OEFinder: a user interface to identify and visualize ordering effects in single-cell RNA-seq data (original) (raw)
Abstract
Summary: A recent article identified an artifact in multiple single-cell RNA-seq (scRNA-seq) datasets generated by the Fluidigm C1 platform. Specifically, Leng et al. showed significantly increased gene expression in cells captured from sites with small or large plate output IDs. We refer to this artifact as an ordering effect (OE). Including OE genes in downstream analyses could lead to biased results. To address this problem, we developed a statistical method and software called OEFinder to identify a sorted list of OE genes. OEFinder is available as an R package along with user-friendly graphical interface implementations which allows users to check for potential artifacts in scRNA-seq data generated by the Fluidigm C1 platform.
Availability and implementation: OEFinder is freely available at https://github.com/lengning/OEFinder
Contact: rstewart@morgridge.org or lengning1@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Single-cell RNA-seq (scRNA-seq) has led to important findings in many fields and is becoming increasingly popular in studies of transcriptome-wide expression (Deng _et al., 2014; Leng et al., 2015; Shalek _et al., 2014; Trapnell et al., 2014; Treutlein et al., 2014). To facilitate scRNA-seq, the majority of studies utilize the Fluidigm C1 platform for cell capture, reverse transcription and cDNA amplification, as this platform allows for rapid and reliable isolation and processing of individual cells. In spite of the advantages, Leng _et al. (2015) identified an artifact in multiple datasets generated by C1 and confirmed that the artifact was present in the cDNA processed by the C1 machine. In particular, in these datasets, there are genes showing higher expression in cells captured in specific capture sites. These capture sites are the ones with small or large plate output IDs. We refer to this artifact as an ordering effect (OE), which has been shown to be independent of organism and laboratory (Leng _et al., 2015). As detailed in Leng _et al. (2015), accurate identification of OE genes is important to ensure unbiased downstream analyses.
Leng _et al. (2015) used an ANOVA-based approach to detect OE genes. The ANOVA-based approach performs well in many cases, but has reduced power when few cells are available. (Note that in empirical data, the cells can be missed owing to the random effects of capture failure—an empty capture site, or doublets—capturing more than one cells in one capture site.) To improve power for identifying OE genes, we developed an approach, OEFinder, based on orthogonal polynomial regression. Results show that OEFinder is less sensitive to sample size and outperforms the ANOVA-based approach when few cells are available.
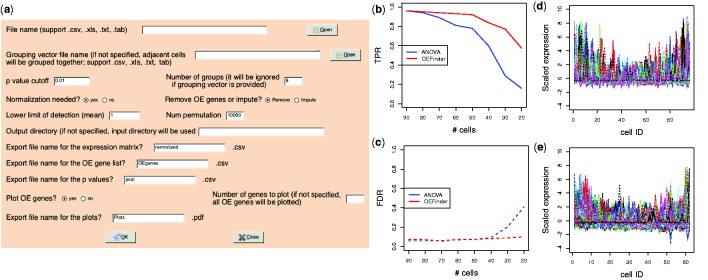
OEFinder is implemented in R, a free and open source language, with a vignette that provides working examples. The graphical user interface (GUI) implementations of OEFinder allow users with little computing background to easily identify and characterize OE genes in scRNA-seq data (Fig. 1a and Supplementary Fig. S4).
Fig. 1.
(a) OEFinder GUI for identifying OE genes (shown is implementation using R/RGtk2 package; an implementation using R/shiny package is also available, see Supplementary Fig. S4). (b, c) Operating characteristics in simulated datasets. The _x_-axes show the number of available cells. The _y_-axis shows TPR and FDR. (d, e) The OE genes identified in the first experiment of Trapnell et al. data and Leng et al. data, respectively. The cells were ordered following the capture site ID. The _y_-axis shows scaled gene expression (_z_-score). Each line represents one OE gene
2 Analysis input and output
2.1 Input
2.1.1 Expression estimates
OEFinder requires a genes-by-cells expression matrix. The expression matrix can be either normalized or unnormalized. If the input matrix is unnormalized, OEFinder applies the Median-by-Ratio normalization method introduced by Anders and Huber (2010) prior to OE detection.
2.1.2 Capture site group definitions
As detailed in Leng _et al. (2015), the capture sites are labeled as A01,…, A12, B01,…, B12,…, H01,…, H12. If the capture site IDs are provided, OEFinder groups cells from sites with the same starting letters. When the capture site information is not available, OEFinder groups cells based on their input order. By default, OEFinder groups cells into eight even-sized groups. The number of groups may be changed by the user.
2.2 Method
The normalized expression values of each gene are scaled to _z_-scores. For each gene, OEFinder applies an orthogonal polynomial regression on _z_-scores against group code. To infer whether gene g follows the OE trend, OEFinder calculates the _P_-value pg,2 of a one-tailed test that tests whether the coefficient of the quadratic term is positive. To account for the goodness of the spline fitting, OEFinder defines an aggregate statistics Sg as −log(pg,2) − log(pmodel), in which pmodel denotes the F test _P_-value of the full model. OEFinder then generates 10 000 simulated genes from permuted data to evaluate the significance of the observed aggregated statistics. By default, genes with permutation _P_-value <0.01 are identified as OE genes. The number of simulated genes and the _P_-value cutoff may be changed by a user (for further details, see Supplementary Section S2).
2.3 Output
2.3.1 List of OE genes
OEFinder outputs two.csv files - one contains a sorted list of OE genes and the other contains p-values for all genes.
2.3.2 Expression matrix for downstream analysis
OEFinder outputs a normalized expression matrix that can be directly input to downstream analyses. The user has the option to choose either removal of the OE genes, or imputation of the OE genes with adjusted values.
2.3.3 Visualization of OE genes
OEFinder generates a .pdf file contains expression plots of the top N OE genes, where N is user specified. An example is shown in Supplementary Fig. S1.
3 Evaluations
3.1 Simulation studies
We conducted eight simulation studies to evaluate the performance of the OE detection algorithms. In each simulation, we generated 5000 expressed genes with 500 OE genes. Expression of OE genes was generated based on expression profiles of OE genes detected in empirical data. (Details of the simulations may be found in Supplementary Section S3.) The eight simulation studies evaluate cases with varying numbers of available cells (20–90 cells). Each simulation study contains 100 repeated simulations.
Figure 1b and c shows the true positive rate (TPR) and false positive rate (FDR) comparing the ANOVA-based method introduced in Leng _et al. (2015) and OEFinder. Results indicate that >60 cells are available, both methods have TPR >90% while FDR is controlled <10%. When fewer cells are available, OEFinder has a higher TPR than the ANOVA-based approach and the FDR is still well controlled. The improved power in OEFinder is likely because the polynomial regression in OEFinder fits the OE trend more specifically than the ANOVA-based approach. Additional simulation results may be found in Supplementary Section S4.
3.2 Case studies
We applied OEFinder on two publicly available datasets with capture site ID information. Trapnell et al. (2014) data and Leng _et al. (2015) data contain four and three experiments, respectively. Figure 1d and e shows 187 and 451 OE genes identified in the first experiment of each dataset. Genes detected by OEFinder show a clear OE pattern. Results of other experiments may be found in Supplementary Section S5.
4 Discussion
We developed an R package OEFinder which can robustly detect OE genes in scRNA data generated by the Fluidigm C1 platform. OEFinder provides user-friendly graphical interface implementations that facilitate use by investigators.
Funding
This work was funded in part by GM102756, 4UH3TR000506, 5U01HL099773, U54 AI117924 Charlotte Geyer Foundation and Morgridge Institute for Research.
Conflict of Interest: none declared.
Supplementary Material
Supplementary Data
References
- Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q. et al. (2014) Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science, 343, 193–196. [DOI] [PubMed] [Google Scholar]
- Leng N. et al. (2015) Oscope: a pipeline for identifying oscillatory genes in unsynchronized single cell RNA-seq experiments. Nat. Methods 12, 947–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A.K. et al. (2014) Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature, 510, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol., 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B. et al. (2014) Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature, 509, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data