Antibiotic-Inducible Promoter Regulated by the Cell Envelope Stress-Sensing Two-Component System LiaRS of Bacillus subtilis (original) (raw)
Abstract
Soil bacteria are among the most prodigious producers of antibiotics. The Bacillus subtilis LiaRS (formerly YvqCE) two-component system is one of several antibiotic-sensing systems that coordinate the genetic response to cell wall-active antibiotics. Upon the addition of vancomycin or bacitracin, LiaRS autoregulates the liaIHGFSR operon. We have characterized the promoter of the lia operon and defined the cis_-acting sequences necessary for antibiotic-inducible gene expression. A survey for compounds that act as inducers of the lia promoter revealed that it responds strongly to a subset of cell wall-active antibiotics that interfere with the lipid II cycle in the cytoplasmic membrane (bacitracin, nisin, ramoplanin, and vancomycin). Chemicals that perturb the cytoplasmic membrane, such as organic solvents, are also weak inducers. Thus, the reporter derived from P_liaI (the liaI promoter) provides a tool for the detection and classification of antimicrobial compounds.
Soil bacteria inhabit a complex and highly variable environment. Bacillus subtilis, a ubiquitously distributed, low G+C gram-positive soil organism, is metabolically versatile and displays sophisticated regulatory responses that allow adaptation to changing environmental conditions (18). Streptomyces coelicolor, the model organism for the actinomycete family, has even greater genomic and regulatory complexity (4, 14). Bacteria from these groups produce large numbers of antibiotics, presumably as a mechanism to suppress the growth of competitors. The ability of these organisms to sense, and respond appropriately to, the presence of antibiotics is crucial to their survival.
We have used DNA microarray analyses to define the genetic responses of B. subtilis to two antibiotics present in its natural habitat, vancomycin and bacitracin (7, 22). The glycopeptide antibiotic vancomycin is produced by Streptomyces toyocaensis and other actinomycetes and binds tightly to D-Ala-D-Ala termini on the pentapeptide side chains of cell wall precursors, thereby inhibiting the formation of peptide cross-bridges by peptidyltransferase (34). While self-resistance of the producing strain has been reported (21), no vancomycin resistance mechanism has so far been described for B. subtilis. The branched cyclic dodecylpeptide antibiotic bacitracin is synthesized by Bacillus licheniformis and some strains of B. subtilis (2, 15). Bacitracin forms a tight complex with the undecaprenyl-pyrophosphate carrier that is released and recycled after the transglycosylation step of peptidoglycan biosynthesis. The presence of this antibiotic is sensed by a specific two-component system (BacRS in B. licheniformis and BceRS in B. subtilis), which in turn strongly induces the expression of a bacitracin-specific ABC transporter that contributes to resistance (22, 25, 26). In other bacteria, reported resistance mechanisms involve the de novo synthesis of undecaprenol-pyrophosphate (6, 8), the target of bacitracin, and exopolysaccharide biosynthesis (27, 31).
Treatment of B. subtilis with sublethal concentrations of vancomycin or bacitracin induces overlapping regulons controlled by alternative sigma factors and two-component systems. Some regulons respond to a very small number of inducers (e.g., BceRS), whereas others (e.g., σM and LiaRS) respond to a variety of agents that perturb cell envelope functions (22). The histidine kinases of the three bacitracin-responsive two-component systems (LiaS, BceS, and YvcQ) share an unusually short sensing domain that is almost completely buried in the cytoplasmic membrane. This led us to hypothesize that they sense perturbations of cell envelope biosynthesis directly through the lipid interface. Therefore, these proteins were named intramembrane-sensing histidine kinases (22). The genes most strongly induced by both antibiotics were liaIH (formerly yvqIH), showing up to 1,000-fold increased induction in the expression level at 5 to 10 min after the addition of bacitracin. This induction was mediated by the response regulator LiaR. The genes for the corresponding two-component system, liaSR, are located directly downstream of the cotranscribed liaIHGF genes (22).
Here we characterize the LiaR-dependent, antibiotic-responsive promoter of the liaIHGFSR locus in B. subtilis. Of all antibiotics tested, this promoter is induced only by bacitracin, nisin, ramoplanin, and vancomycin, i.e., by antibiotics that interfere with the essential undecaprenol cycle in the cytoplasmic membrane. Based on the observed spectrum of its inducers, we renamed the YvqCE two-component system LiaRS (for lipid II cycle interfering antibiotic response regulator and sensor) and the remaining genes of this operon liaIHGF. We demonstrate that the P_liaI_ (liaI promoter) reporter system has potential as a screening tool for antibiotics: it shows a very sensitive, concentration-dependent response to its identified inducers.
MATERIALS AND METHODS
Media and growth conditions.
B. subtilis and Escherichia coli were routinely grown in Luria broth medium at 37°C with aeration. Ampicillin (100 μg/ml) was used for selection of plasmid pJPM122 and its derivatives in E. coli. Kanamycin (100 μg/ml), neomycin (10 μg/ml), chloramphenicol (1 μg/ml), and erythromycin (1 μg/ml) plus lincomycin (25 μg/ml) for macrolide-lincosamide-streptogram resistance, were used for the selection of the B. subtilis mutants used in this study.
Bacterial strains and plasmids.
The strains of E. coli and B. subtilis, as well as the plasmids used, are listed in Table 1. SPβ phages are derivatives of SPβc2Δ2 and were constructed by the integration of a promoter region-cat-lacZ fusion constructed in plasmid pJPM122 into B. subtilis strain ZB307A as described previously (30). SPβ lysates were prepared by heat induction from the lysogenic strains as described (37).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Characteristics or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | φ80_lacZ_Δm15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK−, mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Laboratory stock |
| B. subtilis | ||
| CU1065 | W168 _att_SPβ trpC2 | Laboratory stock |
| ZB307A | W168 SPβ2Δ2::Tn_917_::pSK10Δ6 | 37 |
| BSF2469 | CU1065 liaH::pMUTIN | Zoltan Pragai |
| BSF2470 | CU1065 lial::pMUTIN | Zoltan Pragai |
| HB0933 | CU1065 liaR::kan | 22 |
| HB0940 | W168 SPβ2Δ2::Tn_917_::Φ(P_liaI-29-cat-lacZ_) | This work |
| HB0941 | W168 SPβ2Δ2::Tn_917_::Φ(P_liaI-58-cat-lacZ_) | This work |
| HB0942 | W168 SPβ2Δ2::Tn_917_::Φ(P_liaI-74-cat-lacZ_) | This work |
| HB0943 | W168 SPβ2Δ2::Tn_917_::Φ(P_liaI-83-cat-lacZ_) | This work |
| HB0944 | W168 SPβ2Δ2::Tn_917_::Φ(P_liaI-193-cat-lacZ_) | This work |
| HB0949 | CU1065 SPβ2Δ2::Tn_917_::Φ(P_liaI-58-cat-lacZ_) | This work |
| HB0950 | CU1065 SPβ2Δ2::Tn_917_::Φ(P_liaI-74-cat-lacZ_) | This work |
| HB0952 | CU1065 SPβ2Δ2::Tn_917_::Φ(P_liaI-58-cat-lacZ_), liaR::kan | This work |
| HB0953 | CU1065 SPβ2Δ2::Tn_917_::Φ(P_liaI-74-cat-lacZ_), liaR::kan | This work |
| Plasmids | ||
| pJPM122 | cat-lacZ operon fusion vector for SPβ | 30 |
| pSLZ29 | pJPM122 with P_liaI-29_ | This work |
| pSLZ58 | pJPM122 with P_liaI-58_ | This work |
| pSLZ74 | pJPM122 with P_liaI-74_ | This work |
| pSLZ83 | pJPM122 with P_liaI-83_ | This work |
| pSLZ193 | pJPM122 with P_liaI-193_ | This work |
| Oligonucleotidesa | ||
| 1312 | yvqH fwd: GGAGGAATCAGGTATGG | |
| 1314 | yvqH rev: CTTGACCGCAAATCCTTCC | |
| 1779 | yvqG fwd: CAACTCTTATCGTCAGGCTTCCG | |
| 1311 | yvqH-do rev: CGCTAGATCCCCGCTGTCC | |
| 1503 | PyvqI-559: GGAT_CTGCAG_GGTTTGTGCTGGCGAAAGTCAAGG | |
| 1628 | yvqI-PE: TTAATAAGAATCCGCCTATTG | |
| 1310 | yvqH-do fwd: GCAGACCAGACAAAAGCGGC | |
| 1629 | yvqG-PE: TCCGCTATAATCCGGACATCC | |
| 1506 | PyvqI-193: CCAT_CTGCAG_GCCAAAGCAGAAAGGTCCGACC | |
| 1507 | PyvqI-83: CCAT_CTGCAG_CCGGTGCGAGATACGACTCC | |
| 1508 | PyvqI-74: GGAT_CTGCAG_GATACGACTCCGGTCTTATATAAAAATC | |
| 1509 | PyvqI-58: GGAT_CTGCAG_TATATAAAAATCAATCTCTGATTCG | |
| 1510 | PyvqI-29: GGAT_CTGCAG_GCATATCTTCCAACTTG | |
| 1511 | PyvqI+93: CGAT_GGATCC_TCCTCCAAAAAAGACGGAGATCCC |
DNA manipulations and sequencing.
The preparation of chromosomal DNA, transformation, and SPβ transduction were performed according to standard procedures (9). E. coli plasmid DNA and restriction enzyme fragments were isolated by using the QIAprep spin miniprep and PCR purification kits, respectively (QIAGEN Inc., Chatsworth, Calif.). Restriction endonucleases, DNA ligase (New England Biolabs, Beverly, Mass.), Pfu DNA polymerase (Stratagene, La Jolla, Calif.), and HotStar DNA polymerase (QIAGEN Inc.) were used according to manufacturers' instructions. DNA sequencing was performed with AmpliTaq-FS DNA polymerase and dye terminator chemistry by the DNA services facility of the Cornell New York State Center for Advanced Technology-Biotechnology.
Northern analysis of liaH and liaG.
Total RNA was extracted from 5 ml of B. subtilis CU1065 culture with and without bacitracin (final concentration, 10 μg/ml). Bacitracin was added to the culture at an optical density at 600 nm (OD600) of 0.45 (mid-log phase), and the cultures were incubated for 15 min at 37°C with aeration before the cells were harvested and rapidly frozen at −80°C. RNA was prepared by using the RNeasy kit (QIAGEN) according to the manufacturer's protocol. Internal fragments of liaH and liaG (500- to 750-nucleotide length) were amplified by PCR by using the primer pairs 5′ 1312-3′ 1314 and 5′ 1779-3′ 1311 (Table 1). The PCR fragments were purified by using the QIAGEN PCR purification kit, and 100 ng of each fragment was labeled with [α-32P]dATP (3,000 Ci/mmol, 10 mCi/μl; New England Nuclear) by random oligonucleotide-primed synthesis with the Klenow fragment of DNA polymerase (3′→5′ exo−; New England Biolabs) according to a published procedure (3.5.9-10) (1). Unincorporated [α-32P]dATP was removed by NucAway spin columns (Ambion).
Northern analysis was carried out by using the NorthernMax formaldehyde-based system (Ambion) according to the manufacturer's instruction, with 10 μg of total RNA and Zeta-Probe blotting membrane (Bio-Rad) in a downward transfer setup. After hybridization and washing of the membranes, the blots were wrapped in plastic wrap, exposed for 12 h to a phosphor screen (Molecular Dynamics) and analyzed by a PhosphorImager (Molecular Dynamics).
Primer extension mapping of the liaI promoter site.
For mapping of the liaI promoter, B. subtilis CU1065 cells were grown in LB, and total RNA was isolated from uninduced and bacitracin-induced (final concentration, 10 μg/ml) mid-log phase cultures as described above. Primer extension reactions for liaI were set up as follows: 30 μg of heat-denatured RNA was hybridized at 65°C to ∼2 pmol of end-labeled primer 1628 in buffer containing 60 mM NaCl, 50 mM Tris-HCl (pH 7.9), 10 mM dithiothreitol (DTT), and 40 U of RNasin (Promega) in a total volume of 30 μl. Following hybridization, 50 μl of extension buffer (72 mM NaCl, 50 mM Tris-HCl [pH 7.9], 10 mM DTT, 20 mM MgCl2), 10 mM concentrations of the deoxynucleoside triphosphates, and 2 μl of Superscript II reverse transcriptase (Invitrogen, Carlsbad, Calif.) were added to the mixture, and incubation continued at 37°C for 30 min. The primer extension products were precipitated with ethanol, resuspended in sequence loading buffer, and loaded onto a 6% polyacrylamide sequencing gel. A PCR cycle sequencing kit (Epicentre, Madison, Wis.) was used to generate sequencing ladders corresponding to the liaI promoter region.
Construction of cat-lacZ reporter fusions for P_liaI_ dissection.
For the P_liaI_ -cat-lacZ fusions, promoter fragments of increasing lengths were generated by PCR by using the 5′ primer pair 1506-1510 with the 3′ primer 1511 (Table 1). The PCR was performed in a total volume of 50 μl with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) according to the manufacturer's instruction. The reactions were initially denatured for 2 min at 94°C, followed by 30 cycles of 20 s at 94°C, 30 s at 50°C, 30 s at 72°C, and a final extension of 5 min at 72°C. The resulting PCR products were cloned into pJPM122 (30) as a PstI-to-BamHI fragment (Table 1; restriction sites are underlined in primer sequences), resulting in promoter-cat_-lacZ fusions in plasmids pSLZ29 (as a negative control, lacking the −35 region of P_liaI), pSLZ58 (core promoter), pSLZ74 (single TCCGGT sequence included), pSLZ83 (complete TCCGGT repeat), and pSLZ193 (positive control) (see Fig. 2). The inserts were verified by DNA sequencing at the Cornell BioResource Center. The plasmids were linearized and used to transform ZB307A with neomycin selection to generate strains HB0940 to HB0944. Phages generated from strains HB0941 and HB0942 (SPβ0941 and SPβ0942, respectively) were used to transduce the P_liaI_ -cat-lacZ fusions into B. subtilis strains CU1065 and HB0933, resulting in strains HB0949/0950 and HB0952/0953, respectively (Table 1).
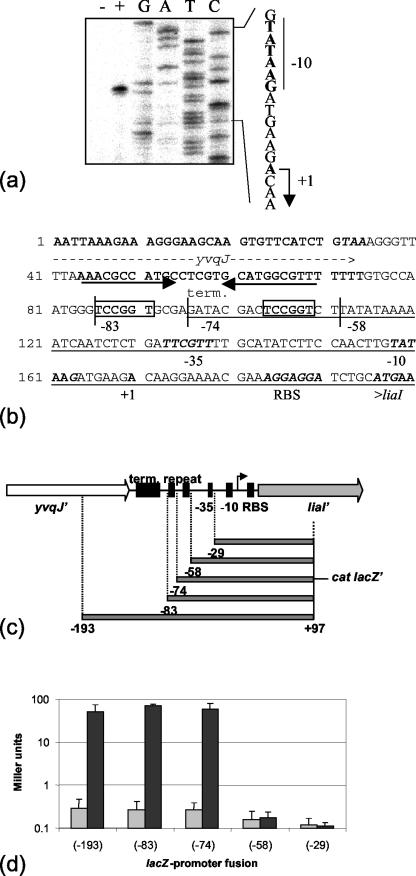
FIG. 2.
Functional dissection of the liaI promoter P_liaI_. (a) Primer extension mapping of the liaI transcriptional start site indicates transcription initiation with the A residue indicated in the sequence to the right. (b) Intergenic squence between yvqJ and liaI. All features are marked underneath the respective lines of the sequence. The end of yvqJ and the beginning of liaI are labeled. The putative yvqJ terminator is indicated by the black arrows. The expression signals for liaI are labeled (−35 and −10 for the promoter and RBS for the putative ribosome binding site). A direct repeat sequence is boxed (see text). The 5′ ends of the fragments used for the promoter dissection are marked and labeled according to their positions relative to the transcriptional start site (see below). The minimal bacitracin-inducible promoter fragment, based on the promoter dissection, is underlined. (c) Graphical representation of the intergenic region and outline of the fragments used for the promoter dissection. The features of the region are represented by black boxes and labeled as above. The arrow indicates the transcriptional start site. term, terminator. (d) β-Galactosidase assay for promoter dissection. Cultures of the P_liaI_ reporter strains HB0940 (−29), HB0941 (−58), HB0942 (−74), HB0943 (−83), and HB0944 (−193) were grown to mid-log phase (OD600 of ∼0.45) and induced by the addition of bacitracin (final concentration, 10 μg/ml). Cells were harvested 30 min postinduction and assayed as described in Materials and Methods. β-Galactosidase activity, plotted on a logarithmic scale for clarity, is expressed in Miller units (23). Dark gray bars represent the induced sample, and light gray bars indicate the uninduced control sample for each strain. Each experiment was performed at least twice, and the standard deviations are indicated by error bars.
P_liaI_ induction assays.
Screening for liaI induction was done by disk diffusion assay essentially as described (7). B. subtilis strains were inoculated from a fresh overnight LB agar plate and grown to mid-log phase (OD600 of ∼0.45) at 37°C with shaking. A total of 20 μl of the culture was mixed with 3 ml of 0.7% soft LB agar (containing 40 μg of XGal per plate) and poured onto the bottom agar. After cooling and drying of the plates (20 min at 37°C), filter paper disks (6-mm diameter) carrying 5 μl of stock solution (antibiotics normally at a concentration of 100 mg/ml each; lysozyme at a concentration of 10 mg/ml; tunicamycin, surfactants, and uncouplers at a concentration of 5 mg/ml each; inhibitors of protein biosynthesis as given in “Media and growth conditions”) were placed on top of the agar. The plates were incubated at 37°C overnight. After incubation for 12 to 24 h, the plates were scored for the appearance of blue rings at or near the edge of the zones of growth inhibition produced by the diffusion of the antibiotics from the filter disks.
For quantitative measurements of β-galactosidase activity, cells were inoculated from fresh overnight LB plates and grown in LB medium at 37°C with aeration until they reached an OD600 of 0.45. A total of 2 ml of the culture was harvested (uninduced control), and the cell pellets were frozen and kept at −80°C. The cultures were induced by the addition of the compound to be tested to the final concentration as described in the individual figure legends and incubated for an additional 30 min at 37°C. Cultures (2-ml) were harvested as described above. The pellets were resuspended in 1 ml of working buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.4 mM DTT) and assayed for β-galactosidase activity as described with normalization to cell density (23). For concentration-dependent induction and killing experiments, cultures of strain BFS2470 were grown in LB medium to mid-log growth phase (OD600 of 0.4 to 0.45), and the antibiotics were added to a final concentration ranging from 0.01 to 100 μg/ml. An uninduced culture was used as a negative control. The cultures were incubated with aeration at 37°C. A sample was taken after 30 min for the β-galactosidase assay, and the turbidity of the remaining culture was measured for at least 5 h to monitor the concentration-dependent effects of the antibiotics on cell growth.
RESULTS
The liaIHGFSR locus forms an operon.
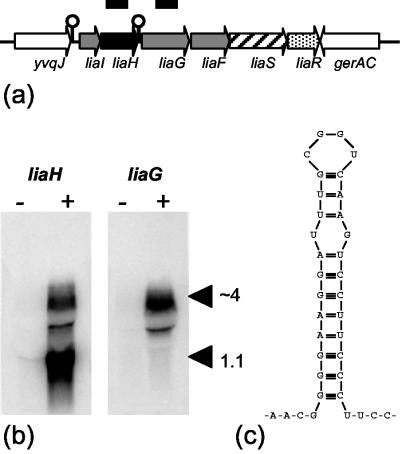
The expression of the liaIHGFSR operon is very tightly regulated and completely LiaR dependent, as shown previously (22). Only a very faint transcript of 1.1 kb, corresponding to liaIH, can be detected by Northern analysis in uninduced mid-log phase cultures (Fig. 1b). Upon the addition of bacitracin, two transcripts are strongly induced, corresponding to liaIH and the complete liaIHGFSR locus (Fig. 1b). This expression pattern may result from transcription termination downstream of liaH (Fig. 1c). The presence of the LiaRS two-component system is a prerequisite for this strong induction.
FIG. 1.
Organization and bacitracin-dependent expression of the liaIHGFSR locus of B. subtilis. (a) Graphic representation of the lia locus including liaS (histidine kinase) and liaR (response regulator). The pspA homolog liaH is shown in black. Genes coding for proteins with unknown function are shown in gray; the genes flanking the lia locus are white. The line corresponds to a size of 7.5 kb. In the course of this work we noticed two errors in the original genome sequence in the noncoding region between liaH and liaG. The corrected sequence resulted in an addition of 150 nucleotides at the 5′ end of liaG. The fragments used as probes for liaH and liaG in the Northern blot (panel b) are represented as thick black bars above the locus. (b) Northern blots showing the bacitracin-dependent expression of liaH and liaG. Expression of each gene was measured by using 10 μg of total RNA from each sample separated on a 1% formaldehyde gel. RNA was transferred to a nylon membrane and hybridized with radiolabeled DNA fragments of the indicated genes. −, uninduced control lane; +, the RNA sample from cultures induced with bacitracin for 15 min (final concentration of 10 μg/ml). The two transcripts are indicated with arrowheads, and the approximate sizes corresponding to liaIH (1.1 kb) and the whole lia locus (∼4 kb) are given. The thin band between the transcripts represents an artifact derived from the quenching of the 4-kb transcript by the abundant 23S rRNA, which can saturate available binding sites on the hybridization membrane. (c) Secondary structure of the stem-loop directly downstream of liaH.
Previous work on the bacitracin stimulon suggested an independent low-level expression of liaGFSR (22). To investigate this further, Northern analysis was performed by using a _liaG_-specific probe. No transcript could be detected in uninduced cultures, and upon bacitracin induction only the full-length transcript appeared (Fig. 1b). Primer extension analysis also failed to detect a transcriptional start site upstream of liaG (data not shown). These results suggest that the _lia_-locus is expressed from the LiaR-dependent promoter upstream of liaI (Fig. 1b). Since LiaRS is required for antibiotic induction, we conclude that it must be expressed even in uninduced cells. Since the number of sensors needed for signal transduction is very low (∼100 molecules of EnvZ per E. coli cell [5]), the lack of a detectable long transcript in uninduced mid-log phase cultures for the lia locus is not too surprising.
Identification of the minimal promoter for the liaI operon.
We mapped the transcriptional start site upstream of liaI by using primer extension analysis (Fig. 2). Transcription initiates at an A residue 25 nucleotides upstream of the liaI start codon. The liaI promoter shows a well-conserved −10 region (TATAAG) and a weakly conserved −35 region (TTCGTT). Response regulators that act as transcriptional activators normally interact with DNA via short, 6- to 10-nucleotide binding sites in close proximity upstream of the promoter. These binding sites can either occur singly or as repeated elements with a spacing of about 8 to 10 nucleotides (16), allowing the response regulator dimer to bind in two neighboring major grooves of the DNA. In the 60 nucleotides between the −35 region and the putative terminator of yvqJ, we identified a 6-nucleotide direct repeat (TCCGGT) with a 12-nucleotide spacing as a candidate site for DNA binding by LiaR.
To analyze P_liaI_ and this putative binding site further, we used a lacZ_-based promoter dissection approach. Fragments of P_liaI ending at position +97 (inside of liaI) and extending to different lengths upstream of the transcriptional start site (Fig. 2c) were used to generate reporter fusions integrated at SPβ. All constructs had very low _lacZ_-expression in the uninduced state (Fig. 2d). The core promoter alone (−58) was not bacitracin inducible and showed only marginally higher expression than the negative control (extending to −29). However, the presence of 16 additional nucleotides (−74), including one copy of the TCCGGT sequence, allowed full induction upon the addition of bacitracin, as also seen with the two larger fragments (extending to −83 and −193). The minimal bacitracin-inducible promoter as defined by these studies is underlined in Fig. 2b. As expected, bacitracin-dependent induction was completely lost in the _liaR_-deletion mutant HB0953 (data not shown).
P_liaI_ is induced by antibiotics that interfere with the lipid II cycle.
P_liaI_ is specifically induced by vancomycin and bacitracin, two antibiotics that interfere with the cycling of lipid II (22). To gain a better understanding of the sensing process, we expanded our analysis to define the range of inducers. Disk diffusion assays were used to screen a variety of antibiotics for the ability to induce β-galactosidase expression from a chromosomal pMUTIN-based liaI′-lacZ fusion in strain BSF2470 (Table 2). The use of pMUTIN leads to an insertion-duplication disruption of a targeted gene, while at the same time putting lacZ under the control of the promoter region of the disrupted gene (33). In addition to the previously identified inducers (bacitracin and vancomycin), the cell wall antibiotics tunicamycin, ramoplanin, nisin, and (weakly) fosfomycin gave a blue ring at the edge of the zone of inhibition, indicating an induction of liaI expression at sublethal concentrations. Negative results were observed for other antibiotics including chloramphenicol, kanamycin, rifampin, spectinomycin, streptomycin, and tetracycline (data not shown).
TABLE 2.
Inducers of liaI expression
| Inducera | Disk diffusion assayb | Concn (μg/ml)c | Increase (_n_-fold) in induction ± SDd | |
|---|---|---|---|---|
| BFS2470 | HB0950 | |||
| Cell wall antibiotics | ||||
| Ampicillin | − | |||
| Bacitracin | + | 10 | 498 ± 92 | 192 ± 16 |
| Cephalosporin | − | |||
| D-cycloserin | − | |||
| Fosfomycin | (+) | 10 | 1.7 ± 0.6 | |
| Moenomycin | − | |||
| Nisin | + | 10 | 423 ± 16 | 156 ± 8.3 |
| Penicillin G | − | |||
| Polymyxin B | − | |||
| Ramoplanin | + | 5 | 422 ± 60 | 144 ± 12 |
| Tunicamycin | + | 50 | 3.1 ± 1.1 | |
| Vancomycin | + | 2 | 63 ± 2.7 | 36 ± 0.2 |
| Organic solvents | ||||
| Diphenyl ether | NT | 10 | 10.8 ± 3.4 | |
| _n_-Hexane | NT | 10 | 7.8 ± 1.2 | |
| Cyclooctane | NT | 10 | 11.6 ± 4.9 | |
| Surfactantse | ||||
| BDMDDA-Br | − | |||
| BDMHDA-Cl | + | 10 | 8.2 ± 1.9 | |
| HDTMA-Br | − |
In quantitative β-galactosidase assays, performed on cells growing in liquid cultures, both nisin and ramoplanin were strong inducers (420-fold increase in induction), as were bacitracin (500-fold increase) and vancomycin (63-fold increase), consistent with earlier studies (22) (Table 2). Equivalent levels of induction were also observed with the P_liaI-74-cat-lacZ_ fragment (in HB0950), albeit with smaller increases (Table 2). While the mode of action is different for these antibiotics, all four interfere with the lipid II cycle that is essential for the biosynthesis of cell envelope polymers (20). Tunicamycin and fosfomycin were weak inducers in the liquid assays (Table 2).
To analyze the specificity of the P_liaI_-lacZ system further, we tested several detergents and surfactants. Detergents like sodium dodecyl sulfate (SDS) and Triton X-100 compromise the integrity of the cytoplasmic membrane. Surfactants serve as emulsifiers by adsorbing and altering the conditions at interfaces due to their amphiphatic nature (24). Detergents did not induce liaI expression in the zone of the inhibition assay and were not further analyzed. Of the three surfactants chosen for our analysis, only benzyldimethylhexadodecylammonium chloride (BDMHDA-Cl) moderately induced liaI expression (Table 2). Organic solvents such as diphenyl ether, _n_-hexane, and cyclooctane also led to an 8- to 12-fold increase in induction. Organic solvents are toxic because they nonspecifically accumulate in and disrupt the cytoplasmic membrane (29).
The liaI and liaH genes code for a small, presumably membrane-bound, protein and a homolog of E. coli pspA (phage-shock protein A), respectively. PspA is involved in the maintenance of cell membrane integrity and proton motive force and is induced by uncouplers such as carbonyl cyanide m-chlorophenylhydrazone and dinitrophenol (36). Although we included these two uncouplers in our screening, neither elevated the level of liaIH expression (data not shown). Lysozyme, which breaks the glycosidic bounds between _N_-acetylmuramic acid and _N_-acetylglucosamine in peptidoglycan, was also ineffective as an inducer (data not shown).
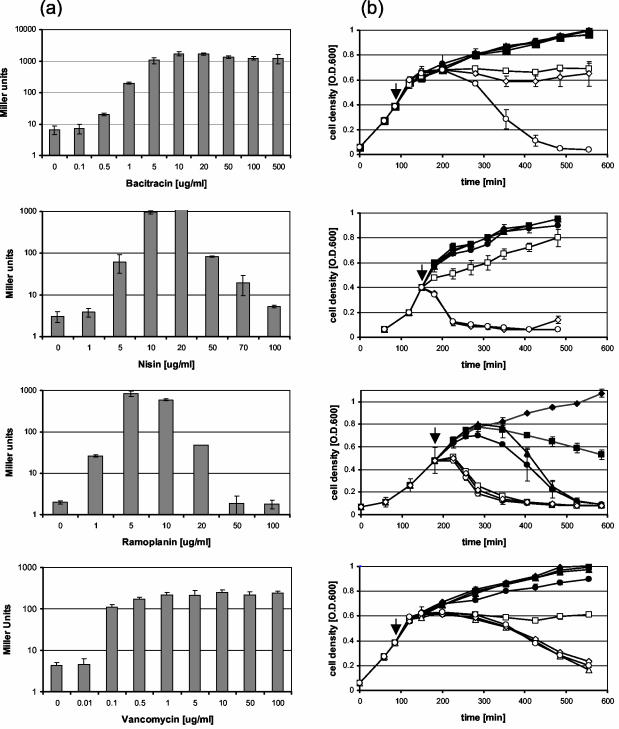
Concentration-dependent induction and killing experiments were performed for bacitracin, nisin, ramoplanin, and vancomycin. In all four cases the induction of liaI occurred in a concentration-dependent manner (Fig. 3a), reaching a maximum at an antibiotic concentration of about 10 μg/ml (1 μg/ml for vancomycin). While the level of lacZ expression remained elevated at higher concentrations of bacitracin and vancomycin, a strong decrease down to uninduced levels was observed for nisin and ramoplanin. This finding correlates well with the bactericidal effects of these antibiotics as inferred from the dramatic decrease in optical density: there is little effect from bacitracin and vancomycin on cell growth during the first 30 to 60 min after the addition of the antibiotics even at high concentrations (Fig. 3b). In contrast, high concentrations of nisin and ramoplanin led to rapid cell lysis, which likely interfered with the induction of β-galactosidase (Fig. 3b).
FIG. 3.
Concentration-dependent induction of liaI and optical density changes for B. subtilis cultures treated with bacitracin, nisin, ramoplanin, and vancomycin. (a) β-Galactosidase assays were performed as described in the legend of Fig. 2. by using strain BFS2470 and different concentrations of the four antibiotics as indicated. Miller units are plotted on a logarithmic scale for clarity. (b) Concentration-dependent killing of B. subtilis by the four antibiotics from the same culture as described in panel a. The times of antibiotic addition are indicated by arrows. The concentrations of bacitracin (□, 50; ◊, 100; and ○, 500 μg/ml), nisin (□, 50; ◊, 70; and ○, 100 μg/ml), ramoplanin (□, 20; ◊, 50; and ○, 100 μg/ml), and vancomycin (□, 5; ⋄, 10; ○, 50; and ▵, 100 μg/ml) that affect the induction of P_liaI_ according to the β-galactosidase assay are indicated.
DISCUSSION
The emergence of antibiotic-resistant pathogens only a few decades after the first clinical use of antibiotics has revived the search for novel antibacterial compounds. The two most common strategies for the identification of new lead compounds involve either target-based approaches or whole-cell screening for bactericidal compounds (28). Target-based approaches focus on essential, conserved genes that are limited to the bacterial world (11). Whole-cell approaches often begin with the high-throughput screening of high-complexity natural product or synthetic chemical libraries. In each case, active compounds must be carefully analyzed to infer the mechanisms of action and to assess toxicity. The prerequisites for efficient screening are unchanged and challenging: assays need to be robust, specific, and sensitive (35). Transcriptional profiling has emerged as a useful tool to identify promoters that respond to specific antibiotic stresses and thereby aid in the functional classification of active compounds (13).
The liaIH (formerly yvqIH) genes are strongly induced in the presence of cell wall antibiotics such as vancomycin and bacitracin (22). This induction depends on the activity of the LiaRS two-component system (formerly YvqCE) (22). In this report we analyzed the regulation and expression of the liaIHGFSR operon in detail. This operon is transcribed from a single LiaR-dependent, σA-type promoter upstream of liaI, and a minimal promoter extending to position −74 relative to the transcription start point was fully inducible (Fig. 2a to c). A stem-loop sequence between liaH and liaG is correlated with the appearance of two different transcripts: a short 1.1-kb large RNA covering liaIH and a long transcript, including the whole locus (Fig. 1). The higher abundance of liaIH relative to liaGFSR mRNA may be due to termination at this stem-loop structure or to stabilization of the smaller transcript against degradation. No terminator structure could be detected downstream of liaR, and its 3′ end is only separated by eight nucleotides from the 3′ end of the convergently transcribed gerAC gene.
The P_liaI_ system described here has several features that recommend it as a useful tool for the detection and classification of compounds eliciting cell envelope stress. Like the VanRS system (32), which is already used in high-throughput screening, the minimal P_liaI_ promoter can be used to construct quantifiable reporter fusions such as the lacZ fusion reported here. The P_liaI_-lacZ reporter fusion has very low basal levels of expression, responds in a concentration-dependent manner to antibiotics, and reaches high levels of induction (Table 2 and Fig. 3a). The dynamic range of P_liaI_ is very high (up to 400-fold increased specific induction compared to a 5- to 10-fold increase reported for VanRS-based reporters in B. subtilis [32]), which suggests that LiaRS-based systems may allow for the sensitive detection of lipid II cycle-interfering antibiotics (Table 2) and discrimination against compounds that nonspecifically perturb the cytoplasmic membrane. In contrast, the VanRS system (32) is induced by a broader range of cell wall antibiotics (S. Donadio, personal communication). The P_liaI_ reporter assay could be further simplified by using the lux reporter (luciferase) instead of lacZ. The lux operon of Photorhabdus luminescens has been adapted and optimized for use in gram-positive bacteria (12). This system does not need an exogenous substrate and produces high levels of light at 37°C, the optimal growth temperature for B. subtilis. It has already been used successfully in Staphylococcus aureus, Streptococcus pneumoniae, and Listeria monocytogenes (12).
We tested a wide range of antibiotics and cell envelope-damaging substances for their ability to induce liaIH gene expression. Based on the nature and strength of the transcriptional response, three groups of inducers can be defined. The strongest inducers (bacitracin, nisin, ramoplanin, and vancomycin) result in a 63- to 400-fold increase in induction of liaIH expression. Indeed, upon induction LiaH is sufficiently abundant to be easily detected in two-dimensional gel electrophoresis and has been assigned as a signature protein for bacitracin-treated cells (3). All four of these antibiotics directly interfere with the essential lipid carrier of cell envelope biosynthesis (see below). The second group consists of weak inducers including some organic solvents and one surfactant (BDMHDA-Cl). Organic solvents accumulate in the hydrophobic interior of the cytoplasmic membrane and may indirectly affect the lipid II cycle. Alternatively, they could interfere with the proposed intramembrane-sensing domain of LiaS (see below). Two of the three surfactants and the two detergents (SDS and Triton X-100) that were tested failed to induce liaIH expression, at least in the plate-based assay. All of these compounds are strong inducers for the VanRS two-component system (32), for which BDMHDA-Cl induces at a concentration 50-fold lower than SDS (S. Donadio, personal communication). We therefore conclude that some as yet unknown properties of BDMHDA-Cl account for the 10-fold increase in induction of liaIH expression. The third group includes some cell wall-active antibiotics that appear to elicit transcriptional induction on plates but for which very weak activity is seen in liquid cultures (Table 2).
Undecaprenol serves as the lipid carrier for cell envelope building blocks which are covalently linked through a phosphoryl group on the cytoplasmic side of the membrane. Once transferred to the extracytoplasmic side of the membrane, lipid II donates the disaccharide-pentapeptide units into the growing peptidoglycan network. Undecaprenol is an essential molecule, and its recycling is a rate-limiting step in cell envelope growth (10). The lipid II cycle is, therefore, an ideal target for antibiotic action. The cell's ability to monitor the state of this crucial process can be viewed as a regulatory device to ensure normal growth and to rapidly detect and respond to perturbations that might result in cell death. The ability of LiaS to sense perturbations of the lipid II cycle is in good agreement with our hypothesis that LiaS, a member of the proposed subfamily of intramembrane-sensing histidine kinases, detects its signal at the membrane interface (22). The LiaRS two-component system may thereby sense a whole group of harmful substances through their effect on a common cellular process.
The importance of this antibiotic-sensing two-component system is stressed by the identification of homologous systems in other gram-positive bacteria. One homologous two-component system, VraSR from S. aureus, is induced by bacitracin and vancomycin and also by other cell wall antibiotics, such as d-cycloserine, imipenem, and ceftizoxime (19). It controls a large regulon, with some target genes having predicted functions linked to cell wall metabolism. Consequently, a VraSR deletion strain shows a significant increase in sensitivity to the antibiotics it senses (19). In contrast, deletion of LiaR target genes does not result in antibiotic sensitivity (22). We suggest that LiaRS and its homologs represent a group of two-component systems that sense damage to the cell envelope. The differences in the spectrum of cell wall antibiotics sensed by these two histidine kinases may be indicative of an evolutionary diversification or specialization within this group of cell wall stress sensors. We note that VraS from S. aureus and Staphylococcus epidermidis shows the maximum phylogenetic distance to LiaS inside the LiaS subgroup of intramembrane-sensing histidine kinases. This difference is especially pronounced in the transmembrane-sensing domain of the proteins (data not shown).
Our investigations so far have only focused on the lia locus itself. A comprehensive microarray analysis of two-component systems in B. subtilis mutants overexpressing the cognate response regulators identified additional putative target genes for LiaRS (17). While we were not able to reproduce all of these authors' findings, there are indications that LiaRS controls more than one target locus and is part of a regulatory cascade (22). An analysis of its regulon is currently under way and will serve as a starting point to identify LiaR-binding sites. The tight regulation and very strong induction of liaIH suggest an important biological function for its gene products, which will be a focus for further studies.
Acknowledgments
We thank S. Donadio (Vicuron) for the generous gift of purified ramoplanin, P. Welzel for the purified moenomycin, and Z. Pragai for the gift of strains BFS2469 and BFS2470. We also thank A Goldborough (RNAworks) for information on quenching of signals by rRNA and S. Donadio for helpful discussions and critical reading of the manuscript.
This work was supported by NIH grant GM-47446 (to J.D.H.).
REFERENCES
- 1.Ausubel, F. M., R. Brent, K. R. E., D. D. Moore, J. G. Seidman, J. A. Smith, and S. K. (ed.). 2003. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Azevedo, E. C., E. M. Rios, K. Fukushima, and G. M. Campos-Takaki. 1993. Bacitracin production by a new strain of Bacillus subtilis. Extraction, purification, and characterization. Appl. Biochem. Biotechnol. 42**:**1-7. [DOI] [PubMed] [Google Scholar]
- 3.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47**:**948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417**:**141-147. [DOI] [PubMed] [Google Scholar]
- 5.Cai, S. J., and M. Inouye. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277**:**24155-24161. [DOI] [PubMed] [Google Scholar]
- 6.Cain, B. D., P. J. Norton, W. Eubanks, H. S. Nick, and C. M. Allen. 1993. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J. Bacteriol. 175**:**3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45**:**1267-1276. [DOI] [PubMed] [Google Scholar]
- 8.Chalker, A. F., K. A. Ingraham, R. D. Lunsford, A. P. Bryant, J. Bryant, N. G. Wallis, J. P. Broskey, S. C. Pearson, and D. J. Holmes. 2000. The bacA gene, which determines bacitracin susceptibility in Streptococcus pneumoniae and Staphylococcus aureus, is also required for virulence. Microbiology 146**:**1547-1553. [DOI] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. VanderHorn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for bacillus. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 10.Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Leeuwenhoek 76**:**159-184. [PubMed] [Google Scholar]
- 11.Donadio, S., L. Carrano, L. Brandi, S. Serina, A. Soffientini, E. Raimondi, N. Montanini, M. Sosio, and C. O. Gualerzi. 2002. Targets and assays for discovering novel antibacterial agents. J. Biotechnol. 99**:**175-185. [DOI] [PubMed] [Google Scholar]
- 12.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68**:**3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, D. 2003. Exploiting genomics, genetics and chemistry to combat antibiotic resistance. Nat. Rev. Genet. 4**:**432-441. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21**:**526-531. [DOI] [PubMed] [Google Scholar]
- 15.Ishihara, H., M. Takoh, R. Nishibayashi, and A. Sato. 2002. Distribution and variation of bacitracin synthetase gene sequences in laboratory stock strains of Bacillus licheniformis. Curr. Microbiol. 45**:**18-23. [DOI] [PubMed] [Google Scholar]
- 16.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5**:**135-141. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183**:**7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390**:**249-256. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49**:**807-821. [DOI] [PubMed] [Google Scholar]
- 20.Lazar, K., and S. Walker. 2002. Substrate analogues to study cell-wall biosynthesis and its inhibition. Curr. Opin. Chem. Biol. 6**:**786-793. [DOI] [PubMed] [Google Scholar]
- 21.Marshall, C. G., I. A. Lessard, I. Park, and G. D. Wright. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42**:**2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50**:**1591-1604. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Neu, T. R. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60**:**151-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumüller, A. M., D. Konz, and M. A. Marahiel. 2001. The two-component regulatory system BacRS is associated with bacitracin “self-resistance” of Bacillus licheniformis ATCC 10716. Eur. J. Biochem. 268**:**3180-3189. [DOI] [PubMed] [Google Scholar]
- 26.Ohki, R., Giyanto, K. Tateno, W. Masuyama, S. Moriya, K. Kobayashi, and N. Ogasawara. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49**:**1135-1144. [DOI] [PubMed] [Google Scholar]
- 27.Pollock, T. J., L. Thorne, M. Yamazaki, M. J. Mikolajczak, and R. W. Armentrout. 1994. Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides. J. Bacteriol. 176**:**6229-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosamond, J., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287**:**1973-1976. [DOI] [PubMed] [Google Scholar]
- 29.Sardessai, Y., and S. Bhosle. 2002. Tolerance of bacteria to organic solvents. Res. Microbiol. 153**:**263-268. [DOI] [PubMed] [Google Scholar]
- 30.Slack, F. J., J. P. Mueller, and A. L. Sonenshein. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J. Bacteriol. 175**:**4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46**:**3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulijasz, A. T., A. Grenader, and B. Weisblum. 1996. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J. Bacteriol. 178**:**6305-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144**:**3097-3104. [DOI] [PubMed] [Google Scholar]
- 34.Walsh, C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, D.C.
- 35.Walters, W. P., and M. Namchuk. 2003. Designing screens: how to make your hits a hit. Nat. Rev. Drug Discov. 2**:**259-266. [DOI] [PubMed] [Google Scholar]
- 36.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. USA 91**:**2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169**:**2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]