Dynamics of Single mRNPs in Nuclei of Living Cells (original) (raw)
. Author manuscript; available in PMC: 2016 Feb 24.
Published in final edited form as: Science. 2004 Jun 18;304(5678):1797–1800. doi: 10.1126/science.1099754
Abstract
Understanding gene expression requires the ability to follow the fate of individual molecules. Here we use a cellular system for monitoring messenger RNA (mRNA)expression to characterize the movement in real time of single mRNA-protein complexes (mRNPs) in the nucleus of living mammalian cells. This mobility was not directed but was governed by simple diffusion. Some mRNPs were partially corralled throughout the nonhomogenous nuclear environment, but no accumulation at subnuclear domains was observed. Following energy deprivation, energy-independent motion of mRNPs was observed in a highly ATP-dependent nuclear environment; movements were constrained to chromatin-poor domains and excluded by newly formed chromatin barriers. This observation resolves a controversy, showing that the energetic requirements of nuclear mRNP trafficking are consistent with a diffusional model.
Recent technological developments have facilitated imaging of single RNA molecules in the cytoplasm of living cells (1). We have developed a cellular system in which the expression of a transgene array can be followed sequentially in single living cells, and we have previously analyzed the particular chromatin-related modifications occurring at this specific locus from a silenced state throughout its transcriptional activation (2). Here we use this system to address the mechanism by which individual mRNA transcripts move within the nucleoplasm after release from the transcription site.
In this system, a genetic locus, its transcribed mRNAs, and the translated protein were rendered visible in cells after electroporation with the cyan fluorescent protein (CFP) or the red fluorescent protein (RFP)–lac repressor protein (marks the genomic locus), yellow fluorescent protein (YFP)–MS2 (labels the mRNA), and pTet-On (for transcriptional induction) (3). Transcriptional activation by doxycycline induced the unfolding of the integrated locus (4) and the recruitment of the YFP-MS2 protein to the locus as a result of the specific labeling of the nascent transcripts bearing the MS2 stem loops. Minutes after induction (15 to 30 min), the MS2 signal began accumulating in the nucleoplasm in a particulate pattern suggestive of mRNA-protein complexes (mRNPs). At later times (1 to 2 hours after induction), mRNPs were detected in the cytoplasm in conjunction with the appearance of cyan fluorescent protein (CFP)–labeled peroxisomes, demonstrating that the tagged RNA was both correctly exported and translated (fig. S1A to D). These components of the gene expression pathway could be detected simultaneously in living cells (3) [Movies S1 to S5 (5)].
The presence of the nascent RNAs at the site of transcription was verified using fluorescent in situ hybridization (FISH) on fixed cells with two different probes either to the MS2 repeats (located in the middle of the transcript) or to the β globin exon (3′ end). Both probes hybridized at the active locus, indicating that the complete pre-mRNA transcripts were retained at the transcription site before release (fig. S2A to D). Colocalization of the mRNA signal (FISH) with the YFP signal demonstrated that the particles visualized in living cells were mRNPs (3). RNA quantification with single-molecule sensitivity was performed on deconvolved FISH images to a single target sequence in the 3′ end of the transcript (6). The majority (~70%) of transcripts hybridized with a single probe, indicating that these nuclear mRNPs represent single RNA transcripts (fig. S2E to I). Imaging of the mRNPs in fixed or living cells showed that mRNPs were excluded from the nucleolus (3) [Movie S5 (5)] and that apart from the transcription site, no site of mRNP accumulation could be detected.
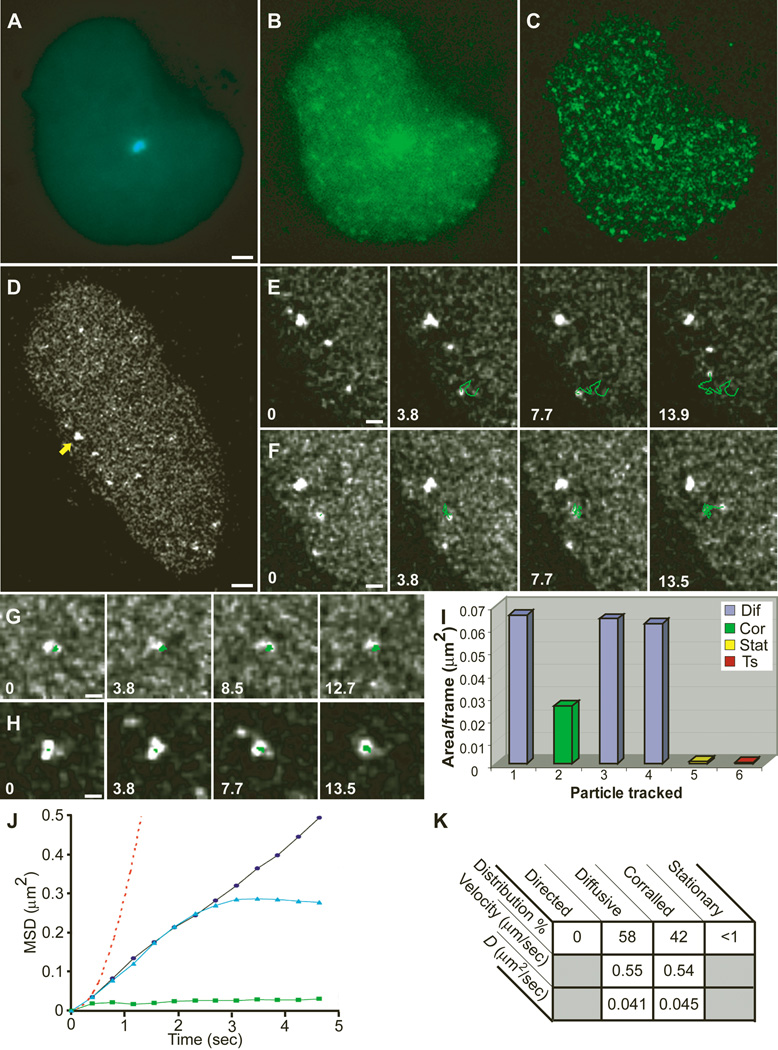
Movements of nuclear mRNPs were followed by sequential imaging of living cells, and the obtained signal was enhanced by deconvolution (Fig. 1A to C). Live cells at early times after induction (up to 30 min) presenting a low concentration of mRNPs were used to continuously track individual particles and study their motion (Fig. 1D). Single-particle tracking (SPT) was performed on mRNPs that remained in focus for a minimum of eight consecutive frames (>3 s) (Fig. 1E to H) [movies S6 to S10 (5)] (3). Total distances traveled for tracked mRNPs were from 2 to 10 µm (mean 5 µm), and mean velocities ranged from 0.3 to 0.8 µm/sec (fig. S3A). These mRNPs displayed a diffusive pattern of movement (Fig. 1E) following a simple diffusion model <r_2> = 4_Dt (Fig. 1J) (3). The diffusion coefficients at 37°C (D_37) ranged from 0.01 to 0.09 µm2/sec (mean 0.04 µm2/sec) (Fig. 1K). The mean square displacement (MSD) plotted over time Δ_t was linear in 58% of the cases, characteristic of simple diffusion (Fig. 1J); for 42% of the mobile mRNPs, the plots began linearly but reached a plateau (Fig. 1J), characteristic of corralled diffusion (Fig. 1F). This result indicated the presence of barriers hindering the movement of the mRNPs. Directed movements in the nucleus were not seen among the tracked particles or observed in the overall population of particles imaged (Fig. 1K), although directed translocations were easily detected for cytoplasmic CFP peroxisomes by using our SPT algorithm (3) (fig. S3C). Extremely confined movements were observed for transcription sites and for some mRNPs (less than 1% of detectable mRNPs) (Fig. 1I and fig. S3B). These mRNPs were confined within small nucleoplasmic volumes. Observations of living cells (3) indicated that once mRNPs were released from the transcription site, their movements through the nucleoplasm on the way to the nuclear pore were rarely hindered by stable interactions with nuclear substructures. Altogether, these data indicated that nucleoplasmic mRNP movement was governed by laws of simple diffusion and was not directional.
Fig. 1.
Live-cell imaging and single-particle tracking of individual mRNPs. Images from time-lapse movies acquired from a cell cotransfected with(A) CFP-lac repressor and (B) YFP-MS2 and induced for more than 30 min. (C) Reduction of noise for tracking of mRNPs was obtained by deconvolution. Bar, 2 µm. (D) Tracking of mRNP particles in a transcriptionally active cell induced for only 20 min (arrow, transcription site) (bar, 2 µm) [Movie S6 (5)] showed (E) diffusing particles [Movie S7 (5)], (F) corralled particles [Movie S8 (5)], (G) stationary particles, and (H) the transcription site. Tracks are marked in green, and time in seconds from the beginning of tracking for eachparticle appears in each frame. Bars, 1 µm. (I) Plot of the area per frame in which each of the tracked particles from this particular cell traveled throughout the tracking period. Diffusive particles are in blue, corralled in green, stationary in yellow, and transcription site in red. Particle 1, particle seen in E; 2, F; 5, G; 6, H. (J) Mean-square displacement (MSD) of tracked nucleoplasmic particles versus time indicated the presence of three types of characterized movements: diffusive (black circles), corralled (blue triangles), and stationary (green squares). Directed movement was never detected (red dotted line; plotted from data of tracked directed cytoplasmic particles). (K) Table summarizing the mean velocities and diffusion coefficients of tracked particles at 37°C. %, percent of tracked particles. _D_diffusion coefficient.
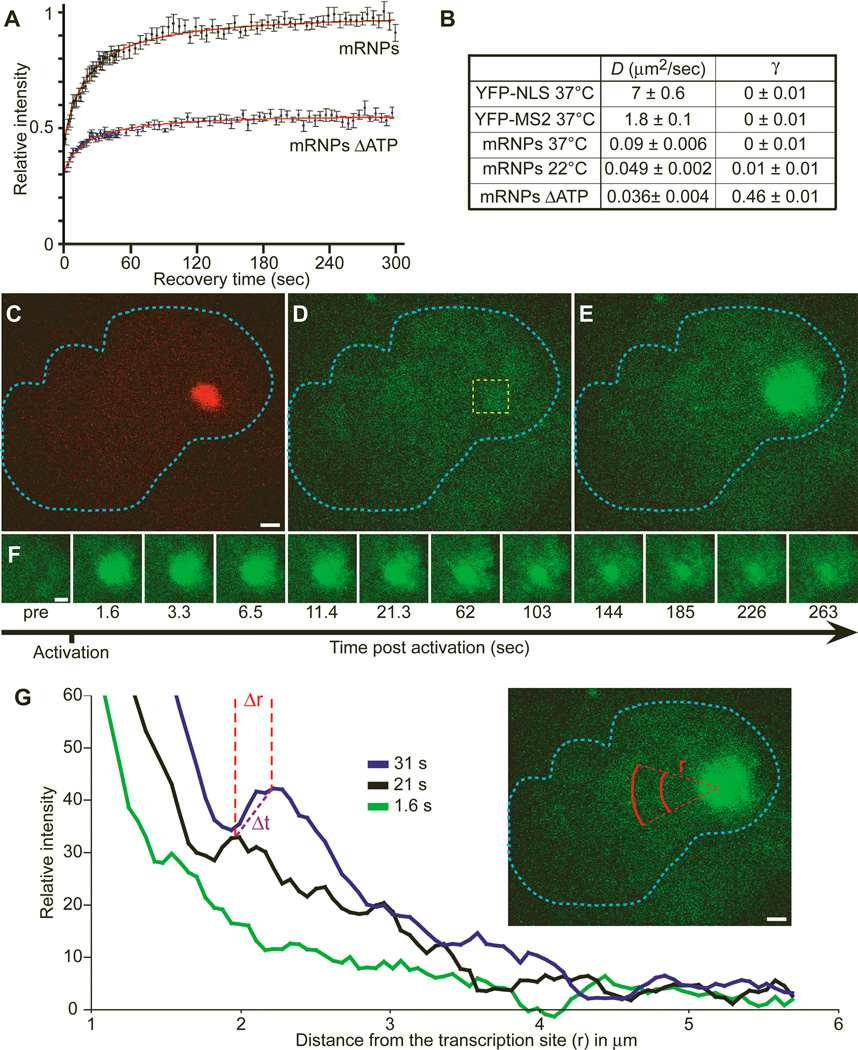
Fluorescence recovery after photobleaching (FRAP) of the tagged mRNPs was followed over time, and the mean diffusion coefficient (_D_37) of these mRNPs was calculated to be 0.09 ± 0.006 µm2/sec (Fig. 2A and B), corroborating the SPT data. As with SPT, an immobile fraction was not observed.
Fig. 2.
FRAP and photoactivation of labeled RNPs. (A) FRAP performed on cells cotransfected with pTet-On and YFP-MS2 and induced for 30 to 60 min with doxycycline. Cells either remained untreated or were ATP depleted for 30 min. Recovery of the bleached signal is shown. (B) Comparison of the calculated diffusion rates (D) and fixed fractions (γ) for free YFP-NLS, free YFP-MS2, mRNPs at different temperatures, and mRNPs in ATP-depleted cells. (C to E) A cell in which the photoactivatable MS2-GFP (MS2-paGFP) gene was stably integrated (2-3-6-PA) was cotransfected with RFP-lac repressor and pTet-On. Transcription was induced for 30 min by doxycycline. The locus was detected (red) prior to photoactivation (C), and the image in GFP before activation was recorded (D). The 405-nm laser was directed at the boxed region of interest (yellow), and the MS2-paGFP was detected at the transcription site 1.635 s after activation (E). Bar, 2 µm. (F) The RNP signal emanating from the transcription site was followed for 262 s (bar, 2 µm) [Movie S11 (5)]. (G) To quantify the movement of the mRNPs, we measured concentric arcs at various distances (r) from the transcription site, as indicated in red, and plotted the normalized (to preactivation value) average intensities per pixel for each arc. The three plots represent three examples of time points (green, 1.635 s; black, 21.255 s; blue, 31.065 s). The distances between the peaks of the signal wave (Δ_r_) were calculated over time (Δ_t_) and the diffusion coefficients determined.
We then addressed the movement of mRNPs present in the vicinity of the transcription site, using a third technique. By use of a photoactivatable form of green fluorescent protein (GFP) (7) fused to the MS2 protein, we followed the mRNPs in a “visible pulse-chase” experiment. A short pulse of activating light (405 nm) confined only to the transcription site fluorescently labeled those RNPs being actively transcribed or just released (Fig. 2C to E). This generated an initial pool of fluorescent mRNAs that, upon release from the transcriptional machinery, diffused away from the transcription site in all directions (Fig. 2F and G) (3) [movie S11 (5)]. Measurements of the area in which these mRNAs moved over time (Fig. 2G) showed a mean diffusion coefficient (_D_22) of 0.034 ± 0.006 µm2/sec, consistent with the range of measurements obtained by the previous two approaches (FRAP yields a _D_22 = 0.049 ± 0.002 µm2/sec, as discussed below and in fig. S4H). In summary, a similar range of diffusion coefficients for nuclear mRNPs was measured with three techniques.
Analysis of the SPT data did not show any characteristics of anomalous diffusion (8), expected if the mRNPs were to exhibit energy-dependent binding, and SPT performed at room temperature showed _D_s of 0.02 to 0.09 µm2/s (mean 0.044 µm2/s) (fig. S3D). According to the Stokes-Einstein equation (3), the diffusion of mRNPs should be affected by the absolute temperature, the radius of the particle, and the viscosity of the nucleoplasm. Because the nucleus is basically an aqueous solution (9), nuclear viscosity decreases in a temperature-dependent manner and should lead in turn to a drop in diffusion coefficients. FRAP experiments (3) (fig. S4) revealed that the _D_s of mRNPs and free YFP-MS2 protein were affected in a linear manner by a reduction in temperature. The linear relationship between D and temperature argues against an energy-driven mechanism for translocation of mRNPs in the nucleus (10). There has been persistent speculation concerning the role of actin and myosin in RNA transport in the nucleus (11, 12); however, inhibitors to known motors did notaffect the motion of the nuclear mRNPs (3).
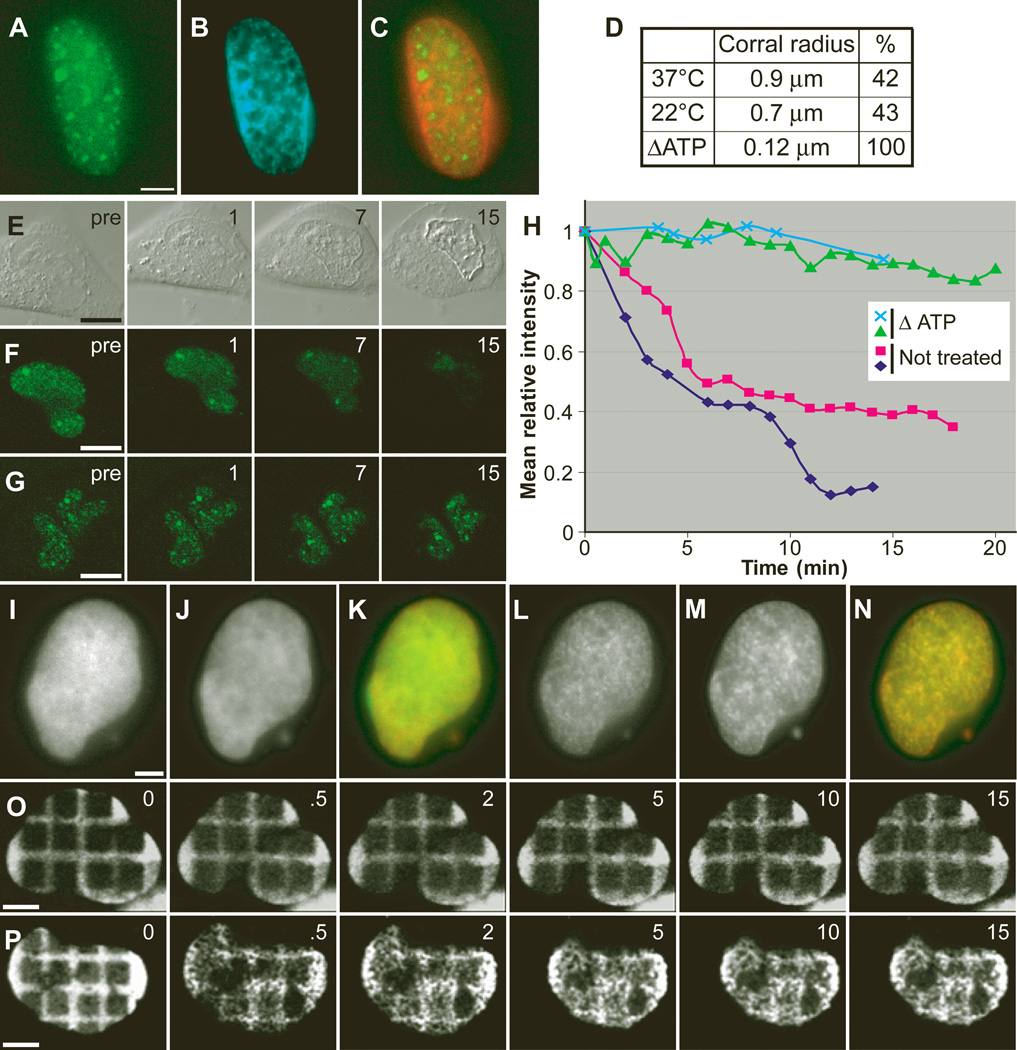
Whether nuclear mRNP movement is an energy-dependent process is controversial (13, 14), and thus far no study has provided an explanation for any energy-dependent effects. To determine whether diffusion was independent of cellular metabolism, we treated cells with the general metabolic inhibitors 2-deoxyglucose (2-DG) and sodium azide. Unexpectedly, the majority of the mRNPs showed measurable decreased mobility throughout the nucleoplasmic space and concentrated in subnuclear regions surrounded by chromatin-dense domains (Fig. 3A to C) (3) [movies S12 to S14 (5)]. Further analysis showed that the mobility was reduced as a result of the constraints imposed by small subnuclear volumes, as determined by the notable decrease in the corralled radius observed by SPT (Fig. 3D). Replenishment of energy levels restored mRNP movement (3). When energy-depleted cells were subjected to mild permeabilization with Triton X-100, mRNPs remained trapped throughout the nucleoplasm, in contrast to undepleted cells where mRNPs rapidly dispersed (Fig. 3E to H). This observation confirmed that under conditions of energy depletion, intranuclear structures constrained mRNP mobility. FRAP of energy-depleted cells showed that 46% of the mRNPs were trapped in the bleached area (Fig. 2A). Direct visualization of the mRNPs (3) [(movies S12 and S14 (5)] showed that, although constrained, the mRNPs moved within areas of confinement. Particles that moved in and out of a bleached area had an apparent diffusion coefficient of 0.036 µm2/s (Fig. 2B). This D was lower than observed for nondepleted cells; however, the extremely corralled nature of the mRNPs under these conditions could explain this difference.
Fig. 3.
Effect of energy depletion on mRNP movement. (A to C) A transfected and transcriptionally induced cell, 10 min after energy depletion. (A) Concentrated YFP-MS2 nuclear mRNPs; (B) Hoechst DNA stain; (C) Merge of YFP-MS2 mRNPs from (A) in green and Hoechst from (B) in red, showing segregation into two domains [movies S12 to S14 (5)]. (D) Table summarizing the corral radius measured for corralled particles tracked at 37°C, 22°C, and ATP-depleted cells. %, percent of corralled particles from total tracked. (E to G) Cells treated with 0.02% Triton (pre, before Triton; 1, 7, and 15 s, after Triton). (E) Differential interference contrast image. (F) YFP-MS2 labeled mRNPs are lost from the nucleus during permeabilization (time in minutes). (G) Same as (F), but the cells were first energy depleted and then permeabilized, and mRNP loss is slower. (H) Plots of the mean fluorescence intensity over time in the nucleus of permeabilized cells, treated as in (F) (red and dark blue lines) or as in (G) (green and light blue lines). (I) Cell expressing H2B-YFP and (J) counterstained with Hoechst. (K) Merge of (I) and (J). (L to N) The same cell 10 min after ATP depletion, showing the formation of nuclear subdomains. Bar, 5 µm. [movies 15 to 17 (5)]. (O) Time course of a cell transfected with H2B-YFP in which squares have been photobleached to form a grid (times are in minutes). (P) Same time course as in O, but ATP was depleted at time 0 min, showing major changes in chromatin structure together with nuclear shrinkage [movie S18 (5)]. Bar, 5 µm.
The stalled mRNPs localized in DNA-poor regions, and the reduction in corral size strongly implied that a change in nuclear substructure obstructed free diffusion of mRNPs under conditions of energy deprivation. Chromatin structure in living cells after energy depletion was then examined. Cells transiently expressing histone H2B-YFP, together with a brief exposure to Hoechst DNA stain, showed the rapid formation (seconds) of dense chromatin regions upon energy depletion (Fig. 3I to N) (3) [movies S15 and S16 (5)], followed by a reduction in nuclear size (3). These effects were reversible when inhibitors were washed away (3) [movie S17 (5)]. A grid pattern generated by photobleaching H2B-YFP demonstrated that the nucleus underwent rapid major restructuring immediately upon energy depletion (Fig. 3O and P) (3) [movie S18 (5)]. Similar changes were also observed by electron microscopy (EM) (fig. S5A and B). The nucleoplasmic signal was diffusely distributed in untreated cells, whereas in the energy-depleted cells subnuclear spaces devoid of electron-dense material could be observed. These spaces were surrounded by electron-dense barriers, reminiscent of the restructured chromatin regions surrounding the mRNPs seen in the fluorescent images (Fig. 3A, B, L, and M). H2B-YFP redistribution compared with specific nuclear markers under conditions of energy depletion (3) showed that the dense chromatin structures were spatially segregated from other nucleoplasmic domains (3) [movies S19 and S20 (5)]. Interestingly, under these conditions, concentrated mRNPs colocalized with speckle domains (3) [movies S21 to S23 (5)]. We conclude that nuclear reorganization observed under energy stress is a general effect influencing both chromatin and interchromatin domains.
A model for free diffusion of nuclear mRNPs was proposed more than a decade ago (15). Other studies using a variety of methods—labeled antisense probes (16–18), GFP-tagged RNA-binding proteins (19), or EM analysis of the distribution of mRNPs (20)—have proposed that a population of RNPs move by diffusion through the nucleoplasmic space and have estimated a wide range of diffusion coefficients for these labeled RNPs [for an extended discussion, see (3)]. In most of these studies, a diffuse fluorescent signal attributed to populations of mRNPs was followed, but the possibility could not be excluded that an exchange of the fluorescent tag was contributing to the measured mobility rates. In contrast, the specific MS2 protein amplifies only on mRNAs containing the MS2 aptamer, thereby forming single mRNPs that can be individually and agnostically tracked. Observations of thousands of mRNP translocations confirmed a strictly diffusion-based model revealing subsets of free diffusing and heretofore undetected corralled and, on rare occasions, constrained states. There were no stable attachments outside the transcription site, nor noticeable accumulations in subnuclear domains. These movements were ATP independent, and the major nuclear reorganization we observed during energy stress may explain the previously described changes in the mobility of various nuclear components [see discussion in (3)]. While the reporter RNP is a bona fide mRNA, it is possible that specific mRNPs or other cell types could have alternative means of nuclear egress.
In summary, this approach allows direct observation of the primary transcript of gene expression and accesses the statistical variance and diversity of molecular events within the population. It provides a means for testing microenvironmental models that act on each molecule. One such model, that of a system directing these transcripts, can be eliminated. Clearly, the complexity of RNA trafficking requires further investigation.
Supplementary Material
Movie S1
Movie S18
Movie S19
Movie S2
Movie S20
Movie S21
Movie S22
Movie S23
Movie S3
Movie S4
Movie S5
Movie S10
Movie S6
Movie S7
Movie S8
Movie S9
Supplemental
Movie S11
Movie S12
Movie S13
Movie S14
Movie S15
Movie S16
Movie S17
Acknowledgments
We thank A. Femino for advice on RNA quantification, B. Hong and M. Cammer for programming assistance, F. Macaluso and L. Cummings for the EM work, B. Ovryn for introducing us to Mathematica software, and T. Meier for useful discussions and critical reading of the manuscript. Supported by NIH EB2060 and DOE63056 to R.H.S and GM42694 to D.L.S.
References and Notes
- 1.Fusco D, et al. Curr. Biol. 2003;13:161. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janicki SM, et al. Cell. 2004;116:683. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Materials, methods, and additional data are available ?as supporting material on Science Online [Google Scholar]
- 4.Tsukamoto T, et al. Nature Cell Biol. 2000;2:871. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- Movies S1 to S23 are available at http://singerlab.org/supplements/science_v304p1797. [Google Scholar]
- 6.Femino AM, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 7.Patterson GH, Lippincott-Schwartz J. Science. 2002;297:1873. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 8.Saxton MJ. Biophys. J. 1996;70:1250. doi: 10.1016/S0006-3495(96)79682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D. J. Cell Sci. 2004;117:2159. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 10.Lang I, Scholz M, Peters R. J. Cell Biol. 1986;102:1183. doi: 10.1083/jcb.102.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rando OJ, Zhao K, Crabtree GR. Trends Cell Biol. 2000;10:92. doi: 10.1016/s0962-8924(99)01713-4. [DOI] [PubMed] [Google Scholar]
- 12.Pestic-Dragovich L, et al. Science. 2000;290:337. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 13.Politz JC, Pederson T. J. Struct. Biol. 2000;129:252. doi: 10.1006/jsbi.2000.4227. [DOI] [PubMed] [Google Scholar]
- 14.Carmo-Fonseca M, Platani M, Swedlow JR. Trends Cell Biol. 2002;12:491. doi: 10.1016/s0962-8924(02)02387-5. [DOI] [PubMed] [Google Scholar]
- 15.Zachar Z, Kramer J, Mims IP, Bingham PM. J. Cell Biol. 1993;121:729. doi: 10.1083/jcb.121.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Politz JC, Browne ES, Wolf DE, Pederson T. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6043. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Politz JC, Tuft RA, Pederson T, Singer RH. Curr. Biol. 1999;9:285. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 18.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. J. Cell Biol. 2004;165:191. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calapez A, et al. J. Cell Biol. 2002;159:795. doi: 10.1083/jcb.200203046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh OP, Bjorkroth B, Masich S, Wieslander L, Daneholt B. Exp. Cell Res. 1999;251:135. doi: 10.1006/excr.1999.4490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1
Movie S18
Movie S19
Movie S2
Movie S20
Movie S21
Movie S22
Movie S23
Movie S3
Movie S4
Movie S5
Movie S10
Movie S6
Movie S7
Movie S8
Movie S9
Supplemental
Movie S11
Movie S12
Movie S13
Movie S14
Movie S15
Movie S16
Movie S17