High density three-dimensional localization microscopy across large volumes (original) (raw)
. Author manuscript; available in PMC: 2016 Sep 7.
Published in final edited form as: Nat Methods. 2016 Mar 7;13(4):359–365. doi: 10.1038/nmeth.3797
Abstract
Extending three-dimensional (3D) single molecule localization microscopy away from the coverslip and into thicker specimens will greatly broaden its biological utility. However, localizing molecules in 3D with high precision in such samples, while simultaneously achieving the extreme labeling densities required for high resolution of densely crowded structures is challenging due to the limitations both of conventional imaging modalities and of conventional labeling techniques. Here, we combine lattice light sheet microscopy with newly developed, freely diffusing, cell permeable chemical probes with targeted affinity towards either DNA, intracellular membranes, or the plasma membrane. We use this combination to perform high localization precision, ultra-high labeling density, multicolor localization microscopy in samples up to 20 microns thick, including dividing cells and the neuromast organ of a zebrafish embryo. We also demonstrate super-resolution correlative imaging with protein specific photoactivable fluorophores, providing a mutually compatible, single platform alternative to correlative light-electron microscopy over large volumes.
Introduction
Localization microscopy methods enable the study of biological structures well below the optical diffraction limit1. However, despite their ability to localize fluorescent molecules in both two and three-dimensions2-4, the majority of applications have focused on the ventral surface or very thin (less than one micron) lamellar regions of adherent cells. Two challenges arise when extending localization microscopy into thicker three-dimensional (3D) samples. First, in order to localize single molecules, background signal from fluorescent emitters residing outside of the focal plane must be reduced either by limiting the excitation volume or by restricting the thickness of the specimen. Confinement at sub 100 nm length scales can be achieved via total internal reflection fluorescence (TIRF) microscopy or by mechanical sectioning of larger samples5. However, these strategies respectively limit observations to immediately adjacent to the coverslip or remove the structure of interest from its native 3D context. At larger length scales, oblique illumination6, multifocal detection7, temporal focusing8, 9, and selective plane illumination (SPIM)10 have all been used to attain high localization precision in thick samples. However, the region of activation and/or excitation in each case is thicker than the depth of focus of detection, leading to out-of-focus background and premature bleaching of activated molecules and degrading both the attainable localization precision and localization density.
This last fact is especially relevant because an often overlooked challenge for high-resolution 3D structural imaging is that the number of molecules required to adequately sample a given spatial frequency and thus resolve a structure of a given size, scales exponentially with the spatial dimension in which those structures are imaged (Supplementary Note 1-5, Supplementary Figs. 1-7, Supplementary Video 1). This is often addressed by analogy to the Nyquist sampling theorem1, 11-19 which links optical resolution, localization density, and spatial dimension in localization microscopy. By the Nyquist criteria, resolving a structure at 20 nm resolution would require the localization of up to 37,500 unique molecules within a 3D (250 × 250 × 600 nm) diffraction limited volume in the worst-case scenario where the volume is filled with structures at this scale. Even if such labeling densities can be achieved with conventional fluorescent proteins or antibodies while avoiding over-expression artifacts and maintaining labeling specificity, the on:off contrast ratio of the activated:non-activated fluorophore would still have to be over 40,000:1 in order to accurately localize the one activated molecule against the background of those remaining, even under perfect optical sectioning illumination. Moreover, because localizations represent a stochastic sampling of the underlying molecular distribution, adequately sampling a given spatial frequency with sufficient signal to noise can require an order of magnitude increase in the number of molecules compared to what is strictly required according to the Nyquist criterion.20
To address the illumination challenge and insure that only molecules near the focal plane are excited, we use lattice light sheet microscopy11 to illuminate an approximately 1.1 μm thick plane that is coincident with and is comparable in thickness to the depth of focus of our detection objective of 1.1 numerical aperture (NA) (Supplementary Note 6). To address the labeling density challenge in thick specimens, we combine this illumination scheme with point accumulation for imaging of nanoscale topography (PAINT) microscopy21. In comparison to methods that label the specimen prior to imaging, PAINT labels continually bind to the specimen throughout the imaging process, providing a continuous source of fluorescent molecules to label the structure (Supplementary Video 2, Supplementary Note 7). We also introduce two new PAINT labels that selectively bind to either intracellular membranes or fluorgenically to DNA (Supplementary Fig. 8, Supplementary Note 8). We use this combination of confined planar illumination, stochastic binding, and novel chemical probes to perform lattice light sheet–PAINT (LLS-PAINT) microscopy at high localization precision and high labeling density, in multiple colors, for samples up to 20 microns thick including dividing cells and at the periphery of small embryos.
Results
The effect of localization density for structural imaging
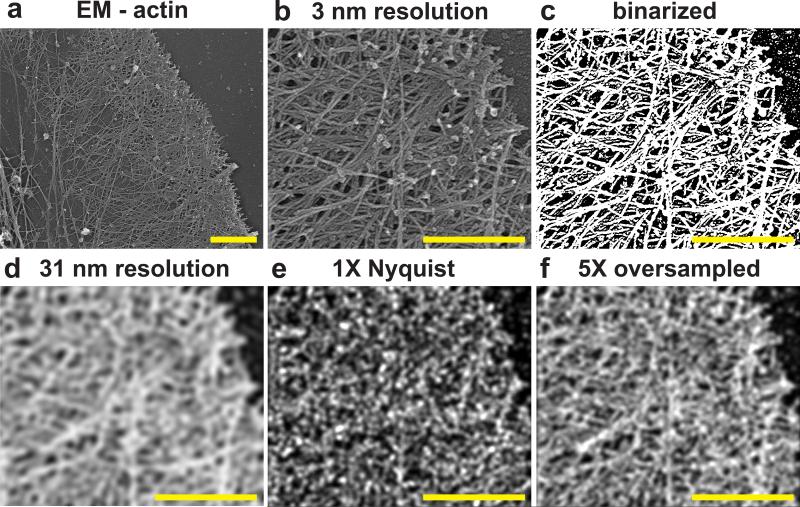
Many studies use the Nyquist criterion, i.e. two unique molecular localizations per unit length to incorporate the effect of localization density on resolution1, 11-19. However, a “sample” in the context of the Nyquist-Shannon sampling theorem is different than a localization event in localization microscopy (Supplementary Note 1): the former relates to a periodic sampling of a scalar valued function, whereas the latter involves stochastic, non-periodic sampling and does not, in a single event, reveal the underlying molecular density at a given location. Rather, each “event” is drawn from a probability distribution that is proportional to the underlying density of fluorophores22, which themselves are usually only proxies for the often incompletely labeled endogenous molecular targets of true biological interest. When used for structural imaging, the task for localization microscopy then is to observe a sufficient number of localization events to resolve the underlying distribution or structure. In light of this distinction, and to determine what density of localized molecules would be required to obtain a given resolution, we utilized both simplified 1D and 2D signals, as well as experimentally relevant images of the actin cytoskeleton23 (Fig. 1, Supplementary Notes 1-5, Supplementary Figs. 1-7). Our results suggest that obtaining localization microscopy data that is qualitatively comparable to electron microscopy (EM) images of well-defined resolution can require a five-fold or greater increase in the number of molecules to be localized than has previously been reported according to the Nyquist criteria. Therefore, in the rest the manuscript, we report resolution using this five-fold oversampling condition. However, we stress the following: 1) these values are inferred lower bounds on the attainable resolution, 2) the resolution varies throughout the cellular volume, and 3) true resolution at any given point will further be influenced by the combination of labeling specificity to the target, linker length from the label to the target, the accuracy of drift correction (see below), the possibility of molecule diffusion within a single camera exposure (especially for PAINT probes), the spatial frequency content of the sample itself22, 24, and the desired level of certainty on underlying molecular density20.
Fig. 1. The effect of labeling density in localization microscopy.
(a) Electron micrograph of actin in the lamellapodia of an adherent cell reproduced with permission from23. (b) Zoomed in region plotted at an inferred resolution of 3 nm. (c) Binarized “ground truth” distribution of actin vs. background. (d) Replotted ground truth image filtered to 31 nm resolution. (e) Simulated localization microscopy image plotted at 1X Nyquist density and 10 nm localization precision. This would be reported as having an identical 31 nm resolution assuming a requirement of 1X Nyquist sampling. (f) Simulated localization microscopy image plotted at five-fold greater localization density than in (e). This would be reported as having a 31 nm resolution assuming a requirement for 5X oversampling compared to Nyquist (c.f. Supplementary Note 1). Scale bars are 1 μm (a), 500 nm (b-f)
3D localization microscopy of intracellular membranes
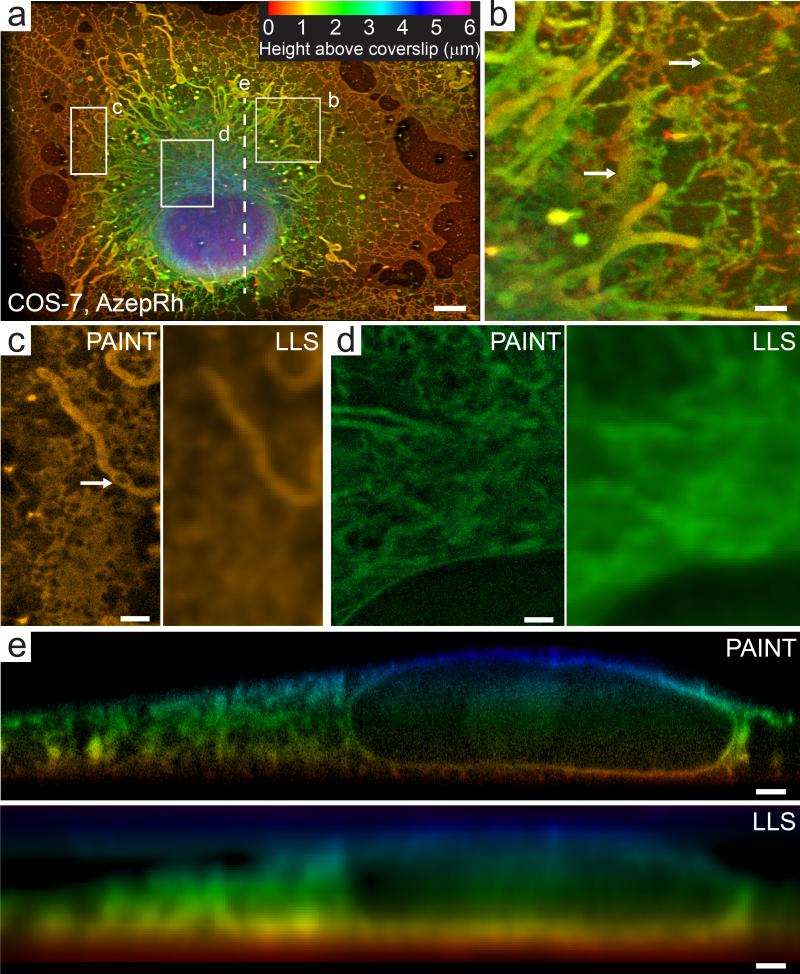
Screening of disparate rhodamine derivatives revealed that azepanyl-rhodamine (AzepRh,25) was an effective membrane stain (Supplementary Fig. 8). We used this probe to preferentially label the intracellular membranes throughout the 3D volume of an adherent, chemically fixed, COS-7 cell (Fig. 2 a, b, Supplementary Video 3, 4). We recorded nearly 150,000 3D volumes comprising over 20 million 2D images acquired in 270 nm steps. From these images, we localized approximately 400 million individual molecules to a median Cramer Rao Lower Bound (CRLB) precision of 8.9 × 9.7 × 41 nm in the lateral and axial directions, corresponding to a 20.9 × 22.8 × 96.4 nm full width at half maximum for a point object (i.e. 2.35 times larger than the CRLB). Note that in lattice light sheet microscopy, the detection axis (denoted z, and which has the poorest resolution) lies oblique to the coverslip (Supplementary Fig. 9). For ease of visualization, most images are displayed in a plane parallel to the coverslip and perpendicular to the coverslip's z (cz) axis; however, we note that this blends both the high precision lateral and low precision axial localizations into the image. The resolution bounds inferred from the localization precision and labeling density varied within different regions of the sample. However, using a density criterion of five-fold oversampling compared to Nyquist, the 23,400 – 6,950 localizations per diffraction limited volume here yields a density-limited lower bound of 40-60 nm resolution throughout much of the cell (Supplementary Video 4). When combined with the anisotropic localization precision noted above, this results in an inferred lower bound on the overall resolution of approximately 45-64 nm in the lateral and 104-114 nm in the axial directions respectively (Eq. 6, Supplementary Note 1). Across the entire six micron depth of the cell, the super-resolved image displayed substantially higher resolution than a deconvolved diffraction limited lattice light sheet image (Fig. 2 c-e, Supplementary Video 3), and several sections reveal features that can be clearly distinguished at ~70 nm in the lateral and ~100 nm in the axial dimensions respectively, (Supplementary Fig. 10), which is consistent with these values.
Fig. 2. Whole cell 3D localization microscopy of intracellular membranes.
(a) Maximum intensity projection of a COS-7 cell imaged with LLS- PAINT microscopy using AzepRh dye. Color encodes the height above the coverslip. Note that this projection mixes the high resolution x-axis with the low resolution z-axis of the microscope (Supplementary Fig. 9), but is more natural for the specimen geometry. (b) Maximum intensity projection of the region outlined in (a). Arrows indicate both sheet-like and reticulated tubule morphology of the endoplasmic reticulum. Larger tubular structures are mitochondria. (c) Single 20 nm thick orthoslice taken approximately 500 nm above the coverslip from the region outlined in (a). White arrow highlights a void in a tubular mitochondria. (d) Single 20 nm thick orthoslice parallel to and approximately 2.5 microns above the coverslip from the perinuclear region outlined in (a). (e) 12.5 nm thick orthoslice oriented perpendicular to the sample scan axis (s axis, Supplementary Fig. 9) along the dotted line indicated in (a). For comparison, (c)-(e) also show orthoslices from a deconvolved diffraction-limited lattice light sheet volume of the same regions (labeled LLS). Scale bars are 5 μm (a), 1 μm (b-e). See Methods for a description of the image post processing. See also Supplementary Videos 3 and 4.
Although AzepRh labels membranes of the endoplasmic reticulum (ER), mitochondria, Golgi, and lysosomes (Supplementary Fig. 11-14), ER and mitochondria could be discriminated through most of the cell by their characteristic morphologies. We observed both reticulated tubules and flattened sheet-like ER structures in the peripheral lamella as well as through the 3D perinuclear volume (Fig. 2 b, white arrows). An extensive tubular ER network was also observed immediately concentric to the outer nuclear envelope. Several mitochondria showed voids, similar to what has been observed previously with 2D, live-cell, localization microscopy and 3D structured illumination microscopy (SIM) using vital dyes17, 26, 27 (Fig. 2 c). Because of its ubiquitous staining pattern, LLS-PAINT datasets using AzepRh resemble the contrast obtained in heavy metal stained electron microscopy (EM) images. However, while lower resolution than EM images in the plane of the coverslip, LLS-PAINT offers sub-diffraction 3D resolution throughout whole cellular volumes without the need for mechanical sectioning.
Multicolor lattice light sheet – PAINT microscopy
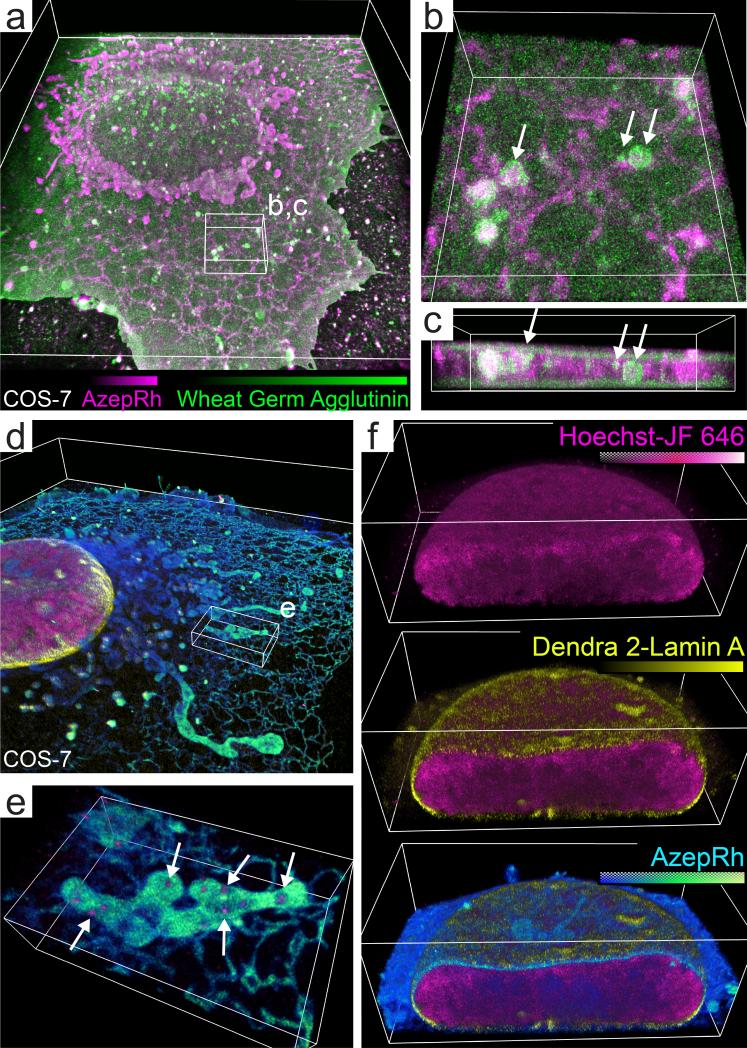
In contrast to EM, PAINT offers multicolor imaging by sequential staining with labels that have affinity to different structures. To demonstrate, we performed sequential imaging of a COS-7 cell with AzepRh (70,000 volumes, 91 million molecules) and wheat germ agglutinin-Alexafluor 555 (45,000 volumes, 34 million molecules) to label intracellular and plasma membranes respectively. This combination allowed visualization of both dorsal and ventral plasma membranes, 3D membrane ruffles at the cell periphery, and internalized plasma-membrane lined vesicles that appeared docked to the tubular network of the endoplasmic reticulum (Fig. 3 a-c, Supplementary Videos 5-6).
Fig. 3. Multicolor lattice light sheet – PAINT microscopy.
(a) Volume rendering of a COS-7 cell that has been sequentially imaged with AzepRh and wheat germ agglutinin – Alexafluor 555 to label intracellular and plasma membranes respectively. (b-c) Magnified perspective and orthogonal view of the region outlined in (a). White arrows indicate internalized vesicles adjacent to the endoplasmic reticulum yet peripherally stained with wheat germ agglutinin. (d) Three color PALM/PAINT imaging of DNA (Hoechst – JF646, PAINT, magenta), the nuclear envelope (Dendra 2 – lamin A, PALM, yellow), and intracellular membranes (AzepRh, PAINT, cyan) in a COS-7 cell. (e) Magnified perspective view of the region outlined in (d). White arrows highlight mitochondrial DNA nucleoids. (f) Cutaway views oriented perpendicular to the s axis through the nucleus in (d). Bounding boxes are 50 × 60 × 6.5 μm (a), 6 × 6 × 2.1 μm (b), 6 × 2.6 × 1.5 μm (c), 60 × 60 × 7.5 μm (d), 7 × 4.4 × 1.6 μm (e), and 21.7 × 13 × 7.5 μm (f). See also Supplementary Videos 5-8.
PAINT microscopy is also compatible with both diffraction-limited fluorescence and super-resolution PALM microscopy, which further extends the range of structures that can be visualized together. We transiently expressed lamin-A coupled to photoconvertable Dendra-2 to label the nuclear envelope. After fixation, lattice light sheet PALM imaging was performed as described previously11. Once the population of Dendra-2 molecules was exhausted (14,000 volumes, 5.7 million molecules), PAINT imaging was performed first with a novel fluorogenic DNA stain, Hoechst-JF646 (Supplementary Fig. 8) (32,800 volumes, 10.6 million molecules), followed by AzepRh (76,000 volumes, 85 million molecules) to label DNA and intracellular membranes respectively (Fig. 3 d-f, Supplementary Videos 7-8). This strategy revealed interactions between organelles and proteins that could not be determined by either method alone. Hoechst labeling revealed heterogeneous density variations within the cell nucleus that was surrounded by the nuclear lamin network. However, we also observed small clusters of Hoechst staining outside the nucleus that, in the context of multicolor imaging with AzepRh, were revealed to reside within the aforementioned mitochondrial voids, suggesting that they are in fact mitochondrial DNA nucleoids (Fig. 3 e, Supplementary Video 7).
3D architecture of membranes and chromosomes in mitosis
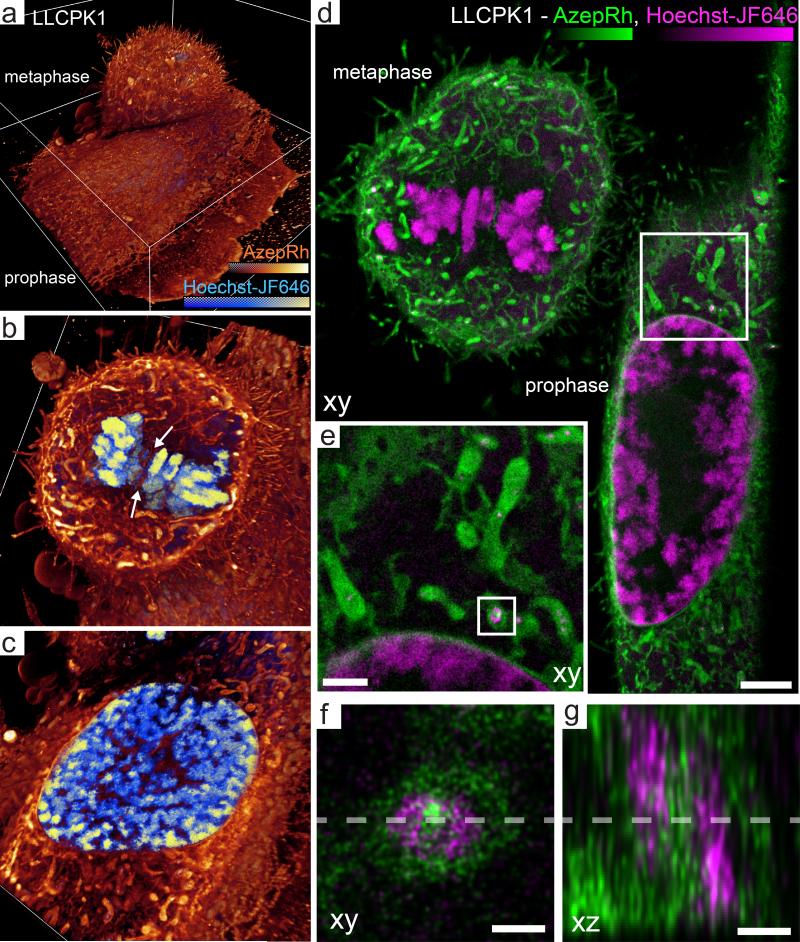
Adherent interphase cells have relatively flat aspect ratios. However, even adherent cells become thicker and more isotropic as they undergo mitosis. Thanks to the planar confinement of the illumination, we could acquire multicolor, 3D LLS-PAINT images of LLCPK1 cells in prophase, metaphase, and telophase with approximate dimensions of 40 × 50 × 7 μm, 20 × 20 × 18 μm and 50 × 25 × 20 μm respectively. Two-color PAINT imaging was performed using either Hoechst-JF646 (80,154 volumes, 57 million molecules) and AzepRh (112,000 volumes, 191 million molecules) (Fig. 4, Supplementary Videos 9-11), or wheat germ agglutinin-AlexaFluor 555 (20,000 volumes, 38 million molecules) and a commercially available BODIPY-TR methyl ester dye (87,000 volumes, 379 million molecules) (Supplementary Fig. 15, Supplementary Videos 12-14) that stained intracellular membranes in a manner similar to the AzepRh label described above (Supplementary Video 15). Throughout the 20 μm thickness of these specimens, the LLS-PAINT images displayed substantially higher resolution than raw or deconvolved diffraction-limited lattice light sheet images of resolution of 260 × 260 × 730 nm (for 560 / 580 nm excitation / emission), as well as lattice light sheet structured illumination microscopy (SIM)-mode images of resolution of 170 × 260 × 320 nm (Supplementary Figs. 9, 16, and Supplementary Videos 9, 12). LLS-PAINT also provided superior images compared to 3D widefield SIM of theoretical resolution 105 × 105 × 330 nm, as estimated by the cutoff of the optical transfer function, applied to a similar mitotic cell or TIRF-SIM of theoretical resolution of 96 × 96 nm applied to the peripheral region of a thin cell (Supplementary Figs. 17-19, Supplementary Note 9).
Fig. 4. 3D architecture of membranes and chromosomes during mitosis.
(a) Volume rendering of two dividing LLCPK1 cells in prophase (bottom) and metaphase (top) labeled with Hoechst – JF646 and AzepRh to visualize DNA and intracellular membranes. (b) Cutaway section midway through the metaphase cell in (a). White arrows highlight tubules of endoplasmic reticulum that intercalate between the condensed chromosomes in the metaphase plate. (c) Cutaway section midway through the nucleus of the prophase cell in (a). (d) 200 nm thick xy orthoslice through the same dataset. (e) Magnified view of the region outlined in (d). (f, g) Further magnified view of the region outlined in (e) showing 10 nm (f, xy) or 5 nm (g, xz) thick orthoslices through a mitochondria adjacent to the nuclear envelope. A ring shaped nucleoid of mitochondrial DNA (magenta) is embedded adjacent to mitochondrial membranes (green). Dashed lines indicate the regions from which corresponding xy or xz orthoslices were made. Cutaway sections in b and c are orientated perpendicular to the cz axis. Bounding boxes are 46 × 47.5 × 18 μm (a). Scale bars are 3 μm (d), 1 μm (e), 200 nm (f,g). See also Supplementary Videos 9-11, 16 and Supplementary Fig. 20.
Dividing LLCPK1 cells were densely covered with small, 1-3 μm long × 0.2-0.4 μm diameter membrane protrusions that labeled at their periphery with wheat germ agglutinin and throughout their volume with AzepRh or BODIPY-TR methyl ester. In some samples, 10-20 μm long retraction fibers anchored the cell to the coverslip (Supplementary Fig. 15). Hoechst-JF646 labeled both nuclear and mitochondrial DNA (Supplementary Video 9, 10). Often, we could observe complex shaped nucleoids such as ring shaped structures that resided within voids within the mitochondria (Fig. 4 d, e). Close inspection revealed clear separation of the DNA and membrane signals at length scales of less than 100 nm (Fig. 4 f, g, Supplementary Fig. 20, Supplementary Video 16). Metaphase chromosomes imaged via LLS-PAINT with Hoechst-JF646 had rough surfaces with many 100 - 200 nm long protrusions. EM images of isolated, pre-swollen chromosome spreads display similar protrusions28, 29, that are thought to be composed of locally decondensed loops of chromatin. However, LLS-PAINT permits direct 3D visualization of these structures in situ in dividing cells, as well as in multicolor imaging with intracellular membranes (Supplementary Fig. 21, Supplementary Video 9-11). ER structures, for example, formed a network of tubules that occasionally intercalated between chromosomes (Fig. 4 b), although this apparent tubular architecture may also result from fixation-induced fenestration of cisternal sheets30.
3D architecture of the zebrafish neuromast sensory organ
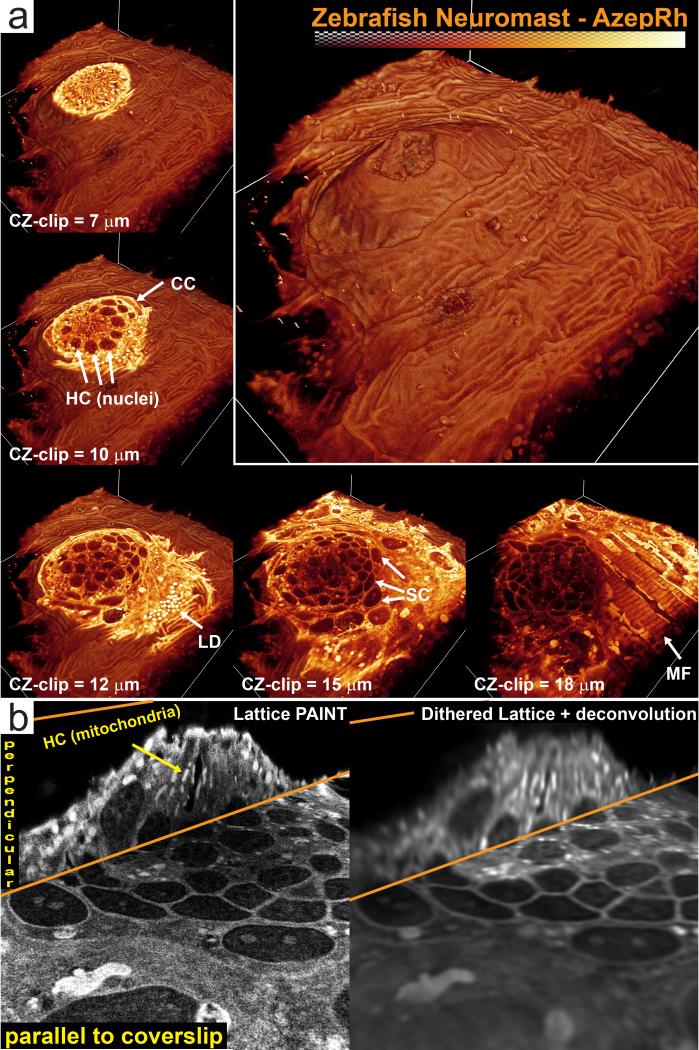
While optical aberrations and label diffusion limit the depth attainable by the LLS-PAINT technique to ~20-30 μm, the microscope geometry does permit the study of the peripheral regions of much larger multicellular specimens to these depths. To illustrate, we performed LLS-PAINT imaging of a neuromast organ at the periphery of an intact zebrafish embryo using the AzepRh dye (95,000 volumes, 1 billion molecules) (Fig. 5, Supplementary Video 17, 18). Each organ consists of a rosette of hair cells surrounded by a layer of support cells. Kinocilia and stereocilia emerge from the hair cells and protrude through a circular opening in the periderm. The LLS-PAINT dataset revealed multiple layers of crosshatched striated muscle peripheral to the neuromast organ and interior to the outer layer of ridged periderm cells. In our example, the kinocilia had flattened against the base, but we could also begin to resolve smaller stereocilia at the neuromast opening. Imaging deeper into the specimen revealed a rosette of approximately 15 hair cells (HC) with dense bundles of mitochondria that extended basally and radially away from the cuticular plate (Fig. 5 a, top). Heterogeneous staining variations in hair cell and support cell nuclei (Fig. 5 a, middle) closely resembled condensed chromatin regions from electron tomography datasets31, although, it is uncertain whether AzepRh directly interacts with chromatin in zebrafish. Surrounding the hair cell rosette was a layer of support cells (SC) (Fig. 5 a, bottom middle). In addition to higher resolution, the LLS-PAINT dataset exhibited improved contrast compared to diffraction-limited deconvolved lattice light sheet (Fig. 5 b), stemming from its superior optical sectioning, defined by the z-localization precision.
Fig. 5. 3D architecture of membranes in the zebrafish neuromast sensory organ.
(a) Volume rendering of a neuromast on the lateral line of a 5 day old zebrafish embryo labeled with AzepRh. Cutaway views oriented perpendicular to the cz axis show the internal architecture at different depths within the organ. Several cell types and features can be identified. A central rosette of hair cells (HC) with dense mitochondrial staining appears immediately basal to the organ opening. This is surrounded by a layer of crescent cells (CC) and support cells (SC). Adjacent to the neuromast, lipid droplets (LD) and myofibrils (MF) are visible. (b) 30 nm thick orthoslices oriented either perpendicular to the cz or s axis through the center of the neuromast. For comparison, deconvolved, diffraction-limited lattice light sheet orthoslices of the same regions are shown at right. Bounding box is 87.8 × 98.7 × 27.5 μm. See Methods for a description of the image post processing. See also Supplementary Videos 17-18.
Compensation of non-linear sample motion
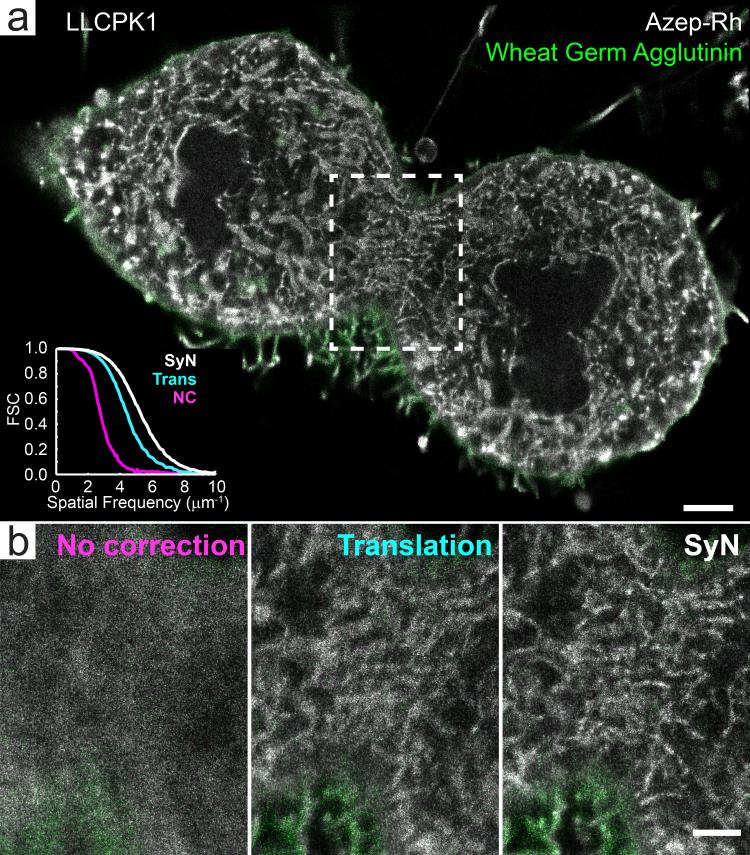
PAINT microscopy is inherently performed under non-equilibrium conditions. If the labels bind permanently rather than transiently32, which is likely the case for the examples shown here, then the sample will be progressively perturbed as additional dye molecules bind. When imaged over many hours, the sample gradually swelled, and if left uncorrected, this swelling reduced the resolution of the final image (Fig 6.). While this swelling is expected to occur with any exogenous labeling strategy, including heavy metal staining for EM, the deformation would not manifest itself during imaging because conventional super-resolution microscopy and EM are performed after labeling. Translational drift correction, common in localization microscopy of thin samples, is not sufficient to correct for the more complex non-linear deformations exhibited by thicker 3D specimens in PAINT. In light of this, we utilized a combination of fiducial-based translational registration and non-linear image transformation. We used a symmetric diffeomorphic image registration (SyN) algorithm, available through the open source Advanced Normalization Tools (ANTs), which is commonly used for template generation and image registration in neuroscience33. With this, we registered sequential images plotted from time-integrated sub-portions of the total data to generate a spatio-temporally varying correction function (Supplementary Note 10, Supplementary Figs. 22-25). Although more pronounced for the thicker 3D specimens, non-linear swelling correction substantially increased image resolution in all datasets when compared to no drift correction or translation only drift correction (Fig. 6, Supplementary Videos 19, 20).
Fig. 6. Compensation for non-linear swelling in lattice light sheet – PAINT datasets.
(a) A 60 nm thick orthoslice oriented perpendicular to the z-axis through the dividing LLCPK1 cell shown in Supplementary Fig. 15 that has been corrected for spatially-invariant translational drift as well as nonlinear swelling using the SyN algorithm. Fourier shell correlation performed within the boxed region of the AzepRh images, shows a shift in the weighting toward higher spatial frequencies for the SyN corrected datasets when compared to no correction and translation only correction. (b) 60 nm thick orthoslices of the boxed region in (a) at various levels of displacement correction. Scale bars are 2 μm (a), 1 μm (b).
Discussion
By limiting the illumination volume and providing high labeling density, LLS-PAINT microscopy allows for sub-100 nm resolution imaging of thick, densely crowded specimens including dividing cells and at the periphery of small embryos. Due to the stochastic nature of localization microscopy, resolving a given spatial frequency can require a five-fold or greater increase in the number of localized molecules compared to what would be required according to the Nyquist criteria. This suggests that previously reported resolution values that assume a requirement for 1X Nyquist density may be overly optimistic1, 11, 14-19, if the expectation is that such values apply to all structures across the field of view in absence of a-priori knowledge about the spatial structure of the sample. It also illustrates the limitations of using fluorescent labels as surrogates to resolve an underlying structure to which the labels are bound. Endogenous expression levels of many target proteins may not support sampling an underlying structure at high spatial resolution, even if every protein molecule of a specific type could be labeled and localized. This may explain why prototypical localization microscopy datasets often focus on a few structural proteins that are normally expressed at or locally condensed at very high densities such as cytoskeletal components (actin, tubulin), focal adhesions (vinculin, paxillin) or endocytic pits (clathrin, caveolin). Further, it suggests that PAINT labels targeted to lipids can enable higher resolution than localization microscopy techniques that target a specific protein, and may point out an intrinsic limitation of protein engineering that would be shared even by EM methods utilizing protein-specific contrast34, 35. Further discussion on the design criteria for new PAINT probes to target additional structures is presented in Supplementary Note 8.
An obvious corollary is that the need for high localization density over a large, densely crowded 3D volume requires an enormous number of localization events that require substantial time to acquire. Many of the datasets presented here are comprised of over 100,000 3D volumes, each consisting of approximately 100 2D image planes. At 25 ms exposure per image, this translates to a volumetric imaging rate of 0.4 Hz, resulting in a total imaging duration of approximately 69 hours. Serial labeling of multiple targets increases this further. Thus it is not uncommon to acquire a single multicolor dataset over approximately 3-10 days. While this is substantially slower than the imaging rates reported for 2D localization microscopy at comparable reported resolution15, it is comparable to the acquisition rate reported elsewhere for high density 3D localization microscopy19 when normalized to the number of focal planes and thus imaging volume acquired. Ultimately, the volumetric imaging rate for LLS-PAINT is limited by the rate of dye diffusion into the specimen (Supplementary Fig. 26).
Despite the long acquisition times, membrane-binding PAINT molecules such as AzepRh and BODIPY-TR methyl ester can play an important role in correlative microscopy when used in conjunction with protein-specific super-resolution methods such as PALM. Although 3D nanoscale membrane-protein correlation has been achieved with combined EM and iPALM to place the protein-specific information in a global context36, heavy metal staining for EM has a deleterious effect on fluorescent labels, so that sample preparation becomes a delicate balancing act of providing enough EM contrast without impacting the labeling density too negatively. Furthermore, 3D EM of samples a micron or more in thickness requires mechanical sectioning which, with the exception of sectioning via focused ion beam milling37, requires careful registration to minimize distortions. Finally, unlike EM-PALM, PAINT-PALM correlation occurs in the same instrument without the need to remove the specimen and relocate the original field of view. Indeed, live cell 3D imaging of cellular dynamics can be recorded with the lattice light sheet immediately prior to in situ fixation and subsequent 3D PAINT and/or PALM, enabling further correlation of dynamics and function with membrane and protein structure, or permitting fixation and subsequent super-resolution imaging to be synchronized to a specific event such as drug delivery or optogenetic stimulation.
Online Methods
Instrumentation
We performed all experiments at room temperature on a previously described lattice light sheet microscope11. We made several modifications to accommodate the 3D PAINT experiments. First, to perform axial localization via astigmatism3, 38, we mounted a 1000 mm focal length cylindrical lens (ThorLabs, J1516RM-A) approximately 40 mm from the camera sensor. Second, we utilized a 2W 560 nm continuous wave laser (MPB Communications, 2RU-VFL-P-2000-560-B1R) to increase illumination intensities at the sample and thus imaging speed. Due to losses within the optical system, incident powers were in the range of 5-25 mW into the back pupil of the excitation objective. Complete experimental details for each dataset including illumination power (as measured at the back pupil of the excitation objective and estimated at the sample), exposure time per 2D image, number of 2D images per volume and the volumes per dataset are available in Supplementary Table 1. Finally, to compensate for evaporation and to maintain dye concentration throughout an experiment, we continuously perfused buffer from an approximately 1-2 L reservoir through the sample bath using a peristaltic pump (Warner Instruments, Model 720). PAINT label concentration varied between samples and labels, but was generally between 100 pM and 1 nM in phosphate buffered saline (PBS). Documentation for the construction of a copy of the lattice light sheet microscope is freely available after execution of a research license with HHMI. Researchers can also apply to access the microscope as visitors through the Advanced Imaging Center at Janelia Research Campus (www.janelia.org/aic).
Sample Preparation
We pre-cleaned 5mm diameter coverslips (Warner Instruments, CS-5R) by immersing in 1 M KOH for 1 hour followed by 1 hour immersion in deionized water, both under sonication. We dried the coverslips and heated to them 100 °C on a hotplate. We then deposited 10 μl of a 1:100 dilution of stock solution of 50 nm gold nano rods (Nanopartz, A12-50-600-25) to 200 proof EtOH onto each coverslip and allowed it to evaporate. We then sputtered 40 nm of SiO2 onto the coverslips using a Denton Explorer (Denton Vacuum LLC). Approximate density of fiducial markers ranged between 5 and 25 per 100 × 100 μm field of view. We obtained COS-7 cells from ATCC. We obtained LLCPK1 cells as a gift from Dr. Mike Davidson at Florida State University. We maintained all cell lines in Dulbecco's Modiified Eagle Medium + 10% fetal bovine serum and verified them to be free of mycoplasma. Prior to plating cells, we UV sterilized coverslips and coated them with 10 μg/ml of fibronectin (Millipore, FC010) for 1 hr to aid adhesion before rinsing with sterile PBS. After 24-48 hours we fixed cells for 1 hr at room temperature using a solution of 4% paraformaldehyde (Electron Microscopy Sciences), 0.1 % gluteraldehyde (Electron Microscopy Sciences) in PHEM buffer (25 mM Hepes, 10 mM EGTA, 2 mM MgCl2, and 60 mM PIPES, all from Sigma Aldrich). Prior to fixation, we transfected the cell from Figure 3 with Dendra2 – Lamin A as described previously11. Where indicated, we purchased BODIPY TR methyl ester from LifeTechnologies (cat# C34556). Zebrafish experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee of the Howard Hughes Medical Institute, Janelia Research Campus. To image the neuromast organ, we anesthetized 5 day old Casper strain zebrafish embryos39 in MS-222 and fixed in 4% paraformaldehyde in PBS over night at 4 °C with gentle agitation on a rocker. After fixation, we embedded the fish in a thin layer of 1% low melting point agarose with a 1:5 dilution of stock solution 50 nm gold nano rods. We aspirated excess agarose to leave only a very thin (~ one micron) layer surrounding the neuromast structures.
Single molecule localization and image characterization
We stored individual volumes as multipage Tiff stacks and imported into Matlab (The Mathworks) for further analysis. We identified fluorescent molecules as local maxima in a Gaussian filtered version of each 2D image and the raw, photon converted data from each molecule was fit to a generalized elliptic 2D Gaussian via Levenberg-Marquardt optimization. We linked sequential localizations of the same molecule into a single higher accuracy localization using a spatio-temporal cutoff filter. We performed all optimization on a graphics processing unit (GPU, NVIDIA GeForce GTX Titan) using custom written CUDA code called via a Matlab mex function. Full details of the numerical procedure are provided in Supplementary Note 11. For each dataset, we report the mean number of photons detected for each molecule, the number of molecules per 100 nm cubic volume, the x, y, and z localization precision, and the lower bounds on the x, y and z image resolution as a function of spatial coordinates in the image. These data represent a thorough characterization of each experiment and are presented in Supplementary Videos 4, 6, 8, 11, 14, and 18. For reference, when these movies are opened in Quicktime, individual frames can be manually selected to more fully view the data. Additional statistics and histograms regarding the localizations for each dataset are provided in Supplementary Fig. 27.
Fourier ring correlation (FRC) and Fourier shell correlation (FSC) represent the relative weighting of spatial frequencies in an image. Thus, if the underlying sample remains the same, a shift in the FRC/FSC of an image toward higher spatial frequencies can reveal the beneficial effects of drift correction. We computed the FSC comparison (in Fig. 6) following previously outlined methods24. We point out here that although this metric is useful for analyzing changes in resolution that occur through transformations to or by incorporating additional localizations within the same sample, care should be taken when using this method as an absolute metric for comparing the spatial resolution between images of different labels or structures (Supplementary Fig. 28). Further discussion is provided in Supplementary Note 4.
Code Availability
All the data in the manuscript are available upon request including high resolution, 3D images as well as the raw localization statistics. Software for identifying, localizing and plotting single molecule data is freely available after execution of a research license with HHMI. The Advanced Normalization Tools Software for image registration is freely available from http://stnava.github.io/ANTs/.
Image rendering
We plotted 3D localization microscopy images either by rendering each individual molecule as a 3D Gaussian probability distribution (for 2D slice representation) or by generating a 3D super-resolution histogram and smoothing the final image with a Gaussian kernel (for volume renderings). For certain datasets, we applied gamma correction to more clearly illustrate fine scale structures. Two datasets also underwent additional rescaling prior to display. First, the fixation buffer used for the COS-7 cell displayed in Figure 2 was contaminated with microbial debris. This debris was visible in the reconstructed dataset and obscured the intracellular structures of interest when displayed as a maximum intensity projection. Therefore, prior to image rendering, we masked structures exterior to the cell with a user generated binary image filter. Second, the periphery of the zebrafish neuromast dataset in Figure 5 showed greater numbers of localizations and thus higher contrast than interior regions. In order to more clearly illustrate features within the hair cell rosette, we normalized the contrast between both interior and peripheral regions using contrast limited adaptive histogram equalization via the CLAHE plugin in Fiji40. For comparison purposes, the resolution characterization images of these datasets in Supplementary Videos 4 and 18 show the full unprocessed data. We plotted final images as 3D matrices in Matlab and exported them in NifTI format using the “NifTI tools” toolbox from the Matlab File Exchange. We then imported NifTI images into Amira (FEI) or Fiji for visualization, image rendering, and Supplementary Video generation.
Supplementary Material
1
vid17
vid18
vid19
vid2
vid20
vid3
vid4
vid5
vid6
vid7
vid1
vid8
vid9
vid10
vid11
vid12
vid13
vid14
vid15
vid16
Acknowledgements
We gratefully acknowledge M. Davidson of the National High Magnetic Field Laboratory and Department of Biological Science, Florida State University, Tallahassee, FL for the cell line stably expressing calnexin and H2B and for the plasmids used for comparative staining, D. Li for discussion of resolution metrics and localization density, B. Hoeckendorf for assistance culturing and mounting zebrafish embryos, and H. White and the shared resource teams at Janelia for assistance with cell culture. W.R.L., L.S., J.B.G., T.A.B, L.D.L, and E.B. are funded by the Howard Hughes Medical Institute (HHMI). B.B.A. acknowledges financial support from NIH grant #K01ES025432-01 (to B.B.A) through NIEHS.
Footnotes
Author Contributions:
E.B. supervised the project; W.R.L, L.D.L, and E.B. conceived the idea; D.E.M, W.R.L, and E.B. developed the instrument control program; W.R.L and L.S. designed the single molecule fitting and plotting software; L.D.L, J.B.G and T.A.B designed and characterized the AzepRh and Hoechst-JF646 PAINT probes; B.B.A developed the SyN algorithm and ANTs software used for non-linear swelling correction; W.R.L built the instrument and performed the imaging experiments; W.R.L and E.B. wrote the paper with input from all co-authors.
Statement of Competing Financial Interests:
Portions of the technology described herein are covered by one or more pending Patent Applications filed by E.B. and W.R.L., or L.D.L. and J.B.G., and assigned to HHMI.
References for Main Text
- 1.Sauer M. Localization microscopy coming of age: from concepts to biological impact. J Cell Sci. 2013;126:3505–3513. doi: 10.1242/jcs.123612. [DOI] [PubMed] [Google Scholar]
- 2.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanguneri S, Flottmann B, Horstmann H, Heilemann M, Kuner T. Three-dimensional, tomographic super-resolution fluorescence imaging of serially sectioned thick samples. PLoS One. 2012;7:e38098. doi: 10.1371/journal.pone.0038098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajj B, et al. Whole-cell, multicolor superresolution imaging using volumetric multifocus microscopy. Proc Natl Acad Sci U S A. 2014;111:17480–17485. doi: 10.1073/pnas.1412396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaziri A, Tang J, Shroff H, Shank CV. Multilayer three-dimensional super resolution imaging of thick biological samples. Proc Natl Acad Sci U S A. 2008;105:20221–20226. doi: 10.1073/pnas.0810636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cella Zanacchi F, et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8:1047–1049. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- 11.Chen BC, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demmerle J, Wegel E, Schermelleh L, Dobbie IM. Assessing resolution in super-resolution imaging. Methods. 2015;88:3–10. doi: 10.1016/j.ymeth.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Deschout H, et al. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat Methods. 2014;11:253–266. doi: 10.1038/nmeth.2843. [DOI] [PubMed] [Google Scholar]
- 14.Holden SJ, Uphoff S, Kapanidis AN. DAOSTORM: an algorithm for high- density super- resolution microscopy. Nat Methods. 2011;8:279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- 15.Huang F, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shim SH, et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci U S A. 2012;109:13978–13983. doi: 10.1073/pnas.1201882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shroff H, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349 doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci U S A. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald JE, Lu J, Schnitzer MJ. Estimation theoretic measure of resolution for stochastic localization microscopy. Phys Rev Lett. 2012;109:048102. doi: 10.1103/PhysRevLett.109.048102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieuwenhuizen RP, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm JB, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–250. 243, 250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MG. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc Natl Acad Sci U S A. 2012;109:5311–5315. doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao L, Kner P, Rego EH, Gustafsson MG. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- 28.Ris H. Stereoscopic electron microscopy of chromosomes. Methods Cell Biol. 1981;22:77–96. doi: 10.1016/s0091-679x(08)61871-3. [DOI] [PubMed] [Google Scholar]
- 29.Harrison CJ, Allen TD, Britch M, Harris R. High-resolution scanning electron microscopy of human metaphase chromosomes. J Cell Sci. 1982;56:409–422. doi: 10.1242/jcs.56.1.409. [DOI] [PubMed] [Google Scholar]
- 30.Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens KN, et al. Ultrastructural analysis of aminoglycoside-induced hair cell death in the zebrafish lateral line reveals an early mitochondrial response. J Comp Neurol. 2007;502:522–543. doi: 10.1002/cne.21345. [DOI] [PubMed] [Google Scholar]
- 32.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci U S A. 2012;109:6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 38.Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
vid17
vid18
vid19
vid2
vid20
vid3
vid4
vid5
vid6
vid7
vid1
vid8
vid9
vid10
vid11
vid12
vid13
vid14
vid15
vid16