Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair (original) (raw)
. Author manuscript; available in PMC: 2017 Jan 28.
Abstract
Mutational processes constantly shape the somatic genome, leading to immunity, aging, and other diseases. When cancer is the outcome, we are afforded a glimpse into these processes by the clonal expansion of the malignant cell. Here, we characterize a less explored layer of the mutational landscape of cancer: mutational asymmetries between the two DNA strands. Analyzing whole genome sequences of 590 tumors from 14 different cancer types, we reveal widespread asymmetries across mutagenic processes, with transcriptional (“T-class”) asymmetry dominating UV-, smoking-, and liver-cancer-associated mutations, and replicative (“R-class”) asymmetry dominating POLE-, APOBEC-, and MSI-associated mutations. We report a striking phenomenon of Transcription-Coupled Damage (TCD) on the non-transcribed DNA strand, and provide evidence that APOBEC mutagenesis occurs on the lagging-strand template during DNA replication. As more genomes are sequenced, studying and classifying their asymmetries will illuminate the underlying biological mechanisms of DNA damage and repair.
Introduction
A thorough understanding of mutational density and patterns in cancer genomes is important for studying the mechanisms of mutagenesis (Pleasance et al., 2010a; Pleasance et al., 2010b), for modeling the evolution of cancer genomes (Alexandrov et al., 2013; Nik-Zainal et al., 2012b) and for identifying cancer genes (Lawrence et al., 2013). In cancer genomes, somatic mutations exhibit heterogeneity in total mutation density, in mutation spectra among tumors and cancer types, and in mutation density along the genome within a given tumor (Lawrence et al., 2013; Pleasance et al., 2010a; Pleasance et al., 2010b). This heterogeneity is caused by underlying mutational processes that reflect different genetic backgrounds and mutagenic exposures, and by a non-uniform epigenomic landscape with variation in DNA replication timing, chromatin structure and gene expression levels across the genome (Lawrence et al., 2013; Pleasance et al., 2010a; Pleasance et al., 2010b; Polak et al., 2015; Polak et al., 2014; Waddell et al., 2015).
One challenge inherent in the analysis of genomic mutations is the loss of strand information that occurs between the initial occurrence of a mutagenic lesion and the ultimate readout by DNA sequencing. For instance, consider a mutational process whose initiating event is oxidative attack on the guanine of a C:G base pair. In principle if we isolated the DNA immediately after such an attack, we could directly observe the lesion; however, in genomic sequencing data we don't encounter mutations until many cell divisions later. The result of such a lesion is generally a A:G mismatch after the first cell division, leading to a stable A:T basepair after an additional round of replication. Since approximately half of C:G base pairs are oriented with the cytosine on the reference (Watson) and half on the anti-reference (Crick) strand, roughly equal numbers of “G→T” and “C→A” mutations are seen. A lesion at the cytosine of a C:G basepair could produce exactly the same result, so working backwards we cannot determine the base of the original DNA damage. This is because using the genomic reference strand as the “frame of reference” for base pair orientation is merely an arbitrary convention.
However, we can recover some strand information by considering a more biologically meaningful reference frame. In regions that undergo DNA transcription, the DNA can be oriented with respect to the transcribed strand. Thus we would consider a C:G→A:T base pair change to be a “C→A” or “G→T” mutation depending on whether the C or the G is in the template strand for transcription. Alternatively we can use DNA replication to define a frame of reference. In this case, whether the C of a C:G base pair is on the leading or the lagging strand of DNA replication would determine the type of mutation. Because replication and transcription are each associated with opportunities for the asymmetric (strand-specific) introduction and repair of DNA damage, they each have the potential to leave their footprints in a patient's mutational profile, in the form of unequal rates and patterns of mutations on the two strands of DNA (Francioli et al., 2015; Green et al., 2003; Lobry, 1996; Lujan et al., 2012; Pleasance et al., 2010a; Pleasance et al., 2010b; Polak and Arndt, 2008; Polak et al., 2010; Shinbrot et al., 2014; Touchon et al., 2005).
Strand asymmetry has already been well studied in the context of transcription. DNA lesions encountered on the transcribed (“template”) strand can stall progression of the RNA polymerase, leading to the recruitment of a nucleotide excision repair (NER) complex that can correct the damage(Donahue et al., 1994; Fousteri and Mullenders, 2008; Hanawalt and Spivak, 2008; Jiang and Sancar, 2006; Mellon et al., 1987; Spivak and Ganesan, 2014). Importantly, higher transcription levels of a gene are associated with more opportunities for transcription-coupled repair (TCR), leading to an inverse correlation between the expression level of a gene and its mutation density (Chapman et al., 2011; Lawrence et al., 2013; Pleasance et al., 2010a). Conversely, damage on the non-template (“sense”) strand may fail to stall the RNA polymerase and therefore could escape repair by TCR. In addition, the non-template strand remains single-stranded during the process of transcription, and is therefore more vulnerable to damage (Jinks-Robertson and Bhagwat, 2014). In combination, these mechanisms lead to differences in mutation densities and spectra on the transcribed and non-transcribed strands (Pleasance et al., 2010a; Pleasance et al., 2010b). Notably, transcriptional strand asymmetry provides information regarding damage and TCR beyond what can be gathered from the correlation of mutational densities with expression, since the latter is convolved with other genomic factors such as chromatin-state- and replication-timing-dependent mismatch repair (MMR)(Supek and Lehner, 2015).
Strand asymmetry can also be viewed in the reference frame of DNA replication. The DNA replication fork is composed of a leading strand, copied in a largely continuous fashion, and a lagging strand, copied as a discontinuous series of Okazaki fragments. DNA polymerases α, δ, and ε work together to replicate the DNA but have distinct roles in synthesis and proofreading. The resulting asymmetry reflects an imbalance in the types of mutations introduced on the leading versus lagging strand, although it is still a matter of debate whether this occurs due to the division of labor of distinct polymerases in DNA synthesis (Miyabe et al., 2011; Nick McElhinny et al., 2008), or due to specialized polymerase proofreading properties (Johnson et al., 2015; Stillman, 2015). Additionally the lagging strand endures longer exposure as single-stranded DNA (ssDNA)(Yu et al., 2014) and as such may be more vulnerable to ssDNA-targeting mutagens. These factors lead to replication-associated mutational asymmetry that flips (i.e. inverts which strand has the higher mutation density) at replication origins. Replication strand asymmetries were observed as local skews in nucleotide composition in the chromosomes of bacterial (Lobry, 1996; McLean et al., 1998) and eukaryotic (Touchon et al., 2005) species, are associated with robustly programmed yeast replication origins (Koren et al., 2010), and have also been experimentally demonstrated in yeast (Lujan et al., 2012; Pavlov et al., 2002).
Results
A framework for analysis of replicative and transcriptional asymmetries
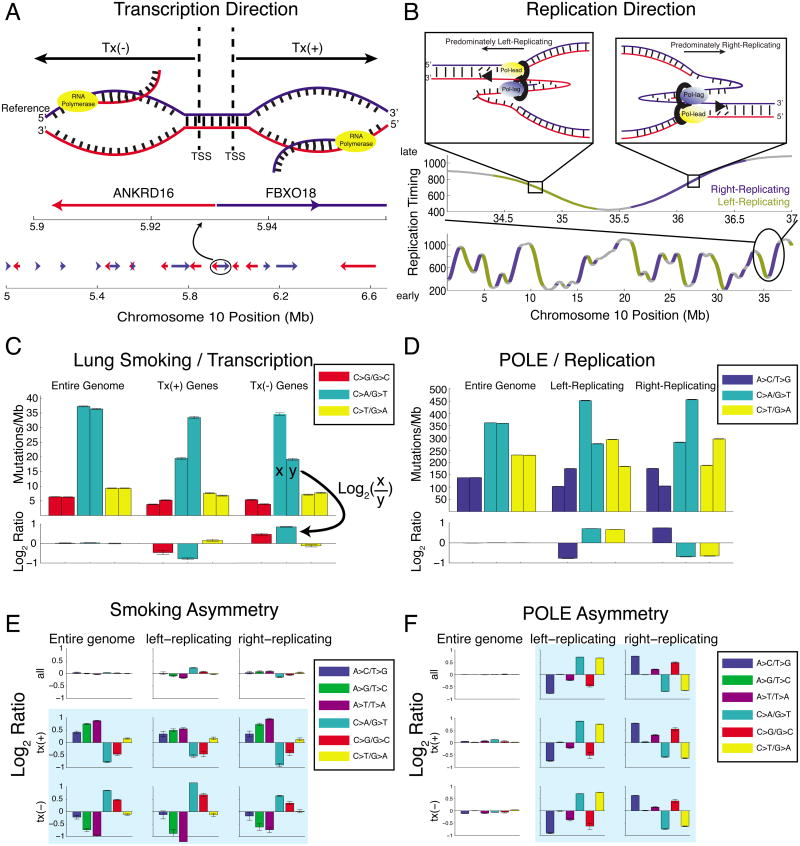
We partitioned the human genome in two ways: first, by transcription direction, using RefSeq gene definitions (Figure 1A). We annotated genomic regions as tx(+) when they encoded genes on the reference strand, and tx(-) when they encoded genes on the complementary strand. We considered the patterns of mutations in smoking-associated lung cancers, combining mutation data from seven lung adenocarcinomas (LUAD) that exhibited a strong smoking signature. Mutational densities of C:G→A:T are highest in both tx(+) and tx(-) genes when the guanine is on the non-transcribed strand (Figure 1C). This is consistent with the known mechanism of the smoking signature, driven by carcinogen attack at guanines (Denissenko et al., 1996). TCR lowers the mutational densities of C:G base pairs where the guanine serves as the transcription template (denoted Cntx:Gtx), relative to IGR regions. In contrast, Gntx:Ctx base pairs do not benefit from this extra opportunity for repair, resulting in undiminshed mutation density of Gntx:Ctx→Tntx:Atx, as shown previously (Pleasance et al., 2010b).
Figure 1. Mutational strand asymmetry associated with transcription (left) and replication (right).
(A) Transcription direction: Tx(+) regions carry the coding sequence of a gene on the genomic reference strand, and Tx(-) regions on the genomic complement strand. (B) Replication direction: positive slope in replication-timing data indicates general rightward movement of the replication complex (“right-replicating”), while negative slope indicates left-replicating. (C) Lung cancers show strong transcriptional (“T-class”) asymmetry. Each pair of bars (upper axis) shows the density of mutations at C:G (left bar) and G:C (right bar) base pairs. When summing across the entire genome, base-pair orientation does not affect mutational densities. In tx(+) regions, G:C base pairs show a higher density of G→T transversions than C:G base pairs; the opposite is true in tx(-) regions. Lower axis shows the log2 ratio of each pair of bars. (D) POLE-mutant cancers (colorectal and endometrial) show strong replicative (“R-class”) asymmetry. Left-replicating regions show a higher density of mutations at C:G base pairs, and right-replicating regions at G:C. (E) Lung cancers show strong T-class asymmetry but little R-class. (F) POLE-mutant cancers show strong R-class strand asymmetry but little T-class.
The second form of genome partitioning was by DNA replication direction. Since the entire genome is replicated every time a cell divides (but only a portion is transcribed), replication direction has the potential to exert larger asymmetries in mutational data. However, determining direction is much more challenging for replication than transcription, since the precise locations of replication origins in the human genome are not known. This has precluded a comprehensive analysis of replicative strand asymmetry thus far.
To enable an analysis of replication direction and strand asymmetry, we utilized high-resolution genomic replication timing data from deep DNA sequencing of S- and G1-phase cells from lymphoblastoid cell lines of six individuals (Koren et al., 2012). This data exhibits peaks and valleys in a timing-vs-location landscape that correspond to the approximate locations of replication origins (or origin clusters) and replication termini (Figure 1B). The regions between valleys and peaks correspond, in principle, to regions that replicate predominantly in a single direction (from origin to termination zone) and for which predominate replication direction can be assigned. This approach has previously been used to reveal compositional skews and asymmetric evolutionary germline mutations in the human genome (Chen et al., 2011). However, there are inherent limitations in the identification of replication origins based on replication timing valleys, and a lack of a gold standard (i.e. a set of replication origins with known locations) with which to benchmark this approach.
The valleys and peaks (constant timing regions) are the source of most tissue-specific variation in the profiles (Rhind and Gilbert, 2013; Ryba et al., 2010), and furthermore present no clear direction of replication. Therefore we excluded these regions from our analysis, and focused on timing transition regions (TTRs), which are highly conserved (Rhind and Gilbert, 2013; Ryba et al., 2010) and have a prominent slope that indicates the general direction of replication, either “left-replicating” or “right-replicating”. (We use the terms “left” and “right” when viewing the DNA in the standard orientation.) While TTRs were first thought to represent regions that are entirely uni-directional in replication (Ryba et al., 2010), it was later suggested that the vast majority of these regions are replicated too quickly for a single replication fork, and are more likely replicated by origins that fire in close succession (Guilbaud et al., 2011; Rhind and Gilbert, 2013). For any pair of sequentially firing origins the greater portion of the inter-origin distance is replicated by the fork originating from the earlier of the two origins. The result is that in aggregate the larger portion of a TTR is synthesized in the early-to-late direction (Figure S1). Thus TTRs have a predominant replication direction given by the sign of their slope. Restricting analysis to these regions enabled us to assign the predominant replication direction of 38% of the genome.
To validate our ability to measure replicative asymmetry using these left- and right-replicating definitions, we considered the one known case of replicative mutational asymmetry: tumors carrying functional mutations in the proofreading exonuclease domain of POLE, the gene encoding polymerase ε (designated as “POLE tumors”)(Shinbrot et al., 2014). The exonuclease domain of polymerase ε is responsible for proofreading during synthesis of the leading strand (Nick McElhinny et al., 2008; Shinbrot et al., 2014), and POLE tumors were previously reported to have high rates of C:G mutations (to A:T or T:A) asymmetrically introduced at cytosines replicated on the leading strand template near three well-characterized origins of replication (Shinbrot et al., 2014). As a consequence, in these tumors we would expect to see predominantly C→A mutations in left-replicating regions and G→T in right-replicating regions, since we hypothesized these regions to be enriched for respectively leading- and lagging-strand synthesis on the reference strand.
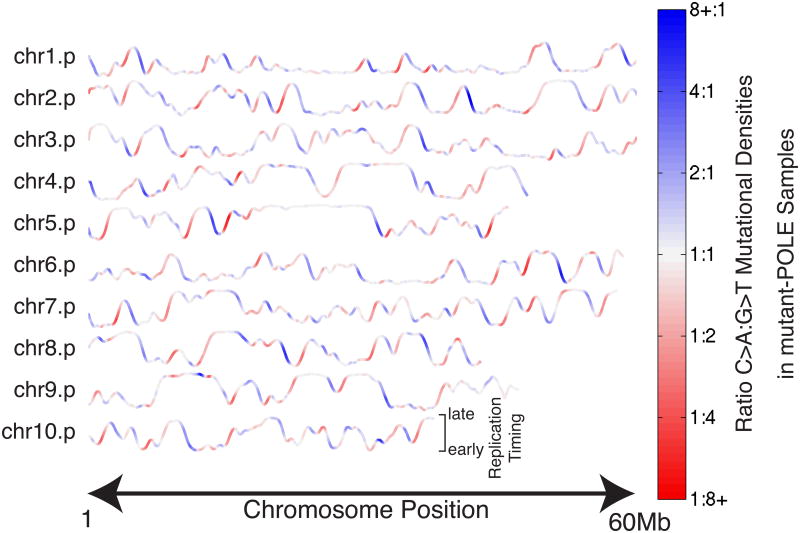
Indeed when asymmetry is visualized along the chromosome, asymmetric C:G→A:T mutations in a pooled cohort of twelve mutant-POLE colorectal and endometrial tumors corresponds strikingly to the slope of the replication timing profile (Figure 2). Higher densities of C→A mutation occur in regions of negative slope, while higher G→T densities occur in regions of positive slope. In TTRs (see Experimental Procedures) the magnitude and direction of this imbalance correlates well with the slope of the profile (R2 = 0.53) while in constant-timing regions no such correlation exists (R2=0.08). Comparing left- and right-replicating regions, we measured a near two-fold enrichment for the expected mutation type (Figure 1D). This is consistent with the recently reported preference for mutations at C:G base pairs where the cytosine is on the leading template strand measured next to 3 well-localized origins of replication (we will denote such base pairs Clft:Grt) (Shinbrot et al., 2014) and validates our ability to extract replication direction from replication-timing profiles. Furthermore we tested our method on replication-timing datasets from various cell types, including embryonic stem cells, induced pluripotent stem cells, neural precursor cells, and lymphoblast cell lines (Figure S2)(Ryba et al., 2010). All yielded very similar patterns of asymmetry, demonstrating the robustness of our method to tissue-specific variations in replication timing profiles.
Figure 2. Strand asymmetry in POLE-mutant cancers reflects directionality of DNA replication timing-transition regions (TTRs).
Replication timing profiles are shown for the p-arms (up to 60Mb) of the first ten chromosomes. Profiles are colored by the local ratio of C→A to G→T mutations in a cohort of 12 mutant-POLE genomes (colorectal and endometrial). Strikingly, late-to-early TTRs (where slope is negative) frequently have a strong bias towards C→A mutations (blue), consistent with leading-strand synthesis using the reference strand as template. Conversely, early-to-late TTRs (positive slopes) show bias towards G→T mutations (red), consistent with lagging-strand synthesis using the reference strand as template(Shinbrot et al., 2014).
Having analyzed each reference frame separately, we jointly considered transcriptional (T-class) and replicative (R-class) asymmetry. By focusing the analysis on regions that are both transcribed and located in TTRs, we can control for potential confounding factors such as chromatin state, since transcribed regions are typically in open chromatin, and TTRs often reside at boundaries between open and closed chromatin (Lawrence et al., 2013). Surprisingly, we observed near-complete mutual exclusivity of R- and T-class asymmetries in the smoking-associated (lung) and POLE-associated (colorectal, endometrial) cohorts. In smoking-associated genomes, the direction of mutational asymmetries flips with transcription direction, but shows little dependence on replication direction, even when controling for transcription direction (Figure 1E). These observations show that smoking-associated lung cancers have a mutational pattern dominated by T-class asymmetry, with very little evidence of R-class asymmetry.
The opposite pattern was seen in POLE-associated cancers, where mutational asymmetries depended entirely on replication direction, and showed little response to change in transcription direction (Figure 1F). Thus, POLE-associated cancers have a mutational pattern dominated by R-class asymmetry, with almost zero T-class asymmetry.
The asymmetry map of cancer genomics
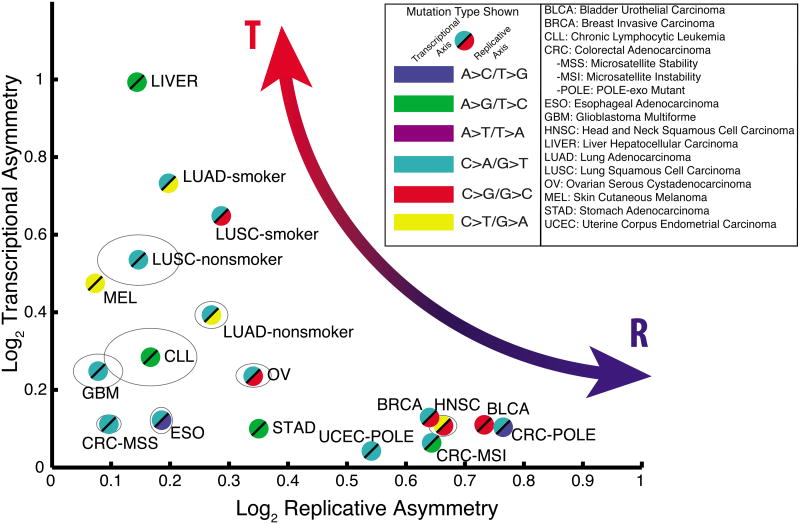
Having established that we can observe and separate transcriptional and replicative strand asymmetries in two well-understood mutational processes, we performed a comprehensive analysis of mutational strand asymmetries across many tumor types. We analyzed somatic mutations in 590 whole-genome sequences across 14 tumor types, partitioned into 18 patient cohorts (separating out POLE and microsatellite-instability (MSI) cases in the colorectal and endometrial cohorts, and separating smokers from non-smokers in the two lung cohorts) (Table S1). For each cohort, we identified the mutation type having the largest asymmetry, with respect to transcription and to replication (Figure 3). This revealed a continuum of tumor types, ranging from tumors with predominant transcriptional (T-class) asymmetry to those with predominant replicative (R-class) asymmetry. For example, the melanoma, liver, and lung cohorts fell on the T-class side of the spectrum, while tumors frequently associated with an APOBEC signature (BLCA, BRCA, and HNSC) or microsatellite instability (CRC-MSI) showed R-class asymmetries at levels comparable to those of POLE tumors (CRC-POLE and UCEC-POLE).
Figure 3. Cancer cohorts vary widely across the asymmetry map.
For each cohort listed, the maximal replicative asymmetry (x-axis) and the maximal transcriptional asymmetry (y-axis) were measured and plotted. Grey ellipses denote 95% confidence intervals for cohorts in which these extend beyond the bounds of the plot symbols.
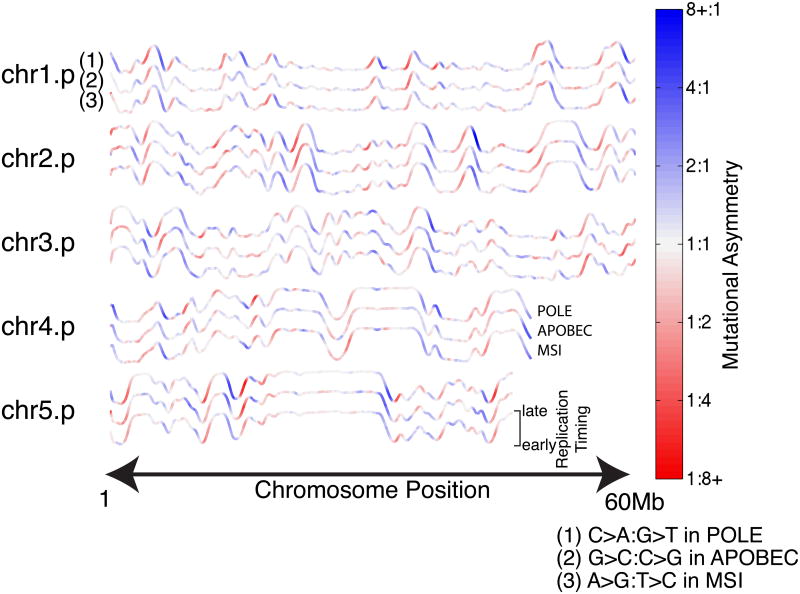
The genomic asymmetry profiles of R-class tumors are strikingly concordant among each other within TTRs (POLE-APOBEC R2=0.50, POLE-MSI R2=0.66, APOBEC-MSI R2=0.42) as well as with the slope of the replication timing profile (POLE R2=0.56, APOBEC R2=0.47, MSI R2=0.49) (Figure 4), a trend robust to substituting replication timing profiles from various cell types (Ryba et al., 2010) (Figure S3,4). Importantly, we were able to detect statistically significant levels of asymmetry in all cohorts in at least one mutation type, and 8/15 showed either T-class or R-class asymmetry with greater than 50% enrichment (>0.58 in Figure 3) for at least one mutation type. Overall, these results demonstrate that mutational strand asymmetries are widespread across cancer.
Figure 4. Replicative asymmetry is concordant across three distinct R-class mutational processes.
Color representing mutational asymmetry is overlaid on replication timing profiles as in Fig. 2. Profiles are shown in triplets colored by: (1) C→A:G→T asymmetry in 12 mutant-POLE colorectal and endometrial genomes; (2) G→C:C→G asymmetry in 22 APOBEC-enriched bladder, breast, and head-and-neck genomes; and (3) A→G:T→C asymmetry in 9 MSI-associated colon genomes.
Trends in mutational asymmetries
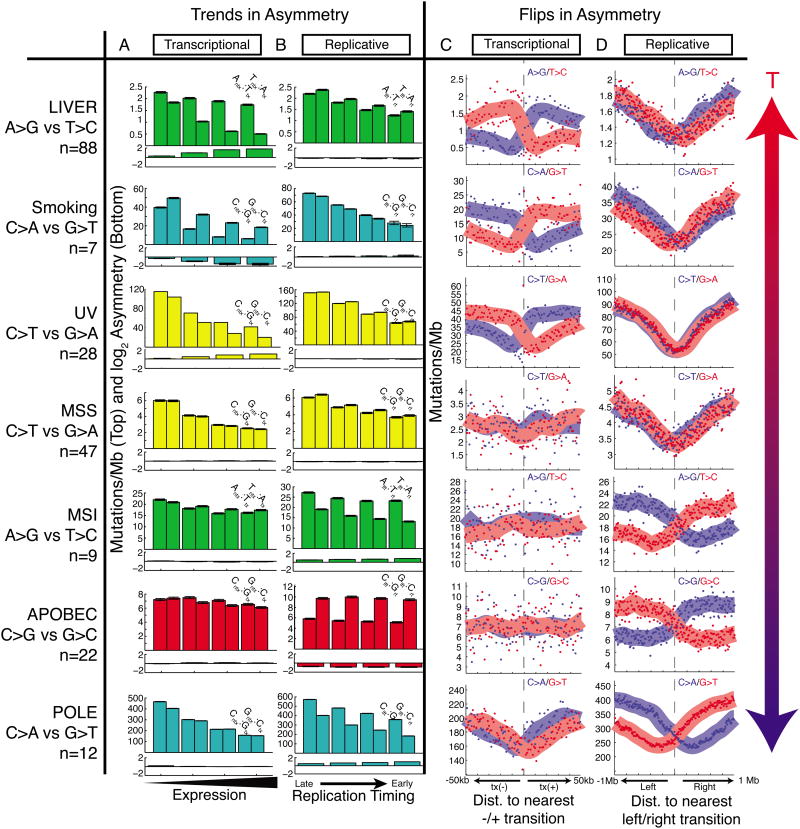
Next, we explored how mutational asymmetries depend on other variables such as expression levels, replication timing, and distance from transitions in replication or transcription direction. We focused on mutational processes that we identified as being the chief sources of asymmetry, and identified the samples in which these processes were the major contributor to the overall mutational burden (Table S2). First, we analyzed transcriptional asymmetry as a function of gene expression level, and replicative asymmetry as a function of DNA replication timing (Experimental Procedures). For most processes, we observed a decrease in mutational burden at higher expression levels (Figure 5A). Transcriptional asymmetry, which reflects TCR activity, was seen in a subset of these cohorts (liver A→G, smoking C→A, and UV C→T) and was maximal in highly expressed regions. In other cohorts (e.g. POLE C→A, microsatellite stable cancers (MSS) C→T), no transcriptional asymmetry was seen, perhaps due to the fact that other covariates (such as replication timing and chromatin state) correlate with expression levels but affect mutational burden via repair mechanisms that are independent of transcription. Similarly, for most processes, we observed a decrease in mutational burden in earliest-replicating regions (Figure 5B). Replicative asymmetry was seen in a subset of cohorts (MSI, APOBEC, POLE), and was strongest in earliest-replicating regions (especially in the case of POLE), but absent in other cohorts. To control for differences in chromatin state of TTRs and transcribed regions, in all of these cohorts we again performed a joint analysis of T- and R-class asymmetries (Figure S5).
Figure 5. Trends and flips in asymmetry.
(A) Transcriptional strand asymmetry measured across four quartiles of expression levels. Total mutation density tends to decrease with expression level, and T-class asymmetry (liver, smoking, UV) is maximal at highest expression. (B) Replicative strand asymmetry measured across four quartiles of replication timing. Total mutational density tends to decrease with earlier replication, and R-class asymmetry (MSI, APOBEC, POLE) is maximal at earliest replication. (C) Strand-specific mutational density measured in the vicinity of bidirectional promoters. T-class asymmetry flips at transitions from tx(-) to tx(+) regions. (D) Strand-specific mutational density measured in the vicinity of replication timing minima. R-class asymmetry flips at these left-to right transitions.
We also analyzed the effect of genomic position with respect to transitions in transcription or replication direction. We examined transcriptional asymmetry around minus-to-plus transcription-direction transitions (Figure 5C), typically representing bidirectional promoters (Trinklein et al., 2004)); and replicative asymmetry around left-to-right replication-direction transitions (Figure 5D), i.e. replication-timing minima (Experimental Procedures). Mutations associated with smoking, UV, and liver cancer showed transcriptional strand asymmetries that flipped sign at transitions in transcription direction. Other cancers maintained balanced mutation densities on both sides of these transitions. Conversely, mutations associated with POLE, MSI, and APOBEC showed replicative strand asymmetries that flipped sign at replication-timing minima. Other cohorts showed no such behavior at changes in replication direction. Exploring each of these asymmetries further can shed light on the operational mechanisms of mutagenesis and repair in these tumors.
Mutational asymmetries reveal mechanisms of mutagenesis
The above analyses lead to insights into the mechanisms of incompletely understood mutational processes, such as the APOBEC and liver signatures. The APOBEC signature consists of C→G and C→T mutations in the context TCW (W = A or T) and is thought to reflect the activity of APOBEC-family cytidine deaminase enzymes (Alexandrov et al., 2013; Lawrence et al., 2013; Roberts et al., 2013). While the precise details of this phenomenon in cancer are not completely understood, a large body of work has characterized many aspects of this form of mutagenesis. APOBEC enzymes target ssDNA (Conticello, 2012), cause mutation clusters termed kataegis (Nik-Zainal et al., 2012a), and do not cause the usual increase in mutational densities in late-replicating, open chromatin, and highly expressed regions (Kazanov et al., 2015). The main occurrences of ssDNA in human cells have been speculated to be at double strand breaks (DSBs), R-loops in transcription bubbles, and the lagging strand of the DNA replication fork. Experiments in model organisms have shown that APOBEC enzymes are indeed capable of inducing mutagenesis at DSBs (Taylor et al., 2013) and transcription bubbles (Lada et al., 2015; Taylor et al., 2014).
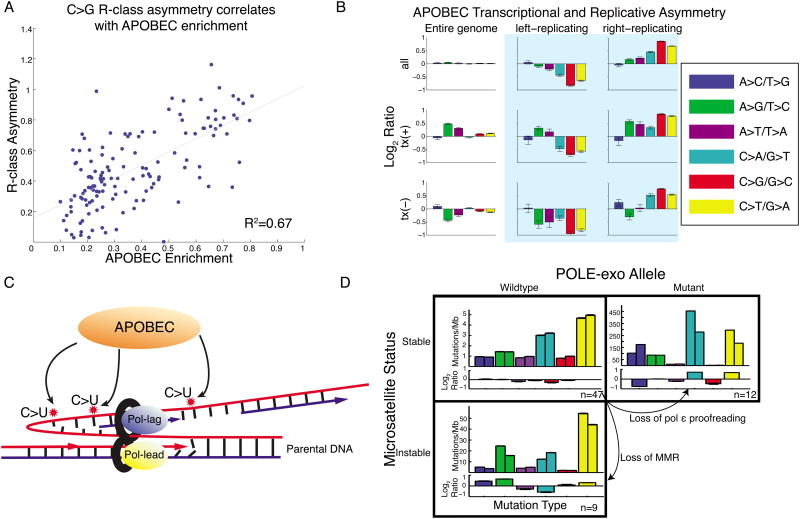
Our results suggest that, in humans, APOBEC mutagenesis primarily occurs on the lagging-strand template during DNA replication. The APOBEC signature shows strong R-class asymmetry, with a higher rate of C→G and C→T mutations in right-replicating regions (Figure 3,5), where reference-strand DNA is predicted to be replicated as the lagging-strand template, exposed as ssDNA between Okazaki segments. The magnitude of this asymmetry increases with enrichment of the APOBEC signature (Figure 6A), and joint analysis of both classes of asymmetry placed APOBEC squarely at the R-class end of the spectrum (Figure 6B). Note that in all breast, bladder and head and neck samples, even when the fraction of APOBEC mutations is low, significant R-class asymmetry is observed, suggesting that it is not merely a property of hypermutation. These findings are further supported by research in model organisms concurrent with this study. Bhagwat et al. (Bhagwat et al., 2016) found that overexpression of APOBEC3G in E. coli leads to a C:G→T:A signature that shows a replicative strand bias consistent with cytosine deamination of the lagging-strand template. Additionally, in a yeast model, Roberts and colleagues (Hoopes et al., 2016) showed that overexpression of APOBEC3A and B produces a similar replicative asymmetry.
Figure 6. R-class asymmetries associated with APOBEC and MSI.
(A) Bladder, breast, and head-and-neck cohorts. Samples with highest enrichment of APOBEC signature show highest replicative asymmetry of C→G mutations. (B) APOBEC-enriched samples are dominated by replicative asymmetry (as Fig. 1E,F) (C) Proposed model: APOBEC deaminates cytosine to uracil on the ssDNA of the lagging-strand template during DNA replication. (D) R-class asymmetry in MSS, MSI, and POLE-mutant cohorts. MSS samples have little asymmetry. Loss of MMR or pol ε proofreading leads to imbalance in mutations between the leading and lagging strands.
Taken together, these findings suggest that the R-class model is the primary mechanism for APOBEC mutagenesis in humans. In this model, APOBEC-family enzymes deaminate cytosines on the lagging-strand template during DNA replication, likely while it is single-stranded (Figure 6C). The resulting uracil is excised, and subsequent replication either incorporates an adenine across from this abasic site, resulting in a C→T mutation, or (mediated by REV1 activity), incorporates a cytosine, resulting in a C→G mutation (Helleday et al., 2014). This model is also supported by the unusual lack of increase in mutational densities in late-replicating regions (Figure 5)(Kazanov et al., 2015). As MMR has been suggested to underlie this variation in mutational densities (Supek and Lehner, 2015), this may imply that APOBEC-associated mutagenesis evades the MMR machinery. This is consistent with the R-class model, in which the lagging-strand template (i.e. the parental strand) is deaminated; MMR, which relies on the parental strand to correct mistakes on the nascent strand, would be unable to correct this error without a correct template. Genome-wide we observed only a small amount of APOBEC T-class asymmetry (Figure 6C), but a previous report showed that overexpressing APOBEC in yeast resulted in mutations that were transcriptionally asymmetric (Lada et al., 2015). Indeed, when we restricted to 5′-UTRs (the regions reported to have the strongest transcriptional asymmetry) we revealed APOBEC T-class asymmetry also in humans (Figure S6). However, in the genome-wide analysis the T-class asymmetry is dwarfed by the contributions from the R-class model.
Intriguingly, we observed a similar APOBEC mutational R-class asymmetry in the human germline. We measured replicative asymmetry in a set of 11,020 de novo germline mutations (Francioli et al., 2015) and found that C→G and C→T mutations showed no significant R-class asymmetry outside the TCW context (1127 C→G/T vs. 1171 G→A/C in the leading-strand reference frame, p = 0.35). When we focused on the TCW context (the preferred target of APOBEC mutagenesis), we were able to detect a significant level of R-class asymmetry (109 TCW→G/T vs. 151 WGA→A/C mutations in the leading-strand reference frame, p = 0.014) (Figure S7). Further studies analyzing a larger number of mutations will be required to fully understand the potential impact of APOBEC enzymes on germline mutagenesis and its evolutionary implications.
A mechanism of transcription-coupled DNA damage
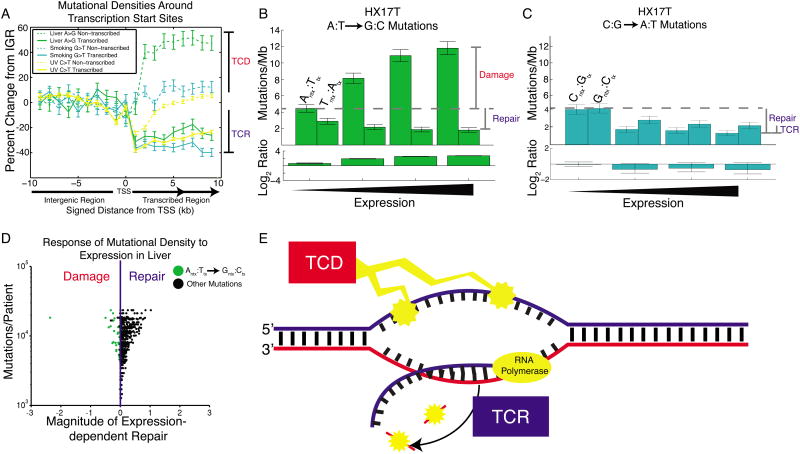
In contrast to APOBEC- and MSI-associated mutations, liver A:T→G:C mutations showed little replicative asymmetry, but instead showed transcriptional asymmetry similar to that seen in lung cancer (LUSC and LUAD in Figure 3; Smoking C→A vs. G→T in Figure 5). Closer inspection of transcriptional strand asymmetry revealed a distinction between the liver A→G signature and the two other T-class examples: UV-associated C→T and smoking-associated G→T. Mutations generated by UV light and smoking are lower in density on the transcribed strand compared to proximal intergenic regions (due to TCR), while mutational densities on the non-transcribed strand remain constant regardless of transcription (Figure 7A). The liver A→G signature also shows the expected TCR effect on the transcribed strand; however, mutational densities of A→G on the non-transcribed strand drastically increase in transcribed regions. This suggests that transcriptional asymmetry in liver is not only due to repair of the transcribed strand, but is also compounded by damage to the complementary non-transcribed strand, a phenomenon we call transcription-coupled damage (TCD).
Figure 7. Transcription-coupled damage in liver cancer.
(A) Mutational densities in the vicinity of promoters. When crossing from non-transcribed regions (IGR) to transcribed regions, mutational densities on the transcribed strand fall, reflecting transcription-coupled repair (TCR). On the non-transcribed strand there is usually little change from IGR levels, with the notable exception of liver cancer, where mutational densities increase from IGR levels, consistent with transcription-coupled damage (TCD). (B) Liver cancer patient HX17T shows a dramatic expression-dependent increase in A→G mutational densities on the non-transcribed strand only. (C) In the same patient, G→T mutational densities show only the usual expression-dependent decrease, on both strands. (D) Most liver patients show dominant TCR. However, for A→G mutations on the non-transcribed strand (green dots), some show the opposite trend, reflecting dominant TCD. The leftmost dot is patient HX17T. (E) TCD damages the non-transcribed strand, exposed as ssDNA during transcription. TCR repairs the transcribed strand. Both of these processes contribute to T-class asymmetry.
At the extreme, we observed one liver cancer sample, HX17T, that showed a 3-fold transcription-dependent increase in A→G mutational densities on the non-transcribed strand (Figure 7B). This is in contrast to the usual trend in which non-transcribed strand mutational densities decrease with expression, due to more effective global genome repair (GGR) and mismatch repair (MMR) in open chromatin and early replicating regions. This effect is unique to the A→G signature; in that same sample, C→A mutational densities (driven by carcinogen attack (Alexandrov et al., 2013)) showed the usual decrease on both strands (Figure 7C). In our cohort of 88 liver cancer samples, we examined the slope of this response of mutational density to expression level, for each of the twelve possible mutation types (Figure 7D). In a two-tailed test we found that 25/88 of the liver patients showed a significant increase in A→G mutational densities on the non-transcribed strand (Experimental Procedures), while only 9/88 showed a significant decrease, showing that, in the majority of samples, the A→G signature does not show the usual repair (Table S3). As mentioned before, the contributions of MMR and GGR are confounding factors when considering the effect of expression levels on mutational densities, since higher expression is correlated with earlier replication timing and more open chromatin state. As a result, on the non-transcribed strand, higher expression could both lead to higher damage by TCD and higher levels of repair by MMR and GGR. Different contributions of these damage and repair processes likely underlie this observed variation across patients.
While the strong transcriptional asymmetry of the A→G signature in liver cancer has been noted (Alexandrov et al., 2013), we propose that this is due to two separate processes operating on different strands -- TCD and TCR (Figure 7E). This explains the extreme transcriptional asymmetry of liver A→G compared to other signatures (Figure 3). Furthermore these results suggest that the A→G signature is caused by a mutational process distinct from typical bulky adduct damage. Finally, we noticed that one colorectal patient (“CRC-8”) from an earlier study of nine colorectal whole genomes (Bass et al., 2011) showed the same signature of TCD. Thus this phenomenon may enriched in liver but not exclusive to it.
Mismatch repair balances mutational asymmetry
Colorectal cancers with functional MMR (i.e. MSS) show little replicative asymmetry of any mutation type (aside from C→G mutations which are in part due to low levels of APOBEC signature). As mentioned above, loss of functional polymerase ε proofreading results in R-class asymmetry. MSI colorectal tumors, typically resulting from damage to the MMR system (Kane et al., 1997; Shinbrot et al., 2014; Vilar and Gruber, 2010), also show replicative asymmetry (Figure 6D). This would suggest that MMR (in addition to exonuclease proofreading) is required to balance mutational asymmetries generated during DNA replication. This phenomenon has also been reported in yeast (Lujan et al., 2012), and our results suggest the same is true in humans.
The implications of this role for MMR reach beyond the realm of cancer research. Without such balancing, asymmetric introduction of germline mutations would result in local depletion of specific nucleotides over evolution. Indeed, a slight replicative imbalance can be detected in the reference genome: Clft:Grt base pairs outnumber Glft:Crt base pairs by 2.1% on average, and Alft:Trt base pairs outnumber Tlft:Art base pairs by 3.7%. This is in line with a previous result measuring a mean compositional skew of 3.72% (Chen et al., 2011). However the relative mildness of these imbalances, compared to the much stronger mutational asymmetries seen in MMR-deficient tumors, suggests that MMR has played an important role through evolution in maintaining genome symmetry.
Discussion
Our results highlight the widespread mutational strand asymmetries observed in cancer genomes, mediated by DNA replication, RNA transcription, and their associated repair pathways. Study of these prominent sources of asymmetry has mostly been performed in model organisms (Lobry, 1996; Lujan et al., 2012; McLean et al., 1998; Pavlov et al., 2002; Touchon et al., 2005), and here we extend this analysis to humans via cancer genomics. Our work addresses several of the most prominent processes in cancer and provides insight into their biological mechanisms. Analysis of asymmetries associated with the growing number of mutational processes discovered by sophisticated signature decoupling approaches (Alexandrov et al., 2013; Kasar et al., 2015; Lawrence et al., 2013) will provide a fuller view of these processes and further illuminate their underlying sources. Our ability to detect mutational asymmetries will improve with higher-resolution replication-timing and transcription maps, and with improving knowledge of human replication origins. Finally, we note that there may be additional useful reference frames for symmetry breaking beyond the two used here.
Classifying patients according to their patterns of mutational strand asymmetry may have clinical relevance. Tumors with defects in DNA repair mechanisms have been shown vulnerable to synthetically lethal therapeutic interventions that further disrupt genome stability (Carreras Puigvert et al., 2015; Curtin, 2012; Middleton et al., 2015). As discussed, R-class asymmetries can be introduced either by asymmetric damage at the replication fork, or by deficiency in the proofreading and repair of DNA synthesis. In the latter case, R-class asymmetry may serve as a proxy for replicative stress and could suggest synthetic lethality as an effective avenue for treatment. Similarly, individual patients of T-class tumor types (such as melanoma) that do not themselves exhibit T-class asymmetry potentially reveal a deficiency of TCR. Thus analyzing asymmetries of both classes may facilitate a better match between patients and treatments.
Additionally, patient-specific responses to classic chemotherapy drugs are often poorly understood and difficult to predict. Analyzing responses to these drugs in conjunction with mutation rates and asymmetries at the replication fork or transcription bubble may provide useful insights into these drugs' functionality. This in turn could allow for more targeted use of the drugs, as well as better control of unintended side effects.
Strand asymmetry may be particularly impactful in the earliest driving events of cancer, due to that defining feature of carcinogenesis, the transformation of cells into an aberrantly proliferative state. Some DNA lesion may push the cell from a resting state (without DNA replication) into active mitosis, and the initial strand hit by that driver lesion is crucial: due to the absence (or infrequence) of DNA replication in these pre-malignant cells, DNA damage of the non-transcribed strand may wait a very long time to be propagated as a mutation to the transcribed strand where it can exert its driving effect.
Beyond cancer, somatic mutational processes play an important role in a broad range of diseases, including aging (Kennedy et al., 2012; Kenyon, 2010), autoimmune disease (Ross, 2014), and neurological disorders (Poduri et al., 2013). Many of the same background mutational processes are active in cancerous and noncancerous cells (such as methylated CpG deamination/“aging”, UV damage, and environmental mutagens) and the lessons learned from the clonal expansion of mutations in cancer will aid in the understanding of these universal processes. Novel mutational and repair processes continue to emerge from cancer genome sequencing studies, and viewing them through the lens of mutational strand asymmetry can provide immediate insights into their molecular mechanisms.
Experimental Procedures
Data Provenance
We assembled a collection of 590 whole genome sequences from 14 tumor types by combining published data from the Cancer Genome Atlas (TCGA) with other published datasets (Alexandrov et al., 2013; Dulak et al., 2013; Lawrence et al., 2014).
Statistical Analysis
MATLAB code to generate asymmetry metrics and figures is available at www.broadinstitute.org/cancer/cga/AsymTools.
Determining transcription and replication direction, and calculating densities
Transcription direction was determined according to the Refseq database. Replication direction was defined using replication timing profiles generated in six lymphoblastoid cell lines, as published in (Koren et al., 2012). We determined left- and right- replicating regions based on the sign of the derivative of the profile (negative is left-replicating and positive is right-replicating). To only define regions in TTRs, we required a slope with a magnitude of at least 250 replication timing units (“rtu”) per Mb. These arbitrary units range from 100 to 1200 denoting the beginning and end of S-phase.
Mutational densities for a given base pair change b1:b2_→_m1:m2 and its complementary mutation b2:b1_→_m2:m1 in a given list of regions were determined by the formula
rb1:b2→m1:m2=nb1→m1p∗Nb1rb2:b1→m2:m1=nb2→m2p∗Nb2
where nb_→_m is the number of observations of b_→_m mutations with respect to the genomic reference strand, Nb is the number of chances for this mutation to happen, i.e. the number of occurrences of the motif b in the given region of the reference genome (on the genomic reference strand), and p is the number of patients analyzed. Asymmetry was then calculated in a given region by
ab→m=log2(rb1:b2→m1:m2rb2:b1→m2:m1)
as seen in Figure 1.
Calculating global mutational asymmetries
When calculating global mutational asymmetries redundant mutations with respect to a given strand are summed together. For example to calculate global genome mutational densities with respect to the leadings strand, we calculate:
rb1:b2→m1:m2=nl,b1→m1+nr,b2→m2p∗(Nl,b1+Nr,b2)rb2:b1→m2:m1=nl,b2→m2+nr,b1→m1p∗(Nl,b2+Nr,b1)ab→m=log2(rb1:b2→m1:m2rb2:b1→m2:m1)
where subscripts “l” and “r” refer to events in left- and right- replicating regions respectively. Essentially this approach distinguishes a b1:b2 base pair by whether base b1 is on the presumed leading/lagging strand rather than the genomic reference. The same approach is used for calculating asymmetry with respect to the sense strand, using tx(+) and tx(-) regions instead of left- and right-replicating respectively.
Correlation of R-class asymmetry with direction of transition-timing regions (TTRs)
Replication timing data and POLE, APOBEC, and MSI asymmetry metrics were aggregated in 100kb bins and smoothed using a moving average over 10 bins. The replication timing data was plotted, and the profiles were colored by the asymmetry metrics in the POLE cohort (Figure 2), or all three R-class cohorts (Figure 4). Correlations restricted to TTRs were calculated by only considering regions with a slope of > 250 rtu/Mb.
Binning by expression and replication timing
Expression profiles were an average of many cell lines, as used in (Lawrence et al., 2013). To perform binning by functional covariates as seen in Figure 5AB, expression and replication timing values were projected onto 20kb intervals. These intervals were then sorted by expression for Figure 5A and replication timing for Figure 5B, separated into bins with an even number of intervals, and mutational rates and asymmetry were calculated for the intervals in each bin.
Identifying transcription and replication direction transitions
Minus- to plus- transcription transitions were identified by taking all bidirectional gene pairs (opposing genes with transcription start sites within 1kb of each other (Trinklein et al., 2004)) in the Refseq database and calculating the midpoint of their transcription start sites. Left-to-right replication transitions were similarly identified by calculating the midpoint between the right and left boundaries of defined left- and right- replicating regions respectively.
Determining response of mutational densities to increasing expression
Response of mutational densities to increasing expression as shown in Figure 7BC was performed by for each patient and for each of the six possible mutations, creating a figure as shown in Figure 7D. Then linear regression was performed separately on each series of bars (the left bars for the non-transcribed strand and the right bars for the transcribed strand). For these two regressions, we calculated a 95% confidence interval for the slope and assessed significance based on whether zero fell inside the interval. Then for each regression we plotted the more conservative bound of the corresponding confidence interval i.e. the value closer to or equal to zero.
Creating a hypothetical replication timing distribution
The hypothetical replication timing curve shown in Figure S1A was creating by first taking its real counterpart in Figure S1B. Then the locations of origins of replication were randomly assigned assuming a rate of one origin per 40kb. From these origins a more detailed profile was drawn by assuming constant polymerase speed, and smoothing the result.
Supplementary Material
1
10
11
2
3
4
5
6
7
8
9
Acknowledgments
We thank Ashok Bhagwat and Steven Roberts and their collaborators for sharing their unpublished data with us, and allowing us to reference them in this work. We thank John Iafrate for valuable insights about the relevance of strand asymmetry in carcinogenesis.
Footnotes
Author Contributions: Conceptualization, M.S.L., P.P., G.G., N.J.H., P.S., D.A.W., E.S., K.R.C., A.Ko., J.K.; Methodology, M.S.L., P.P., N.J.H., P.S.; Software, N.J.H., M.S.L.; Formal Analysis N.J.H., M.S.L., P.P.; Investigation, N.J.H., P.P., M.S.L.; Resources, M.S.L., G.G., A.Ko., D.A.W.; Data Curation, E.R., N.J.H., P.P., M.S.L., A.Ko.; Writing – Original Draft, N.J.H., M.S.L., A.Ko., P.P., G.G.; Writing – Review & Editing, N.J.H., M.S.L., A.Ko., P.P., G.G., D.A.W., P.C.H., E.S., K.R.C., E.R., J.H., P.S., Y.M., A.Ka.,L.B.; Visualization, N.J.H., M.S.L.; Supervision, G.G., M.S.L., P.P., A.Ko., D.A.W., P.C.H.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AS, Hao W, Townes JP, Lee H, Tang H, Foster PL. Strand-biased Cytosine deamination at the Replication Fork causes Cytosine to Thymine Mutations in Escherichia coli (PNAS) 2016 doi: 10.1073/pnas.1522325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras Puigvert J, Sanjiv K, Helleday T. Targeting DNA repair, DNA metabolism and replication stress as anti-cancer strategies. FEBS J. 2015 doi: 10.1111/febs.13574. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Duquenne L, Audit B, Guilbaud G, Rappailles A, Baker A, Huvet M, d'Aubenton-Carafa Y, Hyrien O, Arneodo A, et al. Replication-associated mutational asymmetry in the human genome. Mol Biol Evol. 2011;28:2327–2337. doi: 10.1093/molbev/msr056. [DOI] [PubMed] [Google Scholar]
- Conticello SG. Creative deaminases, self-inflicted damage, and genome evolution. Ann N Y Acad Sci. 2012;1267:79–85. doi: 10.1111/j.1749-6632.2012.06614.x. [DOI] [PubMed] [Google Scholar]
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, Genome of the Netherlands, C. van Duijn CM, Swertz M, Wijmenga C, et al. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Program, N.C.S. Green ED. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- Guilbaud G, Rappailles A, Baker A, Chen CL, Arneodo A, Goldar A, d'Aubenton-Carafa Y, Thermes C, Audit B, Hyrien O. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput Biol. 2011;7:e1002322. doi: 10.1371/journal.pcbi.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes J, Cortez L, Mertz T, Malc EP, Mieczkowski PA, Roberts SA. APOBEC3A and APOBEC3B Deaminate the Lagging Strand Template during DNA Replication (Cell Reports) 2016 doi: 10.1016/j.celrep.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Bhagwat AS. Transcription-associated mutagenesis. Annu Rev Genet. 2014;48:341–359. doi: 10.1146/annurev-genet-120213-092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerase delta in Replication of Both the Leading and Lagging DNA Strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, Lawrence MS, Kiezun A, Fernandes SM, Bahl S, et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat Commun. 2015;6:8866. doi: 10.1038/ncomms9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanov MD, Roberts SA, Polak P, Stamatoyannopoulos J, Klimczak LJ, Gordenin DA, Sunyaev SR. APOBEC-Induced Cancer Mutations Are Uniquely Enriched in Early-Replicating, Gene-Dense, and Active Chromatin Regions. Cell Rep. 2015;13:1103–1109. doi: 10.1016/j.celrep.2015.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133:118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, Sunyaev SR, McCarroll SA. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 2012;91:1033–1040. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Tsai HJ, Tirosh I, Burrack LS, Barkai N, Berman J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 2010;6:e1001068. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lada AG, Kliver SF, Dhar A, Polev DE, Masharsky AE, Rogozin IB, Pavlov YI. Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes. PLoS Genet. 2015;11:e1005217. doi: 10.1371/journal.pgen.1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, Nick McElhinny SA, Kunkel TA. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Middleton FK, Patterson MJ, Elstob CJ, Fordham S, Herriott A, Wade MA, McCormick A, Edmondson R, May FE, Allan JM, et al. Common cancer-associated imbalances in the DNA damage response confer sensitivity to single agent ATR inhibition. Oncotarget. 2015;6:32396–32409. doi: 10.18632/oncotarget.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012a;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012b;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Arndt PF. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Lawrence MS, Haugen E, Stoletzki N, Stojanov P, Thurman RE, Garraway LA, Mirkin S, Getz G, Stamatoyannopoulos JA, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32:71–75. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Querfurth R, Arndt PF. The evolution of transcription-associated biases of mutations across vertebrates. BMC Evol Biol. 2010;10:187. doi: 10.1186/1471-2148-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KA. Coherent somatic mutation in autoimmune disease. PLoS One. 2014;9:e101093. doi: 10.1371/journal.pone.0101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak G, Ganesan AK. The complex choreography of transcription-coupled repair. DNA Repair (Amst) 2014;19:64–70. doi: 10.1016/j.dnarep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- Stillman B. Reconsidering DNA Polymerases at the Replication Fork in Eukaryotes. Mol Cell. 2015;59:139–141. doi: 10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Wu YL, Rada C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife. 2014;3:e03553. doi: 10.7554/eLife.03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Nicolay S, Audit B, Brodie of Brodie, E.B. d'Aubenton-Carafa Y, Arneodo A, Thermes C. Replication-associated strand asymmetries in mammalian genomes: toward detection of replication origins. Proc Natl Acad Sci U S A. 2005;102:9836–9841. doi: 10.1073/pnas.0500577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
10
11
2
3
4
5
6
7
8
9