T cell fate and clonality inference from single cell transcriptomes (original) (raw)
. Author manuscript; available in PMC: 2016 Sep 22.
Published in final edited form as: Nat Methods. 2016 Mar 7;13(4):329–332. doi: 10.1038/nmeth.3800
Abstract
The enormous sequence diversity within T cell receptor (TCR) repertoires allows specific TCR sequences to be used as lineage markers for T cells that derive from a common progenitor. We have developed a computational method, called TraCeR, to reconstruct full-length, paired TCR sequences from T lymphocyte single-cell RNA-seq by combining existing assembly and alignment programs with “combinatorial recombinome” sequences comprising all possible TCR combinations. We validate this method to quantify its accuracy and sensitivity. Inferred TCR sequences reveal clonal relationships between T cells whilst the cells’ complete transcriptional landscapes can be quantified from the remaining RNA-seq data. This provides a powerful tool to link T cell specificity with functional response and we demonstrate this by determining the distribution of members of expanded T cell clonotypes in a mouse Salmonella infection model. Members of the same clonotype span early activated CD4+ T cells, as well as mature effector and memory cells.
INTRODUCTION
T lymphocytes recognise specific peptide–major histocompatibility complex (pMHC) combinations presented on the surface of antigen presenting cells1. This is mediated by the T cell receptor (TCR), an extremely diverse heterodimeric cell-surface protein comprising an α and β chain encoded by genes produced by V(D)J recombination of loci during T cell development2. The DNA sequence diversity of mouse TCRs has been estimated as 5×1021 different paired combinations3 and this enormous space of possible paired TCR sequences allows us to assume that cells with identical paired TCR genes arose from the same T cell clone.
The diversity of individual TCR chains has been used as a proxy for overall clonal diversity within bulk populations of T lymphocytes4–6 but these studies cannot determine the paired chains within each cell. This limits their ability to perform high-resolution determination of clonal relationships between cells and also to draw conclusions about the antigenic specificities of the cells1.
An ability to study paired TCR sequences within individual cells will be extremely powerful in understanding the adaptive immune response in the context of discerning the ‘grammar’ of TCR recognition or designing therapeutic TCR molecules. Furthermore, making a connection between TCR sequence and the transcriptional identity of individual T cells will be crucial in connecting cellular transcriptional fate with antigen specificity, modelling the dynamics of clonal expansion within T cell populations and investigating T cell phenotypic plasticity.
Paired TCR analysis has been performed in individual single-cells using specific amplification or capture of TCR genes7–11 without providing additional information about the cells in question. In addition, biases in PCR primer efficiency prevent accurate determination of TCR expression levels. A method that also amplifies a small set of ‘phenotyping marker’ genes12 provides limited information about cellular phenotype but requires a priori knowledge of informative genes as well as the design and optimisation of multiplexed PCR primers.
Single-cell RNA-seq (scRNA-seq) has already proved valuable in investigating the transcriptional heterogeneity and differentiation processes of cell populations13–19 and has revealed a novel T lymphocyte subset20. Determination of recombined TCR sequences from T cell scRNA-seq datasets has not yet been reported.
Existing computational tools for TCR analysis are designed for experiments that use bulk cell populations and require the targeted amplification of TCR loci during the experimental step21–25. Here, we present a novel method that enables full-length, paired TCR sequences to be reconstructed from single-cell RNA-seq data with high accuracy and sensitivity. Importantly, this method requires no alterations to standard scRNA-seq protocols and so can be applied easily to any species and sample for which scRNA-seq is possible. This novel approach links clonal ancestry and antigen specificity with the comprehensive transcriptomic identity of each studied T cell.
RESULTS
Our method (Supplementary Fig. 1a) extracts TCR-derived sequencing reads for each cell by alignment against ‘combinatorial recombinomes’ comprising all possible combinations of V and J segments (Supplementary Fig. 1b). Reads are then assembled into contiguous sequences which are analysed to find those that represent full-length, recombined TCR sequences. Importantly, the reconstructed recombinant sequences typically contain nearly the complete length of the TCR V(D)J region (Fig. 1) and so allow high-confidence discrimination between closely related and highly-similar gene segments. Here, we use scRNA-seq data generated using the SMART-Seq protocol26 with the Fluidigm C1 microfluidics system. Our method would, however, work with any scRNA-seq data derived from full-length cDNA.
Figure 1.
Distributions of lengths of reconstructed TCR sequences. Reconstructed sequences were trimmed to include the region derived from the V gene, junction and J gene. The lengths of these sequences are plotted as histograms and kernel density estimates for TCRa (upper) and TCRb (lower). Dotted lines represent the interquartile range of lengths of full-length sequences derived from the combinatorial recombinome files.
We have made the TCR reconstruction tool, ‘TraCeR’ (Supplementary Software), available at www.github.com/teichlab/tracer.
We analysed scRNA-seq data from 272 FACS-sorted CD4+ T cells isolated from spleens of C57BL/6N mice (Supplementary Table 1, Supplementary Fig. 2). We detected at least one productive alpha chain in 74%–96% of cells, a productive beta in 88%–96% and paired productive alpha-beta chains in 70%–93% (Table 1, Supplementary Table 2). This compares favourably with previous PCR-based TCR sequencing approaches that detected productive, pairs in 50%–82% of cells7,9,10,12.
Table 1.
TCR reconstruction statistics
| Mouse | TCRα reconstruction | TCRβ reconstruction | Paired productivechains |
|---|---|---|---|
| Uninfected day 0 | 39/50 (78%) | 46/50 (92%) | 37/50 (74%) |
| Day 14 mouse 1 | 68/71 (96%) | 68/71 (96%) | 66/71 (93%) |
| Day 14 mouse 2 | 29/39 (74%) | 35/39 (90%) | 28/39 (72%) |
| Day 49 | 87/112 (78%) | 98/112 (88%) | 78/112 (70%) |
Our method detected two alpha chain recombinants in 42% of cells and two beta chain recombinants in 22% (Supplementary Table 2). We detected two productive alpha chains in 35 out of 188 (19%) cells with at least one productive alpha chain and two productive beta chains in 16 out of 247 (6%). These data are in line with previous observations28. The best-performing PCR-based method did not detect multiple β recombinants in any of the 1268 cells studied12 because it filtered data to remove any TCRβ chains that were represented by less than 85% of reads. In only one cell (0.3%) did we detect two apparently non-productive sequences for a locus and both of these sequences were validated by the PCR-based approach described below.
We compared the TCR sequences reconstructed by our method with those detected by a multiplex PCR-based approach12that we adapted for use with mouse cells (Supplementary Note 1, Supplementary Fig. 3, Supplementary Table 3). We also determined the effects of sequencing depth, read-pairing and read length upon the performance of our method (Supplementary Note 2,Supplementary Figs. 4–5). Our approach provides sequences in good concordance with those generated by the PCR-based method and is able to successfully reconstruct TCR sequences from a variety of sequencing depths and read types.
We also applied TraCeR successfully to over 700 single-cell transcriptomes from a recently published study29 (Supplementary Note 3). As a negative control, we applied our method to 192 scRNA-seq datasets generated from mouse embryonic stem cells27. No TCR sequences were reconstructed from these highly transcriptionally active and promiscuous cells.
Taken together, these data indicate that our method accurately and sensitively determines the sequences of recombined and expressed TCR loci within individual T cells from single-cell RNA-seq data.
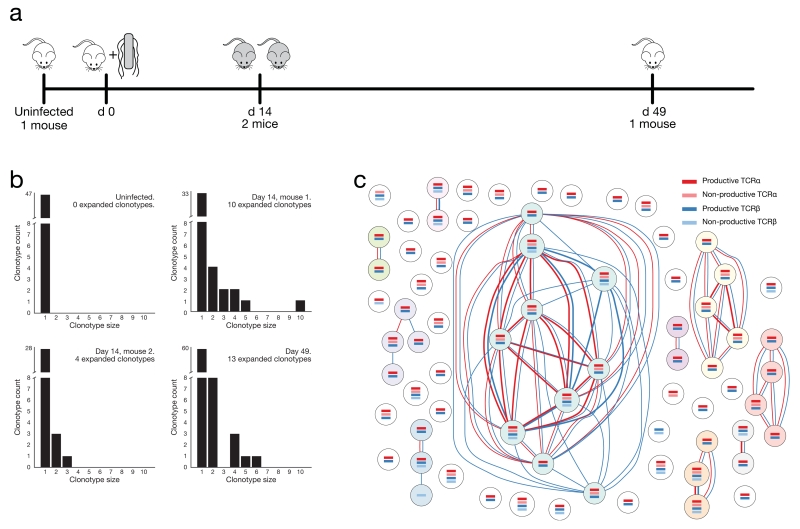
We demonstrated an application of our approach by investigating the CD4+ T lymphocyte clonotypes present within the spleens of mice prior to, during or after a non-lethal infection with Salmonella typhimurium (Fig. 2a, Supplementary Table 1), a bacterium that elicits a strong type-1 CD4+ T cell response. We analysed effector cells (CD4+CD8−TCRB+NK1.1−CD44HighCD62LLow) at day 14 when their relative abundance is close to its maximum, and memory cells (CD4+CD8−TCRB+NK1.1−CD44HighCD62LLowCD127High) at day 49 when the infection has been resolved30 (Supplementary Fig. 2).
Figure 2.
Assessment of clonal CD4+ T cell expansion during Salmonella typhimurium infection. (a) Schematic of timeline for Salmonella infection experiment. (b) Distribution of expanded clonotypes within splenic CD4+ T cell populations analysed by single-cell RNA-seq. The x-axis indicates the number of cells within the expanded clonotypes whilst the y-axis represents the number of clonotypes of each size. (c) Clonotype network graph from day 14, mouse 1. Each node in the graph represents an individual splenic CD4+ T lymphocyte. Coloured bars within the nodes indicate the presence of reconstructed TCR sequences that were detected for each cell. Dark coloured identifiers are productive, light coloured are non-productive. Red edges between the nodes indicate shared TCRα sequences whilst blue edges indicate shared TCRβ sequences. Edge thickness is proportional to the number of shared sequences.
Analysis of the TCR sequences present within the splenic CD4+ T cells enabled us to identify 12 invariant natural killer T (iNKT) cells31 that were excluded from further analyses (Supplementary Note 4).
We compared recombinant identifiers between cells to find clonally-related cells that expressed TCR genes with exactly the same nucleotide sequence. We found no TCR sharing between cells from different mice nor between cells from within the uninfected mouse (Fig. 2b, Supplementary Fig. 6, Supplementary Table 2). This is to be expected given the huge potential diversity of TCR nucleotide sequences.
We saw evidence of clonotype expansion within activated CD4+ T lymphocytes from each mouse at day 14 of Salmonella infection as well as from the mouse at week 7 post-infection (Figs. 2b, 2c and Supplementary Figs. 7–9. Supplementary Table 2). TCR sequences within expanded clonotypes from these mice are likely to be specific for Salmonella antigens. Importantly, we observed multiple cells that share all their detected recombinant sequences including those that are non-productive indicating that our method is detecting the correct combinations of TCR recombinants within the cells. Furthermore, observations of cells that share multiple TCR sequences provide increased confidence that those cells are genuinely clonally related due to the extremely small likelihood that all recombination events would arise identically in two independent cells during development in the thymus.
Developing T lymphocytes in the thymus first perform recombination at the TCRβ locus and undergo proliferation prior to recombining their TCRα loci. Cells generated from a single progenitor by this proliferative expansion will all have the same TCRβ recombinant but will each randomly generate a different α recombinant prior to continuing maturation and entering the periphery. It is therefore possible that a particular single TCRβ chain can be found with multiple partners and we detected cells that share TCRβ sequences but have different TCRα sequences (Supplementary Fig. 10). This illustrates the value of paired TCRαβ sequences when inferring accurate clonal relationships between T cells.
It should be noted that there is no evidence of contamination across microfluidics chip capture or harvest sites or adjacent wells in the 96-well plates used (Supplementary Fig. 11).
Single-cell RNA-seq allows cells to be classified according to their gene expression profiles. To combine this with our knowledge of clonal relationships, we quantified gene expression within each single cell and performed independent component analysis (ICA) to reduce the gene expression space to two dimensions (Fig. 3a). We were able to use 14,889 informative genes for ICA. This is a great deal larger than the 17 phenotyping genes that were used in a previous PCR-based approach to determining clonality and cell fate12.
Figure 3.
Distribution of expanded clonotypes throughout the Th1 response to S. typhimurium infection. (a) Dimensionality reduction of single-cell gene expression data by independent component analysis (ICA). Each single CD4+ T cell is plotted in reduced two-dimensional space according to its gene expression profile. Points are colored according to the timepoint from which they were sampled or according to their expression of marker genes indicative of their phenotype. Where the expression of a set of genes (Th1 genes and proliferation markers) is plotted, this is the sum of TPM values for the genes within the set. (b) Clonotype distribution in gene-expression space. Three representative expanded clonotypes from day 14 mouse 1 are shown as purple points on top of all other cells within the gene expression space.
We analysed the expression of 259 genes that indicate a Th1 cell fate32, Il7r (CD127) which is indicative of effector-memory T cells33, Ccr7 (a marker of central-memory T cells)34 and a set of seven genes that are expressed in proliferating cells35 (Fig 3a). Expression of these genes allowed us to separate the cells into four populations: activated proliferating cells that are differentiating to the Th1 fate, mature differentiated Th1 effector cells, effector memory-like cells and central memory-like cells. Cells from the uninfected mouse are mostly central memory-like, cells from the mouse at day 14 have an activated or Th1 effector phenotype whilst cells from day 49 (sorted to be CD127high, a marker of effector memory fate) are found in the effector-memory region of the ICA gene expression space.
We then determined the distributions of expanded clonotypes within the reduced gene-expression space (Fig 3b and Supplementary Figs. 12-14), excluding those that shared a TCRβ sequence but had different TCRα recombinants. Cells derived from the same progenitor can be seen throughout the activated differentiating, Th1 effector and effector-memory populations. This suggests that, after activation by binding to a Salmonella_antigen–MHC complex, the progeny of a particular CD4+ T cell differentiate asynchronously. Members of one clonotype exist across the full spectrum of proliferation and differentiation states that occur during the_Salmonella response.
DISCUSSION
Here, we present a method for the determination of paired T cell receptor sequences from individual T lymphocytes achieved solely by analysis of standard single-cell RNA-seq datasets. Our method is sensitive, accurate and easy to adapt to any species for which annotated TCR gene sequences are available; it does not need large numbers of multiplexed PCR reactions. We also fully expect our method to be easily extended to the study of the analogous B cell receptor/antibody sequences within B lymphocytes although considerations of clonality would need to take into account the process of somatic hypermutation.
Combining TCR reconstruction with single-cell RNA-seq allows us to assess the cells’ phenotypes using orders of magnitude more genes than existing PCR-based approaches while obviating the need for a priori knowledge of phenotyping genes of interest. This will permit the discovery of novel or poorly-characterised phenotypic subtypes in conjunction with the analysis of their TCR sequences.
Currently, the cost of single-cell RNA-seq is prohibitive for its use in very large-scale surveys of the entire immune repertoire within an organism. However, these methods are practical for the analysis of smaller, selected lymphocyte subsets. In the illustrative example presented here we are able to draw meaningful immunological insights from just 272 cells. A recent study sequenced 722 single T lymphocytes29 and our method found expanded clonotypes likely to provide additional biological insights. It is clear that the throughput of single-cell RNA-sequencing methods is increasing whilst cost decreases and we expect ever larger datasets to become standard.
A combined knowledge of T cell clonal dynamics, TCR specificity and detailed transcriptional phenotype is likely to be of great use in the study of T cell responses to infection, auto-antigens or vaccination and will provide insights into both pathogenic mechanisms and therapeutic approaches.
ONLINE METHODS
Ethics statement
Mice were maintained under specific pathogen-free conditions at the Wellcome Genome Campus Research Support Facility (Cambridge, UK). These animal facilities are approved by and registered with the UK Home Office. Animals were sacrificed by approved animal technicians in accordance with Schedule 1 of the Animals (Scientific Procedures) Act 1986. Oversight of the arrangements for Schedule 1 killing was performed by the Animal Welfare and Ethical Review Body of the Wellcome Genome Campus.
Cell preparation
Female C57BL6/N mice aged 6-8 weeks were infected intravenously with 0.2 ml Salmonella Typhimurium M525 containing 5×105 CFU of bacteria in sterile phosphate buffered saline (PBS, Sigma-Aldrich). At day 14 or 49 post infection (p.i.) mice were sacrificed with spleens and livers being harvested. An uninfected mouse (day 0) was also sacrificed. The sample size of four mice was chosen to provide sufficient example data for application of our method. Mice were randomly chosen to receive infection and randomly assigned to sacrifice at day 14 or day 49. No blinding was performed. Bacteria were enumerated from the livers by serial dilution and plating onto agar plates (Oxoid) to confirm levels of infection. Single-cell suspensions were prepared by homogenising spleens through 70 μm strainers and lysing erythrocytes. Following incubation with CD16/CD32 blocking antibody, the cells from the uninfected mouse and from day 14 p.i. were stained with titrated amounts of fluorochrome conjugated antibodies for CD44(FITC), CD25(PE), CD62L(PE-CF594), TCRβ(PerCP-Cy5.5), CD8α(APC-H7), NK1.1(BV421), and CD4(BV510). The cells from day 49 p.i. were stained with antibodies for CD44(FITC), CD127(PE), CD62L(PE-CF594), TCRβ(PerCP-Cy5.5), NK1.1(APC), CD8α(APC-H7), CD4 (BV510), and Sytox Blue viability stain. Antibody details can be found in Supplementary Table 4. Cell sorting was performed using a BD FACSAria II instrument using the 100 micron nozzle at 20 psi using the single cell sort precision mode. The cytometer was set up using Cytometer Setup and Tracking beads and compensation was calculated using compensation beads (for antibodies, eBioscience UltraComp) and cells (for Sytox Blue) using automated software (FACSDiva v6).
Single-cell RNA-sequencing and gene expression quantification
Capture and processing of single CD4+ T cells was performed using the Fluidigm C1 autoprep system. Cells were loaded at a concentration of 1,700 cells μl−1 onto C1 capture chips for 5-10 μm cells. We used microscopic inspection of the C1 capture sites to determine which contained only a single cell so as to exclude empty wells or those containing multiple cells. From the five chips used in this work we captured 329 single cells out of a possible 480 (68.5%). ERCC (External RNA Controls Consortium) spike-in RNAs (Ambion, Life Technologies) were added to the lysis mix. Reverse transcription and cDNA preamplification were performed using the SMARTer Ultra Low RNA kit (Clontech). Sequencing libraries were prepared using Nextera XT DNA Sample Preparation kit with 96 indices (Illumina), according to the protocol supplied by Fluidigm. Libraries from 303 single cells were pooled and sequenced on Illumina HiSeq2500 using paired-end 100 base reads.
Reads were mapped to the Mus musculus genome (Ensembl version 38.70) concatenated with the ERCC sequences, using GSNAP36 with default parameters. Gene-specific read counts were calculated using HTSeq37. Thirty-one cells (out of 303) with detected transcripts for fewer than 2000 genes, or with more than 10% of measured exonic reads corresponding to genes coded by the mitochondrial genome, were excluded from further analyses. The 272 cells that passed these quality control steps were used in the analyses presented here.
Reconstruction and analysis of TCR sequences from RNA-seq data
Combinatorial recombinome files were separately created for the TCRα and TCRβ chains. To generate these fasta files, nucleotide sequences for all mouse V and J genes were downloaded from The International ImMunoGeneTics information system38 (IMGT, www.imgt.org). Every possible combination of V and J genes was generated for each TCR locus such that each combination was a separate sequence entry in the appropriate recombinome file. Within the recombinome files we did not attempt to encompass all possible sequences that could be generated by junctional diversity during V(D)J recombination. Instead, ambiguous ‘N’ nucleotide sequence characters (not to be confused with ‘N nucleotides’ added by terminal deoxynucleotidyl transferase during recombination within the cell) were introduced into the junction between V and J genes in each sequence entry to improve alignments of reads that spanned diverse junctional sequences (Figure 1b). Seven N nucleotides were used in TCRβ combinations whilst one N nucleotide was used in the TCRα combinations. V gene leader sequences are not well annotated within IMGT and so 20 N nucleotides were added at the 5ʹ end of the V sequence to permit alignment of sequencing reads that included the leader sequence.
TCRα or TCRβ constant region cDNA sequences were downloaded from ENSEMBL and appended to the 3ʹ end of each combined sequence to permit alignment of reads that ran into the constant region. The full-length TCRα constant region was used whilst the only the first 259 nucleotides of the TCRβ constant gene were used since these are identical between both Trbc homologs that are found within the mouse genome. The combinatorial recombinomes used in this work can be found alongside the other tools at www.github.com/teichlab/tracer.
RNA-seq reads from each cell were aligned against each combinatorial recombinome independently using the Bowtie 2 aligner39. Bowtie 2 is ideal for alignment against the recombinomes because it can align against ambiguous N nucleotides within a reference and also introduce gaps into both the reference and read sequences. This allows it to align reads against the variable junctional regions. We used the following Bowtie 2 parameters with low penalties for introducing gaps into either the read or the reference sequence or for aligning against N nucleotides ‘--no-unal -k 1 --np 0 --rdg 1,1 --rfg 1,1’.
For each chain, separately, we used the reads that aligned to the appropriate recombinome as input to the Trinity RNA-seq assembly software40 using its default parameters.
V, D and J gene sequences downloaded from IMGT were used to generate appropriate databases for use with IgBLAST41. Contigs assembled by Trinity were used as input to IgBlast and the resulting output text files were processed with a custom parsing script. Contigs were classed as representing TCR sequences if they contained gene segments from the correct locus (ie TCRα genes for TCRα contigs) and if their reported V and J alignments had E-values below 5×10−3. If multiple contigs within the same cell represented the same recombinant sequence, these were collapsed so that the sequence was only represented once in the cell for subsequent analyses. In some cases where two contigs derived from the same original sequence but one was shorter than the other, IgBLAST assigned different V sequences if the shorter sequence did not provide sufficient information to distinguish between highly similar genes. This typically occurred with V genes that were part of the evolutionary expansion events that caused gene duplication and triplication within the TCRα locus42. In these cases, the sequences were collapsed into a single assignment that used the results from the longest contig. The IgBLAST results for the TCR sequences within each cell were then reduced to an identifying string (Fig. 1a) consisting of the V gene name, the junctional nucleotide sequence and the J gene name (eg. TRBV31_AGTCTTGACACAAGA_TRBJ2-5) which was used for comparisons between sequences within other cells.
It is important to determine whether a particular TCR mRNA sequence is productive and therefore able to be translated to produce a full-length TCR polypeptide chain. To do this for the reconstructed TCR sequences we first converted them to entirely full-length sequences by using full-length V and J gene sequences from IMGT appropriate to the gene segments assigned by IgBLAST. Since sequences from laboratory mouse strains well-characterised and TCRs do not undergo somatic hypermutation we can make the assumption that variations between the RNA-seq derived sequences and the reference sequences outside the junctional region are due to PCR and/or sequencing errors and so can be ignored. We check that these full-length sequences are in the correct reading frame from the start of the V gene to the start of the constant gene and that they lack stop codons. If this is the case, the sequence is classed as productive. For analysis of CDR3 amino-acid sequences we translate the productive recombinants and define the CDR3 as the region flanked by the final cysteine residue of the V gene and the conserved FGXG motif in the J gene as previously described23. Although this approach is likely to be most accurate when analysing data from inbred, well-characterised mouse strains it does risk losing useful sequence information that represents genuine germline polymorphism that may be present when analysing cells from humans or other outbred populations. To address this, it is possible to instruct TraCeR to omit the step that replaces the assembled sequences with reference sequences from IMGT. In this case, productivity is calculated solely from the sequence as assembled by Trinity. For the data presented here, both methods give almost identical assessments of productivity (Supplementary Fig. 14).
Expression levels of the TCR genes found within a cell were quantified by appending that cell’s full-length recombinant sequences to a file containing the entire mouse transcriptome (downloaded from http://bio.math.berkeley.edu/kallisto/transcriptomes/) and then using this file for the generation of an index suitable for use with the pseudoalignment-based Kallisto algorithm43. This index was then used with the RNA-seq reads for the cell as input for Kallisto in quantification mode to calculate transcripts per million (TPM) values for each TCR sequence. If a cell was assigned more than two recombinant sequences for a particular locus (5/272, 1.8% of cells in this study), the sequences were ranked by their TPM values and the two most highly-expressed were used for further analyses. Kallisto’s speed in constructing indices and performing expression quantification makes it ideal for this task.
After assignment of TCR sequences to each cell within an experiment, we used custom Python scripts to compare the recombinant identifiers present in each cell to find cases where multiple cells contained the same identifier. These analyses were used to generate network graphs where each node in the graph represents a single cell and edges between the nodes represent shared TCR sequences.
Code availability
The analyses described above are performed by our tool, TraCeR which is freely available at www.github.com/teichlab/tracer and in Supplementary Software.
PCR-based sequencing of TCR sequences
Primers were designed to amplify all possible recombined TCR sequences from both the TCRα and TCRβ loci (all sequences can be found in Supplementary Table 5). Two constant region primers were designed to be complementary to the Trac or Trbc genes close to their 5ʹ ends. Sets of primers complementary to all TCRα and β V gene sequences downloaded from IMGT were also designed. Primers were designed to regions of homology between V genes and included degeneracy where appropriate so as to minimise the number of primers required. In total, 34 TCRα and 31 TCRβ primers were used. All primers were designed with a Tm of 71–73 °C. All V gene primers were designed with the sequence ACACTCTTTCCCTACACGACGCTCTTCCGATCT at their 5ʹ end to allow amplification by the Illumina PE 1.0 primer (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT) while the constant region primers were designed with the sequence TCGGCATTCCTGCTGAACCGCTCTTCCGATCT at their 5ʹ end so that they could be amplified by barcoding primers containing a unique 11 nt index sequence (Supplementary Table 5). The barcoding primers also contain the Illumina PE 2.0 sequence.
Full-length (oligo-dT primed) cDNA produced from single-cells by the C1 system (Fluidigm, USA) was used as template in two PCR reactions, one for each TCR locus. 0.4 μl of cDNA were used in each reaction along with each V primer at 0.06 μM and the constant primer at 0.3 μM. Phusion DNA polymerase (NEB, USA) was used to perform the amplification in 25 μl final volume. The cycling conditions for this step were 98 °C 30 s; 98 °C 10 s, 60 °C 10s, 72 °C 30 s × 16 cycles; 72 °C 5 min. 4 °C. A 1 μl aliquot of the first reaction was used as template in a second PCR amplification, again using Phusion in a 25 μl reaction volume. Here, the Illumina PE 1.0 primer was used with a barcoding primer unique for each cell and each primer was at 0.4 μM. The cycling conditions for this step were 98 °C 30 s; 98 °C 10 s, 58°C 10s, 72 °C 30 s × 16 cycles; 72 °C 5 min. 4 °C. PCR products of the correct size for sequencing were purified using 0.7 volumes of AMPure XP beads (Beckman Coulter) according to the manufacturer’s instructions. Purified products were pooled and submitted to the Wellcome Trust Sanger Institute (WTSI) Sequencing Facility for sequencing using a MiSeq (Illumina) with 250bp paired-end reads.
Processing PCR data
Reads generated by MiSeq sequencing of PCR products were de-multiplexed by the WTSI Sequencing Facility according to their barcode sequences. Reads were then trimmed to remove low-quality regions and adapter sequences using TrimGalore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The TCR-targeted PCR primers were designed to provide amplicons short enough such that the forward and reverse paired reads would overlap upon sequencing enabling read pairs to be merged using FLASH44. Merged read sequences were then filtered to remove those under 200 nucleotides in length to remove artefactual sequences. Following this step, read sequences for each cell were subsampled where necessary such that there were 50,000 sequences or fewer from each cell. This reduced the computational time and requirements for the next stage whilst still providing sufficient information about the sequences present. As described previously12, we assumed that sequences from an individual cell that had at least 95% sequence identity were derived from the same original cDNA sequence and so these were combined to generate a consensus sequence. The consensus sequences for each cell were analysed by IgBLAST to find sequences that represented recombined TCRs and the number of sequencing reads supporting each TCR were used to filter out background sequences that had few reads.
Comparing PCR and RNA-seq data
For each cell, sequences derived from PCR analysis or reconstructed from RNA-seq data were trimmed to only include the regions assigned by IgBlast as containing V, D or J sequences. This removed any leader sequences or constant regions. Trimmed reconstructed RNA-seq sequences were aligned against the trimmed PCR-derived sequences in a set of pairwise comparisons using BLAST. If an alignment was reported, the number of mismatches across the entire alignment were counted, as were the number of mismatches between the nucleotides that encoded the CDR3 region (defined here as the 30nt following the end of the framework 3 region as annotated by IgBlast). If the CDR3 regions contained any mismatches, the alignment was classed as discordant, otherwise the two sequences were classed as concordant. Sequences from one method (RNA-seq or PCR) that did not align successfully with any sequence from the other method were classed as discordant.
Gene expression quantification and dimensionality reduction
Genes were filtered to remove those expressed (TPM>1) in fewer than three cells. Dimensionality reduction of the remaining gene expression data was performed by independent component analysis (ICA) using the FastICA Python package.
For plotting gene expression for each cell within ICA space, 259 genes indicating a Th1-like fate and seven indicators of proliferation (Mki67, Mybl2, Bub1, Plk1, Ccne1, Ccnd1, Ccnb1) were taken from previous work32,35 and their expression levels (in TPM) were summed for each cell.
Clonotype distribution within gene expression space
Cells that did not appear to be derived from the same progenitor (same TCRβ but differing TCRα chains) were removed from the expanded clonotype groups. Cells belonging to a particular expanded clonotype were then plotted within the ICA reduced gene expression space.
Supplementary Material
Supplementary Figures
Supplementary Figure Captions
Supplementary Software
Supplementary Table 1 & Notes
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
ACKNOWLEDGEMENTS
We thank V. Svensson, T. Hagai, J. Henriksson and other members of the Teichmann laboratory along with G. Lythe for helpful discussions. We thank the Wellcome Trust Sanger Institute Sequencing Facility for performing Illumina sequencing and the Wellcome Trust Sanger Institute Research Support Facility for care of the mice used in these studies. This work was supported by European Research Council grant ThSWITCH (number 260507, SAT) and the Lister Institute for Preventative Medicine (SAT).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
ACCESSION CODES
Sequencing data from the cells described in this manuscript were deposited at ArrayExpress with accession number E-MTAB-3857.
REFERENCES
- 1.Rossjohn J, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 2.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr. Opin. Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieber MR. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- 4.Becattini S, et al. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 5.Mamedov IZ, et al. Quantitative tracking of T cell clones after haematopoietic stem cell transplantation. EMBO Mol. Med. 2011;3:201–207. doi: 10.1002/emmm.201100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas N, et al. Tracking global changes induced in the CD4 T-cell receptor repertoire by immunization with a complex antigen using short stretches of CDR3 protein sequence. Bioinformatics. 2014;30:3181–3188. doi: 10.1093/bioinformatics/btu523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dash P, et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J. Clin. Invest. 2011;121:288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnemann C, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat. Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 9.Turchaninova MA, et al. Pairing of T-cell receptor chains via emulsion PCR. Eur. J. Immunol. 2013;43:2507–2515. doi: 10.1002/eji.201343453. [DOI] [PubMed] [Google Scholar]
- 10.Kim S-M, et al. Analysis of the paired TCR α- and β-chains of single human T cells. PLoS One. 2012;7:e37338. doi: 10.1371/journal.pone.0037338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howie B, et al. High-throughput pairing of T cell receptor α and β sequences. Sci. Transl. Med. 2015;7:301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- 12.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 14.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 20.Mahata B, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7:1130–1142. doi: 10.1016/j.celrep.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolotin DA, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 22.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat. Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 23.Thomas N, Heather J, Ndifon W, Shawe-Taylor J, Chain B. Decombinator: a tool for fast, efficient gene assignment in T-cell receptor sequences using a finite state machine. Bioinformatics. 2013;29:542–550. doi: 10.1093/bioinformatics/btt004. [DOI] [PubMed] [Google Scholar]
- 24.Kuchenbecker L, et al. IMSEQ-a fast and error aware approach to immunogenetic sequence analysis. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv309. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, et al. TCRklass: a new K-string-based algorithm for human and mouse TCR repertoire characterization. The Journal of Immunology. 2015;194:446–454. doi: 10.4049/jimmunol.1400711. [DOI] [PubMed] [Google Scholar]
- 26.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolodziejczyk AA, et al. Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell Stem Cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: an update and reappraisal. The Journal of Immunology. 2010;185:3801–3808. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaublomme JT, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittrücker H-W, Köhler A, Kaufmann SHE. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect. Immun. 2002;70:199–203. doi: 10.1128/IAI.70.1.199-203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 32.Stubbington MJT, et al. An atlas of mouse CD4+ T cell transcriptomes. Biol. Direct. 2015;10:14. doi: 10.1186/s13062-015-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kallies A. Distinct regulation of effector and memory T-cell differentiation. Immunol. Cell Biol. 2008;86:325–332. doi: 10.1038/icb.2008.16. [DOI] [PubMed] [Google Scholar]
- 34.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat. Rev. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 36.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefranc M-P, et al. IMGT®, the international ImMunoGeneTics information system®. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bosc N, Lefranc M-P. The mouse (Mus musculus) T cell receptor alpha (TRA) and delta (TRD) variable genes. Dev. Comp. Immunol. 2003;27:465–497. doi: 10.1016/s0145-305x(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 43.Bray N, Pimentel H, Melsted P, Pachter L. Near-optimal RNA-Seq quantification. 2015 doi: 10.1038/nbt.3519. arXiv [q-bio.QM] at < http://arxiv.org/abs/1505.02710>. [DOI] [PubMed]
- 44.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Captions
Supplementary Software
Supplementary Table 1 & Notes
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5