The Lipid Bilayer Modulates the Structure and Function of an ATP-binding Cassette Exporter (original) (raw)
Abstract
ATP-binding cassette exporters use the energy of ATP hydrolysis to transport substrates across membranes by switching between inward- and outward-facing conformations. Essentially all structural studies of these proteins have been performed with the proteins in detergent micelles, locked in specific conformations and/or at low temperature. Here, we used luminescence resonance energy transfer spectroscopy to study the prototypical ATP-binding cassette exporter MsbA reconstituted in nanodiscs at 37 °C while it performs ATP hydrolysis. We found major differences when comparing MsbA in these native-like conditions with double electron-electron resonance data and the crystal structure of MsbA in the open inward-facing conformation. The most striking differences include a significantly smaller separation between the nucleotide-binding domains and a larger fraction of molecules with associated nucleotide-binding domains in the nucleotide-free apo state. These studies stress the importance of studying membrane proteins in an environment that approaches physiological conditions.
Keywords: ABC transporter, fluorescence resonance energy transfer (FRET), membrane bilayer, multidrug transporter, spectroscopy, FRET, LRET, MsbA, luminescence resonance energy transfer, nanodisc
Introduction
ATP-binding cassette (ABC)2 transporters constitute one of the largest families of membrane proteins and are found in all domains of life (1–3). They can be either importers (most prokaryote ABC transporters) or exporters (most mammal ABC transporters) (1–3). The core structure of ABC proteins consists of two transmembrane domains that form the translocation pathway and two conserved nucleotide-binding domains (NBDs) that bind and hydrolyze ATP (1–3), a process essential for their function. Binding of ATP promotes the formation of an NBD dimer in a head to tail orientation (4, 5). In the resulting structure two ATP molecules are “sandwiched” at the dimer interface, and residues from both NBDs form each of the two nucleotide-binding sites (1, 4, 5). NBD dimerization is essential for ATP hydrolysis and requires ATP binding to both nucleotide-binding sites (6), whereas dissociation of the dimers occurs following hydrolysis at only one of the two sites (7). This ATP-dependent NBDs dimerization/dissociation process is coupled to rearrangements of the transmembrane helices, switching the transporters from an inward-facing conformation (dissociated NBDs) to an outward-facing conformation (dimeric NBDs), with the concomitant translocation of substrate (1, 3).
Two of the most studied ABC exporters are the multidrug resistance protein P-glycoprotein (MDR1 and ABCB1) that plays a role in the resistance to chemotherapy of some forms of cancer (3),and its bacterial homolog, the lipid flippase MsbA (8). MsbA is located in the inner membrane of Gram-negative bacteria, where it transports lipid A from the inner to the outer leaflet (9, 10). MsbA has been crystallized in inward- and outward-facing conformations (11) and has also been extensively studied by different spectroscopic techniques (12–18). Several studies point to large motions between the nucleotide-free “open” inward-facing conformation (NBDs separated by tens of Angstroms) and the ATP-bound “closed” outward-facing conformation (tightly associated NBDs). These observations disagree with evidence suggesting that the NBDs remain in close contact at all times during the transport cycle and that such large conformational changes might not be physiological (19–22).
For simplicity and because of technical limitations, essentially all structural studies of ABC exporters have been performed with the proteins solubilized in detergent, locked in specific conformations and/or at low temperature (11, 12, 14, 18, 23–26). However, understanding of the structure-function of membrane proteins requires their study under more physiological conditions, at a minimum: 1) reconstitution into lipid bilayers, their native environment; 2) physiological temperatures; and 3) functioning conditions, as opposed to specific locked conformations. As part of our efforts to study ABC proteins under more natural experimental conditions, we decided to use luminescence resonance energy transfer (LRET) on MsbA reconstituted in nanodiscs. Nanodiscs are nanometer scale discoidal structures that contain a phospholipid bilayer and are soluble and stable (27). LRET is a spectroscopic technique that displays high sensitivity and allows for studies of proteins while they are functional (18, 28–31). LRET is somewhat similar to the traditional FRET, but it has a number of advantages for studies of membrane proteins. The most significant relates to the properties of the donor (Tb3+ in these studies), which displays atomic-like emission and long lifetime of the excited state (ms versus ns of traditional fluorophores) (31). The sharp Tb3+ emission peaks allow for easy isolation of the acceptor emission, whereas the long donor lifetime makes delayed acquisition possible; i.e. the detector can be gated for acquisition with a delay of hundreds of μs after the excitation pulse. Delayed detector gating minimizes light-scattering artifacts, and the use of the donor with long-lifetime emission makes the calculation of donor-acceptor distances independent of labeling stoichiometry (31).
We found that MsbA reconstituted in nanodiscs, and at physiological temperature, showed major differences with the published crystal structure in the open inward-facing conformation, as well as with double electron-electron resonance (DEER) experiments and LRET experiments performed with MsbA in detergent micelles (11, 13, 14, 18).
Experimental Procedures
Protein Expression and Purification
A Cys-less Salmonella typhimurium MsbA with an N-terminal decahistidine tag (MsbA CL, the designation for MsbA with mutations C88A and C315A) was used as background to generate the single Cys mutant T561C. The Glu-506 to Gln mutation was introduced into T561C to generate the catalytically inactive E506Q/T561C mutant. MsbA was expressed and purified essentially as described (18). Briefly, MsbA expression in BL21 DE3-RILP Escherichia coli cells (Agilent Technologies, Santa Clara, CA) was induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 4 h at 30 °C. Membrane proteins were solubilized for 1 h at room temperature with 2% _n_-dodecyl-β-d-maltopyranoside (dodecylmaltoside) and 0.04% sodium cholate in a buffer containing 100 mm NaCl, 20 mm Tris/HCl, pH 8, 15% glycerol, 0.5 mm Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), and 0.5 mm phenylmethanesulfonyl fluoride. Solubilized MsbA was purified by metal affinity chromatography (Talon Superflow; Clontech) followed by size exclusion chromatography in a P-6DG column (Bio-Rad) equilibrated with storage buffer (100 mm NaCl, 20 mm Tris/HCl, pH 7.5, 0.065% dodecylmaltoside, 0.04% sodium cholate, 15% glycerol, and 0.2 mm TCEP). MsbA was concentrated to >20 μm and stored at −80 °C until use. Protein concentration was determined by the BCA assay (Bio-Rad), and purity was estimated at >95% from SDS-PAGE gels stained with Coomassie Blue.
ATPase Measurements
ATP hydrolysis was measured using a variant of the linked enzyme assay (18, 32).
Protein Labeling
T561C and E506Q/T561C were labeled with thiol-reactive LRET probes. First, storage buffer was exchanged to a glycerol/TCEP-free buffer (100 mm NaCl, 20 mm Tris/HCl pH 7.5, 0.065% dodecylmaltoside, and 0.04% sodium cholate) using a spin column (Zeba; Thermo Fisher Scientific, Rockford, IL). The protein was immediately labeled with both the LRET donor Lanthascreen (thiol-reactive Tb3+-chelate DTPA-cs124-EMPH) and the LRET acceptor Bodipy FL (Bodipy: _N_-(2-aminoethyl) maleimide). Both probes were obtained from Life Technologies. Labeling proceeded with the donor and acceptor probes at a 2-fold molar Cys excess for 1 h at room temperature. Nonspecific labeling evaluated using MsbA CL was negligible, as shown previously (18). For some control experiments, T561C was labeled only with Tb3+ or Bodipy FL under similar conditions. For experiments of MsbA in detergent micelles, the labeled protein was run through a Superdex 200 10/300 gel filtration column (GE Healthcare) equilibrated with 100 mm NaCl, 20 mm Tris/HCl, pH 7.5, 0.065% dodecylmaltoside, 0.04% sodium cholate, and 0.2 mm TCEP) to remove free unreacted LRET probes. For experiments of MsbA in a lipid bilayer the free unreacted LRET probes were removed after labeled MsbA had been reconstituted, as described below. We performed the labeling before reconstitution because in that way we could use the same labeled protein for experiments in detergent or after reconstitution in nanodiscs or liposomes. In a previous publication, we labeled MsbA after reconstitution in liposomes (18), and therefore, there is no indication that the accessibility of Cys-561 is limited after reconstitution.
MsbA Reconstitution
Labeled MsbA was reconstituted in nanodiscs following published methodology (27) with some modifications. The membrane scaffold protein (MSP) E3D1 (27) was expressed in BL21 DE3-RILP E. coli cells transformed with the plasmid pMSP1E3D1 (Addgene, Cambridge, MA). Expression was induced at an _A_600 of 1–2 with 1 mm isopropyl-β-d-thiogalactopyranoside, for 2 h at 37 °C, and MSP was purified by metal affinity chromatography (Talon; Clontech) (27). For some experiments, the His tag of the MSP was removed by digestion with TEV protease, and the nontagged MSP was isolated as the flow through on an immobilized Co2+ column. E. coli total lipid extract in chloroform (Avanti Polar Lipids, Alabaster, AL) was dried overnight and solubilized in nanodisc buffer (100 mm NaCl, 20 mm Tris/HCl, pH 7.5, 0.1 mm TCEP) with 100 mm sodium cholate. The MsbA:MSP:lipid ratio was 1:8:110. The MsbA/MSP/lipid mix was incubated for 1 h at 4 °C with gently rotation, and the detergent was then removed by overnight incubation with Biobeads SM-2 (Bio-Rad). The sample was run through a Superdex 200 10/300 gel filtration column (GE Healthcare) equilibrated in nanodisc buffer to remove free unreacted LRET probes and to collect the fraction enriched in MsbA-containing nanodiscs that was used for the experiments. For some experiments, an additional metal affinity chromatography step was performed to separate the MsbA-containing nanodiscs that bind to the resin from the empty nanodiscs. MsbA concentration in the nanodisc samples was estimated on SDS-PAGE gels stained with Coomassie Blue, using detergent-solubilized MsbA of known concentration as standard. For some control experiments, reconstitution was performed in a mixture of E. coli purified phosphatidylethanolamine, phosphatidyl-glycerol and cardiolipin (Avanti Polar Lipids) at a 70🔞12 ratio instead of the E. coli lipid extract that contains ∼18% of unknown components. The ATPase and LRET results obtained with this mix of lipids were undistinguishable from those obtained with the total lipid extract (not shown). For reconstitution in liposomes, E. coli total lipid extract was used at a protein:lipid ratio of 1:200, and detergent removal was done by either overnight incubation with Biobeads SM-2 or by gel filtration.
LRET
LRET recordings were performed essentially as described (6, 7, 18, 28). Nanodisc samples containing 0.1–0.5 μm of labeled MsbA were analyzed in 3-mm path length quartz cuvettes before and after successive additions of ATP, MgSO4, and sodium orthovanadate (Vi). The different states during the ATP hydrolysis cycle during these LRET experiments are defined as follows: apo, nucleotide-free state obtained in nominally divalent cation-free buffer with 1 mm EDTA to chelate trace divalent cations and prevent ATP hydrolysis; ATP-bound, ATP-bound state obtained after addition of 1 mm NaATP; MgATP, continuous hydrolysis state obtained after addition of 10 mm MgSO4; and MgATP + Vi, high energy posthydrolysis state obtained by addition of 0.25 mm Vi in the presence of MgATP. Emission was measured on either a Photon Technology International spectrometer (QM3SS; PTI, London, Canada) or an Optical Building Blocks phosphorescence lifetime photometer (EasyLife L; OBB, Birmingham, NJ). The emission was recorded with a 200-μs delay from the beginning of a ∼1-μs excitation pulse from a xenon flash lamp. This 200-μs delay allows the selective recording of long lifetime processes (donor emission or sensitized emission from the acceptor) after the short lifetime processes (e.g. acceptor emission resulting from direct excitation, scattering of the excitation pulse) have largely disappeared. Excitation was set to 335 nm, and emission was collected through bandpass filters (Tb3+: 490/10 nm; Bodipy FL: 520/10 nm, Omega Optical, Brattleboro, VT). Control experiments mixing equal proportions of nanodiscs containing MsbA labeled with Tb3+-only and Bodipy FL-only showed essentially not intermolecular LRET. Similar conditions were used for parallel LRET experiments of MsbA in detergent micelles.
LRET Decay Analysis
Distances between the donor and acceptor probes were calculated according to: E = 1 − τDA/τD, and r = _R_0(_E_−1 − 1)1/6, where E is the efficiency of energy transfer, τD is the lifetime of the donor in the absence of acceptor, τDA is the sensitized emission lifetime (lifetime of the acceptor that that arises from LRET), R is the donor-acceptor distance, and _R_0 is the Förster distance (the distance at which E = 0.5). The _R_0 for the Tb3+/Bopidy pair was 41 Å, making this LRET pair very sensitive to distances changes in the ∼25–55 Å range). Lifetimes were determined by fitting of the LRET decays to a multiexponential function, and the goodness of fit was determined from the random residual distribution, which showed no structure and chi-squared values near unity. The decay from Tb3+-only MsbA-nanodisc complexes (recorded at 490 nm) was well fitted by a two-exponential function with lifetimes τD1 = 474 ± 107 μs (5% of the signal) and τD2 = 1,867 ± 41 μs (95% of the signal). This longest lifetime was used as the donor only τD. All Bodipy FL sensitized emission decays (recorded at 520 nm) were well fitted by a three-exponential decay. However, the fastest component (shortest distance; τDA = 100–200 μs) was disregarded for the calculations of distances and percentage of molecules in each conformation because the contribution of this exponential component to the whole signal was ≤5% and did not change appreciably under different experimental conditions, and it is likely the result of a very small fraction of protein aggregates and/or instrument artifacts, as described (18, 28). The number of donor-acceptor pairs that contribute to a distance component (percentage MsbA molecules in each conformation) was estimated from the relative intensity of each lifetime component corrected by the rate of energy transfer (k = 1/τDA − 1/τD) (18, 33). To assess whether LRET intensity decays corresponded to discrete multiexponential decays or a distribution of lifetimes, we analyzed the data by an exponential series method (Felix 32 analysis software; PTI) designed to recover lifetime distributions without any a priori assumptions about their shapes (34, 35). We analyzed the decays using a series of 200 exponentials with fixed, logarithmically spaced lifetimes and variable pre-exponentials.
It is important to mention that our typical LRET distances have standard deviations below 2 Å (18, 28) (see Table 1). This is not the error in the calculated LRET distance versus the distance expected from the crystal structures, but the result of very high experimental reproducibility with atomic resolution. There is an unavoidable uncertainty about the position of the optical probes because of their length, which introduces an error when distances between the probes are assumed to be those between the reacted thiols in the Cys side chains. However, because measurements from others and us have shown remarkably close distances to those expected from crystal structures (18, 28, 30, 36), LRET seems more than adequate to address domain movements in MsbA.
TABLE 1.
Tb3+-Bodipy FL distances during the catalytic cycle of MsbA
The two dominant exponential components of the sensitized emission decay are presented as distances 1 and 2. τDA indicates Bodipy FL-sensitized emission lifetime; R is the calculated donor-acceptor distance; % molecules is the percentage of molecules displaying the corresponding distance. Apo, ATP-bound, MgATP, and MgATP + Vi states were obtained as described under “Experimental Procedures.” The data are presented as means ± S.D., and n is the number of independent experiments in each conformational state.
| State | Distance 1 | Distance 2 | ||||
|---|---|---|---|---|---|---|
| τDA | _R_1 | % molecules | τDA | _R_2 | % molecules | |
| μ_s_ | Å | μ_s_ | Å | |||
| T561C nanodiscs (n = 8) | ||||||
| Apo | 577 ± 16 | 35.9 ± 0.3 | 48 ± 7 | 1313 ± 36 | 47.4 ± 0.7 | 52 ± 7 |
| ATP-bound | 559 ± 9 | 35.6 ± 0.1 | 71 ± 3 | 1204 ± 89 | 45.2 ± 1.4 | 29 ± 3 |
| MgATP | 573 ± 56 | 35.8 ± 0.8 | 51 ± 7 | 1210 ± 85 | 45.5 ± 1.6 | 49 ± 7 |
| MgATP + Vi | 604 ± 21 | 36.3 ± 0.3 | 70 ± 4 | 1232 ± 78 | 45.9 ± 1.5 | 30 ± 4 |
| E506Q/T561C nanodiscs (n = 3) | ||||||
| Apo | 669 ± 1 | 37.2 ± 0.1 | 42 ± 5 | 1345 ± 19 | 48.0 ± 0.4 | 58 ± 5 |
| ATP-bound | 568 ± 2 | 35.7 ± 0.1 | 76 ± 1 | 1179 ± 107 | 44.9 ± 1.8 | 24 ± 1 |
| MgATP | 590 ± 3 | 36.1 ± 0.1 | 75 ± 1 | 1227 ± 143 | 45.8 ± 2.6 | 25 ± 1 |
| MgATP + Vi | 579 ± 1 | 35.9 ± 0.1 | 74 ± 1 | 1160 ± 100 | 44.8 ± 1.4 | 26 ± 1 |
| T561C detergent (n = 6) | ||||||
| Apo | 535 ± 7 | 35.1 ± 0.1 | 7 ± 1 | 1543 ± 18 | 52.5 ± 0.6 | 93 ± 1 |
| ATP-bound | 542 ± 15 | 35.3 ± 0.2 | 63 ± 4 | 1386 ± 118 | 48.7 ± 1.4 | 37 ± 4 |
| MgATP | 550 ± 24 | 35.3 ± 0.4 | 41 ± 7 | 1448 ± 75 | 50.1 ± 2.0 | 59 ± 7 |
| MgATP + Vi | 576 ± 15 | 35.7 ± 0.2 | 61 ± 3 | 1372 ± 97 | 48.3 ± 2.1 | 39 ± 3 |
Stopped Flow LRET Studies
To study the kinetics of the conformational changes that take place after adding ATP or switching from the ATP-bound state to the continuous hydrolysis state (MgATP state), we performed LRET experiments at 37 °C using a stopped flow mixing accessory (RX2000; Applied Photophysics, Surrey, UK) with a 20-μl cell (<10-ms dead time), placed in the sample compartment of the EasyLife L lifetime photometer.
Data Presentation and Statistics
The data are shown as means ± S.E. or S.D., as indicated. Statistical comparisons were performed by the Student's t test for paired or unpaired data or one-way analysis of variance, as appropriate. p < 0.05 in a two-tail analysis was considered significant. The number of experiments (n) corresponds to independent measurements from at least three different protein preparations.
Results
MsbA Reconstitution and ATPase Activity
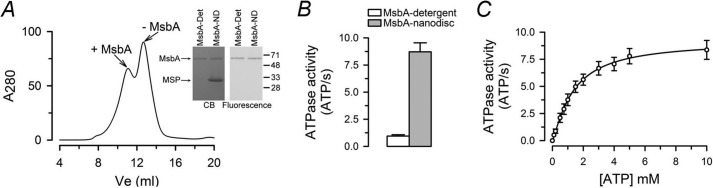
To study the functioning of an ABC transporter in a lipid bilayer and at physiological temperature, we reconstituted detergent-solubilized MsbA in nanodiscs, self-assembled complexes that consist of a lipid bilayer encased within a MSP (27). We used the same single-Cys mutant (T561C) that we recently studied in MsbA-detergent micelles to compare MsbA structure and function in nanodiscs versus detergent. Cys-561 is on the outside of the NBD, readily accessible for labeling by thiol-selective LRET probes, and its ATPase activity is not affected by labeling (18). Fig. 1A shows a typical size exclusion chromatography profile of the MsbA-nanodisc preparation following reconstitution. Fig. 1A also shows a SDS-PAGE of the fraction enriched in MsbA-containing nanodiscs (MsbA-ND, the heavier peak on the left) that confirmed that only MsbA is labeled with the LRET probes, as is the case for MsbA in detergent. The rate of ATP hydrolysis of MsbA-ND was ∼10-fold faster than that of MsbA in detergent (8.7 ± 0.8 versus 0.9 ± 0.1 ATP/s; Fig. 1B), with a Km for MgATP similar to that previously reported (Fig. 1C) (14, 18). This higher ATPase activity for MsbA-ND versus MsbA-detergent is in agreement with previous reports (14, 37) and could result from a stimulatory effect of the lipids. Phospholipids increase the ATPase activity of MsbA in detergent, and the protein reconstituted in liposomes displays higher ATP hydrolysis rate compared with the protein in detergent (10, 17). The rate of ATP hydrolysis by MsbA-ND remained relatively high at room temperature (2.3 ± 0.9 ATP/s versus 0.05 ± 0.01 ATP/s in detergent micelles); this rate is still more than twice the hydrolysis rate of MsbA-detergent at 37 °C, showing that the activity of MsbA in the membrane is less sensitive to temperature.
FIGURE 1.
Activity of MsbA-T561C in nanodiscs. A, gel filtration chromatogram of purified MsbA (T561C mutant) reconstituted in nanodiscs. The heavier peak on the left corresponds to MsbA-nanodisc complexes (+ MsbA), whereas the peak on the right corresponds to empty nanodiscs (− MsbA). _A_280 is the absorbance measured at 280 nm. The panels on the right correspond to purified T561C labeled with Bodipy FL that was either in detergent micelles (MsbA-Det) or reconstituted in nanodiscs (MsbA-ND). The samples were subjected to SDS-PAGE, and the gel was visualized by Coomassie Blue staining (CB) and fluorescence. The positions of molecular mass markers (in kDa) are indicated on the right. The arrows point to purified MsbA and MSP. B, ATPase activity of purified MsbA T561C at 37 °C. The values are presented as means ± S.E. (n = 7 for each condition). The hydrolysis rate of MsbA in nanodiscs was significantly higher than that of MsbA in detergent (p < 0.001). C, dependence of the MsbA-ND ATPase activity on ATP concentration. The measurements were performed at 37 °C, with Mg2+ kept constant at 12 mm. The values are presented as means ± S.E. (n = 4). The solid line is a fit of the Hill equation to the data (_V_max = 9.2 ± 0.2 ATP/s; Km = 1.4 ± 0.1 mm; n = 1.2 ± 0.1).
LRET Spectra and Time Course of Intensity Changes
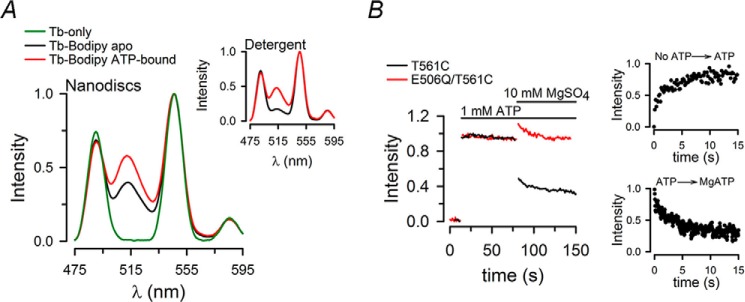
For these experiments, the single-Cys T561C MsbA mutant was labeled with Tb3+ only or with Tb3+ and Bodipy FL. The LRET emission spectra of the labeled MsbA-ND **(**Fig. 2_A_) displayed the typical sharp emission peaks from Tb3+ and a long lifetime emission of the acceptor (peak at ∼515 nm) that arises from LRET (sensitized emission). The ATP-bound protein (red) showed a large LRET sensitized emission, but contrary to what was observed in MsbA-detergent (Fig. 2A, inset), there was also a significant LRET emission in the apo state (black). This increased LRET emission from MsbA-ND in the apo state was the result of a larger proportion of molecules with “associated” NBDs (see below). Fig. 2B shows the time course of the increase in LRET emission after manual addition of NaATP to the active MsbA (T561C, black) or the catalytically inactive mutant E506Q/T561C (red), and the response to later addition of Mg2+ to promote hydrolysis. Fig. 2B also shows the changes induced by ATP and Mg in the active protein after rapid mixing in a stopped flow device. The ATP-induced increase in intensity follows the NBD dimerization process (as more NBDs get closer to each other the average distance between the probes decreases and LRET increases). This occurred in both active and catalytically inactive MsbA because they both bind ATP (18). The subsequent addition of Mg2+ to T561C in the nucleotide-bound conformation resulted in a partial reversal of the increase in intensity (black), which agrees with the presence of a larger fraction of dissociated NBDs after hydrolysis, as reported for MsbA in detergent (18). As expected, the Mg-induced decrease in LRET intensity was not observed in the inactive E506Q/T561C MsbA (red) because dimer dissociation follows ATP hydrolysis (18). These results show that conformational changes of MsbA in nanodiscs can be followed by LRET. In general, the changes in intensity follow a similar pattern to those observed in MsbA-detergent (18). However, the changes in MsbA-ND were faster, perhaps because of a stimulatory effect of the lipids discussed above. The half-life of ATP-induced dimerization in MsbA-ND was ∼5 s (Fig. 2B), whereas for MsbA-detergent it was ∼1 min (18). The transition of the MsbA-ND from the NaATP-bound state to the continuous hydrolysis state (in MgATP) was also fast (Fig. 2B). These faster transitions between states are consistent with the faster rate of ATP hydrolysis in nanodiscs versus detergent. It is important to notice that these LRET experiments have been performed under a saturating ATP concentration (1 mm) because the apparent Kd for ATP induced dimerization of MsbA in nanodiscs was 95 ± 3 μm. Similarly to what has been observed in detergent, the Kd for ATP-induced dimerization of the NBDs is much lower than the Km(ATP) for the ATPase activity, suggesting that dimerization is not the rate-limiting step of the hydrolysis cycle (18). Also, because the [MgATP] in these experiments is close to the Km(ATP), the decrease in LRET sensitized emission in response to Mg2+ may be smaller than the maximal attainable effect. We did not perform experiments at high [ATP] because quenching of the signal by the nucleotide makes the analysis difficult (18).
FIGURE 2.
Effects of ATP on LRET spectra and sensitized emission intensity changes. A, emission spectra from T561C labeled with Tb3+ only in the apo state (Tb-only, green) or Tb3+ and Bodipy FL in the apo (Tb-Bodipy apo, black) and ATP-bound (Tb-Bodipy ATP-bound, red) states. Protein concentration was 0.5 μm, and 1 mm ATP was present for at least 5 min before collecting the spectra. The data were normalized to the 546-nm Tb3+ emission peak. Spectra from MsbA in nanodiscs (main panel) and in detergent (inset) are shown. B, typical changes in Bodipy FL sensitized emission in T561C-nanodisc (T561C, black) and E506Q/T561C-nanodisc (E506Q/T561C, red) complexes in response to sequential additions of ATP and MgSO4. Signals were normalized to the total change elicited by ATP. Stopped flow LRET records are shown in the smaller panels on the right to display changes that were too fast to follow in a standard cuvette (main panel). Records are representative of data from at least five similar experiments. All data were obtained at 37 °C.
LRET Decays during the ATP Hydrolysis Cycle
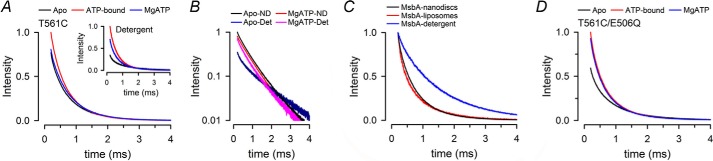
LRET is a very sensitive technique to measure changes in distance between donor-acceptor probes attached to a protein (18, 28, 30, 38). Distances can be calculated from the sensitized emission decay of the acceptor after a single excitation pulse, as described under “Experimental Procedures” and previous publications (18, 28). Fig. 3A shows typical LRET sensitized emission intensity decays from MsbA-ND in the nucleotide-free (Apo, black) and ATP-bound states (red) and under continuous hydrolysis in the presence of MgATP (blue). For comparison, the equivalent intensity decays from MsbA-detergent complexes are also shown (Fig. 3A, inset). There were two noticeable differences between the decays of MsbA-ND and MsbA-detergent: 1) the increase in LRET intensity elicited by shifting MsbA from the apo to the ATP-bound state was significantly smaller when the protein was incorporated in nanodiscs (black versus red lines in Fig. 3A; which is also consistent with the spectra in Fig. 2A); and 2) the LRET intensity decay during continuous ATP hydrolysis in MsbA-ND was closer to that in the apo state (Fig. 3A; black versus blue lines), whereas in MsbA-detergent this decay (blue) was intermediate between those in the apo (black) and ATP-bound states (red) (Fig. 3A, inset). When graphed together in a log scale (Fig. 3B), it was evident that during hydrolysis conditions, the LRET decays of MsbA-ND and MsbA-detergent were not very different; the main difference was the slow decay observed only for the apo MsbA-detergent. The LRET decay in the apo state was also faster when MsbA was reconstituted in unilamellar liposomes (Fig. 3C), indicating that this faster decay was an effect of the lipid bilayer and not the result of reconstitution in nanodiscs. Thus, in the absence of nucleotide, MsbA appears to adopt different conformations when reconstituted in the membrane and when in detergent micelles. The catalytically inactive MsbA mutant E506Q/T561C-ND (Fig. 3D) also showed a large LRET intensity in the apo state. As expected, the LRET intensity increased with ATP but did not decrease by addition of Mg2+ because the Mg-induced NBDs dimer dissociation requires hydrolysis (18). Together, these results suggest that apo state MsbA in a lipid bilayer adopts a conformation(s) closer to that of the ATP-bound protein in detergent than to that of the apo state MsbA in detergent.
FIGURE 3.
Sensitized Bodipy FL emission decays during the ATP-hydrolysis cycle. A, sensitized Bodipy FL emission decays from MsbA in nanodiscs (T561C-ND, main panel) and in detergent (T561C-Det, inset). Black, Apo; red, ATP-bound; blue, MgATP. Intensities were normalized to the ATP intensity at 200 μs. The traces are representative of eight similar experiments. B, semilog graph of selected LRET decays from A. Apo-ND, nucleotide-free T561C in nanodiscs; MgATP-ND, T561C in nanodiscs in the presence of MgATP; Apo-Det, nucleotide-free T561C in detergent; MgATP-ND, T561C in detergent in the presence of MgATP. C, comparison of the sensitized Bodipy FL emission decays from T561C in detergent, nanodiscs and liposomes. The data were obtained in the absence of nucleotides (apo state), and the intensities were normalized to the intensity at 200 μs to emphasize the faster decays in liposomes and nanodiscs, but the intensities in liposomes and nanodiscs were higher than those in detergent as a result of the increased LRET (see A, B, and D). The traces are representative of three similar experiments. D, LRET decays from E506Q/T561C in nanodiscs. See A for details. All experiments were performed at 37 °C.
NBD-NBD Distance Changes during the ATP Hydrolysis Cycle
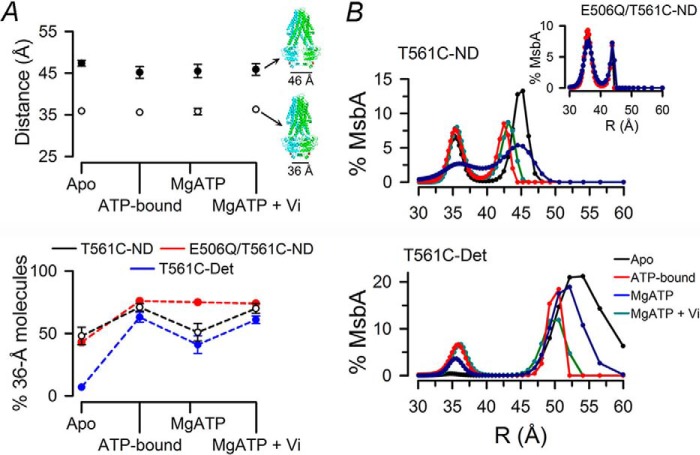
As previously observed for MsbA in detergent micelles (18), the LRET decays from MsbA-ND occurred in a multiexponential fashion (Fig. 3), indicating the existence of more than one donor-acceptor distance in every experimental condition. Fig. 4A and Table 1 show the distances calculated from the two dominant lifetimes of the LRET decays and the percentage of MsbA molecules displaying each distance in the different states. The LRET decays of MsbA-ND were described by two coexistent distances (Fig. 4A, top panel) of ∼36 Å (open circles) and ∼46 Å (closed circles) that remained essentially unchanged during the ATP hydrolysis cycle. The ∼36 Å distance has been observed before in MsbA-detergent (18) and is consistent with the outward-facing, closed conformation structure obtained in the presence of MgAMPPNP (11). However, the longest distance in apo MsbA-ND (47 ± 1 Å) was significantly shorter than that in MsbA-detergent (53 ± 1 Å; Table 1) and much shorter than the distance in the MsbA apo crystal structure (>80 Å) (11). This shorter NBD-NBD distance (represented by a structure with partially opened NBDs: Fig. 4A, top right) was not the result of confinement of MsbA in the nanodiscs, because very similar results were obtained when MsbA was reconstituted in ∼100-nm diameter unilamellar liposomes (18) (Fig. 3C).
FIGURE 4.
Conformational changes during the ATP hydrolysis cycle. A, distances calculated from the LRET sensitized Bodipy FL emission decays and changes in the distribution of molecules during the ATP hydrolysis cycle. Top panel, calculated distances (R) in different states during the hydrolysis cycle in T561C-nanodisc complexes (means ± S.D., n = 8; open and filled circles correspond to _R_1 and _R_2 in Table 1, respectively). Possible MsbA structures associated with each distance are shown on the right. Monomers in the MsbA homodimer are depicted in blue and cyan; Cys-561 is shown in red. The 36 Å structure is represented by the outward-facing conformation (Protein Data Bank code 3B60) with MgAMPPNP bound, and the 46 Å structure was obtained as an intermediate conformation during the morph transition from the open inward-facing (Protein Data Bank code 3B5W) to the outward-facing conformation using PyMOL (Schrödinger). Bottom panel, percentage of MsbA molecules displaying the shorter distance (∼36 Å). The data obtained from T561C in nanodiscs (T561C-ND, black, n = 8), E506Q/T561 in nanodiscs (E506Q/T561C-ND, red, n = 3), and T561C in detergent (T561C-Det, blue, n = 6) are presented as means ± S.D. Percentages of molecules in each conformation were calculated from the fractional intensity contribution of each exponential component divided by the rate of energy transfer (k = 1/τDA − 1/τD). B, distance distributions calculated from the lifetime distributions of LRET sensitized emission intensity decays under different states during the hydrolysis cycle. Black, Apo; red, ATP-bound; blue, Mg-ATP; cyan, MgATP + Vi. The data are from T561C-nanodiscs (top panel) and T561C in detergent (bottom panel). The inset displays distribution of distances from E506Q/T561C-nanodisc complexes. All data were obtained at 37 °C.
Essentially, the results for MsbA-ND can be described by two main conformations: NBDs in close proximity (36 Å distance) or NBDs partially separated (∼46 Å distance). Similar to what has been reported for MsbA in detergent (18), the differences in LRET intensity observed during the hydrolysis cycle were not caused by changes in distances but were mainly the result of changes in the percentage of molecules adopting each conformation (Fig. 4A, bottom panel, and Table 1). Fig. 4A (bottom panel) shows how the proportion of molecules adopting the 36-Å conformation changed during the ATP hydrolysis cycle for MsbA in nanodisc (T561C-ND in black; E506Q/T561C-ND in red) and MsbA in detergent (T561C-Det in blue). From this figure, it is evident that in the absence of nucleotides the proportion of molecules in the 36 Å conformation was significantly higher in MsbA-ND compared with MsbA-detergent (48 ± 7% versus 7 ± 1%; Table 1). This higher proportion of molecules adopting the short distance explains the higher LRET intensity observed for the apo MsbA-ND (Figs. 2A and 3A). In all cases, the percentage of molecules adopting the 36 Å conformation (Fig. 4A, bottom panel) increased in the ATP-bound state, as expected from ATP-induced NBD dimerization. During ATP hydrolysis (in MgATP), this percentage decreased to ∼50% only for the active MsbA (black and blue versus red), as the NBDs dissociate/reassociate during hydrolysis; this steady state where approximately half of the molecules have dimeric NBDs and the other half have dissociated NBDs has been reported before for MsbA in detergent micelles and isolated NBDs, as is the result of continuous NBDs dissociation (after ATP hydrolysis) and redimerization (induced by rebinding of ATP) during the ATP hydrolysis cycle (18, 28, 29). As mentioned above, the percentage of molecules with dissociated NBDs in MgATP is probably underestimated because we performed the experiments at a [MgATP] below saturation for ATP hydrolysis. The equilibrium for the active protein was shifted to a larger proportion of molecules in the 36 Å conformation after ATP hydrolysis was inhibited by Vi. To summarize, the differences in LRET intensities and decays between MsbA-ND and MsbA-detergent can be explained by: 1) a much larger fraction of NBDs associated or in very close proximity in MsbA-ND in the apo state and 2) the substitution of the long distance of fully dissociated NBDs in MsbA-detergent by a shorter distance compatible with loosely associated NBDs or NBDs dissociated but very close to each other.
Fig. 4B shows the typical distribution of distances in the LRET sensitized emission intensity decays analyzed by an exponential series method (see “Experimental Procedures”) (34, 35). This type of analysis can recover lifetime distributions (which can be converted to distance distributions) without assumptions about their shapes, and it can therefore be used to discriminate between continuous distributions and discrete multiexponentials decays (34, 35). Clear peaks centered at the distance values obtained from the multiexponential fits confirmed the presence of discrete conformations in MsbA-ND in the apo, ATP-bound, and Vi-inhibited states (Fig. 4B, top panel). However, in MsbA-ND during continuous ATP hydrolysis (MgATP, blue), the distance distribution was broader, suggesting that in addition to the dominant conformations of ∼36 and ∼46 Å, the protein is adopting a wider range of conformations around the 36 Å distance. This broader distribution was completely reversed by inhibition of the ATP hydrolysis with Vi (cyan). Furthermore, the presence of Mg2+ had no effect on the lifetime distribution of the inactive E506Q/T561C-ND (Fig. 4B, inset), indicating that the widening of distance distribution was dependent on ATP hydrolysis. For MsbA-detergent (Fig. 4B, bottom panel), we observed a different behavior. The peaks of the distance distribution were narrow under all studied conditions, with the exception of the peak corresponding to the longer distance in the apo state, suggesting large flexibility of the nucleotide-free MsbA in detergent micelles.
Discussion
In an attempt to study the structure and activity of a membrane protein in native-like conditions, we have used LRET to measure distance changes between the NBDs of the ABC transporter MsbA incorporated into a nanodisc lipid bilayer at 37 °C. We found that MsbA in nanodiscs was more active and less sensitive to temperature than MsbA in detergent. We also found that the longest measured distance for apo MsbA in nanodiscs was 47 Å, significantly shorter than the large separations of >80 Å in the apo inward-facing crystal structure (Protein Data Bank code 3B5W) (11) and >60 Å from DEER studies of MsbA reconstituted in liposomes or nanodiscs (12–14). Recently we reported a relatively short separation between dissociated NBDs (49 Å between probes bound to Cys-561 of each NBD) for MsbA reconstituted in liposomes; however, light scattering from the liposomes introduced potential errors in the distance calculations and prevented a complete study of the conformational changes during the ATP hydrolysis cycle (18). Here, we took advantage of soluble nanodiscs (27) to study MsbA reconstituted in a lipid bilayer and at physiological temperature. Our results suggest that in these native-like conditions, apo MsbA adopts a conformation with NBDs only partially separated instead of NBDs widely apart.
The shorter LRET distance in nanodiscs (∼36 Å) has also been observed in detergent (18) and is consistent with the closed outward-facing crystal structure of MsbA bound to the nonhydrolyzable ATP analog MgAMPPNP (11). Because we identified two distances under steady state conditions (∼36 and ∼46 Å), they represent distinct MsbA conformations that coexist in equilibrium. The coexistence of two MsbA conformations in equilibrium has been reported previously in LRET (18) and DEER experiments (12). In our experiments, these two distances remained essentially unchanged during the experimental conditions representing different steps during the ATP hydrolysis cycle, but the proportion of MsbA molecules adopting each conformation did change (Fig. 4A). ATP shifted the equilibrium toward a higher proportion of MsbA molecules adopting the short distance, as expected from the ATP-induced dimerization of the NBDs. Promotion of hydrolysis by Mg2+ increased the proportion of molecules adopting the longer distance, as expected from NBDs dimer dissociation following ATP hydrolysis. Under continuous hydrolysis, the dissociated NBDs rebind MgATP and redimerize, leading to an equilibrium with ∼50% of molecules in each conformation. MsbA in detergent displays a very similar behavior (18), confirming that either in detergent micelles or in a lipid bilayer, MsbA transitions between conformations with dimeric and dissociated NBDs. However, the conformational changes occur faster in the reconstituted protein, perhaps by a stimulatory effect of the lipids, which can also result in the wider range of conformations during continuous hydrolysis.
Another important difference found between MsbA in nanodiscs and in detergent was the proportion of molecules adopting the short distance in the apo state (Fig. 4A and Table 1). In detergent, most molecules (∼95%) adopted the longest distance conformation of ∼53 Å, as expected from completely dissociated NBDs. However, in nanodiscs only approximately half of the molecules displayed a long distance of ∼47 Å, which suggests partially dissociated NBDs. The other half of the reconstituted MsbA molecules in the apo state displayed a short distance (∼36 Å). This was unexpected because it is thought that the formation of the NBD dimer requires binding of ATP (1, 3). One possible explanation for this short distance in the absence of nucleotides comes from the published closed inward-facing structure of MsbA (Protein Data Bank code 3B5X). In this structure, the NBDs are in close proximity, and the distance between the residues 561 is ∼36 Å, but the regions that each NBD contributes to form the ATP binding pockets are misaligned (11). Until now, this inward-facing conformation with relatively closed NBDs has been considered a transient structure during the transition from the widely open inward-facing conformation to the closed outward-facing conformation (11). However, this or a related structure can constitute a dominant structure during the catalytic cycle. An electron microscopy study of P-glycoprotein in a lipid bilayer also showed NBDs in close proximity in apo state (39). Additional studies will be needed to provide more information about this apo “closed” structure adopted by MsbA in a lipid bilayer.
The crystal structures of MsbA and P-glycoprotein in the absence of lipids and DEER distances of reconstituted MsbA support the common view of large conformational changes when the exporter switches between the inward- and outward-facing conformations (11–14, 24, 25, 40). The available information from electron microscopy of ABC transporters in a lipid membrane corresponds to snapshots at low resolution that do not provide a unified and definitive picture. For example, a study of BmrA suggests large conformational changes during the hydrolysis cycle (41), whereas a study of P-glycoprotein suggests small conformational changes elicited by substrates and nucleotides (42). In general, available studies of BmrA and MsbA suggest similarity between the conformational changes of these proteins in detergent and liposomes (23, 40, 43). In contrast to the studies described above, our studies allow for a straightforward comparison of the same MsbA preparation at 37 °C in the lipid bilayer versus detergent and clearly point to smaller conformational changes in the bilayer, as well as to a much larger fraction of NBDs in close proximity for reconstituted MsbA in the apo state. The discrepancy with previous studies could be explained by differences in methodologies and experimental conditions, stressing the importance of performing studies in near physiological conditions.
Our conclusion that the catalytic cycle of MsbA in a lipid bilayer proceeds with small conformational changes agrees with a FRET study of P-glycoprotein in liposomes at 37 °C that included single-molecule recordings (22); the results are consistent with a model where the NBDs remain in contact during the hydrolysis cycle or experience only a small separation. Our studies in MsbA differ from the P-glycoprotein FRET studies in that we found fairly well defined conformations as opposed to a broader distribution of distances (22). However, it is presently unclear whether this difference is the result of the difficulty in quantifying distances from the single-molecule FRET data, the fact that the duration of the bursts in single-molecule FRET is significantly shorter than the hydrolysis cycle, and/or that there are important mechanistic differences between MsbA and P-glycoprotein. A DEER spectroscopy and simulation study supports the latter (26), but additional information will be needed to conclude that there are major structural differences between the catalytic cycles of different ABC exporters.
The exponential series analysis of the decays was compatible with two discrete conformations in MsbA-detergent in all studied conditions, with the clear exception of the apo state, where a broader distribution around the 53 Å distance was apparent. The increased sampling distance of MsbA in detergent in the apo state suggests that under certain experimental conditions (e.g. crystallization conditions), it may be possible to lock the protein in a rare conformation where the NBDs are far apart from each other. Two discrete conformations were also observed in MsbA-ND under all conditions, except during hydrolysis, where there was a broader distribution around the 36 Å distance. Perhaps the lipid bilayer stabilizes conformations near the closed NBD dimer that improve the ATP hydrolysis efficiency.
In general, our results suggest that in MsbA reconstituted in a lipid bilayer and at physiological temperature, the conformational changes during the ATP hydrolysis cycle are smaller (NBDs separated at most by ∼10 Å) than those expected from prior studies (11, 14). The studies presented here assess structural changes in an ABC exporter under native-like conditions and show major differences with the widely open inward-facing crystal structure and also with other studies carried out under less physiological conditions. These include our previous LRET studies of MsbA in detergent. The results stress the importance of performing structural/functional studies of membrane proteins under native-like conditions that include the insertion into lipid bilayers and normal temperatures.
Author Contributions
M. E. Z. and G. A. A. conceived and designed the study and analyzed and interpreted the data. M. E. Z. performed all experiments. R. S. C. performed preliminary studies of MsbA in detergent and liposomes and discussed the experimental data with M. E. Z. and G. A. A.
Acknowledgments
We thank Drs. Luis Cuello, Luis Reuss, and Ina Urbatsch and the Center for Membrane Protein Research members for critical discussions.
*
This work was supported by the Cancer Prevention & Research Institute of Texas Grant RP101073. The authors declare that they have no conflicts of interest with the contents of this article.
2
The abbreviations used are:
ABC
ATP-binding cassette
DEER
double electron-electron resonance
LRET
luminescence resonance energy transfer
MsbA-ND
MsbA reconstituted in nanodiscs
MSP
membrane scaffold protein
NBD
nucleotide-binding domain
TCEP
Tris (2-carboxyethyl) phosphine hydrochloride
Vi
sodium orthovanadate.
References
- 1.Al-Shawi M. K. (2011) Catalytic and transport cycles of ABC exporters. Essays Biochem. 50, 63–83 [DOI] [PubMed] [Google Scholar]
- 2.Bouige P., Laurent D., Piloyan L., and Dassa E. (2002) Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3, 541–559 [DOI] [PubMed] [Google Scholar]
- 3.Sharom F. J. (2008) ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics 9, 105–127 [DOI] [PubMed] [Google Scholar]
- 4.Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., and Tainer J. A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 5.Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., and Hunt J. F. (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoghbi M. E., and Altenberg G. A. (2014) ATP binding to two sites is necessary for dimerization of nucleotide-binding domains of ABC proteins. Biochem. Biophys. Res. Commun. 443, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoghbi M. E., and Altenberg G. A. (2013) Hydrolysis at one of the two nucleotide-binding sites drives the dissociation of ATP-binding cassette nucleotide-binding domain dimers. J. Biol. Chem. 288, 34259–34265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King G., and Sharom F. J. (2012) Proteins that bind and move lipids: MsbA and NPC1. Crit. Rev. Biochem. Mol. Biol. 47, 75–95 [DOI] [PubMed] [Google Scholar]
- 9.Doerrler W. T., Gibbons H. S., and Raetz C. R. (2004) MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279, 45102–45109 [DOI] [PubMed] [Google Scholar]
- 10.Doerrler W. T., and Raetz C. R. (2002) ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 [DOI] [PubMed] [Google Scholar]
- 11.Ward A., Reyes C. L., Yu J., Roth C. B., and Chang G. (2007) Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra S., Verhalen B., Stein R. A., Wen P. C., Tajkhorshid E., and McHaourab H. S. (2014) Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife 3, e02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou P., and McHaourab H. S. (2010) Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys. J. 98, L18–L20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou P., and McHaourab H. S. (2009) Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J. Mol. Biol. 393, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchaklian A. H., and Klug C. S. (2005) Characterization of the Walker A motif of MsbA using site-directed spin labeling electron paramagnetic resonance spectroscopy. Biochemistry 44, 5503–5509 [DOI] [PubMed] [Google Scholar]
- 16.Schultz K. M., Merten J. A., and Klug C. S. (2011) Characterization of the E506Q and H537A dysfunctional mutants in the E. coli ABC transporter MsbA. Biochemistry 50, 3599–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckford P. D., and Sharom F. J. (2008) Functional characterization of Escherichia coli MsbA: interaction with nucleotides and substrates. J. Biol. Chem. 283, 12840–12850 [DOI] [PubMed] [Google Scholar]
- 18.Cooper R. S., and Altenberg G. A. (2013) Association/dissociation of the nucleotide-binding domains of the ATP-binding cassette protein MsbA measured during continuous hydrolysis. J. Biol. Chem. 288, 20785–20796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George A. M., and Jones P. M. (2013) An asymmetric post-hydrolysis state of the ABC transporter ATPase dimer. PLoS One 8, e59854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones P. M., O'Mara M. L., and George A. M. (2009) ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 34, 520–531 [DOI] [PubMed] [Google Scholar]
- 21.Jones P. M., and George A. M. (2009) Opening of the ADP-bound active site in the ABC transporter ATPase dimer: evidence for a constant contact, alternating sites model for the catalytic cycle. Proteins 75, 387–396 [DOI] [PubMed] [Google Scholar]
- 22.Verhalen B., Ernst S., Börsch M., and Wilkens S. (2012) Dynamic ligand-induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J. Biol. Chem. 287, 1112–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borbat P. P., Surendhran K., Bortolus M., Zou P., Freed J. H., and McHaourab H. S. (2007) Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5, e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aller S. G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P. M., Trinh Y. T., Zhang Q., Urbatsch I. L., and Chang G. (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Jaimes K. F., and Aller S. G. (2014) Refined structures of mouse P-glycoprotein. Protein Sci. 23, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen P. C., Verhalen B., Wilkens S., McHaourab H. S., and Tajkhorshid E. (2013) On the origin of large flexibility of P-glycoprotein in the inward-facing state. J. Biol. Chem. 288, 19211–19220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., and Sligar S. G. (2009) Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoghbi M. E., Krishnan S., and Altenberg G. A. (2012) Dissociation of ATP-binding cassette nucleotide-binding domain dimers into monomers during the hydrolysis cycle. J. Biol. Chem. 287, 14994–15000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoghbi M. E., Fuson K. L., Sutton R. B., and Altenberg G. A. (2012) Kinetics of the association/dissociation cycle of an ATP-binding cassette nucleotide-binding domain. J. Biol. Chem. 287, 4157–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posson D. J., Ge P., Miller C., Bezanilla F., and Selvin P. R. (2005) Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature 436, 848–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvin P. R. (2002) Principles and biophysical applications of lanthanide-based probes. Annu. Rev. Biophys. Biomol. Struct. 31, 275–302 [DOI] [PubMed] [Google Scholar]
- 32.Urbatsch I. L., Sankaran B., Weber J., and Senior A. E. (1995) P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J. Biol. Chem. 270, 19383–19390 [DOI] [PubMed] [Google Scholar]
- 33.Heyduk T., and Heyduk E. (2001) Luminescence energy transfer with lanthanide chelates: interpretation of sensitized acceptor decay amplitudes. Anal. Biochem. 289, 60–67 [DOI] [PubMed] [Google Scholar]
- 34.James D. R., and Ware W. R. (1986) Recovery of underlying distributions of lifetimes from fluorescence decay data. Chem. Physics Lett. 126, 7–11 [Google Scholar]
- 35.Siemiarczuk A., Wagner B. D., and Ware W. R. (1990) Comparison of the maximum entropy and exponential series methods for the recovery of distributions of lifetimes from fluorescence lifetime data. J. Phys. Chem. 94, 1661–1666 [Google Scholar]
- 36.Posson D. J., and Selvin P. R. (2008) Extent of voltage sensor movement during gating of shaker K+ channels. Neuron 59, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawai T., Caaveiro J. M., Abe R., Katagiri T., and Tsumoto K. (2011) Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS Lett. 585, 3533–3537 [DOI] [PubMed] [Google Scholar]
- 38.Cha A., Snyder G. E., Selvin P. R., and Bezanilla F. (1999) Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402, 809–813 [DOI] [PubMed] [Google Scholar]
- 39.Lee J. Y., Urbatsch I. L., Senior A. E., and Wilkens S. (2002) Projection structure of P-glycoprotein by electron microscopy: evidence for a closed conformation of the nucleotide binding domains. J. Biol. Chem. 277, 40125–40131 [DOI] [PubMed] [Google Scholar]
- 40.Zou P., Bortolus M., and McHaourab H. S. (2009) Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 393, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fribourg P. F., Chami M., Sorzano C. O., Gubellini F., Marabini R., Marco S., Jault J. M., and Lévy D. (2014) 3D cryo-electron reconstruction of BmrA, a bacterial multidrug ABC transporter in an inward-facing conformation and in a lipidic environment. J. Mol. Biol. 426, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 42.Lee J. Y., Urbatsch I. L., Senior A. E., and Wilkens S. (2008) Nucleotide-induced structural changes in P-glycoprotein observed by electron microscopy. J. Biol. Chem. 283, 5769–5779 [DOI] [PubMed] [Google Scholar]
- 43.Mehmood S., Domene C., Forest E., and Jault J. M. (2012) Dynamics of a bacterial multidrug ABC transporter in the inward- and outward-facing conformations. Proc. Natl. Acad. Sci. U.S.A. 109, 10832–10836 [DOI] [PMC free article] [PubMed] [Google Scholar]