV, Egly JM, Melki J. The gene encoding p44, a subunit of the transcription factor TFIIH, is involved in large-scale deletions associated with Werdnig-Hoffmann disease. Am. J. Hum. Genet. 1997;60:72–79. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Supplementary Movie