Modeling Alzheimer's Disease with Induced Pluripotent Stem Cells: Current Challenges and Future Concerns (original) (raw)
Abstract
Alzheimer's disease (AD) is the most prevalent type of dementia and its pathology is characterized by deposition of extracellular β_-amyloid plaques, intracellular neurofibrillary tangles, and extensive neuron loss. While only a few familial AD cases are due to mutations in three causative genes (APP, PSEN1, and PSEN2), the ultimate cause behind the rest of the cases, called sporadic AD, remains unknown. Current animal and cellular models of human AD, which are based on the A_β and tau hypotheses only, partially resemble the familial AD. As a result, there is a pressing need for the development of new models providing insights into the pathological mechanisms of AD and for the discovery of ways to treat or delay the onset of the disease. Recent preclinical research suggests that stem cells can be used to model AD. Indeed, human induced pluripotent stem cells can be differentiated into disease-relevant cell types that recapitulate the unique genome of a sporadic AD patient or family member. In this review, we will first summarize the current research findings on the genetic and pathological mechanisms of AD. We will then highlight the existing induced pluripotent stem cell models of AD and, lastly, discuss the potential clinical applications in this field.
1. Introduction
Since Alzheimer's disease (AD) was first diagnosed by Dr. Alois Alzheimer in 1906 [1], it has become the most prevalent neurodegenerative disease overall. Over 30 million people had suffered from AD worldwide before 2010, and the count number is estimated to double every 20 years to reach 66 million in 2030 and 115 million in 2050 [2] (http://www.alz.co.uk/research/statistics; accessed October 9, 2012).
Clinically, AD is characterized by gradual memory loss and a progressive learning disability and inability to carry out daily tasks. The main pathological hallmarks of AD are thought to be the deposition of extracellular senile plaques composed of insoluble β_-amyloid (A_β) peptide, the formation of intracellular neurofibrillary tangles, and the loss of cholinergic neurons in the basal forebrain, amygdala, hippocampus, and cortical area. However, only the A_β_ and the abnormal truncated and hyperphosphorylated tau hypotheses cannot fully explain all of the symptoms of AD. Indeed, various antiamyloid drugs succeeded in lowering the A_β_ levels in the brain but failed to slow down the cognitive decline in the treated patients [3, 4]. Additionally, antitangle drugs, which target the kinases and activators involved in the hyperphosphorylation of tau (including the GSK-3 inhibitors, Tideglusib and the methylthioninium chloride tau aggregation inhibitor, Rember) were successful in phase II clinical trials [5] but showed imprecise efficacy in larger phase II trials.
Due to AD's multifactorial and heterogeneous features, its ultimate etiology of AD is not thoroughly understood. While mutations of Presenilin 1 (PSEN1), Presenilin 2 (PSEN2), and amyloid precursor protein (APP) account for most of the early-onset familial AD cases [6, 7], the etiology of the remaining 95% sporadic AD patients is complicated, which is due to various factors including aging, gender, education, and genotype of apolipoprotein E (ApoE) [8]. Therefore, there is a pressing need for the emergence of new technologies and models reflecting the progression of AD in patients, confirming the disease pathology, and predicting novel or optimal therapeutic strategies.
Since its creation in 2006 by Yamanaka groups, induced pluripotent stem cell (iPSC) is considered as a potential tool for modeling neurodegenerative diseases [9]. By forced expression of certain genes, including Oct 3/4, Sox2, Klf4, and c-Myc, patients' specific somatic cells are reprogrammed towards their pluripotent state. In this manner, iPSCs are generated artificially and regain the ability to convert into any cell type of the three germ layers: mesoderm, ectoderm, and endoderm. Several preclinical studies, by modeling both familial and sporadic AD, have established promising methods to gather insights into the exact cellular mechanisms, potential therapeutic strategies, and personalized treatments for AD. Here we summarize the current research on the pathogenesis and iPSC-based models of AD and highlight the potential future application of these cells.
2. Genetics and Pathology of AD
Given the fact that most AD cases are sporadic and that the disease occurs at an old age, an increasing evidence indicates that the underlying cellular or molecular pathological process may start early and progress throughout one's life. The early-onset, familial AD (FAD) accounts for less than 5% of all AD sufferers, and the late-onset, sporadic AD (SAD) affects the remaining 95% [10] (http://www.molgen.vib-ua.be/ADMutations/). FAD and SAD appear to share the same clinical and pathological process in a way that both types of AD patients exhibit progressive dementia clinically, extracellular A_β_ plaques, and intracellular accumulation of phosphorylated tau protein. In general, major achievements of understanding to AD came from the study of the familial AD and mostly from FAD patients with disease-causing mutations.
Genetic factor is considered to be among main contributors to the risk of AD. Mutations in disease-causing genes and disease-risk genes have been identified and linked with either early-onset AD (EOAD) or late-onset AD (LOAD) (Table 1). Usually, EOAD is inherited in an autosomal dominant manner, and by linkage analyses three rare forms of EOAD have been identified to be linked to their causative genes which include one that encodes for the amyloid precursor protein (APP) and two coding for presenilin, PSEN1 and PSEN2. Approximately 50% FAD patients carry mutations in the three causative genes. Among them, mutations in PSEN1 that represent the majority comprise the majority (approximately 70–80%) of the mutations in EOFAD, followed by APP mutations (15–20%) and mutations of PSEN2 accounting for less than 5% [10]. The amyloid cascade hypothesis demonstrates the underlying targets of the three causative genes. In central nervous system (CNS), the APP protein functions as a neuron surface receptor and participant in neurite growth, neuronal adhesion, and axonogenesis. Physically, the APP protein is cleaved by α_-, β_-, and γ_-secretase at three major sites, respectively. The α_-secretase (mainly ADAM10, a disintegrin and metalloproteinase 10) mediated cleavage reduces the production of A_β, while β_-secretase (mainly BACE I, β site APP-cleaving enzyme I) and γ_-secretase lead to A_β production [11, 12]. PSEN1 and 2 are transmembrane protein components of the γ_-secretase complex involved in A_β production during APP processing. The A_β clearance pathway includes Neprilysin, IDE (insulin-degrading enzyme), ECE (endothelin-converting enzyme), and ACE (angiotensin-converting enzyme) [13, 14]. Imbalance between production and degradation of A_β (e.g., the mutations of APP, PSEN1, and PSEN2) results in its accumulation and aggregation in the brain. The consequences of A_β accumulation include a series of abnormal cellular responses such as the formation of intracellular neurofibrillary tangles (NFTs) made of abnormal truncated and hyperphosphorylated tau [15, 16], microglial and astrocytic activation, inflammatory response, oxidative stress, mitochondrial dysfunction, and at last neuron loss. Tau is a microtubule-associated protein with function of promoting microtubule assembly and stability that may also be involved in the establishment and maintenance of neuronal polarity, axonal transport, and neurite outgrowth, although there are no known tau mutation in AD. In the AD brain, the principal hallmark of tau pathology is the formation of paired helical filaments (PHFs) and NFTs. Tau hyperphosphorylation is a potent inducer of tau pathology because hyperphosphorylated tau displays an increased propensity to form PHFs. It is possible that A_β peptides that have initially accumulated in the AD brain could activate some tau kinases to promote tau phosphorylation through insulin or wnt pathway [17, 18]. Among these, GSK3_β_ is identified to be able to phosphorylate tau at several sites to form PHFs in neurofibrillary tangles distributed in AD brain [19]. In some PSEN1 mutation cases, GSK3 also became active with the existence of A_β_ peptide [20]. On the other hand, tau is also a substrate for various proteases. Truncations of tau protein at aspartic acid 421 (D421) and glutamic acid 391 (E391) residues by several caspases are associated with NFTs in the brains of AD patients [21, 22]. In vitro A_β_ treatment produces a 17 kDa fragment (tau 45–230), and overexpression of it induces neuronal apoptosis [23]. Additionally, Calpains, thrombin, and cathepsins are also involved in tau truncation apart from caspase [24–26]. However, more tau fragments found in AD brain are not well investigated and their production and impact remain to be identified.
Table 1.
Causative and risk variants of AD.
| Gene | Variant | Effect allele frequency | Odds ratio | Function | AD-related pathways |
|---|---|---|---|---|---|
| APP | — | — | — | A_β_ peptide precursor, neurite outgrowth, adhesion, and axonogenesis | APP processing, produce A_β_ |
| PSEN1 | — | — | — | Component of γ_-secretase complex that cleaves APP into A_β fragments | APP amyloidogenic pathway, cleaves APP |
| PSEN2 | — | — | — | Component of γ_-secretase complex that cleaves APP into A_β fragments | APP amyloidogenic pathway, cleaves APP |
| Frequent variants | |||||
| APOE | — | — | — | Component of lipoproteins, transports lipids and cholesterols, mediates synaptogenesis, synaptic plasticity, and neuroinflammation | A_β_ clearance |
| CR1 | rs6656401 | 0.197 | 1.18 | Bind C3b and C4b, and moderate the activity of the complement system | A_β_ clearance |
| BIN1 | rs6733839 | 0.409 | 1.22 | Participant in Clathrin-mediated endocytosis, intracellular trafficking, apoptosis, and interacting with the microtubule cytoskeleton | Mediate tau toxicity |
| CD2AP | rs10948363 | 0.266 | 1.10 | Cytoskeletal organization, endocytosis | Mediate tau toxicity |
| EPHA1 | rs11771145 | 0.338 | 0.9 | Mediate brain development, particularly axonal guidance | Immune response, neural development |
| CLU | rs9331896 | 0.375 | 0.86 | extracellular chaperone, inhibits formation of amyloid fibrils by APP | A_β_ clearance |
| MS4A6A | rs983392 | 0.403 | 0.9 | Signal transduction | unknown |
| PLCALM | rs10792832 | 0.358 | 0.87 | Clathrin-mediated endocytosis | A_β_ clearance |
| CD33 | rs3865444 | 0.307 | 0.94 | Inhibition of cell activity | A_β_42 uptake |
| HLA-DRB5–HLA-DRB1 | rs9271192 | 0.276 | 1.11 | Histocompatibility antigen, peptide antigen binding | Immune response |
| PTK2B | rs28834970 | 0.366 | 1.10 | Induce long term potentiation in hippocampus | Synapse function and neural development |
| SLC24A4and RIN3 | rs10498633 | 0.217 | 0.91 | Calcium transport, brain and neural development | Neural development |
| DSG2 | rs8093731 | 0.017 | 0.73 | Component of intercellular desmosome junctions | unknown |
| INPP5D | rs35349669 | 0.488 | 1.08 | Regulate cell proliferation and survival | Immune response |
| MEF2C | rs190982 | 0.408 | 0.93 | Participant in hippocampal-dependent learning and memory by suppressing the number of excitatory synapses,and neuronal development and distribution | Neural development, synapse function |
| NME8 | rs2718058 | 0.373 | 0.93 | Spermatogenesis, ciliary functions | unknown |
| ZCWPW1and NYAP1 | rs1476679 | 0.278 | 0.91 | Epigenetic regulation (ZCWPW1); brain and neural development (NYAP1) | Neural development |
| CELF1 | rs10838725 | 0.316 | 1.08 | mRNA splicing | unknown |
| FERMT2 | rs17125944 | 0.092 | 1.14 | Cell adhesion, cell shape and Wnt signaling pathway | Mediates tau toxicity |
| CASS4 | rs7274581 | 0.083 | 0.88 | cell adhesion and cell spreading | Cytoskeleton and axonal transport |
| SORL1 | rs11218343 | 0.039 | 0.77 | Lipoprotein uptake, APP trafficking to and from Golgi apparatus | APP trafficking |
| ABCA7 | rs115550680 | 0.09 | 1.79 | lipid metabolism, phagocytosis of apoptotic cells | A_β_ clearance |
| Rare variants | |||||
| ADAM10 | Q170H and R181G | — | — | Constitute and regulate _α_-secretase activities | APP nonamyloidogenic pathway |
| APP | rs63750847 | 0.0045 | 0.24 | Inhibit _β_-secretase activities | APP processing |
| TREM2 | rs75932628 | 0.0063 | 2.26 | A_β_ clearance | Immune response |
| UNC5C | rs137875858 | 0.0003298 | 2.15 | Increase susceptibility to neuronal cell death | Inflammatory response |
| PLD3 | rs145999145 | 0.003077 | 2.1 | APP trafficking and cleavage | APP processing |
| AKAP9 | rs144662445 | 0.0006298 | 2.75 | unknown | unknown |
| rs149979685 | 0.000432 | 3.61 |
Due to the multifactorial and heterogeneous nature of AD, genetic counseling of SAD is empiric and relatively nonspecific. It is often speculated that SAD is the combined action of unknown environmental factors and multiple susceptibility genes. Among them, frequent variations of apolipoprotein E (APOE) are the only well-documented association with SAD. APOE is a component of several lipoproteins consisting of 3 isoforms determined by cysteine-to-arginine substitutions at positions 112 and 158 of the amino acid sequence [27]. Individuals with heterozygous APOE _ε_4 are 4 times more likely to develop AD while homozygous for APOE _ε_4 are 8 times relative to individuals without APOE _ε_2 and APOE ε_3 allele [28]. In CNS, APOE is thought to facilitate clearance of A_β, and the APOE ε_4 allele seems to have the lower ability to clear A_β resulting in a high risk of developing AD. APOE _ε_4 is also identified with smaller gray matter volumes and accelerated brain aging [29, 30].
With the application of Genome-Wide Association Study (GWAS) since 2005, next generation whole exome (WGS), and whole-genome sequencing (WES) to identify common and rare variations, a series of genes or locus are proposed to increase the risk of AD. These include the identified common risk variation of CR1, CLU [31, 32], PICALM [32], BIN1 [33], HLA-DRB5/DRB1 [34], CD2AP, MS4A, ABCA7, CD33 [35, 36], INPP5D, MEF2C, SLC24A4-RIN3, CASS4, NME8, ZCWPW1, PTK2B, CELF1, and SORL1 [34]. Their functions vary from brain development, guiding neural plasticity, cytoskeletal organization, cell apoptosis, and lipid metabolism to A_β_ uptake and microtubule cytoskeleton interaction (Reviewed in [37]). While these frequent variations are responsible for risk of AD, rare variation detected from WES/WGS might have larger effect sizes than the common variations. A rare variant of Triggering Receptor Expressed on Myeloid Cells 2 (TREM2) gene, the rs75932628 (R47H) mutation was confirmed to increase the age of onset in LOAD patients [38]. Further studies on the TREM2_-associated risk of AD indicated that it is the recessive loss-of-function mutations in TREM2 that were responsible for early-onset dementia [39]. Generally speaking, TREM2 is expressed by microglial cells in CNS and was found to be presented with amyloid plaques in the brain of AD mice, suggesting that TREM2 may play a role of A_β clearance. The presence of TREM2 R47H variant was also confirmed in population from French [40], Spanish [41], Catalan [42], and Belgian [43]; however, in a study involving 1133 patients and 1157 subjects from China, the R47H variant was not detected [44]. In our study with 360 AD cases and 400 controls of Chinese population, the rs201280312-T (A130V) variant was detected in two of the AD cases [45], suggesting the genetically heterogeneous nature of TREM2 mutations.
Other rare variations were identified in genes coding for Netrin receptor, UNC5C [46], phospholipase D3 (PLD3) [47], ATP-binding cassette transporter (ABCA1) [48], and disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) [49, 50], although their functional participation in AD occurrence needs further investigation. Elucidating the genetic contribution is a major concern in understanding SAD while there is neither animal models nor proper cell models of SAD to modeling SAD in a dish.
3. iPSCs Reprogramming and the Basis of Alzheimer's Disease Modeling
Both embryonic stem (ES) cells and induced pluripotent stem cells (iPSCs) have the ability to self-renew and differentiate into all three germ layers, thereby endowing us the possibility of reconstructing all types of cells, tissues, and even organs. However the applications for human ES (hES) are limited by several challenging problems such as allogeneic immune rejection, potential tumor formation, and ethical issues concerning the utility of human embryo [51, 52]. Derived from human fibroblast, iPSC was first generated by Yamanaka groups in 2007 [53]. With the similar ability of differentiation with ES cells, but without the concerns of immune rejection problems or ethical issues, iPSC soon gained worldwide attention.
3.1. Introduction of iPSCs Technology
The iPSCs were first generated from mouse fibroblasts through the retroviral-mediated introduction of four transcription factors (OCT4, SOX2, KLF4, and c-MYC) by Takahashi and Yamanaka in 2006 [9]. They found that forced expression of the four extrinsic factors was sufficient to return somatic fibroblasts into a pluripotent state within a few weeks. As soon as a year after this breakthrough, the technique was applied to human fibroblasts [53, 54]. The induced pluripotent stem cells showed similar colony morphology, gene expression, cell surface marker expression, and the ability to self-renew and differentiation as embryonic stem cells (ESC). Since then, more combinations of transcription factors emerged, such as forced expression of OCT4, SOX2, NANOG, and LIN28 mediated via lentiviral vector to reprogram human fibroblasts into an undifferentiated state [55]. One of the concerns for iPSCs methodology is that the insertion of retrovirus vectors into human genome might become a potential threat to troublesome changes such as tumor-genesis. To avoid shortcomings brought by viral vector interaction, nonintegrating viruses have been applied for generation of the iPSCs, including Adenovirus [56] and Sendai Virus [57–60]. However, these methods are either of low efficiency or technological immaturity. Thus, more alternatives for nonintegrating methods were invented such as transfection miRNA for transcription factors [61], episomal plasmids, three oriP/EBNA plasmids (a kind of plasmid vector that may express for a long period of time) harboring either an Oct4, Sox2, Nanog, and Klf4, an Oct4, Sox2, and SV40 large Tantigen, or a c-myc and Lin28 combination. This way, human foreskin fibroblasts were reprogrammed into iPSCs as soon as 20 days after transfection [62].
Efforts were also directed at improving the reprogramming efficiency as the original method of reprogramming by Yamanaka achieved an efficiency of only ~0.02% at ~30 days after retroviral transduction [53] and the mRNA expression method mentioned above was able to raise the efficiency to 1.4% within 20 days [61]. It has been found that by shifting the culture condition to 5% O2 and adding valproic acid into the cell culture medium, the efficiency could be increased to 4.4% [61]. miRNA has been believed to be another promising factor that could increase the reprogramming efficiency with or without Yamanaka factors since some miRNAs are upregulated in both iPSCs and hESCs. For instance, with the presence of four Yamanaka factors, miR-302b and/or miR-372 could increase the efficiency of reprogramming in MRC5 and BJ-1 fibroblasts from 10- to 15-fold compared with the four factors alone [63], while expression of miR302/367 only could transform ~10% of the BJ-1 fibroblasts into iPSCs after 12–14 days after infection [64].
In addition to seeking safer reprogramming factors and improving the reprogramming efficiency, searching for the proper cell sources for reprogramming represents another important strategy. Although skin fibroblasts are considered as the traditional, classical cell source for iPSCs generation, one must undergo a rather invasive procedure for donating samples. The collection of cells from other lower yield sources can be far less invasive, including the collection of mononuclear cells from peripheral blood [65], or hair follicles [66], or even the exfoliated renal epithelial cells from urinary sediments [67]. However, due to the inherent inefficiency of iPSC generation, a large amount of somatic cells is required. Furthermore, the culture potential of the primary cells often varies according to the donors' ages, physical conditions, and long-term drug use. Consequently, it is urgent to decipher the primary underlying cause of the differences between various cell sources and to find an easier method for isolating enough somatic cells in the least invasive manner.
3.2. Application of iPSC in Alzheimer's Disease
In light of the important benefits conferred by their self-renewal and multidirectional differentiation capacity, iPSCs are valued in the context of regenerative medicine and disease modeling, especially for neurodegenerative disorders. Indeed, despite the limitations imposed by the low-efficiency and time-consuming nature of the reprogramming process, iPSCs remain a relevant tool to study the fundamental etiology of neurological diseases and perform high-throughput drug screening for CNS disorders. In fact, several neurological diseases have been modeled using iPSCs, such as monogenic disorders and versions of complex diseases caused by known mutations have been modeled by iPSCs. These disorders include, among others, Parkinson's disease (PD) with SNCA, PINK1, PARK2, GBA1, and LRRK2 mutations [68–70], amyotrophic lateral sclerosis (ALS) with TDP43 mutation [71], Huntington's disease (HD) with HTT mutation [72], and Spinal muscular atrophy (SMA) with SMN1 mutation [73]. Similarly, AD is another slow progressing disease with a poorly understood etiology and a lack of efficient therapeutic strategies. Therefore, it is of the utmost importance for the AD patients' unmet clinical needs that we identify suitable, disease-relevant cell models to solve these problems.
iPSCs can be directionally differentiated into neurons using a specific array of protocols. First, neural stem/progenitor cells are generated from iPSCs with the presence of the neuroectoderm inducer, retinoic acid [74–76]. A similar outcome can also be achieved by inhibiting the bone morphogenetic protein (BMP) and the transforming growth factor-β (TGF_β_) superfamily signal transduction pathways [77, 78], both of which are capable of directing epidermal or mesodermal differentiation. Then, these neural stem cells could be further exposed to certain growth factors to direct differentiation into specific neuronal subtypes. To model AD, induced cholinergic neurons can be generated using a combination of Compound E, 2S-2-N-propanamide (Calbiochem), and Compound W, 3,5-bis(4-nitrophenoxy)benzoic acid, to activate specific intracellular signaling pathways targeting the repressor element 1-silencing transcription factor (REST) and its corepressor (CoREST) [79, 80]. These induced neurons are further selected by specific markers expressed on the endogenous neurons. Upon transplantation into animal models of neurodegenerative diseases, these neurons function well and contribute to the recovery of several neurological deficits.
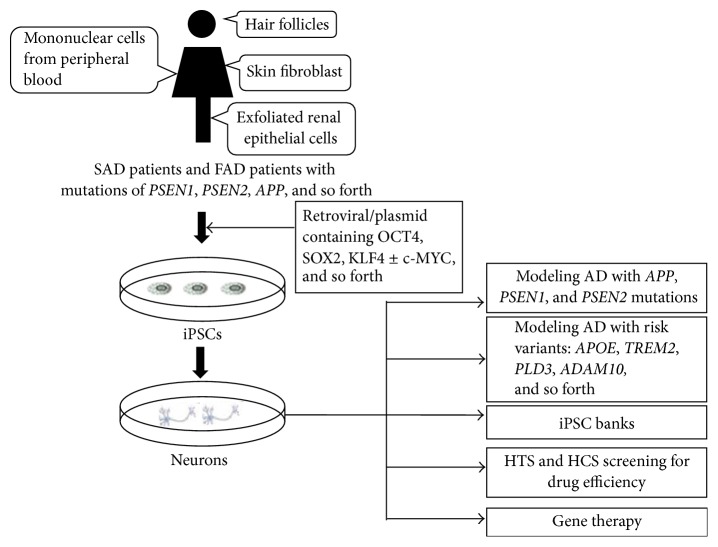
In general, isolated somatic cells undergo a series of reprogramming and neural differentiation procedures to generate a large number of induced AD patient-specific neurons for both research and transplantation purposes (Figure 1). However, there are some hurdles to be overcome before attaining the stage of clinical application. First, even though iPSCs and induced neurons retain the original patients' specific genome, some random DNA alterations and epigenetic changes cannot be avoided during either the reprogramming or the differentiation processes [81]. The potential alterations in DNA splicing or gene expression may induce clonal heterogeneity within the iPSCs and result in cellular functional changes. Second, neuroglia cells participate in the induction of immune responses and A_β_ peptide clearance in AD pathogenesis, and the participation of astrocytes and microglia cells may have an important influence on our understanding and interpretation of the figuring out of the AD-specific cellular phenotypes and drug efficiency. Actually, GWAS analyses have yielded a pattern of common cellular pathways involved in AD patients carrying certain risk variants. In the future, it will not be enough to induce the formation of cholinergic neurons only. Instead, three-dimensional human neural cell culture models will be essential for accurate AD modeling [82]. Third, before reaching the stage of clinical application, it will be essential to determine the optimal and safest iPSC generation and neuron differentiation protocols to use. Indeed, some protocols using integrative viruses and the culture media or feeder cell layers containing animal components constitute potential health threats due to the potential for unwanted immune responses and tumor-genesis.
Figure 1.
The generation of iPSCs and iPSC-derived neurons from various sources of somatic cells and application of iPSC in Alzheimer's disease.
4. Specific Cellular Phenotypes and Processes in the iPSC-Based Models of AD
Modeling AD using iPSCs was initiated from the modeling of familial cases with mutations in disease-causing genes including APP, PSEN1, and PSEN2. Until now, five out of eight publications reported reprogramming of iPSCs-derived cholinergic neurons from patients with FAD (summarized in Table 2), indicating that modeling AD using iPSCs is still in its infancy. However, these studies, which are seeking to find AD-specific unique cell phenotype and AD-related cellular processes, appear as the first step in gaining insights into the genetic contributions to AD.
Table 2.
Human somatic cell reprogramming-based neuronal models of Alzheimer's disease.
| Disease | Genetic defect | Outcome | Drug test | Reference |
|---|---|---|---|---|
| Azheimer's disease | PSEN1 A264E; PSEN2 N141I | Increase secretion of A_β_1–42 in neurons with mutations | _γ_-secretase inhibitors | Yagi et al., 2011 [80] |
| Duplication of APP; Sporadic | Increase secretion of A_β_1–42 and phosphorylated tau (Thr231) in neurons with mutations | _β_-secretase inhibitors | Israel et al., 2012 [84] | |
| APP E693Δ; APPV717L; Sporadic | Increase of intracellular A_β_ olimgo in neurons with APPE693Δ; | DHA | Kondo et al., 2013 [86] | |
| Asymptomatic and symptomatic APP V717I | Increase secretion of A_β_1–42 and A_β_1–38 in neurons with mutations | A_β_ antibody | Muratore et al., 2014 [85] | |
| PSEN1 A246E; PSEN1 M146L | Gene expression differences between neurons with mutations of PSEN1 and controls | no | Sproul et al., 2014 [83] | |
| Sporadic | Changes in gene expression as well as the inducible subunits of the proteasome complex associated with AD in AD-iPS derived neuronal cells | _γ_-secretase inhibitors | Hossini et al., 2015 [87] |
4.1. FAD Disease Modeling with iPSC
iPSCs have been generated from patients with several mutations of PSEN1, PSEN2, and APP by five groups. Two groups analyzed the production of A_β_ peptides and the accumulation of phosphorylated tau protein. Both PSEN1 A246E- and PSEN2 N141I-expressing mutant neurons showed an increased ratio of A_β_42 to A_β_40 compared with control neurons, but the ratio in iPSCs lines was very low, indicating that the secretion of A_β_ peptides varies during differentiation. In addition, no accumulation of tau protein was observed in this type of FAD-derived neurons [80]. However, the PSEN1 A246E mutant was further analyzed during the differentiation and observed an increase in the ratio of A_β_42 to A_β_40 in both fibroblasts, neural progenitor cells (NPCs) and early neurons [83], which is somewhat conflicting with the result of Takuya Yagi groups [80].
Induced neurons that carry a duplication of APP exhibited a higher level of A_β_40 but not A_β_42. In fact, the A_β_42 and A_β_38 levels were completely lower than the detection range of the assay [84]. Through fluorescence-activated cell sorting, researchers were able to isolate a more than 95% pure culture of induced neurons. Purified neurons also exhibited higher levels of phosphorylated tau (Thr 231) and active GSK-3_β_. Compared with control neurons, RAB5-positive early endosomes were enlarged in the neurons from patients with duplication of APP, suggesting that early endosomes may regulate APP processing to result in the increased level of phospho-tau, neurofibrillary tangles, synaptic loss, and apoptosis. Muratore et al. found that iPSCs and neurons harboring the APP (V717I) mutation showed a twofold increase in the production of A_β_42 and a slight increase in A_β_40 [85]. The A_β_38 level and the calculated A_β_38/40 ratio were also significantly increased compared with control neurons [85]. Furthermore, FAD neurons secreted a lower ratio of APPs_α_/APPs_β_. The APPs_β_ production showed a 1.4-fold increase compared with controls [85], suggesting that the V717I mutation may primarily alter the initial epsilon site of cleavage within APP. In another study in patients with APP E693Δ mutation [86], the A_β_ oligomers accumulated in the iPSCs-derived neurons and astrocytes with APP E693Δ. The hall markers of ER stress and oxidative stress, including BiP, cleaved caspase-4, PRDX4-coding antioxidant protein peroxiredoxin-4, and ROS levels, were also increased in the FAD neurons. Therefore, there was a possibility that the intracellular A_β_ oligomers may provoke an antioxidant stress response resulting in increased ROS levels. These results supported the hypothesis that oxidative stress participates in the pathogenesis of AD.
4.2. SAD Disease Modeling with iPSCs
Primary cells from SAD patients have also been used for reprogramming studies and were mostly compared with iPSC originating from cells donated by FAD patients. By researching into the SAD case, Hossini et al. were able to draw an AD-related protein interaction network composed of APP and GSK3_β_ among others [87]. In Israel et al.'s study, relative to nondemented controls, both iPSCs and neurons generated from mutation of the APP gene and SAD patients showed elevated levels of A_β_ peptides, hyperphosphorylation of tau, and GSK3_β_. While neurons from only one of two SAD patients exhibited increased levels of intracellular A_β_ aggregates, similar to the cells derived from the _APP_-E693Δ FAD patients. It is possible that some underlying de novo acquired genes may also participate in the pathogenesis of SAD cases, reflecting the inherent variability of iPSCs. A recent gene expression study in neurons derived from an 82-year-old SAD patient revealed significant gene expression changes between primary cells and induced neurons [87]. The iPSCs technique offers an opportunity to study the underlying molecular events leading to SAD without interference of environmental contributions, allowing the identification of novel AD-associated networks of regulated genes. However, one concern related to the heterogeneous nature of SAD is that the iPSCs-derived neurons from AD patients without inheritance need to expand until cells are produced to enable statistically meaningful analyses.
4.3. Using iPSC-Derived Models to Screen Novel Drugs for AD
Novel treatments targeting the amyloid cascade, APP processing, and ER stress have been tested on the iPSC-derived models of both FAD and SAD. Some of them exhibited significant efficiency on the cell-based models and may become candidate drugs to cure AD patients in the future. _γ_-secretase inhibitors were first screened in iPSCs and neurons carrying PSEN1 A246E and PSEN2 N141I mutations [80]. In the presence of compound E, a potent _γ_-secretase inhibitor, both A_β_42 and A_β_40 decreased sharply in a dose-dependent manner in FAD neurons. Another _γ_-secretase substrate, the Notch intracellular domain, was also inhibited in a dose-dependent manner, suggesting that both PSEN1 and PSEN2 iPSCs-derived neurons respond to the γ_-secretase inhibitor treatment. In Muratore et al.'s study, both FAD neurons with APP V717I mutation and SAD neurons responded to DAPT, another γ_-secretase inhibitor, and the induced neurons exhibited an inhibited production of A_β_38, 40, and 42 when treated with 5 μ_m DAPT for 48 hours [85]. A_β antibody that binds and sequesters the A_β peptides was able to prevent the increase in the total tau levels in APP V717I neurons, suggesting a crosstalk between the amyloid cascade and tau hyperphosphorylation in the AD brains [85]. However, in the clinical trials, although the antibody succeeded at lowering the levels of A_β peptides, it failed at slowing down the progression of the cognitive impairment. Docosahexaenoic acid (DHA) has been reported to improve ER stress or to inhibit ROS generation [88]. In _APP_-E693Δ neurons, DHA could significantly decrease the BiP protein, cleaved caspase-4, and peroxiredoxin-4 levels, as well as reduce the ROS production, ER stress, and oxidative stress markers. As a consequence, the neurons with an APP_-E693Δ mutation survived longer after DHA treatment for 16 days compared with the SAD and control neurons [86]. Considering that the levels of A_β oligomers, which trigger the ER and oxidative stress, remained unchanged, this indicates that DHA treatment may be considered as a symptom relief invention but not as a preventive/curative therapy. Taken together, these results show that iPSCs-derived models allow screening for proper treatment strategies under specific individual genomes.
5. Challenges and Concerns
The potential applications of the reprogramming technology provide a promising approach to generate accurate human cell models of neurodegenerative disorders. To a certain extent, it now becomes possible to aim at recapitulating AD “in a dish.” While iPSCs-derived cell models allowed the identification of factors associated with the disease phenotypes and the screening of various novel potential therapies, for example, the _β_- and _γ_-secretase inhibitors on the basis of the amyloid cascade hypothesis, there are still more discoveries to be made by using iPSC-derived models of AD (Figure 1).
5.1. Investigation of Novel Mutations in APP, PSEN1, and PSEN2
So far the iPSCs generated from fibroblasts with several FAD mutations, including PSEN1 A264E, L166P and M146L, PSEN2 N141I, APP V717I, E693Δ, and duplication of APP are summarized in Table 2. Compared with neurons induced from control fibroblasts, FAD presenilin and APP mutant neurons exhibited not only an increased A_β_42/40 ratios, an elevated level of phosphorylated tau protein, and an increased activated GSK3_β_. Furthermore, they also displayed abnormal endosomes, indicating a novel dysregulated pathway alongside of APP processing and tau phosphorylation. Therefore, more iPSCs with additional FAD mutations should be investigated to confirm the existing conclusions and help identify other underlying mechanisms behind the disparate phenotypes of the patients.
5.2. Investigation of AD Risk Variants Identified by GWAS, WES, and WGS
Whole-genome sequencing and large scale genome-wide association studies aimed at elucidating the factors that result in SAD has brought to light a variety of rare and frequent variants that may predispose or to protect from AD. However, the processing of these data represents a major challenge. For example, screening for these variants in diverse disease populations is already planned, but the related underlying molecular and biochemical mechanisms as well as the impact of the different genetic variants on the neuronal phenotype and AD risk still remain an unsolved puzzle. Actually, several common cellular pathways are associated with variations identified as GWAS, such as inflammation and immune response, endocytosis, and lipid metabolism. The application of the iPSCs technology to study those potential AD risk variants maybe an efficient way to identify the ultimate impact of these genes on the pathology of SAD, to distinguish between the real and the false positive variations and to find out novel pathways associated with the pathogenesis of AD.
5.3. Construction of the iPSCs Bank for AD
Although cord blood bank has been initiated in many countries, given the expensive procedure and the limited availability at the moment of birth only, iPSCs bank would be a promising way alternative to the cord blood bank to restore and recycle cells from patients with different phenotype and mutations because the iPSCs technology could generate a self-renew, stable progenitor population. Public banks of diseased fibroblasts from patients with genetic mutations responsible for certain neurodegenerative disorders already exist. The existence of these banks allows for categorizing sporadic cases and familial cases with different mutations for personalized medicine purposes, investigating the typical phenotype of each individual's unique genetic background, and may ultimately provide a potential treatment means for regenerative medicine.
5.4. Novel Drug Testing: High-Throughput Screening (HTS) and High Content Screening (HCT)
Since iPSCs could retain the patients' genotype and enable us to recapitulate AD in a dish, neurons derived from the disease-specific iPSCs have been used to test several candidate drugs, such as the γ_- and β_-secretase inhibitors and A_β antibody. The five studies based on iPSCs-derived models of AD patients harboring the mutations of PSEN1, PSEN2, and APP point mutation as well as APP duplication, respectively, all report decreasing A_β peptide levels in iPSCs-derived neurons treated with a γ_-secretase inhibitor, an A_β antibody, and DHA. The reduction in phosphorylated tau and GSK-3_β_ levels was observed in the neurons treated with a β_-secretase inhibitor. Likewise, the nonsteroidal anti-inflammatory drug (NSAID) sulindac sulfide has been proposed to become one of the novel strategies for AD therapy by inhibition of the A_β production [89]. The stem cell-derived neurons expressing wild-type PSEN1 treated with NSAID exhibited a decrease in A_β_42 level. However the therapeutic effect was absent in cells harboring the PSEN1 L166P mutation [90]. These studies constitute the primary attempts at screening potential treatment strategies in human genotype- or disease-specific cells. Using iPSCs, HTS, and HCT could allow for rapid analysis for thousands of compounds and disease hallmarks, as well as various cellular contexts affecting drug efficiency, or chemical toxicity, for example. Indeed, by accessing the induced neurons from a cohort of patients and controls in a 96-well format, researchers could rapidly analyze a substantial number of drugs and chemicals for endpoints such as A_β_ peptides levels or phosphorylated tau levels. Using the HTS technique as a prescreening method would streamline the time-consuming preclinical animal studies and potentially reduce the number of failing clinical trials. After target drugs or compounds identification using HTS, HCS would enable the subsequent analysis of relevant cellular signals and pathways. Highly efficient screening based on iPSC-derived neurons could become a routine drug discovery pathway.
5.5. Gene Therapy and iPSCs Transplantation
The potential iPSCs-based regeneration medicine and gene therapy on AD include gene correction and iPSCs induced neurons transplantation. Gene correction has already been conducted in HD by using homologous recombination in the iPSCs stage, leading to normalized pathogenic HD signaling pathways, including cadherin, TGF-β, BDNF, and caspase activation in neural stem cells [72]. On the basis of the cellular endogenous recombination mechanism, zinc-finger nucleases (ZFNs) and tal-effector nucleases (TALENs) are emerging as engineered nucleases usable to modify individual genomes [91, 92]. First, a wild-type nucleotide sequence binds into the FOK1 nuclease fused to arrayed domains triggering a DNA double-strand break. Then, the endogenous recombination machinery emerged induces DNA homologous recombination and nonhomologous end-joining. Finally, the disease-causing mutations are corrected into the wild-type genomic sequence. Genetically corrected iPSCs or induced neurons transplantation in CNS is still at the stage of animal testing. In 1985, researchers successfully transplanted embryonic cholinergic neurons into an AD rat model. The procedure resulted in memory improvement, suggesting that cholinergic cells transplantation may induce functional recovery in the rodent brains [93]. With the generation of the first iPSCs in 2006, the idea of autologous transplantation emerged with the advantage of reducing immunoreactions usually associated with heterologous transplantation. A recent study on the autologous transplantation of iPSC-derived dopamine neurons in a cynomolgus monkey (CM) PD model demonstrated that after iPSC-derived dopamine neurons injection on one side of midbrain, the CM exhibited motor improvement on the transplanted side without a need for immunosuppression. This study indicated a progression that is a step closer to human clinical applications [94]. However, safety is an essential problem concerning insertional mutagenesis and tumorigenesis prior to clinical use. The amount of induced neurons required for functional improvement is another hurdle, an improvement in the iPSCs/neurons generation protocols is indispensable to get equivalent or greater neuron survival.
After all, the iPSCs technology provides a potential therapy for monogenic disorders, as to AD patients with mutations in APP, PSEN1, and PSEN2, shedding light on ultimate therapy of FAD by correcting these mutation. This strategy also holds great promise for complex diseases like SAD. For example, correcting the mutations of risk genes selectively enables a direct comparison between the iPSC lines derived from WT and mutant cells under the same genome circumstance, significantly reducing the intrinsic genome variability existing in different patients.
Competing Interests
The authors declare that there are no competing interests regarding the publication of this paper.
Authors' Contributions
Weiwei Zhang, Miaojin Zhou, and Tao Zhou conceived and wrote the paper, the table, and the imaging; Bin Jiao and Lu Shen critically edited the paper.
References
- 1.Berchtold N. C., Cotman C. W. Evolution in the conceptualization of dementia and Alzheimer's disease: Greco-Roman period to the 1960s. Neurobiology of Aging. 1998;19(3):173–189. doi: 10.1016/s0197-4580(98)00052-9. [DOI] [PubMed] [Google Scholar]
- 2.Barnes D. E., Yaffe K. The projected effect of risk factor reduction on Alzheimer's disease prevalence. The Lancet Neurology. 2011;10(9):819–828. doi: 10.1016/s1474-4422(11)70072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan H. I., Jacobsen J. S., Rutkowski J. L., Feuerstein G. Z. Translational medicine lessons from flurizan's failure in Alzheimer's disease (AD) trial: implication for future drug discovery and development for AD. Clinical and Translational Science. 2009;2(3):242–247. doi: 10.1111/j.1752-8062.2009.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith A. D. Why are drug trials in Alzheimer's disease failing? The Lancet. 2010;376(9751):p. 1466. doi: 10.1016/s0140-6736(10)61994-0. [DOI] [PubMed] [Google Scholar]
- 5.Navarrete L. P., Pérez P., Morales I., Maccioni R. B. Novel drugs affecting tau behavior in the treatment of Alzheimer's disease and tauopathies. Current Alzheimer Research. 2011;8(6):678–685. doi: 10.2174/156720511796717122. [DOI] [PubMed] [Google Scholar]
- 6.Hardy J., Gwinn-Hardy K. Genetic classification of primary neurodegenerative disease. Science. 1998;282(5391):1075–1079. doi: 10.1126/science.282.5391.1075. [DOI] [PubMed] [Google Scholar]
- 7.Campion D., Dumanchin C., Hannequin D., et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. The American Journal of Human Genetics. 1999;65(3):664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borenstein A. R., Copenhaver C. I., Mortimer J. A. Early-life risk factors for Alzheimer disease. Alzheimer Disease and Associated Disorders. 2006;20(1):63–72. doi: 10.1097/01.wad.0000201854.62116.d7. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Guerreiro R. J., Gustafson D. R., Hardy J. The genetic architecture of Alzheimer's disease: beyond APP, PSENS and APOE. Neurobiology of Aging. 2012;33(3):437–456. doi: 10.1016/j.neurobiolaging.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Götz J., Lim Y.-A., Ke Y. D., Eckert A., Ittner L. M. Dissecting toxicity of tau and β-amyloid. Neurodegenerative Diseases. 2010;7(1–3):10–12. doi: 10.1159/000283475. [DOI] [PubMed] [Google Scholar]
- 12.Borchelt D. R., Thinakaran G., Eckman C. B., et al. Familial Alzheimer's disease-linked presenilin I variants elevate aβ1–42/1–40 ratio in vitro and in vivo. Neuron. 1996;17(5):1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 13.Hardy J., Selkoe D. J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 14.Tanzi R. E., Moir R. D., Wagner S. L. Clearance of Alzheimer's Aβ peptide: the many roads to perdition. Neuron. 2004;43(5):605–608. doi: 10.1016/j.neuron.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 15.Wischik C. M., Novak M., Edwards P. C., Klug A., Tichelaar W., Crowther R. A. Structural characterization of the core of the paired helical filament of Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(13):4884–4888. doi: 10.1073/pnas.85.13.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goedert M., Spillantini M. G., Cairns N. J., Crowther R. A. Tau proteins of alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron. 1992;8(1):159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 17.Magdesian M. H., Carvalho M. M. V. F., Mendes F. A., et al. Amyloid-β binds to the extracellular cysteine-rich domain of frizzled and inhibits Wnt/β-catenin signaling. The Journal of Biological Chemistry. 2008;283(14):9359–9368. doi: 10.1074/jbc.m707108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Townsend M., Mehta T., Selkoe D. J. Soluble Aβ inhibits specific signal transduction cascades common to the insulin receptor pathway. The Journal of Biological Chemistry. 2007;282(46):33305–33312. doi: 10.1074/jbc.m610390200. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez G., Muñoz-Montaño J. R., Satrústegui J., Avila J., Bogónez E., Díaz-Nido J. Lithium protects cultured neurons against β-amyloid-induced neurodegeneration. FEBS Letters. 1999;453(3):260–264. doi: 10.1016/s0014-5793(99)00685-7. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez F., Lucas J. J., Avila J. GSK3 and tau: two convergence points in Alzheimer's disease. Journal of Alzheimer's Disease. 2013;33(supplement 1):S141–S144. doi: 10.3233/jad-2012-129025. [DOI] [PubMed] [Google Scholar]
- 21.Basurto-Islas G., Luna-Muñoz J., Guillozet-Bongaarts A. L., Binder L. I., Mena R., García-Sierra F. Accumulation of aspartic acid421- and glutamic acid 391-cleaved tau in neurofibrillary tangles correlates with progression in Alzheimer disease. Journal of Neuropathology and Experimental Neurology. 2008;67(5):470–483. doi: 10.1097/NEN.0b013e31817275c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamblin T. C., Chen F., Zambrano A., et al. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S.-Y., Ferreira A. The generation of a 17 kDa neurotoxic fragment: an alternative mechanism by which tau mediates β-amyloid-induced neurodegeneration. The Journal of Neuroscience. 2005;25(22):5365–5375. doi: 10.1523/jneurosci.1125-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai T., Guo J.-P., McGeer P. L. Proteolysis of non-phosphorylated and phosphorylated tau by thrombin. The Journal of Biological Chemistry. 2005;280(7):5145–5153. doi: 10.1074/jbc.m409234200. [DOI] [PubMed] [Google Scholar]
- 25.Yang A. J., Chandswangbhuvana D., Margol L., Glabe C. G. Loss of endosomal/lysosomal membrane impermeability is an early event in amyloid Aβ1-42 pathogenesis. Journal of Neuroscience Research. 1998;52(6):691–698. doi: 10.1002/(sici)1097-4547(19980615)52:6<691::aid-jnr8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Kenessey A., Nacharaju P., Ko L.-W., Yen S.-H. Degradation of tau by lysosomal enzyme cathepsin D: implication for Alzheimer neurofibrillary degeneration. Journal of Neurochemistry. 1997;69(5):2026–2038. doi: 10.1046/j.1471-4159.1997.69052026.x. [DOI] [PubMed] [Google Scholar]
- 27.Zlokovic B. V. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurology. 2013;70(4):440–444. doi: 10.1001/jamaneurol.2013.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirier J., Bertrand P., Poirier J., et al. Apolipoprotein E polymorphism and Alzheimer's disease. The Lancet. 1993;342(8873):697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 29.Davies G., Harris S. E., Reynolds C. A., et al. A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Molecular Psychiatry. 2014;19(1):76–87. doi: 10.1038/mp.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean D. C., III, Jerskey B. A., Chen K., et al. Brain differences in infants at differential genetic risk for late-onset alzheimer disease: a cross-sectional imaging study. JAMA Neurology. 2014;71(1):11–22. doi: 10.1001/jamaneurol.2013.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert J.-C., Heath S., Even G., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nature Genetics. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 32.Harold D., Abraham R., Hollingworth P., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature Genetics. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seshadri S., Fitzpatrick A. L., Ikram M. A., et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. The Journal of the American Medical Association. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert J. C., Ibrahim-Verbaas C. A., Harold D., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature Genetics. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naj A. C., Jun G., Beecham G. W., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nature Genetics. 2011;43(5):436–443. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollingworth P., Harold D., Sims R., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nature Genetics. 2011;43(5):429–436. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chouraki V., Seshadri S. Genetics of Alzheimer's disease. Advances in Genetics. 2014;87:245–294. doi: 10.1016/b978-0-12-800149-3.00005-6. [DOI] [PubMed] [Google Scholar]
- 38.Jonsson T., Stefansson H., Steinberg S., et al. Variant of TREM2 associated with the risk of Alzheimer's disease. The New England Journal of Medicine. 2013;368(2):107–116. doi: 10.1056/nejmoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerreiro R., Wojtas A., Bras J., et al. TREM2 variants in Alzheimer's disease. The New England Journal of Medicine. 2013;368(2):117–127. doi: 10.1056/nejmoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pottier C., Wallon D., Rousseau S., et al. TREM2 R47H variant as a risk factor for early-onset alzheimer's disease. Journal of Alzheimer's Disease. 2013;35(1):45–49. doi: 10.3233/JAD-122311. [DOI] [PubMed] [Google Scholar]
- 41.Benitez B. A., Cooper B., Pastor P., et al. TREM2 is associated with the risk of Alzheimer's disease in Spanish population. Neurobiology of Aging. 2013;34(6):1711.e15–1711.e17. doi: 10.1016/j.neurobiolaging.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz A., Dols-Icardo O., Bullido M. J., et al. Assessing the role of the TREM2 p.R47H variant as a risk factor for Alzheimer's disease and frontotemporal dementia. Neurobiology of Aging. 2014;35(2):444.e1–444.e4. doi: 10.1016/j.neurobiolaging.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Cuyvers E., Bettens K., Philtjens S., et al. Investigating the role of rare heterozygous TREM2 variants in Alzheimer's disease and frontotemporal dementia. Neurobiology of Aging. 2014;35(3):726.e11–726.e19. doi: 10.1016/j.neurobiolaging. [DOI] [PubMed] [Google Scholar]
- 44.Yu J.-T., Jiang T., Wang Y.-L., et al. Triggering receptor expressed on myeloid cells 2 variant is rare in late-onset Alzheimer's disease in Han Chinese individuals. Neurobiology of Aging. 2014;35(4):937.e1–937.e3. doi: 10.1016/j.neurobiolaging.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 45.Jiao B., Liu X., Tang B., et al. Investigation of TREM2, PLD3, and UNC5C variants in patients with Alzheimer's disease from mainland China. Neurobiology of Aging. 2014;35(10):2422.e9–2422.e11. doi: 10.1016/j.neurobiolaging.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 46.Wetzel-Smith M. K., Hunkapiller J., Bhangale T. R., et al. A rare mutation in UNC5C predisposes to late-onset Alzheimer's disease and increases neuronal cell death. Nature Medicine. 2014;20(12):1452–1457. doi: 10.1038/nm.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cruchaga C., Karch C. M., Jin S. C., et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2014;505(7484):550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupton M. K., Proitsi P., Lin K., et al. The role of ABCA1 gene sequence variants on risk of Alzheimer's disease. Journal of Alzheimer's Disease. 2014;38(4):897–906. doi: 10.3233/JAD-131121. [DOI] [PubMed] [Google Scholar]
- 49.Cai G., Atzmon G., Naj A. C., et al. Evidence against a role for rare ADAM10 mutations in sporadic Alzheimer disease. Neurobiology of Aging. 2012;33(2):416–417.e3. doi: 10.1016/j.neurobiolaging.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim M., Suh J., Romano D., et al. Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate α-secretase activity. Human Molecular Genetics. 2009;18(20):3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 52.Tachibana M., Amato P., Sparman M., et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153(6):1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi K., Tanabe K., Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nature Protocols. 2007;2(12):3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 55.Yu J., Vodyanik M. A., Smuga-Otto K., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 56.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy Series B: Physical and Biological Sciences. 2009;85(8):348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seki T., Yuasa S., Oda M., et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7(1):11–13. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Zhou H., Wu S., Joo J. Y., et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D., Kim C.-H., Moon J.-I., et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warren L., Manos P. D., Ahfeldt T., et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu K., Yu J., Suknuntha K., et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117(14):e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanyam D., Lamouille S., Judson R. L., et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnology. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anokye-Danso F., Trivedi C. M., Juhr D., et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seki T., Yuasa S., Fukuda K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nature Protocols. 2012;7(4):718–728. doi: 10.1038/nprot.2012.015. [DOI] [PubMed] [Google Scholar]
- 66.Aasen T., Belmonte J. C. I. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nature Protocols. 2010;5(2):371–382. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- 67.Zhou T., Benda C., Dunzinger S., et al. Generation of human induced pluripotent stem cells from urine samples. Nature Protocols. 2012;7(12):2080–2089. doi: 10.1038/nprot2012115. [DOI] [PubMed] [Google Scholar]
- 68.Byers B., Cord B., Nguyen H. N., et al. SNCA triplication parkinson's patient's iPSC-Derived DA neurons accumulate α-synuclein and are susceptible to oxidative stress. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0026159.e26159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soldner F., Hockemeyer D., Beard C., et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136(5):964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seibler P., Graziotto J., Jeong H., Simunovic F., Klein C., Krainc D. Mitochondrial parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. The Journal of Neuroscience. 2011;31(16):5970–5976. doi: 10.1523/jneurosci.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimos J. T., Rodolfa K. T., Niakan K. K., et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 72.An M. C., Zhang N., Scott G., et al. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11(2):253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebert A. D., Yu J., Rose F. F., Jr., et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaeger I., Arber C., Risner-Janiczek J. R., et al. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development. 2011;138(20):4363–4374. doi: 10.1242/dev.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erceg S., Ronaghi M., Zipancic I., et al. Efficient differentiation of human embryonic stem cells into functional cerebellar-like cells. Stem Cells and Development. 2010;19(11):1745–1756. doi: 10.1089/scd.2009.0498. [DOI] [PubMed] [Google Scholar]
- 76.Amoroso M. W., Croft G. F., Williams D. J., et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. Journal of Neuroscience. 2013;33(2):574–586. doi: 10.1523/jneurosci.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou J., Su P., Li D., Tsang S., Duan E., Wang F. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28(10):1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maroof A. M., Keros S., Tyson J. A., et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12(5):559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ooi L., Wood I. C. Chromatin crosstalk in development and disease: lessons from REST. Nature Reviews Genetics. 2007;8(7):544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 80.Yagi T., Ito D., Okada Y., et al. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Human Molecular Genetics. 2011;20(23):4530–4539. doi: 10.1093/hmg/ddr394.ddr394 [DOI] [PubMed] [Google Scholar]
- 81.Check Hayden E. Stem cells: the growing pains of pluripotency. Nature. 2011;473(7347):272–274. doi: 10.1038/473272a. [DOI] [PubMed] [Google Scholar]
- 82.Choi S. H., Kim Y. H., Hebisch M., et al. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515(7526):274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sproul A. A., Jacob S., Pre D., et al. Characterization and molecular profiling of PSEN1 familial alzheimer's disease iPSC-Derived neural progenitors. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0084547.e84547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Israel M. A., Yuan S. H., Bardy C., et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muratore C. R., Rice H. C., Srikanth P., et al. The familial Alzheimer's disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Human Molecular Genetics. 2014;23(13):3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kondo T., Asai M., Tsukita K., et al. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12(4):487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 87.Hossini A. M., Megges M., Prigione A., et al. Induced pluripotent stem cell-derived neuronal cells from a sporadic Alzheimer's disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genomics. 2015;16(1, article 84) doi: 10.1186/s12864-015-1262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Begum G., Kintner D., Liu Y., Cramer S. W., Sun D. DHA inhibits ER Ca2+ release and ER stress in astrocytes following in vitro ischemia. Journal of Neurochemistry. 2012;120(4):622–630. doi: 10.1111/j.1471-4159.2011.07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yahata N., Asai M., Kitaoka S., et al. Anti-Aβ drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer's disease. PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0025788.e25788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koch P., Tamboli I. Y., Mertens J., et al. Presenilin-1 L166P mutant human pluripotent stem cell-derived neurons exhibit partial loss of γ-secretase activity in endogenous amyloid-β generation. The American Journal of Pathology. 2012;180(6):2404–2416. doi: 10.1016/j.ajpath.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 91.Cathomen T., Keith Joung J. Zinc-finger nucleases: the next generation emerges. Molecular Therapy. 2008;16(7):1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 92.Mussolino C., Cathomen T. TALE nucleases: tailored genome engineering made easy. Current Opinion in Biotechnology. 2012;23(5):644–650. doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 93.Fine A., Dunnett S. B., Bjorklund A., Iversen S. D. Cholinergic ventral forebrain grafts into the neocortex improve passive avoidance memory in a rat model of Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(15):5227–5230. doi: 10.1073/pnas.82.15.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hallett P. J., Deleidi M., Astradsson A., et al. Successful function of autologous iPSC-derived dopamine neurons following transplantation in a non-human primate model of Parkinson's disease. Cell Stem Cell. 2015;16(3):269–274. doi: 10.1016/j.stem.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]