Sigma-1 Receptor as a Pluripotent Modulator in the Living System (original) (raw)
. Author manuscript; available in PMC: 2017 Apr 1.
Published in final edited form as: Trends Pharmacol Sci. 2016 Feb 9;37(4):262–278. doi: 10.1016/j.tips.2016.01.003
Abstract
The sigma-1 receptor (Sig-1R) is an endoplasmic reticulum (ER) protein resides specifically at the interface between ER and mitochondria, called the MAM, where the Sig-1R is recently reported to be involved in certain CNS diseases. In addition to being able to translocate to the plasma membrane to interact with ion channels and other receptors, the Sig-1R is found to exist at the nuclear envelope where it recruits chromatin-remodeling factors to affect the transcription of genes. As well, thorough experimental and bioinformatic means, Sig-1Rs are reported to interact with other membranous or soluble proteins at other loci, including the cytosol. We propose that the Sig-1R is a pluripotent modulator with resultant multiple functional manifestations in the living system.
Keywords: Sigma-1 receptor, Pluripotent modulator, Diseases
The Sigma-1 Receptor: Brief History and Current Status
Martin et al. [1] hypothesized the existence of multiple opioid receptors to mediate different pharmacological effects of morphine and it various structural analogs. These receptors and their prototypic ligands and pharmacological effects are respectively: mu opioid receptor (MOR) for morphine-induced analgesia, kappa opioid receptor for ketocyclazocine-induced dysphoria, and sigma “opioid” receptor for SKF-10047 (N-normetazocine)-induced psychotomesis. Influenced by the multiple opioid receptor hypothesis, Su [2] demonstrated the existence of a “sigma receptor” that however differs from Martin’s sigma “opioid” receptor in that the sigma receptor discovered by Su has very low affinity for naltrexone which is a universal high-affinity blocker for all subtypes of opioid receptors as hypothesized by Martin et al..
The sigma receptor discovered by Su therefore is a receptor on its own and is not a subtype of opioid receptors. The sigma receptor identified by Su was unfortunately mistermed as the sigma “opioid” receptor in its original publication [2] but was later correctly called the sigma receptor [3]. The sigma receptor identified by Su was later recognized as the sigma-1 receptor (Sig-1R) when two subtypes of the sigma receptor were identified as Sig-1R and Sig-2R [4]. The Sig-1R has been cloned and was found to be an ER protein (Box 1) [5]. The Sig-2R has not been cloned so far. Although the progesterone receptor membrane component 1 was suggested to be the Sig-2R [6], the final identification remains to be fully clarified and awaits the confirmation by successful cloning and sequencing of the Sig-2R in the future.
Box 1.
The sigma-1 receptor (Sig-1R) exists at the endoplasmic reticulum(ER)-mitochondrion interface called the MAM where the Sig-1R promotes cellular survival by (1) ensuring Ca2+ signaling from the ER into mitochondria by chaperoning the IP3 receptor; (2) enhancing the ER to nucleus signaling for antioxidant power by chaperoning the ER stress sensor IRE1; and (3) attenuating free radical damage through the Nrf2 signaling.
The Sig-1R can also translocate, upon the stimulation by agonists, from the MAM to the plasma membrane where Sig-1Rs interact with and affect the function of many other receptors, ion channels, and kinases. The Sig-1R can also translocate to the nuclear envelope where Sig-1Rs recruit chromatin-remodeling factors to affect the transcriptional regulation of genes. In addition, Sig-1Rs have been reported to interact with many other membranous and soluble functional proteins in other parts of cell including cytosol. The Sig-1R may thus represent a pluripotent modulator in the living system.
The Sig-1R has been shown to relate to many diseases including Alzheimer’s disease, Parkinson’s disease, Amyotrophic lateral sclerosis, Huntington’s disease, stroke/ischemia, pain/neuropathic pain, certain psychiatric disorders, cocaine addiction, myocardial hypertension, and cancer. Inasmuch as the Sig-1R acts as a pluripotent modulator, the dysfunction of the Sig-1R may thus play a role in those diseases. Thus pharmacological or cellular engineering targeting the Sig-1R, the pluripotent modulator, may provide therapeutic opportunities to treat those diseases.
The Sig-1R is mainly an ER protein where it resides specifically at the ER-mitochondrion interface, referred to as the MAM (mitochondrion-associated ER membrane) [7]. A the MAM, the Sig-1R acts as a molecular chaperone and sustains the proper conformation of the inositol triphosphate (IP3) receptor type 3 to ensure proper Ca2+ signaling from the ER into mitochondria to facilitate the production of ATP [7–9]. At the MAM, the Sig-1R also chaperones an ER stress sensor, inositol-requiring enzyme 1 (IRE1), to ensure the proper transmission of ER stress into the nucleus to call for the enhanced production of anti-stress and antioxidant proteins [10]. Sig-1Rs also attenuate the formation of reactive oxygen species (ROS) by enhancing the signaling of Nrf2 [11].
The Sig-1R can, upon the stimulation of agonists or stress, translocate to the plasma membrane to interact with ion channels, receptors, and kinases [12, 13]. The Sig-1R is also found to translocate to the envelope of the nucleus [14, 15], where the Sig-1R interacts with a nuclear envelope-resident protein emerin and recruits therein a series of chromatin-remodeling factors to regulate the gene transcription [15]. Recently, many studies via experimental or bioinformatics means have identified or proposed the interaction of the Sig-1R with many other functional proteins in the plasma membrane, ER, mitochondria, and even the cytosol.
The above results, when taken together, suggest that the Sig-1R acts as a pluripotent modulator in the living system that may thus affect many human diseases. In this opinion, we report those proteins that have been experimentally shown to interact with the Sig-1R (Figure 1), and other proteins that were reported to link to the Sig-1R via bioinformatics information (Figure 2). We also show in particular the CNS diseases that have been reported to relate to the Sig-1R (Table 1). Collectively, current research findings suggest that the Sig-1R, as a pluripotent modulator via its interactions with diverse classes of other proteins, may play important physiological roles in the living system and that the dysfunction of the Sig-1R may contribute to certain human diseases.
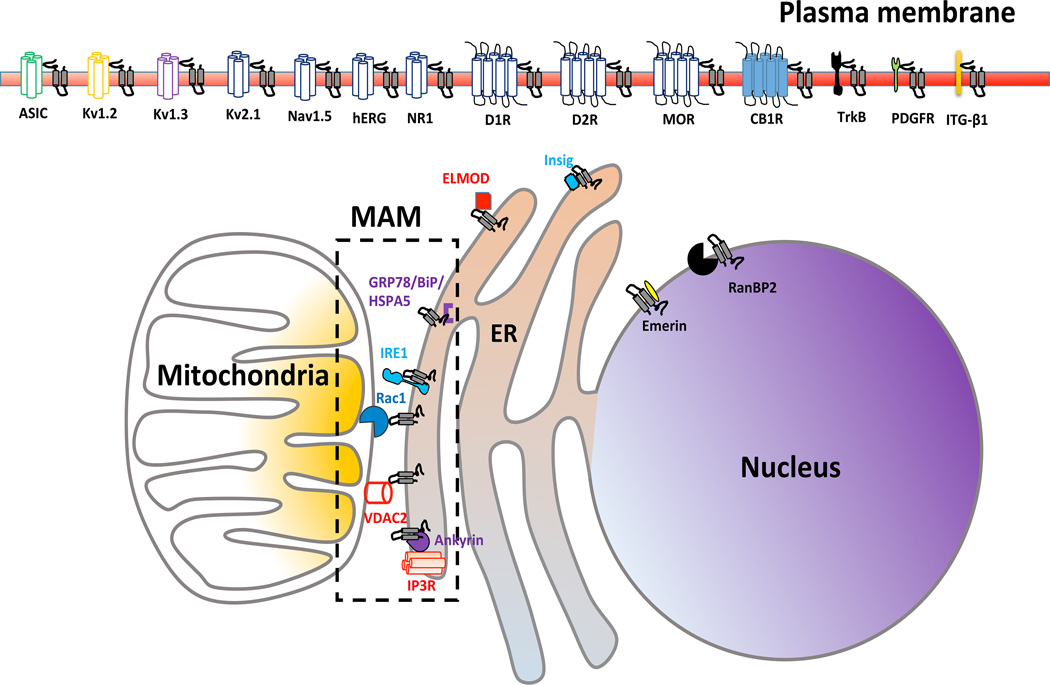
Figure 1. The sigma-1 receptor-interacting proteins as determined via experimental means.
The sigma-1 receptor (Sig-1R) is shown as a two-transmembrane protein in black. ER, endoplasmic reticulum; MAM, the mitochondrion-associated ER membrane. Abbreviations for Sig-1R–interacting proteins are: (A) At the plasma membrane: ASIC, acid-sensing ion channel; Kv1.2, Kv1.3, and Kv2.1, voltage-gated potassium channel; Nav1.5, voltage-gated sodium channel; hERG, voltage-gated potassium channel hERG (human either-à-gogo related gene); NR1, NMDA receptor subunit 1; D1R, dopamine receptor 1; D2R, dopamine receptor 2; MOR, mu opioid receptor; CB1R, cannabinoid receptor 1; TrkB, Tropomyosin receptor kinase B for brain-derived neurotrophic factor; PDGFR, platelet-derived growth factor receptor. (B) In the cytosol, general endoplasmic reticulum membrane, or mitochondrial outer membrane: ELMOD, Cell engulfment and motility domain; Rac1, Ras-related C3 botulinum toxin substrate (Rac)-GTPase; Insig, insulin-induced gene. (C) At the endoplasmic reticulum-mitochondrion contact region called the MAM: GRP78/BiP/HSPA5, glucose response protein/immoprotein binding protein/heat shock protein A5; IRE1, inositol-requiring enzyme 1; VDAC, voltage-dependent anion channel 2; IP3R, inositol trisphosphate receptor. (D) At the nuclear envelope: RanBP2, Ran-binding protein2.
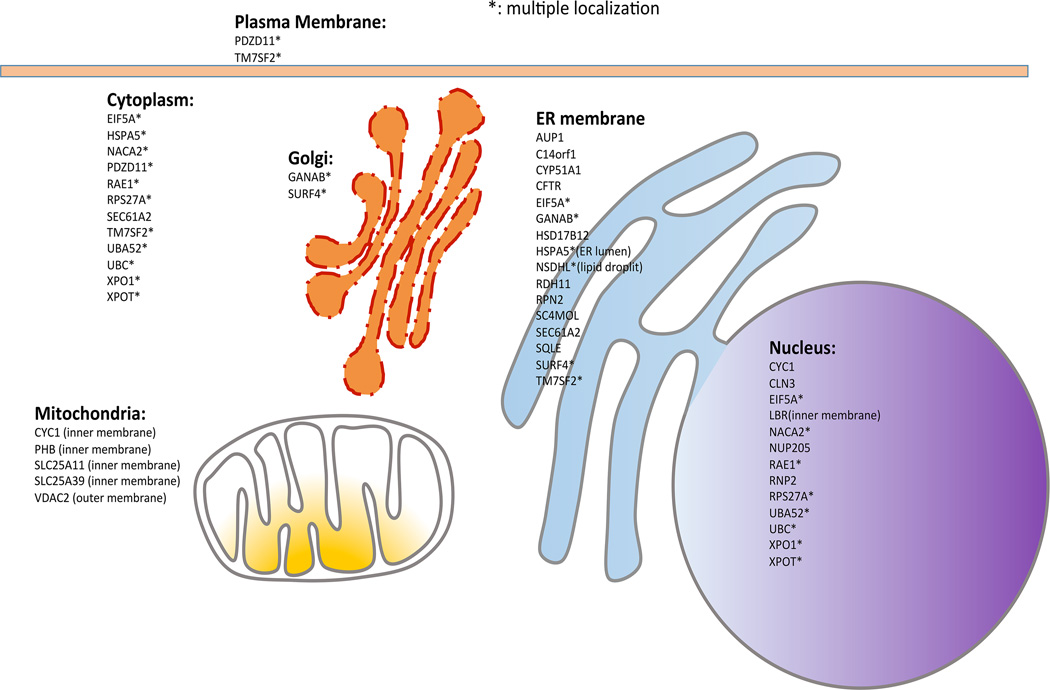
Figure 2. The proposed sigma-1 receptor-interacting proteins through a search created by one of the bioinformatics report [61].
Abbreviations (in alphabetic order) are: AUP1 (ancient ubiquitous protein 1), C14orf1 (chromosome 14 open reading frame 1), CYC1 (cytochrome c-1), CYP51A1 (cytochrome P450, family 51, subfamily A, polypeptide 1), EIF5A (eukaryotic translation initiation factor 5A), GANAB (glucosidase, alpha; neutral AB), HSD17B12 (hydroxysteroid (17-beta) dehydrogenase 12), HSPA5 (heat shock 70kDa protein 5; glucose-regulated protein, 78kDa; BIP), LBR (lamin B receptor), NACA2 (nascent polypeptide-associated complex alpha subunit 2), NSDHL (NAD(P) dependent steroid dehydrogenase-like), NUP205 (nucleoporin 205kDa), PHB (prohibitin), PDZD11 (PDZ domain containing 11), RAE1 (RAE1 RNA export 1 homolog), RDH11 (retinol dehydrogenase 11 (all-trans/9-cis/11-cis)), RPS27A (ribosomal protein S27a), RPN2 (ribophorin II), SC4MOL (sterol-C4-methyl oxidase-like), SEC61A2 (Sec61 alpha 2 subunit (S. cerevisiae)), SLC25A11 (solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11), SLC25A39 (solute carrier family 25, member 39), SQLE (squalene epoxidase), SURF4 (surfeit 4), TM7SF2 (transmembrane 7 superfamily member 2), UBA52 (ubiquitin A-52 residue ribosomal protein fusion product 1), UBC (ubiquitin C), VDAC2 (voltage-dependent anion channel 2), XPO1 (exportin 1 (CRM1 homolog, yeast)), XPOT (exportin, tRNA (nuclear export receptor for tRNAs)).
TABLE 1.
Sigma-1 receptor-associated CNS diseases
| Disorders | Potential locationsand interactingproteins | References |
|---|---|---|
| Amyotrophic LateralSclerosis (ALS)/MotorNeuron Disorders | BiP (ER)Insig (ER)RanBP2 (NE)mAchR (PM) | [75–88][65, 66, 89–93] |
| Alzheimer’s Disease(AD) | Rac-GTPase(mitochondria),BiP (ER andMitochondria)IP3R (MAM)Insig (ER) | [57, 67, 94, 95][73, 96, 97] |
| HIV | PDGFR (PM) | [37, 98][99–103] |
| Huntington’s Disease(HD) | BiP (ER)D2R (PM)Emerin (NE) | [104] [105] [106][69, 107] |
| Pain/Spinal Cord Injury | MOR (PM),NR1 (PM)CB1R (PM)TrKB (PM) | [25] [26, 27, 108] [109] [110] [111][112] [113] [114] [115] [116] [117][118] [119] [120] [121, 122] |
| Parkinson’s Disease(PD) | TrKB (PM)Emerin (NE)IP3R (MAM)Insig (ER) | [70, 123] [124] |
| Psychiatric Disorders(Schizophrenia andDepression) | BiP (ER)TrkB (PM)IP3R (MAM) | [125] [126] [127] [128] [129] [130][131] [132] [133] [134] [135] [36,136] |
| Stroke and Ischemia | BiP (ER)IP3R (MAM) | [137] [123, 138] [139] [140] |
| Traumatic Brain Injury(TBI) | BiP (ER)IP3R (MAM) | [141] |
Sig-1R–interacting proteins per experimental demonstrations including co-immunoprecipitation and proximity assessment experimentations
At the Plasma Membrane
Regardless of its predominant ER membrane expression pattern, many reports demonstrate that the Sig-1R also regulates plasma membrane proteins (Figure 1). The following section briefly reviews all plasma membrane proteins that have been reported so far to interact directly with the Sig-1R.
Acid-sensing ion channels (ASICs)
ASICs are proton-gated cation channels expressed in the peripheral and central nervous neurons and can be activated by acidic pH conditions that occur for example during ischemia.
Sig-1Rs can modulate ASICs via the activation of Sig-1Rs by ligands and inhibits thus ASIC1a–induced calcium influx in rat cortical neurons [16]. However, direct interaction between Sig-1Rs and ASIC1a was later demonstrated by atomic force microscopy imaging. The analysis of the Sig-1R binding to ASIC1a was carried out in cells coexpressing ASIC1a and FLAG/His6-tagged Sig-1R. The results of the stoichiometry suggested that the Sig-1R associates with the trimeric ASIC1a subunit with a 3-fold symmetry [17]. However, opinions differ in those two types of studies in whether the interaction of the Sig-1R and the ASIC1a takes place in lipid rafts.
Dopamine receptors
Dopamine receptors (DR) play crucial roles in many neurological processes, including motivation, cognition, memory, and motor function. There are at least five subtypes of dopamine receptors: D1, D2, D3, D4, and D5. Moreover, those receptors are grouped as D1-like receptor (D1 and D5) and D2-like receptor (D2, D3, and D4), which respectively stimulate or inhibit adenylyl cyclase.
Navarro and colleagues [18] demonstrated the heterodimerization of Sig-1R and dopamine D1 receptor (D1R) in living cells using bioluminescent resonance energy transfer saturation experiments. Colocalization of Sig-1R and D1R was also identified by immunostaining studies. The Sig-1R-D1R interaction was later extended to animal tissue, where Sig-1R-D1R-Histamine H3 receptor complexes were detected in the rat striatum by energy transfer experiments and proximity ligation assays [19]. A similar approach was applied to establish the functional interaction of Sig-1R and D2R. The data suggest that cocaine binds to Sig-1R-D2R heteromers and inhibits the downstream extracellular-signal-regulated kinase-MAPK signaling pathway [20]. These findings suggest that the Sig-1R binds D1R and D2R and thus differentially associates and modulates the D1R and D2R downstream signaling when neurons are stimulated by cocaine.
Muscarinic acetylcholine receptor (mAchR)
mAchRs are G protein-coupled receptors for acetylcholine that plays important roles in the control of motor neurons in the brain.
In a ventral horn motorneuron model, Mavlyutov et al. [21] examined the distributions of Sig-1Rs at the synaptic contact site. Immunoelectron microscopy revealed that Sig-1Rs are located in the postsynaptic densities and are juxtapositional to the metabotropic acetylcholine receptor (mAchR) M2 in very close proximity. The ultrastructure visualization of Sig-1Rs indicated that Sig-1Rs are excluded from the PM; rather, they are primarily located in the subsurface ER cisternae. Since the interaction between Sig-1R and mAchRM2 may promote the survival or proper functioning of motor neurons, the close proximity of the Sig-1R to mAchRM2 suggests a role of the Sig-1R in amyotrophic lateral sclerosis whose hallmark is the motor neuron degeneration.
Mu-opioid receptor (MOR)
The MOR is a subtype of opioid receptors that mediates morphine-induced analgesia. Although the Sig-1R is not a subtype of opioid receptors, as stated in the introduction, numerous studies have been focused on the endogenous Sig-1R as the negative modulator of opioid analgesia [22–26]. As such, Sig-1R antagonists would potentiate morphine-induced analgesia. The functional and physical association of Sig-1R with MOR was recently assessed by the guanosine 5′-O-(3-[35S]thio)triphosphate ([35S]GTPγS) binding and by coimmunoprecipitation experiments using epitope-tagged receptors [27]. Interestingly, this study also showed that, in mouse brain membrane preparations, Sig-1R-selective antagonists could potentiate both opioid receptor and muscarinic acetylcholine receptor-mediated stimulation of [(35)S]GTP gamma S binding. These results suggest a broader role for Sig-1Rs in modulating G-protein-coupled receptor signaling [27].
The Sig-1R also has been demonstrated to modulate opioid analgesia through the Sig-1R′s interaction with the NMDA receptor NR1 subunit (GluN1) [28] (see next section). The potential role of Sig-1R in the cross-regulation between MOR and NMDARs was demonstrated by using peptide interference assay and immunohistochemistry in the mouse mesencephalic periaqueductal grey matter. Those results suggest that the Sig-1R-MOR-GluN1 trimeric complex may play a role in nociception. However, further investigation is needed to totally clarify the relation between the trimeric complex and MOR-induced analgesia.
N-methyl-D-aspartate receptor (NMDAR) and Cannabinoid Receptor 1 (CB1R)
The NMDA receptor is an ion channel type of receptor for glutamate and consists of a heterotetramer between two GluN1 and two GluN2 subunits. The NMDAR controls certain neuronal functions including synaptic plasticity and memory. The CB1R is a G protein-coupled receptor for endocannabinoids such as anandamide and 2-arachidonoylglycerol and is expressed at presynaptic neurons where the CB1R modulates the release of neurotransmitter glutamate.
The Sig-1R has been extensively studied in cognitive function, particularly in psychiatric disorders. Sig-1R agonists have shown to enhance NMDAR functionality [29, 30]. Combining the atomic force microscopy imaging study and the in situ proximity ligation assay, Balasuriya et al. [31] demonstrated a direct interaction between the Sig-1R and NMDAR. The Sig-1R bound directly to the NMDAR subunits, GluN1/GluN2A heterotetramers, specifically via the interaction with the GluN1 subunit. Interestingly, the Sig-1R agonist administration caused an upregulation of GluN2A and GluN2B expression in the synaptosomal fraction [32]. The Sig-1R antagonist abolished the agonist-induced increase of synaptosomal expression of GluN2A and GluN2B and their associated trafficking to the plasma membrane. Coimmunoprecipitation studies also revealed an increased interaction between Sig-1Rs and GluN2 subunits by Sig-1R agonist [32]. A recent study suggested that the Sig-1R functions as a safety switch to control CB1-NMDAR interaction to prevent CB1R–provoked NMDAR hypofunction [33]. The interactions of Sig-1Rs with CB1R, GluN1 and the histidine triad nucleotide-binding protein 1 (HINT1) were visualized by bimolecular fluorescence complementation assay. The data suggested that the assembly of the CB1-HINT1-GluN1 protein complex is critically regulated by the Sig-1R. These findings indicate that the Sig-1R regulates synaptic plasticity perhaps via dynamic protein associations. The main function of the Sig-1R in this regard is suggested to allow for the restoration of the hypo-functional NMDAR that was caused by the interacting CB1R. The authors proposed that this action of the Sig-1R regulates the homeostasis between opposite effects of CB1R and NMDAR in the context of analgesia and certain psychiatric disorders like schizophrenia. The hypo-functional NMDAR has been implicated in schizophrenia.
Tropomyosin receptor kinase B (TrkB)
The TrkB is cell surface tyrosine kinase receptor for brain derived neurotrophic factor (BDNF) and neurotrophin 4. TrkB plays important roles in the brain including synaptic transmission, neurogenesis, learning, and cognition.
Sig-1R agonists exert anti-depressant-like effects and neuroprotective effects via the upregulation of BDNF [34] or the enhanced post-translational processing of BDNF [35]. In addition, Kimura et al. [36] reported that the Sig-1R interacts with the BDNF receptor TrkB in cerebellar granule neurons and promotes the neurite elongation. The report also demonstrated that the coimmunoprecipitation of Sig-1R and TrkB was apparently strengthened by the Sig-1R agonist PRE-084.
Platelet-derived growth factor receptor (PDGFR)
The PDGFR is cell surface tyrosine kinase receptor for PDGF and plays a role in the regulation of cell proliferation, cellular differentiation, cell growth, and development, and relates to many diseases including cancer.
It has been known that the Human immunodeficiency virus (HIV)-associated increase in monocyte adhesion and trafficking is exacerbated by cocaine abuse. One of the underlying mechanisms involves cocaine-mediated upregulation of cell adhesion molecules that result in subsequent disruption of the blood-brain barrier. PDGFR is known to cause the transcriptional increase of an adhesion molecule called ALCAM by activating the transcription factor nuclear factor-kB. However, the exact relationship between the HIV-related action of cocaine and PDGFR remained unknown until the study below was published.
In human brain microvascular endothelial cells, the Sig-1R can interact with the PDGFR and the interaction was intensified by cocaine as a result of cocaine causing the translocation of the Sig-1R from the MAM to the plasma membrane [37]. This interaction of the Sig-1R with PDGFR is important for cocaine to enhance the transmigration or infiltration of leukocytes across the blood-brain barrier by increasing the expression of ALCAM. Further, as nuclear factor -kB also mediates inflammation, the Sig-1R-PDGFR interaction plays an important role in the HIV-induced inflammation which is also known to be exacerbated by cocaine.
Integrin-β1
Integrins are transmembrane receptors for cell adhesion molecules including fibronectin and collagen, and are important for the metastasis of cancer cells. Integrin is a heterodimers consisting of α and β subunits.
The Sig-1R has been reported to interact with integrin-β1 which facilitates cell adhesion [38]. Interestingly, the interaction between Sig-1R and integrin-β1 was blocked by the Sig-1R ligand SKF-10047, which is a Sig-1R agonist. Further, the silencing of Sig-1Rs by siSig-1R attenuated cell adhesion. Those two seemingly contradictory results, agonist producing the same effect as that from siRNA treatment, need to be clarified in the future. Nevertheless, the interaction between Sig-1R and integrin-β1 suggests a role of the Sig-1R in cell adhesion and perhaps the progression of cancer cells.
Voltage-gated potassium channels (Kv)
Kvs are on the plasma membrane that plays an important role for returning the depolarized cell to a resting state during action potentials.
By reconstituting responses seen in Xenopus oocytes, two separate groups showed that Sig-1Rs regulate voltage-gated potassium channels Kv1.3 and Kv1.4. Aydar and colleagues [39] demonstrated a functional interaction between the Sig-1R and Kv1.4 in the absence of ligands. A decade later, Kinoshita et al [40] revealed that Sig-1R interacts at the transmembrane domain of the Kv1.3 channels and alters their kinetics. In contrast to the study led by Aydar et al., Kinoshita and colleagues claimed that Sig-1R ligands are not required to alter (or block) the interactions between Kv1.3 channels and the Sig-1R. The interactions and dynamics of Sig-1R with the voltage gated potassium channel Kv1.2 were later identified and established in the animal model. In this report, cocaine exposure induces Sig-1R translocation to the plasma membrane and shapes intrinsic plasticity via the persistent association of Sig-1R and Kv1.2 in the nucleus accumbens shell medium spiny neuron [41]. Additionally, a recent study using confocal imaging revealed the colocalization of Sig-1R and Kv2.1 channel in the C-terminals of motor neurons [21]. The relationship between the Sig-1R and other ion channels were extended to a cardiac voltage-gated potassium channel hERG (human either-à-gogo related gene). The study was carried out in the leukemic K562 cell line to explore the potential pharmacological targets to reduce cancer progression. Electrophysiological data shows that the Sig-1R modulates the hERG current density in the presence of ligands [42]. Biochemical approaches including the co-immunoprecipitation study suggest that the Sig-1R expression potentiates the hERG subunit’s ER/Golgi translocation and maturation [42]. Atomic force imaging and homogenous time-resolved fluorescence approaches later identified that the Sig-1R interacts with hERG with a four-fold symmetry. The authors clarify that the direct interaction between the Sig-1R and hERG in the plasma membrane is not Sig-1R ligand-dependent but is reduced by cholesterol depletion, suggesting that Sig-1R may bind to hERG in the ER and facilitate hERG assembly and trafficking perhaps in a lipid-raft related fashion [43]. The findings of the Sig-1R interacting with hERG in the ER and potentiating hERG maturation and translocation to the plasma membrane suggest that the Sig-1R may exert chaperoning activities in the ER to facilitate proper protein sorting to their final destinations. Thus, this relationship between the Sig-1R and hERG may apply to other Sig-1R-interacting partners as well.
Voltage-gated sodium channels (Nav)
Nav on the plasma membrane are responsible for action potential initiation and propagation in excitable cells including nerve, muscle, and neuroendocrine cells.
The Sig-1R has been reported to modulate several sodium channels including Nav1.2, Nav1.4 and Nav1.5 [44–46]. Those studies investigated the modulation of the voltage-gated sodium channels by the Sig-1R by using various Sig-1R ligands. Results indicate that Sig-1R agonists exert inhibitory action on the Na+ current that was in turn blocked by the Sig-1R antagonist progesterone [47]. The atomic force microscopy imaging of the co-isolated Sig-1R and Nav1.5 demonstrated that the Sig-1R binds to Nav1.5 with a 4-fold symmetry [44].
Notes on the interaction of the Sig-1R with proteins at the plasma membrane
Increasing reports are adding to the list of the ER Sig-1R chaperone associating partners. While the majority of the findings are based on the assumption that Sig-1R forms physical interactions with these proteins and regulates their activities at the plasma membrane, one needs to note that, due to the lack of a high-affinity Sig-1R antibody as well as the interfering signal from the control IgG which has the same molecular weight as the Sig-1R, immnuoprecipitation and the subsequent western blotting of endogenous Sig-1Rs remains technically challenging. Thus, most of the studies were carried out in the overexpression system, in which Sig-1Rs are expressed together with tagged proteins and are thus usually over-saturated in the cellular compartments that may lead to aberrant protein localizations. Therefore, it is conceivable that the over-expressed tagged Sig-1Rs may associate with some proteins that the endogenous Sig-1R may not associate with. Further, the electron microscopy study by Mavlyutov and coworkers demonstrated that the Sig-1R indeed could be localized in the proximity of plasma membrane. Thus, the Sig-1R may interact with the plasma membrane proteins via Sig-1R’s proximity to the plasma membrane. Finally, a recent report suggested that the Sig-1R activation inhibits store-operated Ca2+ (SOCE) entry effects in rat brain microvascular endothelial cells [48]. However, the physical interaction, if any, between the Sig-1R and the SOCE protein complex ORAI1 or STIM1 has yet to be established.
In the cytosol
Cell engulfment and motility domain (ELMOD)
ELMOD proteins are GTPase-activating proteins (GAPs) for the ADP Ribosylation Factors (ARFs) and ARF-likes (ARLs) and can bind to the activated form of the GTPase (e.g. GTP-ARFs) to speed up the rate of hydrolysis of GTP and consequently inactivate the associated signaling.
Distinct GTPases control a variety of cross-talk signaling pathways, which require specific regulators, guanine-nucleotide exchange factors (GEFs) and GAPs. Those GTPases coupled with and controlled by GEFs or GAPs can be activated or inhibited respectively depending on their roles in the signaling pathway. Thus, investigation of the specificities and binding partners of GEFs and GAPs is essential to the construction of integrated models of cell signaling. Ivanova et al. [49] reported that ELMOD proteins are GAPs for the ARF family with links to deafness. According to their results, the Sig-1R acts as a new effector of the GAP activity of ELMOD1–3 proteins because direct binding of Sig-1R to either ELMOD1 or ELMOD2 results in the loss of GAP activity. This observation opens up a new link between the Sig-1R and GTPase (see below).
Ras-related C3 botulinum toxin substrate (Rac)-GTPase (Rac-GTPase)
Rac-GTPase is a subfamily of the Ras homolog gene (Rho) family of GTPases and is known to regulate tumor-cell migration, dendritic growth, and dendritic spine maturation.
Tsai et al. discovered that the Sig-1R regulates neuritogenesis and spine maturation via a signaling pathway involving Rac1-GTPase and its regulator TIAM1 [50]. Recently, Natsvlishvili et al. [51] reported that, by using immunoprecipitation, the Sig-1R not only directly interacts with Rac1-GTPase, but also forms complexes with IP3R, BiP, and Bcl2, in the brain mitochondria. Interestingly, the ligand-specific assembly complex relies on the Sig-1R agonist/antagonist and the presence of GTP/GDP. The author concluded that the Sig-1R-induced Rac1 signaling would trigger mild oxidative stress and the mild oxidative stress impacts neuroplasticity as well as prevents apoptosis and autophagy.
At the ER-mitochondrion interface and mitochondria
Binding Immunoglobulin Protein (BiP)
BiP, also known as 78 kDa glucose-regulated protein or heat shock 70 kDa protein 5, is a constitutively expressed ER protein that functions as a molecular chaperone. Bip is involved in the folding, translocation and assembly of proteins. It was reported that the Sig-1R forms a Ca2+-sensitive chaperone machinery with BiP under normal physiological conditions [7, 52]. However, the lowering of the local Ca2+ concentration such as the efflux of Ca2+ from IP3R causes the Sig-1R to dissociate from BiP. Further, independent from the effect of local Ca2+, the Sig-1R agonist like cocaine or (+)pentazocine can also cause the dissociation of the Sig-1R from BiP [7, 13]. It is known that this action of the Sig-1R agonist causes the Sig-1R to translocate from the MAM to the plasma membrane and nucleus.
Inositol 1,4,5- trisphosphate receptor type 3 ( IP3R3), Ankyrin
In general, the IP3R plays an important role in the generation, propagation and regulation of cytoplasmic Ca2+ signals that regulate numerous physiological and pathophysiological processes. The IP3R3, a subtype of IP3Rs localizes mainly at the MAM and when activated by its agonist IP3 allows Ca2+ to efflux from the ER into mitochondria. However, after allowing for the Ca2+ efflux, IP3Rs’ conformation is altered and is subjected to proteasomal degradation. The IP3R’s degradation posed a question that puzzled researchers for many years: How does a cell restore the conformation of IP3R3 to ensure the proper Ca2+ signaling from the ER into mitochondria? The answer is: nature has provided the Sig-1R to come to rescue.
After dissociating from BiP, as stated above, the Sig-1R begins to bind IP3R3 at the MAM and chaperone IP3R3 [7]. As such, the Sig-1R ensures proper Ca2+ efflux from the ER into mitochondria via IP3R3 at the MAM and sustains/enhances cellular survival. Interestingly, it was demonstrated by Boehning et al. [8] that cytochrome c can bind IP3R3 and regulate Ca2+ signaling. Therefore, the possibility exists that the trimeric complex of Sig-1R-IP3R3-cytochrome c may play an important role in the homeostatic regulation of ER-mitochondrion Ca2+ signaling.
Ankyrin, a family of cytoskeletal adaptor proteins, has been known to inhibit Ca2+ efflux from intracellular organelles by interacting with IP3Rs at the ER [53]. It was demonstrated that the Sig-1R agonist can cause the dissociation of ankyrin from the IP3R to open up the IP3R for Ca2+ efflux from the ER into the cytosol [54]. However, the exact relationship among the Sig-1R-BiP-ankyrin-IP3R remains to be totally clarified.
Voltage-dependent Anion Channels (VDAC)
VDAC localizes at the outer mitochondrial membrane (OMM) and forms a complex with IP3R that resides at the ER. The VDAC-IP3R complex facilitates transfer of Ca2+ from the ER to mitochondria.
Marriott et al. [55] reported that the Sig-1R can interact with VDAC and contributes to the regulation of mitochondrial pregnenolone synthesis. Studies have shown that the VDAC on the outer mitochondrial membrane may interact with steroidogenic acute regulatory protein [56], another exclusive outer mitochondrial membrane protein, and enhances the cholesterol import. The first report also revealed that deletion of Sig-1R disrupts the bridge formed via the Sig-1R association with VDAC2, resulting in an inhibition of cholesterol influx into the mitochondria. This Sig-1R-VDAC interaction may play an even more important role in the ER-mitochondrion cross-talk that has yet to be totally clarified. Further, it may be possible that this Sig-1R-VDAC interaction is partly responsible for maintaining the integrity of the MAM. In fact, losing the integrity of the MAM by the Sig-1R knockdown has been speculated to relate to the Alzheimer’s disease [57].
Inositol-requiring enzyme 1 (IRE1)
IRE1 is an ER stress sensor that can splice the messenger RNA (mRNA) of X-box binding protein 1 (XBP1) to allow for expression of functionally active transcription factor XBP1 that in turn translocate into nucleus to induce the upregulation of several ER chaperones and antioxidant proteins or enzymes.
The ER provides an exclusive environment for protein synthesis and folding, which is vital to the cellular function. Under normal conditions, the synthesis and degradation of proteins remain in balance. Yet, numerous external insults could break the balance, causing the accumulation of undegraded or misfolded proteins. Thus, cells rely on a system, the unfolded protein response, which regulates the homeostasis of the ER by signaling the protein-handling problem to the nucleus via three ER stress sensors: IRE1, PERK, and ATF6. Mori et al. reported [10] that IRE1, but not PERK or ATF6, resides mainly at the MAM and that the mitochondria-derived ROS can preferentially activate IRE1 at the MAM. Further, the Sig-1R interacts only with IRE1 and not PERK or ATF6. The Sig-1R can thus stabilize IRE-1 to ensure a proper mitochondrion-ER-nucleus signaling axis for cellular survival by eventually prolonging the activation of the IRE1-XBP1 signaling pathway.
Insulin-induced gene (Insig)
Insig is an ER protein that plays an important role in the ER control of the cholesterol and lipid homeostasis by affecting the degradation of lipid-synthesizing enzymes via the ER-associated degradation system (ERAD).
Sig-1R plays a key role in oligodendrocyte differentiation by facilitating degradation of the enzyme Ceremide galatosyl transferase (CGalT, which synthesizes galactosylceremide to negatively affect oligodendrocyte differentiation [58]. The Sig-1R does so by forming a complex with Insig at the ER that is part of the ERAD that degrades the CGalT enzyme [58]. The Sig-1R agonist promotes the formation of the Sig-1R-Insig complex presumably to increase the degradation of CGalT to reduce the production of the negative regulator galactosylceramide. This study suggests that the Sig-1R is an important member of the ERAD system that degrades misfolded proteins immediately outside of the ER.
At the Nuclear Envelope: Emerin
The nuclear envelope consists of the outer nuclear membranes and the inner nuclear membranes. It is a highly organized dynamic barrier that separates the nucleus from the cytosol. It is known that the ER and nuclear envelope form a continuous network since both the inner and outer nuclear membranes are contiguous lipids from rough ER membrane. Dussossoy et al. [59] first reported the colocalization of the Sig-1R and sterol isomerase at the nuclear envelope and the ER. In the Sig-1R-EYFP overexpressing NG108 cells, Sig-1R agonists caused the Sig-1R to translocate toward the nuclear envelope and to the tip of the neurite [60]. Recently, an electron microscopy study revealed the precise subcellular distribution of Sig-1R in retinal neurons [14]. The study confirms that Sig-1Rs are localized in the inner and outer membrane of the nuclear envelope. These observations led to the important discovery that the Sig-1R is a transcriptional regulator at the nuclear envelope. Tsai and Chuang et al [15] discovered that the Sig-1R translocates from the ER to the outer nuclear membrane and interacts with the inner nuclear membrane protein emerin and likely the nuclear pore complex protein Ran-binding protein2 (RanBP2). Confocal imaging and coimmunostaining examinations demonstrated the interaction of Sig-1R and emerin at the nuclear envelope and the subsequent recruitment of barrier-to-autointegration factor, histone deacetylase 2, and the specific transcription factor 3 to the promoter of the monoamine oxidase B (MAOB) to suppress the gene transcription of MAOB. Cocaine acts as the Sig-1R agonist to intensify the complex formation. This finding opens a new chapter on how cocaine or other drugs may change the drug reward system. It is a widely accepted concept that cocaine and other drugs, such as methamphetamine, alter the DA levels in the synaptic cleft by blocking the DA reuptake and hijacking the brain reward system. This latest report demonstrating that cocaine, via the Sig-1R, is able to reduce the MAOB level in the NAc in a dopamine transporter-independent manner provides a new mechanism to understand the complexity of addictive processes.
Sig-1R–interacting proteins from bioinformatics (figure 2)
Proteomic and bioinformatics analyses have become important methods to predict potential protein-protein interactions. Many bioinformatics methods have been developed using different data mining routes and criteria (e.g., [61–63]). We looked into their respective web sites from those studies and chose to use one from the study by Schmitt et al ((funcoup.sbc.su.se) [61]) to look for predicted Sig-1R–interacting proteins partly because the confidence level of each predicted protein was easily accessed. We arbitrarily chose a confidence level of 0.948 for potential candidates because one of nuclear proteins of our interest was at this confidence level.
The predicted Sig-1R-interacting proteins in human are (in alphabetic order): AUP1 (ancient ubiquitous protein 1), C14orf1 (chromosome 14 open reading frame 1), CYC1 (cytochrome c-1), CYP51A1 (cytochrome P450, family 51, subfamily A, polypeptide 1), EIF5A (eukaryotic translation initiation factor 5A), GANAB (glucosidase, alpha; neutral AB), HSD17B12 (hydroxysteroid (17-beta) dehydrogenase 12), HSPA5 (heat shock 70kDa protein 5; glucose-regulated protein, 78kDa; BIP), LBR (lamin B receptor), NACA2 (nascent polypeptide-associated complex alpha subunit 2), NSDHL (NAD(P) dependent steroid dehydrogenase-like), NUP205 (nucleoporin 205kDa), PHB (prohibitin), PDZD11 (PDZ domain containing 11), RAE1 (RAE1 RNA export 1 homolog), RDH11 (retinol dehydrogenase 11 (all-trans/9-cis/11-cis)), RPS27A (ribosomal protein S27a), RPN2 (ribophorin II), SC4MOL (sterol-C4-methyl oxidase-like), SEC61A2 (Sec61 alpha 2 subunit (S. cerevisiae)), SLC25A11 (solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11), SLC25A39 (solute carrier family 25, member 39), SQLE (squalene epoxidase), SURF4 (surfeit 4), TM7SF2 (transmembrane 7 superfamily member 2), UBA52 (ubiquitin A-52 residue ribosomal protein fusion product 1), UBC (ubiquitin C), VDAC2 (voltage-dependent anion channel 2), XPO1 (exportin 1 (CRM1 homolog, yeast)), XPOT (exportin, tRNA (nuclear export receptor for tRNAs)).
The information from bioinformatics provides insight toward the potential proteins that may interact with the Sig-1R in theory. However, more experimental evidence is certainly needed to validate these results. For example, only two (HSPA5/BiP and VDAC2) of 22 Sig-1R-interacting proteins in figure 1 are correctly predicted by the bioinformatic means to be high-potential interactors at the confidence level of 0.948. The other 19 proteins in figure 1 are not predicted as potential interactors even at the confidence level of 0.30 except for the mouse Rac1 which has a confidence level 0.302. On the contrary, a predicted high-potential interactor (LBR: lamin B receptor) did not co-immunoprecipitate with the Sig-1R in an experiment [64]. In addition, a low-potential protein below a confidence level of 0.30, the lamin A receptor, was shown to co-immunoprecipitate with the Sig-1R [64]. This pattern of discordance between the experimental result and bioinformatic prediction concerning the Sig-1R interactors applies to those in human, mouse, or rat. Discordance of similar patterns was also found when searching the potential Sig-1R interactors by using tools from other two reports [62, 63].
The above discordance notwithstanding, it is important to point out that the predicted Sig-1R’s interactors from bioinformatics may in fact be indirect “functional interactors” and not direct physical interactors. Further, many of the high-potential interactors from bioinformatic predictions have not been tested in the co-immunoprecipitation or other proximity assessment assays. The discordance as stated above may be reconciled in future studies.
CNS diseases reported to associate with the Sig-1R: speculation on the role of the Sig-1R in complex with respective interacting protein partner
The Sig-1R has been reported to be involved with certain CNS diseases (Table 1). Although the exact Sig-1R-interacting protein(s) which may relate to each disease is not totally clear at present, we tentatively place those proteins in the table simply to indicate the possibility as such. Nevertheless, the potential roles of the Sig-1R in interaction with its respective partners in some CNS diseases are speculated as follows.
Amyotrophic lateral sclerosis (ALS or Lou Gehrig’s disease)
The loss of function of Sig-1Rs in the motor neuron disrupted the ER-mitochondrion contact, caused a reduction of Ca2+ signaling, and stunned the axon extension, leading to amyotrophic lateral sclerosis in animals [65]. Thus, the action of Sig-1R–IP3R–VDAC in maintaining an intact contact between ER and mitochondria may play an important role in this disease. Further, the close proximity of the Sig-1R to the muscarinic acetylcholine receptor at the plasma membrane [21] may also be involved in this disease. In addition, the Sig-1R-Insig interaction may participate in the etiology of this disease because Insig was reported to be important in this disease by reducing the glutamate-induced excitotoxicity [66].
Alzheimers’ disease
The Sig-1R plays an important role in maintaining the structural integrity of the MAM presumably through the tethering of the Sig-1R with IP3R and VDAC. The ER-mitochondrion cross-talk via the Sig-1R-IP3R-VDAC linkage at the MAM has recently been implicated to play an important role in the pathogenesis of Alzheimer’s disease [57]. Knockdown of Sig-1R caused neurodegeneration. The level of Sig-1Rs is reduced in the brain of human Alzheimer’s patients [57]. In addition, the Sig-1R-Insig interaction may play a role in this disease because Insig has been shown to be involved in the progress of the disease as it affects the cholesterol synthesis [67].
Huntington’s disease
The “dopamine system stabilizer” class of drugs [68] have been suggested as potential agents to treat Huntington’s disease as the drug can act as either a functional agonist or a functional antagonist depending on the initial levels of dopamine. One of those drugs is 4- [3-(methylsulfonyl)phenyl]-1-propylpiperidine (pridopidine) . Pridopidine, in addition to binding to dopamine D2R, binds the Sig-1R with an affinity 20 times higher than that at D2R in a PET imaging study [69], suggesting that pridopidine may work through its duel actions at D2R and Sig-1R [69]. As the Sig-1R has been shown to interact directly with D2R [20], it is tempting to speculate that the Sig-1R-D2R interaction may play an important role in the action of pridopidine. The effect of pridopidine against Huntington’s disease may involve the drug’s ability, like cocaine does [15], to translocate Sig-1Rs to the nuclear envelope to recruit chromatin-remodeling factors to suppress the gene expression of MAOB, thus increasing dopamine level in the brain. This possibility remains to be tested in the future.
Pain or neuropathic pain
The Sig-1R per se or its agonists was demonstrated to be involved in the attenuation of morphine-induced analgesia [25–27]. These results suggest the Sig-1R as an endogenous pain modulator in the CNS. Although the exact molecular mechanism remains to be totally clarified, the Sig-1R was shown to co-immunoprecipitate with the MOR in HEK cells [27]. Further, as stated above, the Sig-1R also co-immunoprecipitates with the NMDA receptor subunit GluN2 [32] as well as with CB1R [33], both of which are known to be involved in pain perception. Thus, the Sig-1R antagonists combined with morphine are being developed as an analgesic agent to reduce the dose of morphine while still maintaining effective analgesia.
Parkinson’s disease
A Sig-1R agonist PRE-084 was found to induce functional neurorestoration in experimental Parkinsonism in that density of dopaminergic fibres is increased and a modest recovery of dopamine level is seen [70]. Further, this agonist treatment also causes a wider intracellular distribution of Sig-1Rs [70], presumably due to the agonist-induced Sig-1R translocation. As such, although the Sig-1R-interacting partner protein was not identified in this report, it is tempting to speculate that the Sig-1R agonist PRE-084 may cause the translocation of Sig-1Rs to the nuclear envelope to bind emerin which in turn recruits chromatin-remodeling factors to suppress the gene expression of MAOB [64], thus reducing the dopamine degradation and causing an increase of dopamine in the brain. However, another possibility for the involvement of the Sig-1R in this disease is that the Sig-1R can interact directly with TrKB [36] to enhance the receptor binding and/or signaling of the BDNF that is known to promote the survival of neurons.
Depression
BDNF, a neurotrophic factor, is known to play an important role in the action against depression because BDNF causes increases of dendritic spines and axon elongation for enhanced communications between neurons [71, 72]. BDNF does so via its receptor TrkB at the plasma membrane. The Sig-1R has been shown to be involved in depression partly through the Sig-1R’s action in stabilizing the post-translational processing of mature BDNF [35]. Interestingly, the Sig-1Rwas also shown to co-immunoprecipitate with TrkB [36], suggesting that one of the antidepressive actions of the Sig-1R may be due to its ability to bind the BDNF receptor TrKB and enhance thereof the downstream signaling of TrkB.
Cocaine addiction
The Sig-1R-Kv1.2 interaction has been shown to shape neuronal and behavioral responses to cocaine [41]. This study demonstrated that cocaine “hijacks” Sig-1R to increase the interaction between Sig-1R and Kv1.2 potassium channel to decrease the intrinsic excitability of GABAergic neurons, thus leading to cocaine-induced behavioral sensitization. In addition, cocaine also causes the translocation of Sig-1R from the ER to the nuclear envelope to interact with emerin to suppress the gene transcription of dopamine-metabolizing enzyme MAOB to facilitate the action of cocaine [64].
Concluding Remarks
Thus, because the Sig-1R represents a new type of functional protein in the living system in that it can bind and modulate many different classes of functional proteins in many parts of the cell, we suggest to call the Sig-1R a pluripotent modulator (see Outstanding Questions box). It is perhaps because of this unique action of the Sig-1R that the receptor is involved in affecting or regulating so many different physiological and pathological conditions.
Outstanding Questions.
What is the structural basis that allows the sigma-1 receptor to be able to interact with so many different classes of proteins with diverse structure and functions?
Why would the nature design such a protein to exist in the living system?
Is the sigma-1 receptor the only protein with this unique pluripotent action or is there any other member of this kind which has yet to be discovered?
Are all of the actions of the sigma-1 receptor described in this article relating to the chaperone nature of the sigma-1 receptor or is it some other as yet unknown nature of the sigma-1 receptor?
Given that the sigma-1 receptor can interact with so many functional proteins in the living system, how can we design drugs to target the specific protein that the sigma-1 receptor interacts with without affecting other interacting proteins?
At the MAM, the interaction between the Sig-1R and other proteins apparently encompasses functional sequelae for cellular survival because the Sig-1R: (1) Chaperones client proteins to maintain proper Ca2+ signaling from ER into mitochondria to ensure mitochondrial ATP production for bioenergetics; (2) Attenuates free radical generation by ensuring proper mitochondrion-ER-nucleus signaling; (3) Maintains the structural integrity of the contact between ER and mitochondria to facilitate the functional cross-talk of these two critical organelles; (4) Serves as a member of the ER-associated degradation system to regulate homeostasis of functional proteins; (5) Serves as the carrier of signaling lipid, specifically myristic acid [73], for proper functionality of neurons.
Outside of the MAM, the functional sequelae of the Sig-1R-target protein interaction may not be involved in cellular survival but in general relates to positive or negative modulation of the function of the target protein as indicated in the main portion of this opinion.
Although we have speculated above the potential role of the Sig-1R-target protein interaction in certain CNS diseases, questions in this regard remain: (1) What is the molecular basis of signaling that may relate the disease, in particular neurodegenerative diseases, to the Sig-1R at the MAM? If a functionally aberrant protein causes a disease, how does the cell signal the aberrance of that protein to the Sig-1R at the MAM? (2) If the disease for some reason causes the Sig-1R to translocate from the MAM, does the Sig-1R translocate to all other parts of the cell where the Sig-1R has been described? (3) Does the Sig-1R affect only the dysfunctional proteins? Does the Sig-1R do anything to a functionally normal protein? (4) Does the Sig-1R regulate the function of the interacting protein partner only by chaperoning the partner’s conformation or by other as-yet-unknown actions?
Several questions also remain concerning the Sig-1R-related therapeutic agents for disease treatment: (1) Sig-1R agonists that may be effective in treating certain neurodegenerative diseases may also cause the Sig-1R to translocate from the MAM. What would the consequence be in terms of treatmetnefficacy? (2) Does the Sig-1R agonist continue to bind the Sig-1R after causing the translocation of the Sig-1R from the MAM? (3) What is the action of the Sig-1R agonist if it is translocated together with the Sig-1R to for example, the plasma membrane? Does the agonist serve to enhance the chaperone activity or other as yet unknown activity of the Sig-1R at the destined location? (4) Can the Sig-1R antagonist block the action of the Sig-1R even after the Sig-1R form a complex with target protein at the plasma membrane or nuclear envelope? (5) In the case of the hERG to which the Sig-1R binds at the ER and then co-translocates to the plasma membrane, how do we design drugs to facilitate or break up the interaction at the desired loci in a cell?
The reason why the Sig-1R can interact with so many structurally diverse proteins can only be speculated at present. Although numbers of chaperone proteins in the living system are limited, those chaperones have to maintain or help degrade thousands of other proteins. Thus, it is understandable that a chaperone has to interact and chaperone with many client proteins. So is the Sig-1R. However, the Sig-1R differs from other chaperones in that the Sig-1R has two transmembrane regions whereas none of other chaperones, as far as we know, has a transmembrane region. A particularly interesting question concerning the Sig-1R having two transmembrane regions is whether the transmembrane regions play any unique role in the action of the Sig-1R as a chaperone. This question deserves to be answered in the future in the light that most of the Sig-1R-interacting proteins are transmembrane proteins. Also, the Sig-1R differs from other chaperones in that while most of the action of other chaperones requires ATP, the Sig-1R does not require ATP in chaperoning the target protein [7].
Resent publications reporting the existence of Sig-1Rs in equilibrium as monomers, dimers and higher oligomeric forms may provide some answers, at least in part, as to why Sig-1Rs may bind so many target proteins or even ligands [74] [75] [13]. The formation of oligomers does not involve disulfide bonds. The Sig-1R agonist seems to favor monomers and dimers while the antagonist favors oligomers. Interestingly, certain ligands abolished the monomeric Sig-1R interactions with the plasma membrane ion channels. Thus, the ligand-gated oligomer/monomer equilibrium state of Sig-1Rs may attribute to Sig-1Rs being able to bind many different classes of ligands, target diverse proteins, and exert different chaperoning activities [13] [74, 75].
Whether the Sig-1R is the only member in this newly termed “pluripotent modulator” remains to be totally clarified in the future. Also unknown is the exact relationship between the ligand-induced oligomerization of Sig-1Rs and all of the actions of Sig-1Rs described above including these in the disease states. More investigations are certainly warranted to advance our understanding on this unique pluripotent modulator protein.
Lastly, although the Sig-2R has not been cloned, it is not reasonable to speculate that the Sig-2R could also be a chaperone with however potentially different interacting partners from those of the Sig-1R. This speculation is made because many studies so far have reported close and overlapping pharmacological and biochemical properties between the Sig-1R and Sig-2R.
Trends Box.
- ■
The sigma-1 receptor (Sig-1R) exists at the endoplasmic reticulum(ER)-mitochondrion interface called the MAM (mitochondrion-associated ER membrane), where the Sig-1R promotes cellular survival - ■
The Sig-1R can, upon the stimulation of agonists or stress, translocate to the plasma membrane to interact with ion channels, receptors, and kinases - ■
Experimental or bioinformatics studies have identified interactions between the Sig-1R and many other functional proteins in the plasma membrane, ER, mitochondria, and even the cytosol. - ■
CNS diseases have been reported to relate to the Sig-1R, including Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, Huntington’s disease, stroke/ischemia, pain/neuropathic pain, and certain psychiatric disorders - ■
Pharmacological or cellular engineering targeting the Sig-1R may provide therapeutic opportunities to treat those diseases
Acknowledgments
This work is supported by the Intramural Research Program of the National Institute on Drug Abuse. Tzu-Chieh Su is supported in part by the Dragon Gate Program of the Ministry of Science and Technology of Taiwan (MOST #105-2911-I-038 -503). Yoki Nakamura is supported in part by the Japanese Society for Promotion of Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin WR, et al. The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. The Journal of pharmacology and experimental therapeutics. 1976;197:517–532. [PubMed] [Google Scholar]
- 2.Su TP. Evidence for sigma opioid receptor: binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. The Journal of pharmacology and experimental therapeutics. 1982;223:284–290. [PubMed] [Google Scholar]
- 3.Su TP, et al. Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science. 1988;240:219–221. doi: 10.1126/science.2832949. [DOI] [PubMed] [Google Scholar]
- 4.Hellewell SB, Bowen WD. A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain research. 1990;527:244–253. doi: 10.1016/0006-8993(90)91143-5. [DOI] [PubMed] [Google Scholar]
- 5.Hanner M, et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, et al. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nature communications. 2011;2:380. doi: 10.1038/ncomms1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 8.Boehning D, et al. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nature cell biology. 2003;5:1051–1061. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
- 9.Tagashira H, et al. Fluvoxamine rescues mitochondrial Ca2+ transport and ATP production through sigma(1)-receptor in hypertrophic cardiomyocytes. Life sciences. 2014;95:89–100. doi: 10.1016/j.lfs.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Mori T, et al. Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PloS one. 2013;8:e76941. doi: 10.1371/journal.pone.0076941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, et al. Sigma 1 receptor regulates the oxidative stress response in primary retinal Muller glial cells via NRF2 signaling and system x, the Na-independent glutamate-cystine exchanger. Free radical biology & medicine. 2015;86:25–36. doi: 10.1016/j.freeradbiomed.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su TP, et al. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends in pharmacological sciences. 2010;31:557–566. doi: 10.1016/j.tips.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu U, Ruoho AE. Biochemical Pharmacology of the Sigma-1 Receptor. Molecular pharmacology. 2015 doi: 10.1124/mol.115.101170. [DOI] [PubMed] [Google Scholar]
- 14.Mavlyutov TA, et al. Subcellular localization of the sigma-1 receptor in retinal neurons - an electron microscopy study. Scientific reports. 2015;5:10689. doi: 10.1038/srep10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SA, et al. Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1518894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrera Y, et al. sigma-1 receptor modulation of acid-sensing ion channel a (ASIC1a) and ASIC1a–induced Ca2+ influx in rat cortical neurons. The Journal of pharmacology and experimental therapeutics. 2008;327:491–502. doi: 10.1124/jpet.108.143974. [DOI] [PubMed] [Google Scholar]
- 17.Carnally SM, et al. Demonstration of a direct interaction between sigma-1 receptors and acid-sensing ion channels. Biophysical journal. 2010;98:1182–1191. doi: 10.1016/j.bpj.2009.12.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro G, et al. Direct involvement of sigma-1 receptors in the dopamine D1 receptor-mediated effects of cocaine. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18676–18681. doi: 10.1073/pnas.1008911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno E, et al. Cocaine disrupts histamine H3 receptor modulation of dopamine D1 receptor signaling: sigma1-D1-H3 receptor complexes as key targets for reducing cocaine’s effects. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:3545–3558. doi: 10.1523/JNEUROSCI.4147-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro G, et al. Cocaine inhibits dopamine D2 receptor signaling via sigma-1-D2 receptor heteromers. PloS one. 2013;8:e61245. doi: 10.1371/journal.pone.0061245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavlyutov TA, et al. The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience. 2010;167:247–255. doi: 10.1016/j.neuroscience.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien CC, Pasternak GW. Sigma antagonists potentiate opioid analgesia in rats. Neuroscience letters. 1995;190:137–139. doi: 10.1016/0304-3940(95)11504-p. [DOI] [PubMed] [Google Scholar]
- 23.Chien CC, Pasternak GW. Selective antagonism of opioid analgesia by a sigma system. The Journal of pharmacology and experimental therapeutics. 1994;271:1583–1590. [PubMed] [Google Scholar]
- 24.Chien CC, Pasternak GW. Functional antagonism of morphine analgesia by (+)-pentazocine: evidence for an anti-opioid sigma 1 system. European journal of pharmacology. 1993;250:R7–R8. doi: 10.1016/0014-2999(93)90650-7. [DOI] [PubMed] [Google Scholar]
- 25.Mei J, Pasternak GW. Sigma1 receptor modulation of opioid analgesia in the mouse. The Journal of pharmacology and experimental therapeutics. 2002;300:1070–1074. doi: 10.1124/jpet.300.3.1070. [DOI] [PubMed] [Google Scholar]
- 26.Mei J, Pasternak GW. Modulation of brainstem opiate analgesia in the rat by sigma 1 receptors: a microinjection study. The Journal of pharmacology and experimental therapeutics. 2007;322:1278–1285. doi: 10.1124/jpet.107.121137. [DOI] [PubMed] [Google Scholar]
- 27.Kim FJ, et al. Sigma 1 receptor modulation of G-protein-coupled receptor signaling: potentiation of opioid transduction independent from receptor binding. Molecular pharmacology. 2010;77:695–703. doi: 10.1124/mol.109.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Munoz M, et al. The sigma1 receptor engages the redox-regulated HINT1 protein to bring opioid analgesia under NMDA receptor negative control. Antioxidants & redox signaling. 2015;22:799–818. doi: 10.1089/ars.2014.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergeron R, et al. Potentiation of neuronal NMDA response induced by dehydroepiandrosterone and its suppression by progesterone: effects mediated via sigma receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16:1193–1202. doi: 10.1523/JNEUROSCI.16-03-01193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martina M, et al. The sigma-1 receptor modulates NMDA receptor synaptic transmission and plasticity via SK channels in rat hippocampus. The Journal of physiology. 2007;578:143–157. doi: 10.1113/jphysiol.2006.116178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balasuriya D, et al. The sigma-1 receptor interacts directly with GluN1 but not GluN2A in the GluN1/GluN2A NMDA receptor. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:18219–18224. doi: 10.1523/JNEUROSCI.3360-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pabba M, et al. NMDA receptors are upregulated and trafficked to the plasma membrane after sigma-1 receptor activation in the rat hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:11325–11338. doi: 10.1523/JNEUROSCI.0458-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Blazquez P, et al. The calcium-sensitive Sigma-1 receptor prevents cannabinoids from provoking glutamate NMDA receptor hypofunction: implications in antinociception and psychotic diseases. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2014;17:1943–1955. doi: 10.1017/S1461145714000029. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto K. Sigma-1 receptor chaperone and brain-derived neurotrophic factor: emerging links between cardiovascular disease and depression. Progress in neurobiology. 2013;100:15–29. doi: 10.1016/j.pneurobio.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Fujimoto M, et al. Sigma-1 receptor chaperones regulate the secretion of brain-derived neurotrophic factor. Synapse. 2012;66:630–639. doi: 10.1002/syn.21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura Y, et al. Sigma-1 receptor enhances neurite elongation of cerebellar granule neurons via TrkB signaling. PloS one. 2013;8:e75760. doi: 10.1371/journal.pone.0075760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao H, et al. Cocaine hijacks sigma1 receptor to initiate induction of activated leukocyte cell adhesion molecule: implication for increased monocyte adhesion and migration in the CNS. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:5942–5955. doi: 10.1523/JNEUROSCI.5618-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer CP, et al. Sigma-1 receptors bind cholesterol and remodel lipid rafts in breast cancer cell lines. Cancer research. 2007;67:11166–11175. doi: 10.1158/0008-5472.CAN-07-1771. [DOI] [PubMed] [Google Scholar]
- 39.Aydar E, et al. The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron. 2002;34:399–410. doi: 10.1016/s0896-6273(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 40.Kinoshita M, et al. Sigma-1 receptor alters the kinetics of Kv1.3 voltage gated potassium channels but not the sensitivity to receptor ligands. Brain research. 2012;1452:1–9. doi: 10.1016/j.brainres.2012.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kourrich S, et al. Dynamic interaction between sigma-1 receptor and Kv1.2 shapes neuronal and behavioral responses to cocaine. Cell. 2013;152:236–247. doi: 10.1016/j.cell.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crottes D, et al. Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. The Journal of biological chemistry. 2011;286:27947–27958. doi: 10.1074/jbc.M111.226738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balasuriya D, et al. A direct interaction between the sigma-1 receptor and the hERG voltage-gated K+ channel revealed by atomic force microscopy and homogeneous time-resolved fluorescence (HTRF(R)) The Journal of biological chemistry. 2014;289:32353–32363. doi: 10.1074/jbc.M114.603506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balasuriya D, et al. The sigma-1 receptor binds to the Nav1.5 voltage-gated Na+ channel with 4-fold symmetry. The Journal of biological chemistry. 2012;287:37021–37029. doi: 10.1074/jbc.M112.382077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao XF, et al. Sigma-1 receptor agonists directly inhibit Nav1.2/1.4 channels. PloS one. 2012;7:e49384. doi: 10.1371/journal.pone.0049384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johannessen M, et al. Voltage-gated sodium channel modulation by sigma-receptors in cardiac myocytes and heterologous systems. American journal of physiology Cell physiology. 2009;296:C1049–C1057. doi: 10.1152/ajpcell.00431.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johannessen M, et al. Antagonist action of progesterone at sigma-receptors in the modulation of voltage-gated sodium channels. American journal of physiology Cell physiology. 2011;300:C328–C337. doi: 10.1152/ajpcell.00383.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brailoiu GC, et al. Cocaine Inhibits Store-Operated Ca2+ Entry in Brain Microvascular Endothelial Cells: Critical Role for Sigma-1 Receptors. The Biochemical journal. 2015 doi: 10.1042/BJ20150934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanova AA, et al. Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. The Journal of biological chemistry. 2014;289:11111–11121. doi: 10.1074/jbc.M114.548529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai SY, et al. Sigma-1 receptors regulate hippocampal dendritic spine formation via a free radical-sensitive mechanism involving Rac1xGTP pathway. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22468–22473. doi: 10.1073/pnas.0909089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Natsvlishvili N, et al. Sigma-1 receptor directly interacts with Rac1-GTPase in the brain mitochondria. BMC biochemistry. 2015;16:11. doi: 10.1186/s12858-015-0040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortega-Roldan JL, et al. Characterization of the human sigma-1 receptor chaperone domain structure and binding immunoglobulin protein (BiP) interactions. The Journal of biological chemistry. 2013;288:21448–21457. doi: 10.1074/jbc.M113.450379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bourguignon LY, Jin H. Identification of the ankyrin-binding domain of the mouse T-lymphoma cell inositol 1,4,5-trisphosphate (IP3) receptor and its role in the regulation of IP3-mediated internal Ca2+ release. The Journal of biological chemistry. 1995;270:7257–7260. doi: 10.1074/jbc.270.13.7257. [DOI] [PubMed] [Google Scholar]
- 54.Hayashi T, Su TP. Regulating ankyrin dynamics: Roles of sigma-1 receptors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:491–496. doi: 10.1073/pnas.98.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marriott KS, et al. sigma-1 receptor at the mitochondrial-associated endoplasmic reticulum membrane is responsible for mitochondrial metabolic regulation. The Journal of pharmacology and experimental therapeutics. 2012;343:578–586. doi: 10.1124/jpet.112.198168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad M, et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) regulates steroidogenic activity via steroidogenic acute regulatory protein (StAR)-voltage-dependent anion channel 2 (VDAC2) interaction. The Journal of biological chemistry. 2015;290:2604–2616. doi: 10.1074/jbc.M114.605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hedskog L, et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7916–7921. doi: 10.1073/pnas.1300677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi T, et al. The lifetime of UDP-galactose:ceramide galactosyltransferase is controlled by a distinct endoplasmic reticulum-associated degradation (ERAD) regulated by sigma-1 receptor chaperones. The Journal of biological chemistry. 2012;287:43156–43169. doi: 10.1074/jbc.M112.380444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dussossoy D, et al. Colocalization of sterol isomerase and sigma(1) receptor at endoplasmic reticulum and nuclear envelope level. European journal of biochemistry / FEBS. 1999;263:377–386. doi: 10.1046/j.1432-1327.1999.00500.x. [DOI] [PubMed] [Google Scholar]
- 60.Hayashi T, Su TP. Intracellular dynamics of sigma-1 receptors (sigma(1) binding sites) in NG108-15 cells. The Journal of pharmacology and experimental therapeutics. 2003;306:726–733. doi: 10.1124/jpet.103.051292. [DOI] [PubMed] [Google Scholar]
- 61.Schmitt T, et al. FunCoup 3.0: database of genome-wide functional coupling networks. Nucleic acids research. 2014;42:D380–D388. doi: 10.1093/nar/gkt984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic acids research. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai SY, et al. Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E6562–E6570. doi: 10.1073/pnas.1518894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernard-Marissal N, et al. Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain : a journal of neurology. 2015;138:875–890. doi: 10.1093/brain/awv008. [DOI] [PubMed] [Google Scholar]
- 66.Taghibiglou C, et al. Sterol regulatory element binding protein-1 (SREBP1) activation in motor neurons in excitotoxicity and amyotrophic lateral sclerosis (ALS): Indip, a potential therapeutic peptide. Biochemical and biophysical research communications. 2011;413:159–163. doi: 10.1016/j.bbrc.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Pierrot N, et al. Amyloid precursor protein controls cholesterol turnover needed for neuronal activity. EMBO molecular medicine. 2013;5:608–625. doi: 10.1002/emmm.201202215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamminga CA, Carlsson A. Partial dopamine agonists and dopaminergic stabilizers, in the treatment of psychosis. Current drug targets CNS and neurological disorders. 2002;1:141–147. doi: 10.2174/1568007024606195. [DOI] [PubMed] [Google Scholar]
- 69.Sahlholm K, et al. Pridopidine selectively occupies sigma-1 rather than dopamine D2 receptors at behaviorally active doses. Psychopharmacology. 2015;232:3443–3453. doi: 10.1007/s00213-015-3997-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Francardo V, et al. Pharmacological stimulation of sigma-1 receptors has neurorestorative effects in experimental parkinsonism. Brain : a journal of neurology. 2014;137:1998–2014. doi: 10.1093/brain/awu107. [DOI] [PubMed] [Google Scholar]
- 71.Duman CH, et al. A role for MAP kinase signaling in behavioral models of depression and antidepressant treatment. Biological psychiatry. 2007;61:661–670. doi: 10.1016/j.biopsych.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 72.Liu RJ, et al. Brain-derived neurotrophic factor Val66Met allele impairs basal and ketamine-stimulated synaptogenesis in prefrontal cortex. Biological psychiatry. 2012;71:996–1005. doi: 10.1016/j.biopsych.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsai SY, et al. Sigma-1 receptor regulates Tau phosphorylation and axon extension by shaping p35 turnover via myristic acid. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6742–6747. doi: 10.1073/pnas.1422001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gromek KA, et al. The oligomeric states of the purified sigma-1 receptor are stabilized by ligands. The Journal of biological chemistry. 2014;289:20333–20344. doi: 10.1074/jbc.M113.537993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mishra AK, et al. The sigma-1 receptors are present in monomeric and oligomeric forms in living cells in the presence and absence of ligands. The Biochemical journal. 2015;466:263–271. doi: 10.1042/BJ20141321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Al-Saif A, et al. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Annals of neurology. 2011;70:913–919. doi: 10.1002/ana.22534. [DOI] [PubMed] [Google Scholar]
- 77.Belzil VV, et al. Genetic analysis of SIGMAR1 as a cause of familial ALS with dementia. European journal of human genetics : EJHG. 2013;21:237–239. doi: 10.1038/ejhg.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casas C, et al. Early presymptomatic cholinergic dysfunction in a murine model of amyotrophic lateral sclerosis. Brain and behavior. 2013;3:145–158. doi: 10.1002/brb3.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukunaga K, et al. The role of SIGMAR1 gene mutation and mitochondrial dysfunction in amyotrophic lateral sclerosis. Journal of pharmacological sciences. 2015;127:36–41. doi: 10.1016/j.jphs.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 80.Iguchi Y, et al. Amyotrophic lateral sclerosis: an update on recent genetic insights. Journal of neurology. 2013;260:2917–2927. doi: 10.1007/s00415-013-7112-y. [DOI] [PubMed] [Google Scholar]
- 81.Kim HJ, et al. Mutations in UBQLN2 and SIGMAR1 genes are rare in Korean patients with amyotrophic lateral sclerosis. Neurobiology of aging. 2014;35:e1957–e1958. doi: 10.1016/j.neurobiolaging.2014.03.001. 1957. [DOI] [PubMed] [Google Scholar]
- 82.Liu ZJ, et al. Identify mutation in amyotrophic lateral sclerosis cases using HaloPlex target enrichment system. Neurobiology of aging. 2014;35:e2811–e2885. doi: 10.1016/j.neurobiolaging.2014.07.003. 2881. [DOI] [PubMed] [Google Scholar]
- 83.Mancuso R, et al. Lack of synergistic effect of resveratrol and sigma-1 receptor agonist (PRE-084) in SOD1G(9)(3)A ALS mice: overlapping effects or limited therapeutic opportunity? Orphanet journal of rare diseases. 2014;9:78. doi: 10.1186/1750-1172-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mancuso R, et al. Sigma-1R agonist improves motor function and motoneuron survival in ALS mice. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2012;9:814–826. doi: 10.1007/s13311-012-0140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mavlyutov TA, et al. Lack of sigma-1 receptor exacerbates ALS progression in mice. Neuroscience. 2013;240:129–134. doi: 10.1016/j.neuroscience.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mavlyutov TA, et al. Role of the Sigma-1 receptor in Amyotrophic Lateral Sclerosis (ALS) Journal of pharmacological sciences. 2015;127:10–16. doi: 10.1016/j.jphs.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ono Y, et al. SA4503, a sigma-1 receptor agonist, suppresses motor neuron damage in in vitro and in vivo amyotrophic lateral sclerosis models. Neuroscience letters. 2014;559:174–178. doi: 10.1016/j.neulet.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 88.Peviani M, et al. Neuroprotective effects of the Sigma-1 receptor (S1R) agonist PRE-084, in a mouse model of motor neuron disease not linked to SOD1 mutation. Neurobiology of disease. 2014;62:218–232. doi: 10.1016/j.nbd.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 89.Prause J, et al. Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Human molecular genetics. 2013;22:1581–1600. doi: 10.1093/hmg/ddt008. [DOI] [PubMed] [Google Scholar]
- 90.Tagashira H, et al. Methyl pyruvate rescues mitochondrial damage caused by SIGMAR1 mutation related to amyotrophic lateral sclerosis. Biochimica et biophysica acta. 2014;1840:3320–3334. doi: 10.1016/j.bbagen.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 91.Tsai SY, et al. Sigma-1 receptor chaperones in neurodegenerative and psychiatric disorders. Expert opinion on therapeutic targets. 2014;18:1461–1476. doi: 10.1517/14728222.2014.972939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ullah MI, et al. In silico analysis of SIGMAR1 variant (rs4879809) segregating in a consanguineous Pakistani family showing amyotrophic lateral sclerosis without frontotemporal lobar dementia. Neurogenetics. 2015;16:299–306. doi: 10.1007/s10048-015-0453-1. [DOI] [PubMed] [Google Scholar]
- 93.Vollrath JT, et al. Loss of function of the ALS protein SigR1 leads to ER pathology associated with defective autophagy and lipid raft disturbances. Cell death & disease. 2014;5:e1290. doi: 10.1038/cddis.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Behensky AA, et al. Afobazole activation of sigma-1 receptors modulates neuronal responses to amyloid-beta25–35. The Journal of pharmacology and experimental therapeutics. 2013;347:468–477. doi: 10.1124/jpet.113.208330. [DOI] [PubMed] [Google Scholar]
- 95.Furuse T, Hashimoto K. Sigma-1 receptor agonist fluvoxamine for delirium in patients with Alzheimer’s disease. Annals of general psychiatry. 2010;9:6. doi: 10.1186/1744-859X-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lahmy V, et al. Blockade of Tau hyperphosphorylation and Abeta(1)(−)(4)(2) generation by the aminotetrahydrofuran derivative ANAVEX2-73, a mixed muscarinic and sigma(1) receptor agonist, in a nontransgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:1706–1723. doi: 10.1038/npp.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin J, et al. Sigma-1 (sigma(1)) receptor deficiency reduces beta-amyloid(25–35)-induced hippocampal neuronal cell death and cognitive deficits through suppressing phosphorylation of the NMDA receptor NR2B. Neuropharmacology. 2015;89:215–224. doi: 10.1016/j.neuropharm.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 98.Yao H, et al. Molecular mechanisms involving sigma receptor-mediated induction of MCP-1: implication for increased monocyte transmigration. Blood. 2010;115:4951–4962. doi: 10.1182/blood-2010-01-266221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buch S, et al. Cocaine and HIV-1 interplay in CNS: cellular and molecular mechanisms. Current HIV research. 2012;10:425–428. doi: 10.2174/157016212802138823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim SG, et al. Cocaine exposure enhances permissiveness of quiescent T cells to HIV infection. Journal of leukocyte biology. 2013;94:835–843. doi: 10.1189/jlb.1112566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y, et al. Sigma-1 receptor agonists provide neuroprotection against gp120 via a change in bcl-2 expression in mouse neuronal cultures. Brain research. 2012;1431:13–22. doi: 10.1016/j.brainres.2011.10.053. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, et al. Involvement of sigma-1 receptor in astrocyte activation induced by methamphetamine via up-regulation of its own expression. Journal of neuroinflammation. 2015;12:29. doi: 10.1186/s12974-015-0250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang X, et al. Cocaine enhances HIV-1 gp120-induced lymphatic endothelial dysfunction in the lung. Physiological reports. 2015;3 doi: 10.14814/phy2.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hyrskyluoto A, et al. Sigma-1 receptor agonist PRE084 is protective against mutant huntingtin-induced cell degeneration: involvement of calpastatin and the NF-kappaB pathway. Cell death & disease. 2013;4:e646. doi: 10.1038/cddis.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miki Y, et al. Sigma-1 receptor is involved in degradation of intranuclear inclusions in a cellular model of Huntington’s disease. Neurobiology of disease. 2015;74:25–31. doi: 10.1016/j.nbd.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 106.Squitieri F, et al. Pridopidine, a dopamine stabilizer, improves motor performance and shows neuroprotective effects in Huntington disease R6/2 mouse model. Journal of cellular and molecular medicine. 2015;19:2540–2548. doi: 10.1111/jcmm.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Squitieri F, de Yebenes JG. Profile of pridopidine and its potential in the treatment of Huntington disease: the evidence to date. Drug design, development and therapy. 2015;9:5827–5833. doi: 10.2147/DDDT.S65738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roh DH, et al. Sigma-1 receptor-induced increase in murine spinal NR1 phosphorylation is mediated by the PKCalpha and epsilon, but not the PKCzeta, isoforms. Neuroscience letters. 2010;477:95–99. doi: 10.1016/j.neulet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 109.Yoon SY, et al. An increase in spinal dehydroepiandrosterone sulfate (DHEAS) enhances NMDA-induced pain via phosphorylation of the NR1 subunit in mice: involvement of the sigma-1 receptor. Neuropharmacology. 2010;59:460–467. doi: 10.1016/j.neuropharm.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 110.Tseng LF, et al. (+)-Morphine attenuates the (−)-morphine-produced tail-flick inhibition via the sigma-1 receptor in the mouse spinal cord. Life sciences. 2011;89:875–877. doi: 10.1016/j.lfs.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nieto FR, et al. Role of sigma-1 receptors in paclitaxel-induced neuropathic pain in mice. The journal of pain : official journal of the American Pain Society. 2012;13:1107–1121. doi: 10.1016/j.jpain.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 112.Romero L, et al. Pharmacological properties of S1RA, a new sigma-1 receptor antagonist that inhibits neuropathic pain and activity-induced spinal sensitization. British journal of pharmacology. 2012;166:2289–2306. doi: 10.1111/j.1476-5381.2012.01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bangaru ML, et al. Sigma-1 receptor expression in sensory neurons and the effect of painful peripheral nerve injury. Molecular pain. 2013;9:47. doi: 10.1186/1744-8069-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi SR, et al. Spinal sigma-1 receptors activate NADPH oxidase 2 leading to the induction of pain hypersensitivity in mice and mechanical allodynia in neuropathic rats. Pharmacological research. 2013;74:56–67. doi: 10.1016/j.phrs.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 115.Moon JY, et al. Sigma-1 receptor-mediated increase in spinal p38 MAPK phosphorylation leads to the induction of mechanical allodynia in mice and neuropathic rats. Experimental neurology. 2013;247:383–391. doi: 10.1016/j.expneurol.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 116.Moon JY, et al. sigma1 receptors activate astrocytes via p38 MAPK phosphorylation leading to the development of mechanical allodynia in a mouse model of neuropathic pain. British journal of pharmacology. 2014;171:5881–5897. doi: 10.1111/bph.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pan B, et al. Sigma-1 receptor antagonism restores injury-induced decrease of voltage-gated Ca2+ current in sensory neurons. The Journal of pharmacology and experimental therapeutics. 2014;350:290–300. doi: 10.1124/jpet.114.214320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Roh DH, Yoon SY. Sigma-1 receptor antagonist, BD1047 reduces nociceptive responses and phosphorylation of p38 MAPK in mice orofacial formalin model. Biological & pharmaceutical bulletin. 2014;37:145–151. doi: 10.1248/bpb.b13-00690. [DOI] [PubMed] [Google Scholar]
- 119.Sanchez-Fernandez C, et al. Modulation of peripheral mu-opioid analgesia by sigma1 receptors. The Journal of pharmacology and experimental therapeutics. 2014;348:32–45. doi: 10.1124/jpet.113.208272. [DOI] [PubMed] [Google Scholar]
- 120.Vidal-Torres A, et al. Effects of the selective sigma-1 receptor antagonist S1RA on formalin-induced pain behavior and neurotransmitter release in the spinal cord in rats. Journal of neurochemistry. 2014;129:484–494. doi: 10.1111/jnc.12648. [DOI] [PubMed] [Google Scholar]
- 121.Moon JY, et al. Spinal sigma-1 receptor activation increases the production of d-serine in astrocytes which contributes to the development of mechanical allodynia in a mouse model of neuropathic pain. Pharmacological research. 2015;100:353–364. doi: 10.1016/j.phrs.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 122.Mori T, et al. Effects of (+)-pentazocine on the antinociceptive effects of (−)-pentazocine in mice. Synapse. 2015;69:166–171. doi: 10.1002/syn.21799. [DOI] [PubMed] [Google Scholar]
- 123.Ruscher K, et al. The sigma-1 receptor enhances brain plasticity and functional recovery after experimental stroke. Brain : a journal of neurology. 2011;134:732–746. doi: 10.1093/brain/awq367. [DOI] [PubMed] [Google Scholar]
- 124.Hong J, et al. Sigma-1 receptor deficiency reduces MPTP-induced parkinsonism and death of dopaminergic neurons. Cell death & disease. 2015;6:e1832. doi: 10.1038/cddis.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kishi T, et al. Association analysis of SIGMAR1 with major depressive disorder and SSRI response. Neuropharmacology. 2010;58:1168–1173. doi: 10.1016/j.neuropharm.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 126.Nakano M, et al. Fluvoxamine and sigma-1 receptor agonists dehydroepiandrosterone (DHEA)-sulfate induces the Ser473-phosphorylation of Akt-1 in PC12 cells. Life sciences. 2010;86:309–314. doi: 10.1016/j.lfs.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 127.Chevallier N, et al. Behavioural phenotyping of knockout mice for the sigma-1 (sigma(1)) chaperone protein revealed gender-related anxiety, depressive-like and memory alterations. Journal of psychopharmacology. 2011;25:960–975. doi: 10.1177/0269881111400648. [DOI] [PubMed] [Google Scholar]
- 128.Elfverson M, et al. Chronic administration of the anabolic androgenic steroid nandrolone alters neurosteroid action at the sigma-1 receptor but not at the sigma-2 or NMDA receptors. Neuropharmacology. 2011;61:1172–1181. doi: 10.1016/j.neuropharm.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 129.Hayashi T, et al. Targeting ligand-operated chaperone sigma-1 receptors in the treatment of neuropsychiatric disorders. Expert opinion on therapeutic targets. 2011;15:557–577. doi: 10.1517/14728222.2011.560837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sabeti J. Ethanol exposure in early adolescence inhibits intrinsic neuronal plasticity via sigma-1 receptor activation in hippocampal CA1 neurons. Alcoholism, clinical and experimental research. 2011;35:885–904. doi: 10.1111/j.1530-0277.2010.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Villard V, et al. Pharmacological interaction with the sigma1 (sigma1)-receptor in the acute behavioral effects of antidepressants. Journal of pharmacological sciences. 2011;115:279–292. doi: 10.1254/jphs.10191fp. [DOI] [PubMed] [Google Scholar]
- 132.Albayrak Y, Hashimoto K. Beneficial effects of the sigma-1 agonist fluvoxamine for tardive dyskinesia in patients with postpsychotic depressive disorder of schizophrenia: report of 5 cases. The primary care companion for CNS disorders. 2012;14 doi: 10.4088/PCC.12br01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fu Y, et al. Fluvoxamine increased glutamate release by activating both 5-HT(3) and sigma-1 receptors in prelimbic cortex of chronic restraint stress C57BL/6 mice. Biochimica et biophysica acta. 2012;1823:826–837. doi: 10.1016/j.bbamcr.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 134.Ito K, et al. Decreased brain sigma-1 receptor contributes to the relationship between heart failure and depression. Cardiovascular research. 2012;93:33–40. doi: 10.1093/cvr/cvr255. [DOI] [PubMed] [Google Scholar]
- 135.Niitsu T, et al. A randomized, double-blind, placebo-controlled trial of fluvoxamine in patients with schizophrenia: a preliminary study. Journal of clinical psychopharmacology. 2012;32:593–601. doi: 10.1097/JCP.0b013e3182664cfc. [DOI] [PubMed] [Google Scholar]
- 136.Moriguchi S, et al. Stimulation of the sigma-1 receptor by DHEA enhances synaptic efficacy and neurogenesis in the hippocampal dentate gyrus of olfactory bulbectomized mice. PloS one. 2013;8:e60863. doi: 10.1371/journal.pone.0060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Allahtavakoli M, Jarrott B. Sigma-1 receptor ligand PRE-084 reduced infarct volume, neurological deficits, pro-inflammatory cytokines and enhanced anti-inflammatory cytokines after embolic stroke in rats. Brain research bulletin. 2011;85:219–224. doi: 10.1016/j.brainresbull.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 138.Ruscher K, et al. Effects of the sigma-1 receptor agonist 1-(3,4-dimethoxyphenethyl)-4-(3-phenylpropyl)-piperazine dihydro-chloride on inflammation after stroke. PloS one. 2012;7:e45118. doi: 10.1371/journal.pone.0045118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sato S, et al. Antidepressant fluvoxamine reduces cerebral infarct volume and ameliorates sensorimotor dysfunction in experimental stroke. Neuroreport. 2014;25:731–736. doi: 10.1097/WNR.0000000000000162. [DOI] [PubMed] [Google Scholar]
- 140.Urfer R, et al. Phase II trial of the Sigma-1 receptor agonist cutamesine (SA4503) for recovery enhancement after acute ischemic stroke. Stroke; a journal of cerebral circulation. 2014;45:3304–3310. doi: 10.1161/STROKEAHA.114.005835. [DOI] [PubMed] [Google Scholar]
- 141.Dong H, et al. Sigma-1 Receptor Modulates Neuroinflammation After Traumatic Brain Injury. Cellular and molecular neurobiology. 2015 doi: 10.1007/s10571-015-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]