GREAT improves functional interpretation of cis-regulatory regions (original) (raw)
. Author manuscript; available in PMC: 2016 Apr 22.
Published in final edited form as: Nat Biotechnol. 2010 May 2;28(5):495–501. doi: 10.1038/nbt.1630
Abstract
We developed the Genomic Regions Enrichment of Annotations Tool (GREAT) to analyze the functional significance of _cis_-regulatory regions identified by localized measurements of DNA binding events across an entire genome. Whereas previous methods took into account only binding proximal to genes, GREAT is able to properly incorporate distal binding sites and control for false positives using a binomial test over the input genomic regions. GREAT incorporates annotations from 20 ontologies and is available as a web application. Applying GREAT to data sets from chromatin immunoprecipitation coupled with massively parallel sequencing (ChIP-seq) of multiple transcription-associated factors, including SRF, NRSF, GABP, Stat3 and p300 in different developmental contexts, we recover many functions of these factors that are missed by existing gene-based tools, and we generate testable hypotheses. The utility of GREAT is not limited to ChIP-seq, as it could also be applied to open chromatin, localized epigenomic markers and similar functional data sets, as well as comparative genomics sets.
The coupling of chromatin immunoprecipitation with massively parallel sequencing, ChIP-seq, is ushering in a new era of genome-wide functional analysis1–3. Thus far, computational efforts have focused on pinpointing the genomic locations of binding events from the deluge of reads produced by deep sequencing4–8. Functional interpretation is then performed using gene-based tools developed in the wake of the preceding microarray revolution9–11. In a typical analysis, one compares the total fraction of genes annotated for a given ontology term with the fraction of annotated genes picked by proximal binding events to obtain a gene-based P value for enrichment (Fig. 1 and Online Methods).
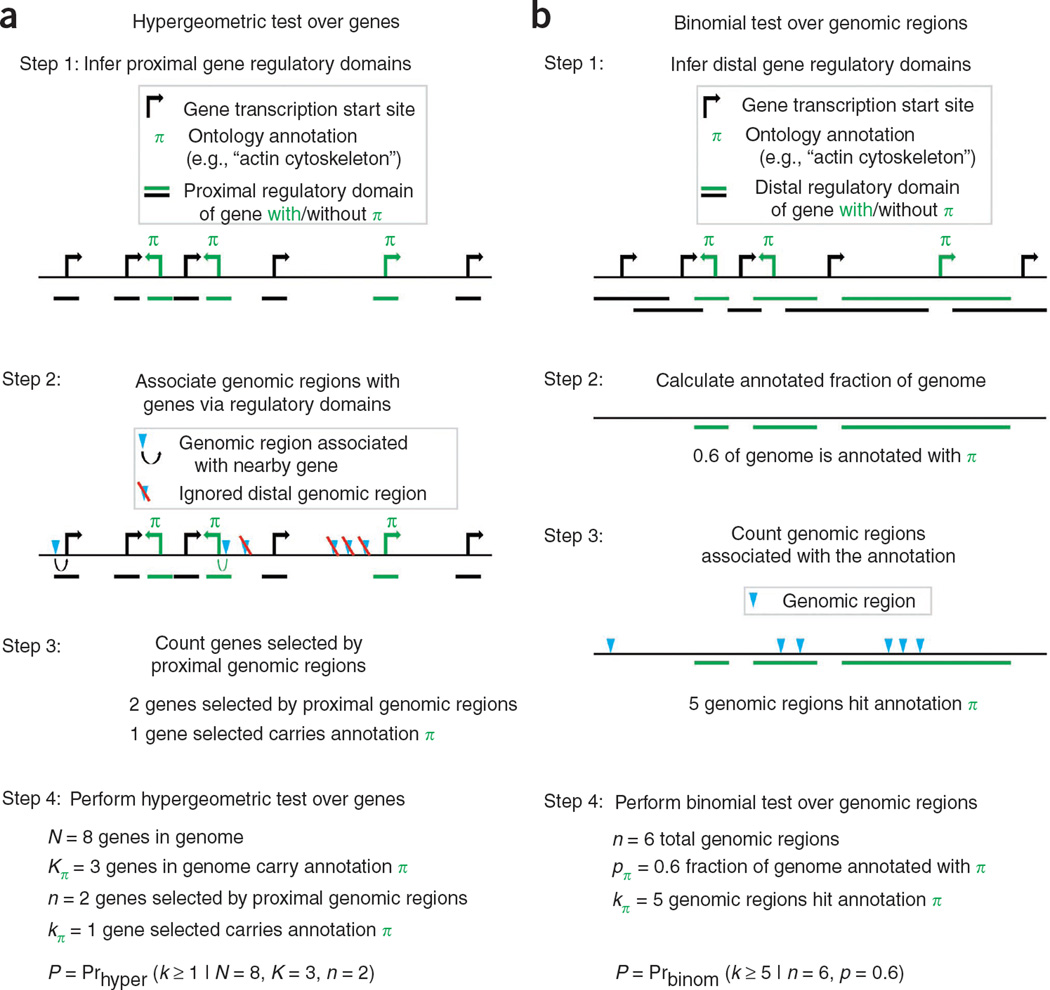
Figure 1.
Enrichment analysis of a set of _cis_-regulatory regions. (a) The current prevailing methodology associates only proximal binding events with genes and performs a gene-list test of functional enrichments using tools originally designed for microarray analysis. (b) GREAT’s binomial approach over genomic regions uses the total fraction of the genome associated with a given ontology term (green bar) as the expected fraction of input regions associated with the term by chance.
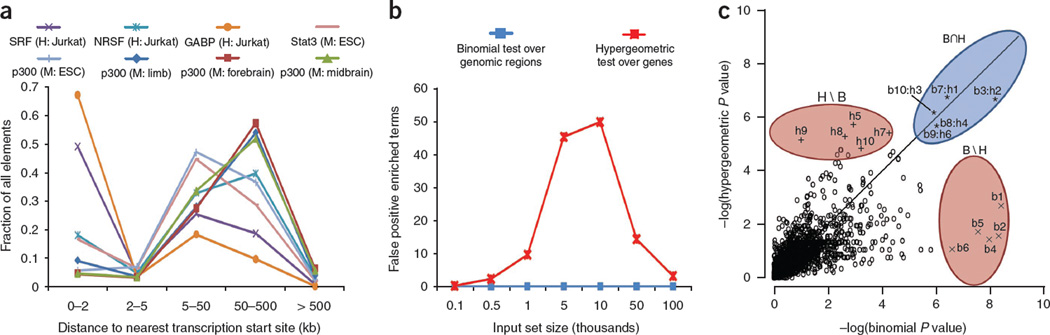
This procedure has a fundamental drawback: associating only proximal binding events (for example, under 2–5 kb from the transcription start site) typically discards over half of the observed binding events (Fig. 2a). However, the standard approach to capturing distal events—associating each binding site with the one or two nearest genes—introduces a strong bias toward genes that are flanked by large intergenic regions12,13. For example, though the Gene Ontology14 (GO) term ‘multicellular organismal development’ is associated with 14% of human genes, the ‘nearest genes’ approach associates over 33% of the genome with these genes. This biological bias results in numerous false positive enrichments, particularly for the input set sizes typical of a ChIP-seq experiment (Fig. 2b and Supplementary Fig. 1). Building on our experience in addressing these pitfalls12,15,16, we have developed a tool that robustly integrates distal binding events while eliminating the bias that leads to false positive enrichments.
Figure 2.
Binding profiles and their effects on statistical tests. (a) ChIP-seq data sets of several regulatory proteins show that the majority of binding events lie well outside the proximal promoter, both for sequence-specific transcription factors (SRF and NRSF, ref. 8; Stat3, ref. 43) and a general enhancer-associated protein (p300, refs. 33,43). Cell type is given in parentheses: H, human; M, mouse. (b) When not restricted to proximal promoters, the gene-based hypergeometric test (red) generates false positive enriched terms, especially at the size range of 1,000–50,000 input regions typical of a ChIP-seq set. Negligible false positive enrichment was observed for the region-based binomial test (blue). For each set size, we generated 1,000 random input sets in which each base pair in the human genome was equally likely to be included in each set, avoiding assembly gaps. We calculated all GO term enrichments for both hypergeometric and binomial tests using GREAT’s 5+1 kb basal promoter and up to 1 Mb extension association rule (see Results). Plotted is the average number of terms artificially significant at a threshold of 0.05 after application of the conservative Bonferroni correction. (c) GO enrichment P values using the genomic region-based binomial (x axis) and gene-based hypergeometric (y axis) tests on the SRF data8 with GREAT’s 5+1 kb basal promoter and up to 1 Mb extension association rule (see Results). b1 through b10 denote the top ten most enriched terms when we used the binomial test. h1 through h10 denote the top ten most enriched terms when we used the hypergeometric test. Terms significant by both tests (B ∩ H) provide specific and accurate annotations supported by multiple genes and binding events (Table 3). Terms significant by only the hypergeometric test (H\B) are general and often associated with genes of large regulatory domains, whereas terms significant by only the binomial test (B\H) cluster four to six genomic regions near only one or two genes annotated with the term (Supplementary Table 46).
RESULTS
Here we describe GREAT, which analyzes the functional significance of sets of _cis_-regulatory regions by explicitly modeling the vertebrate genome regulatory landscape and using many rich information sources.
A binomial test for long-range gene regulatory domains
GREAT associates genomic regions with genes by defining a ‘regulatory domain’ for each gene in the genome. Each genomic region is associated with all genes in whose regulatory domains it lies (Fig. 1b). High-throughput chromosomal conformation capture (3C) approaches such as 5C (ref. 17), Hi-C (ref. 18) or enhanced ChIP-4C (ref. 19) are providing first glimpses of actual gene regulatory domains. Because we still lack precise empirical maps, however, GREAT assigns each gene a regulatory domain consisting of a basal domain that extends 5 kb upstream and 1 kb downstream from its transcription start site (denoted below as 5+1 kb), and an extension up to the basal regulatory domain of the nearest upstream and downstream genes within 1 Mb (GREAT allows the user to modify the rule and distances). GREAT further refines the regulatory domains of a handful of genes, including several global control regions20, by using their experimentally determined regulatory domains. Our tool can also incorporate additional locus-based and genome-wide data as they become available (Supplementary Fig. 2 and Online Methods).
Given a set of input genomic regions and an ontology of gene annotations, GREAT computes ontology term enrichments using a binomial test that explicitly accounts for variability in gene regulatory domain size by measuring the total fraction of the genome annotated for any given ontology term and counting how many input genomic regions fall into those areas (Fig. 1b and Online Methods). In the example above, GREAT expects 33% of all input elements to be associated with ‘multicellular organismal development’ by chance, rather than the 14% of input elements that a gene-based test assumes. The binomial test integrates distal binding events in a way that remains robust regardless of erroneous assignments of genomic regions to genes. Namely, the longer the regulatory domain of any gene—and, by extension, of any ontology term—the greater the expected number of regions associated with this term by chance. Indeed, the binomial statistic markedly reduces the number of false positive enriched terms even when very large regulatory domains are used (Fig. 2b and Supplementary Fig. 1). The binomial test treats each input genomic region as a point-binding event, making it most suitable for testing targets with localized binding peaks. The binomial test also highlights cases in which a single gene attracts an unlikely number of input genomic regions. To separate these biologically interesting gene-specific events from term-derived enrichments that are distributed across multiple genes, we perform both the binomial test and the traditional hypergeometric gene-based test. In doing so, we highlight ontology terms enriched by both tests (term-derived enrichment) separately from those enriched by only the binomial test (gene-specific enrichment) or the hypergeometric test (regulatory domain bias) (Fig. 2c and Supplementary Fig. 3).
GREAT supports direct enrichment analysis of both the human and mouse genomes. It integrates 20 separate ontologies containing biological knowledge about gene functions, phenotype and disease associations, regulatory and metabolic pathways, gene expression data, presence of regulatory motifs to capture cofactor dependencies, and gene families (Supplementary Tables 1–3 and Online Methods). Core computations are performed by the GREAT server while subsequent browsing is executed on the user’s machine. An overview of the tool’s functionality and options when analyzing data is given in Table 1, and its current web interface is shown in Supplementary Figure 4.
Table 1.
GREAT parameters, filters and options, and their effects
| Parameter | Effect |
|---|---|
| Region-gene association rule | Determines how gene regulatory domains are calculated. When we allowed for distal associations, the sets we examinedremained robust regardless of the exact choice of association rule. Our default rule (basal and extension; see Results)models a current hypothesis of gene regulatory domains. |
| Region-gene association rule parameters | Determine the length of each inferred gene regulatory domain. As we show, when the right statistical model is used,including distal associations of up to 1 Mb can strongly increase biological signals. |
| Statistical significance visual filter | Highlights statistically significant results in bold font. Multiple test correction options and thresholds for significancecan be modified. |
| Binomial fold enrichment filter | Complements P value by requiring that statistically significant terms have strong biological effects. Often filters generalontology terms that apply to thousands of genes. |
| Observed gene hits filter | Shows only enriched terms for which input regions select at least this many genes. Helps avoid enrichments owing to numerous regions selecting a small number of genes. |
| Minimum annotation count threshold | Increases statistical power by reducing the number of tests performed, by testing only ontology terms associated a priori with at least this many genes. |
| Display type | Summary display shows only terms statistically significant by both binomial and hypergeometric tests. Full display ignores the statistical significance filter and shows terms that meet all other criteria. |
| Export | Export tables individually or in batches into a file of tab-separated values or publication-ready HTML. |
| UCSC custom tracks | Clicking a specific region from within a term details page opens the University of California Santa Cruz Genome Browser44focused on that region, with two custom tracks automatically loaded—one for the total set of input regions and anotherfor the subset of regions associated with the chosen term. |
Comparison of enrichment tests and regulatory domain ranges
To demonstrate the utility of our approach, we compared GREAT results to previously published gene-based analyses as well as to enrichments from the Database for Annotation, Visualization, and Integrated Discovery (DAVID)21. Most gene-based tools assess enrichments in a very similar manner; we chose DAVID as a representative gene-based tool owing to its popularity and its ability to test a breadth of data sources similar to that of GREAT (Supplementary Table 4).
We analyzed eight ChIP-seq data sets from a range of human and mouse cells and tissues (Supplementary Table 5), each with a different distribution of proximal and distal binding events (Fig. 2a). We tested each data set in six different ways: (i) by reproducing the original study’s list of enrichments, or if the original study did not report enrichments, by using DAVID on the set of genes with binding events within 2 kb of the transcription start site; (ii) by using GREAT with the default regulatory domain definition (basal promoter 5+1 kb and extension up to 1 Mb); (iii) by using GREAT’s hypergeometric test on the set of genes with binding events within 2 kb of the transcription start site, to control for the different gene mappings and ontologies in DAVID and GREAT; (iv) by using GREAT with a 5+1 kb basal promoter and a more limited 50 kb extension; and (v, vi) by using GREAT with either one (v) or two (vi) nearest genes up to 1 Mb (Tables 2 and 3, and Supplementary Tables 6–44, indexed in Supplementary Table 45).
Table 2.
Gene-based ontology enrichments regions bound by SRF in human Jurkat cells
| Term | P value |
|---|---|
| Nucleus | 5.18 × 10−70 |
| Protein binding | 2.16 × 10−50 |
| Cytoplasm | 6.67 × 10−27 |
| Transcription | 4.13 × 10−26 |
| Nucleotide binding | 1.04 × 10−23 |
| Metal ion binding | 1.92 × 10−22 |
| Zinc ion binding | 5.76 × 10−20 |
| RNA binding | 3.38 × 10−18 |
| Regulation of transcription, DNA-dependent | 1.15 × 10−15 |
| ATP binding | 4.84 × 10−15 |
Table 3.
GREAT ontology enrichments for regions bound by SRF in human Jurkat cells
| Ontology | Term | Binomial P value | Binomial foldenrichment | Hypergeometric P value | Distal bindinga | Experimental support |
|---|---|---|---|---|---|---|
| GO: cellular component | Actin cytoskeleton | 6.91 × 10−9 | 3.05 | 2.22 × 10−7 | 38.9% | Ref. 23 |
| Cortical cytoskeleton | 4.03 × 10−6 | 5.90 | 5.41 × 10−4 | 54.5% | Ref. 23 | |
| GO: molecular function | Actin binding | 5.21 × 10−5 | 2.03 | 2.74 × 10−5 | 51.4% | Ref. 23 |
| Transcription factor targets | SRF targets (Jurkat,T/G HA-VSMC, Be(2)-C) | 4.97 × 10−76 | 13.22 | 9.79 × 10−68 | 14.3% | Positive control |
| YY1 targets (HeLa) | 1.45 × 10−6 | 2.09 | 0.0084 | 20.4% | Ref. 26b | |
| E2F4 and p130(T98G, U2OS) | 0.0047 | 2.01 | 0.0027 | 44.4% | Novelc | |
| E2F4 (WI-38) | 0.0194 | 2.08 | 0.0031 | 36.4% | Novelc | |
| Predicted promoter motifs | SRF variants | 4.54 × 10−28 to4.19 × 10−12 | 3.69 to 15.46 | 1.71 × 10−25 to2.04 × 10−9 | 17.4% to28.6% | Positive controls |
| GABPA or GABPB | 4.20 × 10−9 | 3.67 | 6.68 × 10−6 | 27.6% | Novelc | |
| Motif NGGGACTTTCCA | 1.02 × 10−4 | 2.12 | 8.30 × 10−5 | 20.0% | Novelc | |
| EGR1 | 1.71 × 10−4 | 2.03 | 0.0013 | 46.9% | Novelc | |
| Pathway commons | TRAIL signaling pathway | 2.37 × 10−7 | 2.45 | 1.71 × 10−5 | 46.3% | Ref. 29 |
| Class I PI3K signaling events | 9.92 × 10−7 | 2.56 | 4.45 × 10−5 | 44.1% | Ref. 30 | |
| TreeFam | FOS family | 9.66 × 10−9 | 27.89 | 1.21 × 10−6 | 28.6% | Ref. 22d |
GREAT invariably revealed strong enrichments for experimentally validated functions of the specific factors, as well as for testable—and, to our knowledge, novel—functions. It also implicated subsets of regulatory regions in driving the assayed developmental processes and in activating key signaling pathways. In a majority of data sets, distal binding events were essential to recover known functions, strongly suggesting that many of the distal associations are biologically meaningful (see below). Furthermore, in most sets, restricting regulatory domain extension to 50 kb retains many enriched terms but omits roughly half of both the binding events and the genes implicated using the full 1-Mb extension. Although including distal associations is crucial, the exact distal association rule is not—the default rule, the nearest-gene rule, and the two-nearest-genes rule (tests ii, v and vi, respectively) behaved very similarly. Additionally, inclusion of the small set of experimentally determined gene regulatory domains we curated from the literature made very little difference in the rankings of any of the sets (data not shown). We present the analysis of four ChIP-seq data sets below and discuss the remainder in the Supplementary Note.
Serum response factor binding in human Jurkat cells
First, we analyzed a set of genomic regions bound by the serum response factor (SRF) in the human Jurkat cell line, identified via ChIP-seq and mapped to the genome using the quantitative enrichment of sequence tags (QuEST) ChIP-seq peak-calling tool8. This data set’s authors applied existing gene-based enrichment tools, which did not discern specific functions of SRF from the set of regions it binds8, and concluded that SRF is a regulator of basic cellular processes with no specific physiological roles (results reproduced in Table 2). Although SRF is indeed a regulator of basic cellular functions, numerous studies have implicated SRF in more specific biological contexts. SRF is a key regulator of the Fos oncogene22 and has also been described as a “master regulator of actin cytoskeleton”23. Neither FOS nor actin appeared in the top ten hypotheses generated by the previous study (Table 2). The same was true when we used GREAT with only proximal (2 kb) associations (Supplementary Table 6).
However, GREAT analysis of the most significant SRF ChIP-seq peaks8 (QuEST score > 1; n = 556) using the default settings (5+1 kb basal, up to 1 Mb extension) prominently highlights the key observation that gene-based analyses were unable to reveal: SRF regulates genes associated with the actin cytoskeleton23 (Table 3). As postulated above, using both binomial and hypergeometric enrichment tests does highlight informative GO terms more effectively than using either test alone (Fig. 2c and Supplementary Table 46). Moreover, when extension of regulatory domains is limited to 50 kb, one-third of the supporting regions and associated genes are lost, and actin-related terms drop in rank (Supplementary Table 7).
Coupling distal (up to 1 Mb) associations with the many additional ontologies available within GREAT provides a wealth of enrichments for specific known functions of SRF. An enrichment analysis of TreeFam gene families24 shows that SRF binds in proximity to five of six members of the FOS family. Two genes within the Fos family, Fos and Fosb, are previously known targets of SRF (ref. 22). The Transcription Factor Targets ontology25 has compiled data from ChIP experiments that link transcription factor regulators to downstream target genes. GREAT shows that many genes proximal to SRF binding events (in Jurkat cells) are also proximal to YY1 binding events (in HeLa cells), consistent with experiments showing that SRF acts in conjunction with YY1 to regulate Fos (ref. 26). The top six hits in the Predicted Promoter Motifs ontology27 are all variants of the SRF motif generated from different experiments and thus serve as strong positive controls of our method. Using the Pathway Commons ontology28, GREAT predicts that SRF regulates components of the TRAIL signaling pathway and the class I PI3K signaling pathway. Previous experimental work has demonstrated that there is an association between SRF and TRAIL signaling29 and that SRF is needed for PI3K-dependent cell proliferation30.
In addition to rediscovering and expanding specific known functions of SRF, GREAT produces testable hypotheses even for this well-studied transcription factor. The Transcription Factor Targets ontology indicates that SRF binds near genes regulated by E2F4 (in T98G, U2OS and WI-38 cells; Table 3). SRF and E2F4 have not been shown to co-regulate target genes; however, both SRF and E2F4 are known to interact with Smad3 (refs. 31,32), and they may thus be co-regulators of a common set of genes. The Predicted Promoter Motifs ontology reveals additional potential cofactors and co-regulators. It is particularly useful given that many more genes have characterized binding motifs than have genome-wide ChIP data available. In this case, it shows enrichment for SRF binding near genes containing GABP motifs in their promoters. Notably, an independent experiment measuring GABP-bound regions of the genome in Jurkat cells has found that 29% of SRF peaks occur within 100 bp of a GABP peak, suggesting that SRF and GABP may indeed work together8. We were able to generate this same hypothesis using GREAT, without observing the GABP ChIP-seq data.
P300 binding in the developing mouse limbs
Second, we analyzed a recent ChIP-seq data set comprising 2,105 regions of the mouse genome bound by the general enhancer-associated protein p300 in embryonic limb tissue33. Of 25 such regions tested in transgenic mouse assays, 20 showed reproducible enhancer activity in the developing limbs33. Our analysis shows that GREAT identifies functions of enhancers active during embryonic development that gene-based tools do not detect. DAVID analysis of the genes with proximal p300 limb binding events produces only enrichments associated with transcription and involvement in organ morphogenesis, with the closest enrichments being the much broader terms ‘organ development’ and ‘anatomical structure morphogenesis’ (Supplementary Table 10a). In contrast, GREAT analysis of the 2,105 p300 limb peaks using the default settings (5+1 kb basal, up to 1 Mb extension) produces overwhelming support for their putative functional role in limb development (Supplementary Table 10b).
GO enrichments highlight the regulation of transcription factors involved specifically in embryonic limb morphogenesis. The Mouse Phenotype ontology34 points to the developing limbs and skull, hinting at the remarkable overlap of signaling processes involved in head and limb development35. The p300 limb peaks are enriched near genes in the TGF-β signaling pathway, which is known to be involved in limb development36, and the InterPro ontology highlights genes in the Smad family containing the Dwarfin-type MAD homology-1 protein domain (Supplementary Table 10b), which is known to mediate and regulate TGF-β signaling37.
Perhaps the strongest validation for the GREAT methodology comes from the MGI Expression: Detected ontology38. Notably, the enrichments highlighted most prominently by GREAT pinpoint the exact tissue and time point at which the experiment in ref. 33 was performed, providing unique large-scale evidence for the relevance of p300-bound regions to limb gene regulation. The top two ontology terms suggest limb-specific expression during Theiler stage 19 (TS19), which corresponds precisely with embryonic day 11.5, the time point at which the p300 limb peaks were assayed in ref. 33 (Supplementary Table 10b). In contrast, GREAT run with proximal (2 kb) associations retrieves only weak enrichments for limb-associated genes and limb TS19, implicating 7-fold fewer genes and 16-fold fewer p300 limb peaks as being involved in TS19 limb expression than GREAT run with the default association rule (Supplementary Table 11). Moreover, GREAT run with proximal associations completely misses genes with crucial roles in limb development such as Gli3, Grem1 and Wnt7a (ref. 39).
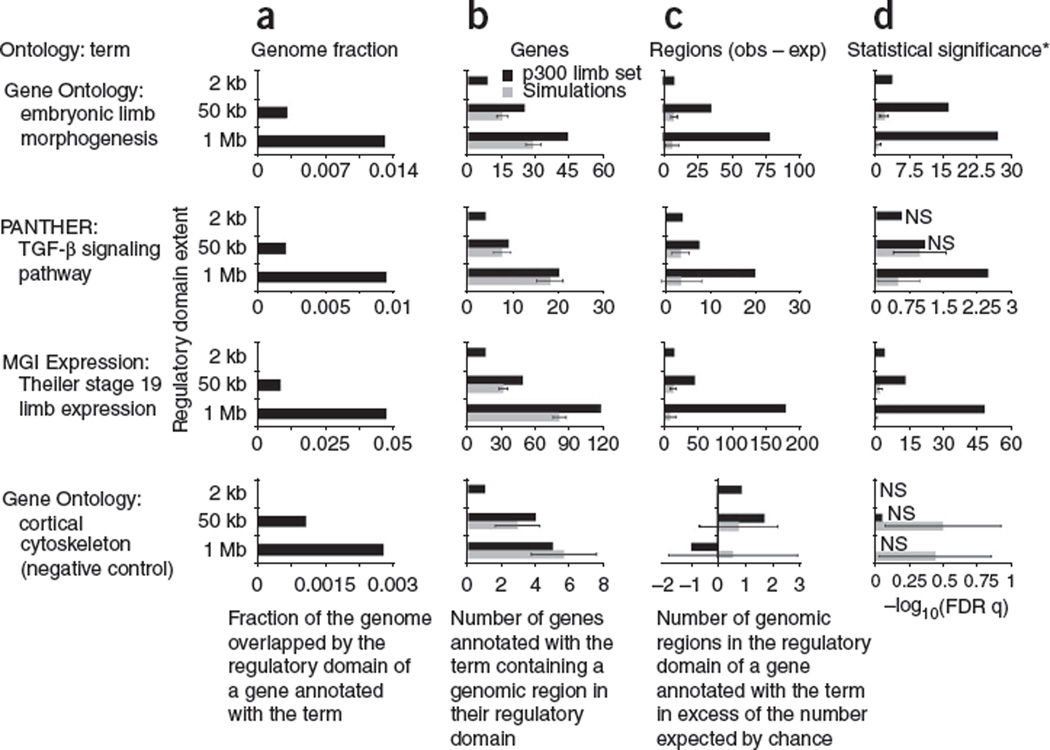
When GREAT’s regulatory domains are extended up to 50 kb, it correctly recovers limb terms, but still implicates only half the genes found with the default association rule and yields P values many orders of magnitude weaker (Fig. 3 and Supplementary Table 12). By extending regulatory domains, we increase both the number of limb-related genes containing one or more p300 limb peaks within their regulatory domains and the number of p300 limb peaks associated with limb-related genes (Fig. 3). When regulatory domains are further extended from 50 kb to 1 Mb, they include even more p300 limb peaks than expected by chance (Fig. 3c), providing strong evidence that many of these distal associations are biologically meaningful.
Figure 3.
Distal binding events contribute substantially to accurate functional enrichments of p300 limb peaks. We examined properties of the 2,105 p300 mouse embryonic limb peaks33 in the context of three known limb-related terms and a negative control term (GO cortical cytoskeleton). Three different association rules were used (see Results): a gene-based GREAT analysis using only peaks within 2 kb of the nearest transcription start site (labeled 2 kb), an analysis with 5+1 kb basal and up to 50 kb extension (50 kb), and an analysis with 5+1 kb basal and up to 1 Mb extension (1 Mb). For each term, we examined the relevance of distal binding peaks by comparing the experimental results (black bars) to the average values of 1,000 simulated data sets (gray bars) in which the 192 proximal ChIP-seq peaks within 2 kb of the nearest transcription start site were fixed and the 1,913 distal peaks were shuffled uniformly within the mouse genome, avoiding assembly gaps and proximal promoters. By design, simulation results for proximal, 2-kb GREAT are identical to the actual data and are thus omitted. (a) Lengthening a 2-kb proximal promoter to a 50-kb extension, expected to increase genome coverage per term (_p_π in Fig. 1b) by 25-fold, causes an actual increase of 19- to 24-fold; in contrast, lengthening a 50-kb extension rule to a 1-Mb extension rule, expected to raise genome coverage 20-fold, leads to an actual increase of only 2.5- to 6-fold because regulatory domains are not extended through neighboring genes. (b) As regulatory domains increase in length from only the proximal 2 kb up to 50 kb and 1 Mb, the number of relevant genes with a p300 limb peak in their regulatory domain increases. The added genes selected only by distal associations are typically enriched for limb functionality compared to simulated data. (c) As regulatory domains increase in length, the number of p300 limb peaks associated with a relevant gene in excess of the number expected by chance increases for all limb-related terms. (d) As in c, the inclusion of distal peaks markedly increases the statistical significance of the correct terms alone. *Statistical significance is measured using the hypergeometric test over genes for 2 kb to mimic current gene-based approaches, and using the binomial test over genomic regions for 50 kb and 1 Mb. Error bars indicate s.d.; NS, not significant at a threshold of 0.05 after false discovery rate multiple test correction; obs, observed; exp, expected. Note scale changes on x axes.
P300 binding in the developing mouse forebrain and midbrain
Finally, we analyzed two ChIP-seq data sets comprising regions bound by p300 in mouse embryonic forebrain and midbrain tissue33. Using the 2,453 forebrain peaks, DAVID correctly highlights forebrain and general brain development (0.004 < P < 0.05), but with terms implicating fewer than ten genes (Supplementary Table 15a). GREAT run with proximal regulatory regions (2 kb) ranks forebrain development higher and is able to implicate additional genes and regions using its unique phenotype and expression ontologies (Supplementary Table 16). Using up to 50 kb extension adds additional related terms and raises the number of genes associated with each term (Supplementary Table 17). This trend continues when the extension is increased to up to 1 Mb, and only this inclusion of distal binding allows detection of significant associations (P = 0.001) with Wnt signaling genes that have known roles in forebrain development40 (Supplementary Table 15b).
When run on the 561 midbrain p300 peaks, DAVID does not yield significant results (P > 0.05; Supplementary Table 20a) and proximal (2 kb) GREAT performs only slightly better, offering three relevant terms associated with very few genes from our unique ontologies (Supplementary Table 21). In contrast, GREAT with up to 1 Mb extension highlights twelve brain-specific enriched terms (Supplementary Table 20b). Many GREAT enriched terms are shared between the forebrain and midbrain peaks (as discussed in Supplementary Note), but GREAT correctly identifies midbrain-specific enrichments such as the GO term ‘compartment specification’. Compartment specification is of interest, as within this tissue at this developmental age, Fgf8 induces Wnt (also enriched within this set) to set up a gene network that establishes the boundary between the midbrain and hindbrain compartments41. GREAT with up to 50 kb extension is able to highlight many of the same terms, but loses roughly half the associated genes and regions and the Wnt enrichment (Supplementary Table 22).
DISCUSSION
GREAT is a new-generation tool aimed at the interpretation of genome-wide _cis_-regulatory data sets. It explicitly models the vertebrate _cis_-regulatory landscape through the use of long-range regulatory domains and a genomic region–based enrichment test, allowing analyses that take into consideration the large number of binding events that occur far beyond proximal promoters. By accounting for the length of gene regulatory domains, GREAT is able to highlight biologically meaningful terms and their associated _cis_-regulatory regions and genes, in a manner that remains robust if there are false associations between input regions and genes. Moreover, these regulatory-domain definitions can naturally incorporate future results from three-dimensional conformation capture studies17–19, radiation hybrid maps42 and other emerging approaches for measuring the regulatory genome in action. By coupling this methodology with many ontologies that span a wealth of biological information types, GREAT produces specific, accurate enrichments that provide insight into the biological roles of _cis_-regulatory data sets of interest.
We comprehensively tested GREAT on multiple ChIP-seq data sets and found that it is able to reproduce many known biological facts that existing methods do not detect, as well as suggest novel hypotheses for further experimental characterization. In particular, our analysis shows that ignoring distal binding events often leads to missing target gene associations, to obtaining weaker P values or even to completely omitting relevant enrichment terms. Besides ChIP-seq data, GREAT can also be applied to the analysis of any data set thought to be enriched for localized _cis_-regulatory regions. This includes functional genomic data sets of open chromatin, localized epigenomic markers, and comparative genomic sets. GREAT may thus prove invaluable in elucidating the _cis_-regulatory functions encoded in genomes.
GREAT is available online (http://great.stanford.edu/); also provided is a means for direct submission from other applications such as genome portals and peak calling tools.
ONLINE METHODS
Gene set definition
Statistical enrichment of ontology terms is dependent upon the genome-wide gene set used in the analysis. GREAT currently supports testing of human (Homo sapiens NCBI Build 36.1, or UCSC hg18) and mouse (Mus musculus NCBI Build 37, or UCSC mm9). To limit the gene sets to only high-confidence genes and gene predictions, we use only the subset of the UCSC Known Genes45 that are protein coding, are on assembled chromosomes and possess at least one meaningful GO annotation14. GO is an ontological representation of information related to the biological processes, cellular components and molecular functions of genes. We rely on the idea that if a gene has been annotated for function it should be included in the gene set, and if no function has been ascribed to a gene its status may be unclear and thus it is best omitted from the gene set. In GREAT version 1.1.3, we use GO data downloaded on 5 March 2009 for human and 23 March 2009 for mouse, leading to gene sets of 17,217 and 17,506 genes for human and mouse, respectively.
A single gene may have multiple splice variants. As annotations are generally given at the gene level, GREAT uses a single transcription start site (TSS) to specify the location of each gene. The TSS used is that of the ‘canonical isoform’ of the gene as defined by the UCSC Known Genes track45.
Association rules from genomic regions to genes
For each gene, we define a ‘regulatory domain’ such that all noncoding sequences that lie within the regulatory domain are assumed to regulate that gene. GREAT currently supports three different parametrized association rules to define gene regulatory domains (Supplementary Fig. 2). The default ‘basal plus extension’ association rule assigns a ‘basal regulatory region’ irrespective of the presence of neighboring genes that extends (using default parameters) 5 kb upstream and 1 kb downstream of the TSS (Supplementary Fig. 2a). Each gene’s regulatory domain is then extended up to the basal regulatory region of the nearest upstream and downstream genes, but no longer than 1 Mb in each direction. The choice of basal regulatory region size and placement was motivated by the location of histone modifications and measures of chromatin accessibility near the TSS of genes46, and the maximum extension distance is based upon work showing that long-range distal enhancers can regulate expression of target genes up to 1 Mb away47,48. All three parameters (basal upstream, basal downstream and maximum extension distance) can be set by the user.
The ‘two nearest genes’ association rule extends each gene’s regulatory domain from the TSS of the canonical isoform to the nearest upstream and downstream TSS (Supplementary Fig. 2b), up to 1 Mb in each direction. This association rule stipulates that each base pair cannot be assigned to more than two genes.
The ‘single nearest gene’ association rule extends each gene’s regulatory domain from the TSS of the canonical isoform in each direction to the midpoint between the TSS and the nearest adjacent TSS (Supplementary Fig. 2c), up to 1 Mb in each direction. This association rule stipulates that each base pair cannot be assigned to more than one gene.
For well-studied genes with experimentally detected distal regulatory elements (reviewed in ref. 20), we manually override the computationally defined regulatory domains. GREAT version 1.1.3 uses experimentally validated regulatory domains for _SHH_47, genes in the β-globin locus49, and KIAA1715, EVX2, HOXD10, HOXD11, HOXD12 and HOXD13 (ref. 50). Future releases of the tool will continue to refine regulatory domains as technological advances, including three-dimensional conformation capture studies17–19 and radiation hybrid maps42, further elucidate interactions between regulatory DNA and its target genes.
Hypergeometric test over genes
The hypergeometric test over genes identifies all genes whose regulatory domains possess one or more genomic regions from the input set and calculates enrichments over the genes with respect to the defined gene set using a hypergeometric distribution. More formally, the hypergeometric test is executed separately for each ontology term π and is defined by four parameters:
- N is the total number of genes in the genome.
- _K_π is the number of genes in the genome that possess ontology annotation π.
- n is the number of genes selected because one or more input genomic regions resides in their regulatory domains.
- _k_π is the number of selected genes that possess ontology annotation π.
The test calculates the P value of the observed enrichment for term π as the fraction of ways to choose n genes without replacement from the entire group of N genes such that at least _k_π of the n possess ontology annotation π, using the formula below.
| ∑i=kπmin(n,Kπ)(Kπi)(N−Kπn−i)(Nn) | (1) |
|---|
In particular, the hypergeometric test counts every gene only once even if it was picked by multiple genomic regions. Terms enriched by the hypergeometric test thus indicate a high ‘term coverage’, where a larger fraction of all genes annotated with the term are selected by the input genomic regions than expected by chance.
Binomial test over genomic regions
To account for the length variability within gene regulatory domains, we implemented a binomial test over genomic regions that uses the fraction of the genome associated with each ontology term as the probability of selecting the term. The binomial test is executed separately for each ontology term π and is defined by three parameters:
- n is the total number of genomic regions in the input set.
- _p_π is the a priori probability of selecting a base pair annotated with π when selecting a single base pair uniformly from all non–assembly gap base pairs in the genome.
- _k_π is the number of genomic regions in the input set that cause annotation π to be selected.
The test calculates the P value of the observed enrichment for term π as the probability of selecting annotation π at least _k_π times in n attempts using the formula below.
| ∑i=kπn(ni)pπi(1−pπ)n−i | (2) |
|---|
The binomial test first maps each input genomic region to the left median base pair in its span, making it most appropriate for assessing enrichment of factors with narrow, precise peaks. The value of _p_π is calculated for each ontology annotation π as the fraction of non–assembly gap base pairs in the genome associated with annotation π. Each input genomic region can then be thought of as a ‘dart’ thrown at the genome, counting as a hit if the left median base pair is annotated with ontology term π. In this test, the length of each gene’s regulatory domain is explicitly accounted for in the calculation of _p_π. This explicit use of regulatory domain size in the significance calculation provides a proper assessment of the enrichment for ontology terms by noncoding sequences. Notably, as the binomial test incorporates the fraction of the genome assigned to each gene in the calculation of statistical significance, it is robust regardless of variation in association rules and occasional incorrect assignments of genomic regions to distal target genes. Ontology terms assigned to genes that have large regulatory domains are inherently weighted such that each binding event associated with the term contributes less to the resulting enrichment than binding events associated with terms assigned to genes with small regulatory domains. However, enrichments under the binomial test may arise from clusters of noncoding regions all near one or a few genes with a particular ontology annotation, as well as from noncoding regions associating with many genes that possess a particular ontology annotation. The hypergeometric test over genes (described above) provides a measure of ‘term coverage’ that can be used to identify terms significant by the binomial test that have many annotated genes selected as well.
Foreground/background hypergeometric test over genomic regions
When a set of input genomic regions is selected from a superset of ‘background genomic regions’ (for example, the repetitive elements that have been exapted into functional roles selected from all repetitive elements in the genome12), one should consider whether the input genomic regions differ in functional composition from the entire set of background genomic regions as a whole. The foreground/background hypergeometric test over genomic regions poses this statistical question by mapping all ontology annotations of each gene to all background genomic regions that lie within its regulatory domain; it then calculates enrichments over the input genomic regions with respect to the superset of background genomic regions using a hypergeometric distribution. Formally, the foreground/background hypergeometric test over genomic regions is executed separately for each ontology term π and is defined by four parameters:
- N is the number of genomic regions in the background set.
- _K_π is the number of genomic regions in the background set that lie within the regulatory domain of some gene annotated with term π.
- n is the number of genomic regions in the foreground set.
- _k_π is the number of genomic regions in the foreground set that lie within the regulatory domain of some gene annotated with term π.
The test calculates the P value of the observed enrichment for term π using the hypergeometric equation shown above, equation (1).
GREAT software
The GREAT core calculation engine is implemented in C and the source code is publicly available for download (http://great.stanford.edu/).
Supplementary Material
Supplemental
Acknowledgments
We thank M. Sirota for an early survey of ontologies, F. Sathira for developing an intermediary core calculation engine, T. Capellini for critical reading of the manuscript, M. Davis and S. Gutierrez for system administration and the communities of ontology developers and curators for providing invaluable data sources. C.Y.M. is supported by a Bio-X graduate fellowship. M.H. is supported by a German Research Foundation Fellowship (Hi 1423/2-1) and the Human Frontier Science Program (fellowship LT000896/2009-l). S.L.C. is a Howard Hughes Medical Institute Gilliam Fellow. A.M.W. is supported by a Stanford Graduate Fellowship. G.B. is a Packard Fellow, Searle Scholar, Microsoft Research Faculty Fellow and an Alfred P. Sloan Fellow. Research was also supported by an Edward Mallinckrodt, Jr. Foundation junior faculty grant and US National Institutes of Health grant 1R01HD059862 to G.B.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
C.Y.M. developed the core calculation engine, processed ontologies, analyzed data sets and co-wrote the manuscript. D.B. designed and developed the web application. M.H. added key ontologies and calculated ontology statistics. S.L.C. performed and wrote the SRF analysis. B.T.S. contributed to data set analysis and manuscript writing. A.M.W. guided website design and wrote user documentation. G.B. and C.B.L. devised the different enrichment tests and developed early core calculation engines. G.B. supervised the project and co-wrote the manuscript. All authors edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 2.Mardis ER. ChIP-seq: welcome to the new frontier. Nat. Methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 3.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 11.Dopazo J. Functional interpretation of microarray experiments. OMICS. 2006;10:398–410. doi: 10.1089/omi.2006.10.398. [DOI] [PubMed] [Google Scholar]
- 12.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc. Natl. Acad. Sci. USA. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taher L, Ovcharenko I. Variable locus length in the human genome leads to ascertainment bias in functional inference for non-coding elements. Bioinformatics. 2009;25:578–584. doi: 10.1093/bioinformatics/btp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 16.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 17.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitz F, Duboule D. Global control regions and regulatory landscapes in vertebrate development and evolution. Adv. Genet. 2008;61:175–205. doi: 10.1016/S0065-2660(07)00006-5. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai J, Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J. Physiol. Pharmacol. 2002;53:147–157. [PubMed] [Google Scholar]
- 23.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 2007;292:70–81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 24.Ruan J, et al. TreeFam: 2008 update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linhart C, Halperin Y, Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–1189. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol. Cell. Biol. 1995;15:5975–5982. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami EG, Bader GD, Gross BE, Sander C. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertolotto C, et al. Cleavage of the serum response factor during death receptor-induced apoptosis results in an inhibition of the c-FOS promoter transcriptional activity. J. Biol. Chem. 2000;275:12941–12947. doi: 10.1074/jbc.275.17.12941. [DOI] [PubMed] [Google Scholar]
- 30.Poser S, Impey S, Trinh K, Xia Z, Storm DR. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J. 2000;19:4955–4966. doi: 10.1093/emboj/19.18.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HJ, et al. SRF is a nuclear repressor of Smad3-mediated TGF-beta signaling. Oncogene. 2007;26:173–185. doi: 10.1038/sj.onc.1209774. [DOI] [PubMed] [Google Scholar]
- 32.Chen CR, Kang Y, Siegel PM, Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 33.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blake JA, et al. The Mouse Genome Database genotypes:phenotypes. Nucleic Acids Res. 2009;37:D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat. Rev. Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- 36.Capdevila J, Izpisúa Belmonte JC. Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- 37.Kretzschmar M, Massagué J. SMADs: mediators and regulators of TGF-beta signaling. Curr. Opin. Genet. Dev. 1998;8:103–111. doi: 10.1016/s0959-437x(98)80069-5. [DOI] [PubMed] [Google Scholar]
- 38.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niswander L. Pattern formation: old models out on a limb. Nat. Rev. Genet. 2003;4:133–143. doi: 10.1038/nrg1001. [DOI] [PubMed] [Google Scholar]
- 40.Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- 42.Park CC, et al. Fine mapping of regulatory loci for mammalian gene expression using radiation hybrids. Nat. Genet. 2008;40:421–429. doi: 10.1038/ng.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 44.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 45.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 46.The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 48.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 49.Levings PP, Bungert J. The human beta-globin locus control region. Eur. J. Biochem. 2002;269:1589–1599. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 50.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental