Mitochondrial RNA granules: Compartmentalizing mitochondrial gene expression (original) (raw)
Abstract
In mitochondria, DNA replication, gene expression, and RNA degradation machineries coexist within a common nondelimited space, raising the question of how functional compartmentalization of gene expression is achieved. Here, we discuss the recently characterized “mitochondrial RNA granules,” mitochondrial subdomains with an emerging role in the regulation of gene expression.
Introduction
RNA granules represent a diverse range of ribonucleoparticles that lack membrane delineation and are often visible by light microscopy. They have been observed in the nucleus and cytosol of somatic cells, neurons, and germ cells and are implicated in a variety of different functions. Nuclear RNA granule structures include the Cajal bodies, speckles, and nucleoli, sites of snRNP biogenesis and maturation, RNA splicing, and ribosomal RNA (rRNA) transcription, respectively. Cytosolic stress granules (SGs) and processing bodies (PBs) contain polyadenylated mRNAs and RNA-related enzymes and are induced under conditions in which translation is inhibited, though PBs also exist under normal conditions. SGs are rich in preinitiation complexes and contain mRNAs that can reenter polyribosomes, whereas PBs contain decapping and deadenylation factors, the RISC complex, and the nonsense-mediated decay machinery and are thought to be mostly involved in mRNA degradation (Kedersha et al., 2005). RNA granules have also been observed within organelles. In chloroplasts, SGs are induced by a variety of stimuli and like cytosolic SGs are rich in preinitiation complexes (Uniacke and Zerges, 2008). RNA granules also exist in mitochondria but appear to play a different role, as they are not induced by stress, and processes such as mRNA splicing and decapping that occur in other types of RNA granules are not relevant to gene expression in mammalian mitochondria.
The human mitochondrial genome is a highly compact circular molecule of ∼16.5 kb that encodes 37 genes, all of which are required for oxidative phosphorylation, including 13 highly hydrophobic membrane components of the electron transport chain, two rRNAs, and 22 tRNAs that provide the specialized translational machinery required to express these proteins. Both strands of the mitochondrial DNA (mtDNA) are almost completely transcribed from a single promoter region, giving rise to polycistronic precursors that are then processed into mono- or bi-cistronic mRNAs, in most cases by excision of flanking tRNAs interspersed throughout the genome. As with nuclear RNA, mitochondrial RNA is further modified during or after transcription by polyadenylation, methylation, aminoacylation, or pseudouridinylation. However, in contrast to nuclear gene expression, which occurs in a well-defined step-wise manner consisting of capping, splicing, polyadenylation, and nuclear export, very little is known about the spatiotemporal organization of gene expression in mitochondria. Currently it is unclear how immature polycistronic RNAs are prevented from interacting with the translation machinery, where and how posttranscriptional modification or mitoribosome assembly occur, or what governs RNA stability. In the absence of a clear membrane delineation within the mitochondrial matrix, mitochondria appear to have developed specialized RNA-rich domains, in which these complex processes are compartmentalized and which likely represent key sites in the organization of mitochondrial gene expression. The composition and function of these mitochondrial RNA granules (MRGs) will be the focus of this paper.
MRGs
Iborra et al. (2004) first described newly made RNAs as punctate mitochondrial structures, which could be labeled with the uridine analogue 5-bromouridine (BrU). We and others later reported a specific colocalization of MRGs with GRSF1, a protein involved in mitochondrial RNA processing and translation (Antonicka et al., 2013; Jourdain et al., 2013). GRSF1 was found to interact with the mitochondrial RNase P complex, and accordingly GRSF1 depletion reduced mature RNAs, with an accumulation of RNA precursors (Jourdain et al., 2013). Not surprisingly, RNase P subunits were also enriched in MRGs, further implicating MRGs as centers of RNA processing. BrU pulse-chase experiments supported this notion, as removal of either GRSF1 or RNase P prevented the transit of newly made RNA through MRGs (Jourdain et al., 2013). Other RNA processing and maturation factors have now been shown to be present within MRGs, such as members of the FASTK family, the mitochondrial poly(A)-polymerase mtPAP, methyltransferases, RNA helicases, and the hSuv3p–PNPase complex or degradosome, which plays a central role in mitochondrial RNA degradation (Wang and Bogenhagen, 2006; Vilardo et al., 2012; Lee et al., 2013; Borowski et al., 2013; Jourdain et al., 2013, 2015; Bogenhagen et al., 2014; Wilson et al., 2014; Antonicka and Shoubridge, 2015; Tu and Barrientos, 2015; Table 1). Although some of these components are found in almost all MRGs, others are only found in a subpopulation of granules. Thus, MRGs are probably dynamic structures, which provide a platform for the spatiotemporal regulation of the numerous processes required for mitochondrial gene expression, including RNA processing, maturation, ribosome assembly, and translation initiation (Fig. 1). Their protein and RNA composition may change continuously depending on the progress of RNA processing and mitoribosome assembly.
Table 1. Composition of the MRGs.
| Gene name | Function | Conserved domains (PFAM) | Reference |
|---|---|---|---|
| DDX28 | Mitoribosome assembly | DEAD; Helicase_C | Antonicka and Shoubridge, 2015; Tu and Barrientos, 2015 |
| DHX30 | Mitoribosome assembly | DEAD; HA2; Helicase_C; OB_NTP_bind | Wang and Bogenhagen, 2006; Antonicka and Shoubridge, 2015 |
| FASTK | RNA processing | FAST_1; FAST_2; RAP | Jourdain et al., 2015 |
| FASTKD2 | RNA processing; mitoribosome assembly | FAST_1; FAST_2; RAP | Antonicka and Shoubridge, 2015; Jourdain et al., 2015 |
| FASTKD5 | RNA processing; mitoribosome assembly | FAST_1; FAST_2; RAP | Antonicka and Shoubridge, 2015 |
| FTSJ2a (MRM2) | rRNA methylation | FtsJ | Lee et al., 2013 |
| GRSF1 | RNA processing; mitoribosome assembly | RRM6 (3x) | Antonicka et al., 2013; Jourdain et al., 2013 |
| HSD17B10 (MRPP2) | RNAse P - 5′ tRNA processing | adh_short | Jourdain et al., 2013; Bogenhagen et al., 2014 |
| KIAA0391 (MRPP3) | RNAse P - 5′ tRNA processing | PRORP | Jourdain et al., 2013; Bogenhagen et al., 2014 |
| MRM1a | rRNA methylation | SpoU_methylase; SpoU_sub_bind | Lee et al., 2013 |
| MTPAP (PAPD1) | Poly(A) polymerase | PAP_assoc | Wilson et al., 2014 |
| PNPT1 (PNPase) | RNA degradation; RNA processing | KH_1; PNPase; RNase_PH; RNase_PH_C; S1 | Borowski et al., 2013 |
| RNMTL1a (MRM3) | rRNA methylation | SpoU_methylase; SpoU_sub_bind | Lee et al., 2013 |
| SUPV3L1 (hSuv3p) | RNA degradation; RNA processing | Helicase_C; SUV3_C | Borowski et al., 2013 |
| TRMT10C (MRPP1) | RNAse P - 5′ tRNA processing; tRNA methyltransferase | tRNA_m1G_MT | Jourdain et al., 2013; Bogenhagen et al., 2014 |
Figure 1.
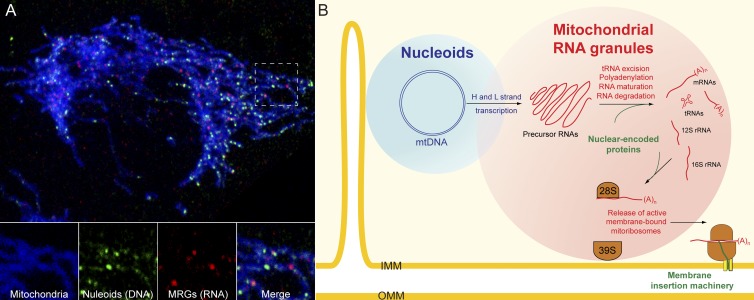
MRGs and nucleoids. (A) Confocal image of a HeLa cell expressing a blue fluorescent protein in mitochondria (mitoBFP), incubated with 2.5 mM BrU for 1 h and labeled with antibodies to mitochondrial SSB (nucleoid marker, green) or BrU (MRG marker, red; Jourdain et al., 2013). (B) Model of the composition and function of MRGs and their interaction with the nucleoids.
All work on MRGs to date has been performed using human or mouse cells, and thus it is legitimate to ask whether they also exist in other species. From the original genome of the ancestral endosymbiotic bacteria, the structural organization and content of the mitochondrial genome has undergone major changes during the course of evolution. In certain species such as green plants or Saccharomyces cerevisae, large mitochondrial genomes encode multiple promoters and introns, whereas in other experimentally tractable organisms such as Schizosaccharomyces pombe, Caenorhabditis elegans, or Drosophila melanogaster, mtDNA is shorter and, as in humans, appears to be transcribed into long polycistronic precursors that are then processed according to the so-called “tRNA punctuation” model. In D. melanogaster, proteins related to known MRG components have been found, including members of the FASTK family, hnRNPs, and mitochondrially-targeted helicases. However, it is currently unknown whether MRGs exist in nonmammalian species, although their existence seems likely in those species with similar mtDNA architecture, mode of DNA replication, and pattern of transcription.
Dynamics of MRGs
Iborra et al. (2004) initially classified the BrU foci into two classes: those detectable after a short pulse of BrU, which colocalize with the mitochondrial nucleoids, and those detectable after a longer pulse of BrU, which are stable over time and do not colocalize with the mitochondrial genome. About 10–20% of MRGs belong to the former class and are intimately associated with nucleoids (Fig. 1 A; Iborra et al., 2004; Jourdain et al., 2013). Moreover, because this subpopulation of MRGs also contains the most rapidly labeling RNA (Iborra et al., 2004), they are most likely associated with transcriptionally active nucleoids, allowing efficient transfer of the primary transcripts. We envisage that within each MRG two distinct pathways occur in parallel (Fig. 1 B). The first involves processing and maturation of the genome-length polycistronic heavy and light strand transcripts to generate the mature mRNAs that encode the subunits of the electron transport chain, requiring tRNA excision, elimination of the noncoding “mirror” RNAs, and selective polyadenylation and posttranscriptional modifications. The second involves excision and maturation of the 12S and 16S rRNAs from the heavy strand transcript and association with nuclear-encoded MPRS and MPRL proteins, resulting in assembly of the 28S and 39S mitoribosomal subunits, respectively. In their initial work on BrU foci, Iborra et al. (2004) reported that BrU-positive foci (i.e., MRGs) are distributed in the vicinity of active cytosolic ribosomes on the mitochondrial surface and the protein import machinery, which would allow for coordinated assembly of mitoribosomes using both nuclear and mitochondrial gene products. Based on this and recent structural analyses of the mitoribosome (Greber et al., 2014; Amunts et al., 2015), the 39S subunit likely provides a membrane anchorage function, which allows release of fully assembled, active mitoribosomes from the MRGs and presentation of the nascent hydrophobic peptides to the associated membrane insertion machinery of the inner mitochondrial membrane (Fig. 1 B). According to this scheme, the compact MRG structure would provide the proximity and high concentration of reactants, together with the organizational framework and access to nuclear encoded gene products, to ensure efficient RNA processing, maturation, and mitoribosome assembly crucial for mitochondrial gene expression.
As MRGs exist independently from the nucleoids, this raises the interesting question of their dynamics and inheritance during cycles of mitochondrial fusion and fission. Nucleoids are found in close proximity to sites of fission, presumably facilitating the segregation of mtDNA throughout the mitochondrial network. During the normal cycle of mitochondrial fusion/fission, and especially during acute perturbations of mitochondrial dynamics, nucleoids may become unevenly distributed, resulting in mitochondria that lack mtDNA. Such mitochondria either re-fuse with mtDNA-containing mitochondria and reacquire the genome, or lose membrane potential and are degraded by mitophagy (Twig et al., 2008). Though MRG dynamics have not yet been investigated by live-cell imaging, mitochondria lacking nucleoids could use nonnucleoid-associated MRGs as a stock of RNA able to sustain translation temporarily and thus extend the survival time of isolated mitochondria until fusion can occur. A similar RNA storage function could be important during mtDNA replication or when transcriptional activity is otherwise decreased. Thus, MRGs could play a buffering role in mitochondrial gene expression, maintaining RNA availability during transient decreases in mitochondrial transcription and/or nucleoid loss.
Structure and formation of MRGs
How is the structural integrity of MRGs maintained? In the more highly studied cytoplasmic granules, proteins containing prion-like domains, Sm folds, or other motifs have been found to be essential in maintaining structural identity. In each case, deletion of the aggregation domain leads to disassembly of the granule. By analogy with the cytoplasmic granules, one or more of the protein components of MRGs might contain a similar aggregation domain. Because the only means currently known to disassemble MRGs is removal of RNA (Iborra et al., 2004; Jourdain et al., 2013), the putative scaffolding protein might contain an RNA-binding motif, in addition to other protein-interacting domains and functions. Intriguing data from Kato et al. (2012) have shown that addition of a biotinylated isoxazole compound to complex cellular extracts selectively precipitates a subset of aggregation-prone, RNA-binding proteins highly enriched in low-complexity (LC) regions, which the authors argue could provide the basis for differential assembly of various RNA granules. Searches for LC domains within known MRG proteins have so far failed to reveal clear candidates; thus, if the mode of MRG assembly resembles that of cytosolic RNA granules, the LC domain–containing proteins required to maintain the MRG architecture have yet to be identified.
Another possible mechanism of MRG assembly is suggested from a recent study of nucleoids in which assembly was shown to occur via a multistep mechanism relying on the cross-strand binding of the mitochondrial transcription factor A (TFAM) to mtDNA (Kukat et al., 2015). TFAM binding induces loop formation in mtDNA and increasing concentrations of TFAM drive binding to multiple and sometimes distant sites, ultimately leading to compaction of the mtDNA into nucleoid-like structures. A similar mechanism could also take place in MRGs, with the putative scaffolding protein binding newly made RNA, leading to formation of foci. Site-specific binding to unprocessed RNA junctions or noncoding RNA could then ensure the controlled release of mature mRNAs to the mitoribosomes and thence to the mitochondrial matrix.
Identification of such factors that coordinate and maintain MRG architecture would be an important step forward in investigating the function of these RNA granules.
Concluding remarks
MRGs are emerging as discrete subcompartments within the mitochondrion, which partition the mitochondrial matrix and enable the posttranscriptional events necessary for mitochondrial gene expression. We propose that the MRGs are assembled around the newly synthesized primary transcripts in close proximity to active nucleoids with which they are transiently associated and that subsequent events in mitochondrial gene expression take place as the MRGs become detached and take up independent locations within the inner mitochondrial matrix. We view MRGs as dynamic structures whose composition changes as the primary transcript is posttranscriptionally modified, processed to mature RNAs, and assembled into mitoribosome complexes for oriented translation and insertion of the mitochondrially encoded proteins into the inner mitochondrial membrane. The changing composition of MRGs, their architecture, and their patterns of segregation during the cycles of mitochondrial fission and fusion are intriguing topics for the future that will give us greater insight into MRG function.
Acknowledgments
The authors thank Dr. Tom Bender for help with artwork and Sofia Zaganelli for help with the fluorescent imagery.
This work was supported by the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Förschung (310030B_160257/1), iGE3, and the State of Geneva.
The authors declare no competing financial interests.
References
- Amunts A., Brown A., Toots J., Scheres S.H., and Ramakrishnan V.. 2015. The structure of the human mitochondrial ribosome. Science. 348:95–98. 10.1126/science.aaa1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H., and Shoubridge E.A.. 2015. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Reports. [DOI] [PubMed] [Google Scholar]
- Antonicka H., Sasarman F., Nishimura T., Paupe V., and Shoubridge E.A.. 2013. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 17:386–398. 10.1016/j.cmet.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F., Martin D.W., and Koller A.. 2014. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 19:618–629. 10.1016/j.cmet.2014.03.013 [DOI] [PubMed] [Google Scholar]
- Borowski L.S., Dziembowski A., Hejnowicz M.S., Stepien P.P., and Szczesny R.J.. 2013. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 41:1223–1240. 10.1093/nar/gks1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B.J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N., Aebersold R., and Ban N.. 2014. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 515:283–286. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Kimura H., and Cook P.R.. 2004. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2:9 10.1186/1741-7007-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., and Martinou J.C.. 2013. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 17:399–410. 10.1016/j.cmet.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain A.A., Koppen M., Rodley C.D., Maundrell K., Gueguen N., Reynier P., Guaras A.M., Enriquez J.A., Anderson P., Simarro M., and Martinou J.C.. 2015. A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Reports. 10:1110–1121. 10.1016/j.celrep.2015.01.063 [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., and Anderson P.. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. (published erratum appears in J. Cell Biol. 2005. 170:847) 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C., Davies K.M., Wurm C.A., Spåhr H., Bonekamp N.A., Kühl I., Joos F., Polosa P.L., Park C.B., Posse V., et al. 2015. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA. 112:11288–11293. 10.1073/pnas.1512131112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.W., Okot-Kotber C., LaComb J.F., and Bogenhagen D.F.. 2013. Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are positioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J. Biol. Chem. 288:31386–31399. 10.1074/jbc.M113.515692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y.T., and Barrientos A.. 2015. The human mitochondrial DEAD-Box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., and Zerges W.. 2008. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 182:641–646. 10.1083/jcb.200805125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., and Rossmanith W.. 2012. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40:11583–11593. 10.1093/nar/gks910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., and Bogenhagen D.F.. 2006. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 281:25791–25802. 10.1074/jbc.M604501200 [DOI] [PubMed] [Google Scholar]
- Wilson W.C., Hornig-Do H.T., Bruni F., Chang J.H., Jourdain A.A., Martinou J.C., Falkenberg M., Spåhr H., Larsson N.G., Lewis R.J., et al. 2014. A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression. Hum. Mol. Genet. 23:6345–6355. 10.1093/hmg/ddu352 [DOI] [PMC free article] [PubMed] [Google Scholar]