NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment (original) (raw)
Abstract
Osmotic stress responses are critical not only to the survival of unicellular organisms but also to the normal function of the mammalian kidney. However, the extent to which cells outside the kidney rely on osmotic stress responses in vivo remains unknown. Nuclear factor of activated T cells 5 (NFAT5)/tonicity enhancer binding protein (TonEBP), the only known osmosensitive mammalian transcription factor, is expressed most abundantly in the thymus and is induced upon lymphocyte activation. Here we report that NFAT5/TonEBP is not only essential for normal cell proliferation under hyperosmotic conditions but also necessary for optimal adaptive immunity. Targeted deletion of exons 6 and 7 of the Nfat5 gene, which encode a critical region of the DNA-binding domain, gave rise to a complete loss of function in the homozygous state and a partial loss of function in the heterozygous state. Complete loss of function resulted in late gestational lethality. Furthermore, hypertonicity-induced NFAT5/TonEBP transcriptional activity and hsp70.1 promoter function were completely eliminated, and cell proliferation under hyperosmotic culture conditions was markedly impaired. Partial loss of NFAT5/TonEBP function resulted in lymphoid hypocellularity and impaired antigen-specific antibody responses in viable heterozygous animals. In addition, lymphocyte proliferation ex vivo was reduced under hypertonic, but not isotonic, culture conditions. Direct measurement of tissue osmolality further revealed lymphoid tissues to be hyperosmolar. These results indicate that lymphocyte-mediated immunity is contingent on adaptation to physiologic osmotic stress, thus providing insight into the lymphoid microenvironment and the importance of the NFAT5/TonEBP osmotic stress response pathway in vivo.
Unicellular organisms use well defined osmotic stress response pathways to compensate for the osmotic loss of intracellular water that occurs upon exposure to a hypertonic environment (1, 2). These pathways couple upstream osmotic sensor proteins with transcription factors that regulate the expression of genes that function to increase the concentration of nonperturbing or compatible intracellular organic osmolytes (3, 4), thereby allowing for normalization of the concentration of water within the cell through osmosis. In the yeast Saccharomyces cerevisiae, the high-osmolarity glycerol pathway links upstream and putative osmotic sensor proteins Sln1p, Sho1p, and Msb2p to the evolutionarily conserved mitogen-activated protein kinase Hog1p. Hog1p in turn activates transcriptional responses mediated by Sko1p, Hot1p, and Msn2p/Msn4p, resulting in the induction of genes involved in glycerol synthesis and thus an increase in the concentration of compatible osmolytes within the cell (2).
The mammalian adaptive osmotic stress response utilizes nuclear factor of activated T cells 5 (NFAT5)/tonicity enhancer binding protein (TonEBP), hereafter referred to as NFAT5 (refs. 5–8), a transcription factor that contains the rel DNA-binding domain (DBD) also found in rel/Dorsal/NF-κB and NFATc proteins (9), to control the transcription of genes that similarly function to increase the concentration of intracellular osmolytes (10). These include the genes encoding aldose reductase, the sodium/myoinositol cotransporter, and the sodium/chloride/betaine cotransporter, which increase the intracellular concentration of sorbitol, inositol, and betaine, respectively (11). In addition, NFAT5 plays a role in the hypertonicity-dependent induction of the molecular chaperone HSP70 encoded by the hsp70.1 gene (12). However, in contrast to unicellular organisms that are directly exposed to the external environment, mammalian cells are not thought to be normally subjected to extremes of hypertonic stress because of highly sensitive and dynamic regulatory mechanisms that maintain body fluid homeostasis (11, 13). The renal medulla represents a clear exception because of the urine-concentrating mechanisms of the kidney, which can generate an extremely hypertonic environment (11). Thus, as expected, complete loss of NFAT5 function results in marked atrophy of the renal medulla (14). However, NFAT5 protein also is expressed in the thymus (7, 15), brain (16, 17), and liver (16), and is induced upon activation of quiescent T lymphocytes (7) and expressed in various tissues of the developing embryo (18). These observations give rise to the hypothesis that the NFAT5 osmotic stress response pathway plays a critical role in enabling mammalian cells to adapt to osmotic stress that occurs physiologically in tissues other than the kidney.
This study tests this hypothesis through the introduction of a mutation into the mouse genome that results in either complete or partial loss of the function of NFAT5 as a DNA-binding transcription factor. Whereas complete loss of function resulted in late gestational lethality and severe impairment of cell proliferation under hyperosmotic culture conditions, animals with partial loss of function were viable and exhibited defects in adaptive immunity. Furthermore, direct measurements revealed lymphoid tissues to be hyperosmolar relative to serum. These results not only demonstrate that NFAT5 represents a critical component of the mammalian osmotic stress response but also provide important insight into the lymphoid tissue microenvironment that illustrates the broader biologic significance of osmotic stress and the NFAT5 osmotic stress response pathway in vivo.
Materials and Methods
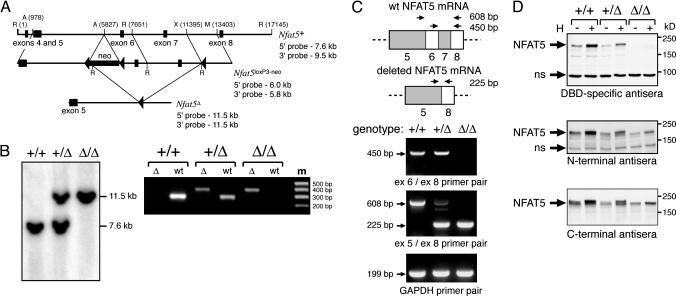
Targeted Disruption of the Nfat5 Gene. The Nfat5 gene was targeted for homologous recombination in R1 (S129/SvJ) embryonic stem (ES) cells by using a gene-targeting vector containing three loxP recombination sites and a neomycin-selectable marker within a 12.4-kb genomic region encompassing exons 5–7 (Fig. 1_A_). Deletion of exons 6 and 7 was achieved by transfecting correctly targeted _Nfat5_loxP3-neo ES cells with a cre recombinase expression vector (provided by J. Marth, University of California at San Diego) and screening for cre-mediated recombination by Southern blotting with both 5′ and 3′ external probes. Chimeric mice derived from blastocyst injections of gene-targeted Nfat5+/Δ ES cells were crossed to C57BL/6, and germline transmission was assessed by coat color (see Supporting Experimental Procedures, which is published as supporting information on the PNAS web site).
Fig. 1.
The targeted deletion of exons 6 and 7 of the Nfat5 gene results in expression of a mutant NFAT5 protein. (A) Gene-targeting strategy for deletion of exons 6 and 7 of the Nfat5 gene by homologous recombination in ES cells (see Materials and Methods). A, _Avr_II; R, _Eco_RI; M, _Msc_I; X, _Xho_I. (B) Targeted deletion within the Nfat5 gene was verified by PCR genotyping with two sets of primers that span the site of insertion of the downstream loxP site in both the wild-type and knockout alleles (Left), and by Southern blot analysis with a 5′ probe located outside of the targeted genomic sequence (Right). (C) RNA isolated from SV40 T antigen-immortalized MEF cell lines was subjected to RT-PCR analysis with primers positioned as indicated relative to that portion of the NFAT5 mRNA encoded by exons 5–8. (D) Whole-cell extracts from MEF cell lines cultured under either standard or hypertonic (H; complete medium plus additional 120 mM NaCl) culture conditions for 16 h were subjected to immunoblot analysis with the indicated antisera. The indicated nonspecific (ns) bands function as internal controls for equal protein loading and transfer.
Generation of MEF Cell Lines. Primary mouse embryonic fibroblasts (MEFs) were obtained from embryonic day 13.5 embryos by using standard methods. Immortalized MEF cell lines were generated by using transfecting primary MEFs using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals) with an SV40 T antigen expression plasmid that confers resistance to neomycin (provided by S. Dowdy, University of California at San Diego) and culturing the cells in medium containing 1 mg/ml G418. The cell lines were genotyped by using PCR and Southern blotting.
Osmolality Measurements. Tissue osmolality was measured by using a vapor pressure osmometer (Model 5520, Wescor, Logan, UT) as described in refs. 19 and 20. Tissue obtained from anesthetized, 6- to 8-week-old C57BL/6 mice was placed in a screw-cap microfuge tube and snap frozen in a dry-ice/methanol bath. No more than two tissues were obtained per animal. Blood samples were obtained by retroorbital venipuncture. Osmolality measurements were made from filter discs absorbed with tissue fluid from frozen tissue that had been fragmented. Proper calibration of the instrument was verified by measurement of standards immediately prior and subsequent to a tissue osmolality measurement. In control experiments, as reported in refs. 19 and 20, measurement of either tissue fluid absorbed onto a filter disk or tissue slices resulted in essentially identical results, and no differences were seen between fresh and frozen tissue. Osmolality measurements of whole blood were identical to those of serum. Culture medium osmolality also was measured by using the previously described method.
Cell Stimulation and Immunoblotting. Splenocyte cell suspensions were cultured as described in ref. 15. The cells were stimulated at 1 × 106 per ml with 1 μg/ml anti-T-cell-receptor antibody (CD3ε chain; clone 145-2C11), 1 μg/ml anti-CD28 antibody (clone 37.51; BD Biosciences, San Diego), or 25 μg/ml (bacterial) lipopolysaccharide (LPS) (Calbiochem). The cells were pulsed by using [3H]thymidine [0.5 μCi per well (1 Ci = 37 GBq); Amersham Biosciences] during the final 12 h of a 72-h culture. Whole-cell extracts were subjected to SDS/PAGE, transferred to poly(vinylidene difluoride) membranes, and probed with rabbit polyclonal antisera directed against the DBD (7), the N terminus (5), or the C terminus (provided by H. M. Kwon, University of Maryland, Baltimore) of the NFAT5 protein.
In Vivo Immune Response. Nfat5+/Δ mice and Nfat5+/+ littermate controls (5–7 weeks old) were immunized s.c. with 50 μg of ovalbumin (Sigma) plus 5 μg of LPS emulsified in incomplete Freund's adjuvant (Sigma). Serum samples were obtained prior to and 3 weeks after immunization. Antigen-specific immunoglobulin (Ig) was measured by ELISA with an alkaline phosphatase-conjugated goat antimouse Igκ secondary antibody (BD Biosciences) detected in a fluorescence-based assay. Total serum IgM and IgG were measured by ELISA.
Reporter Gene Analysis. Reporter studies were performed by using FuGENE 6 transfection of MEF cell lines with an NFAT5-responsive luciferase reporter gene containing two tandem hTonE sites within a minimal promoter derived from the human IL2 gene (7) or with a luciferase reporter gene in which transcription is directed by a 4.1-kb fragment of the mouse hsp70.1 gene promoter (12) (also referred to as HSP70-2). A constitutive alkaline phosphatase reporter was cotransfected to normalize for transfection efficiency.
Results
Complete and Partial Loss of NFAT5 Function by Deletion of Exons Encoding the N-Terminal Portion of the DBD. To define the biologic function of the NFAT5 transcription factor in vivo, exons 6 and 7 of the murine Nfat5 gene were deleted through insertion of loxP recombination sites by homologous recombination followed by cre recombinase-dependent excision in ES cells (Fig. 1 A). These exons encode amino acid residues 254–380, which comprise the N-terminal portion of the DBD. This region mediates critical base-specific contacts with DNA and also forms one of the two interfaces for dimerization within the DBD (21). The targeted deletion thus eliminates a region of the NFAT5 protein that is essential for its function as a site-specific DNA-binding transcription factor (Fig. 6, which is published as supporting information on the PNAS web site). Germline transmission of the allele bearing the exon 6 and 7 deletion (hereafter referred to as _Nfat5_Δ) was verified by using PCR and Southern genotyping (Fig. 1_B_). Analysis of NFAT5 RNA and protein expression in Nfat5+/+, Nfat5+/Δ, and _Nfat5_Δ/Δ MEF cell lines demonstrated that the _Nfat5_Δ allele encodes a mutant protein containing an internal deletion of amino acid residues encoded by exons 6 and 7 (NFAT5Δ254–380), as expected based on the contiguous reading frame maintained between exons 5 and 8 (Fig. 1 C and D). NFAT5 immunoreactivity in _Nfat5_Δ/Δ cells was essentially completely eliminated by using antisera directed against the DBD, and both constitutive and hypertonicity-induced expression of wild-type NFAT5 expression were significantly reduced in Nfat5+/Δ cells (Fig. 1_D_ Top). Moreover, antisera directed against either the N or C terminus demonstrated that upon hypertonic stimulation, the mutant protein, although induced in level of expression, did not undergo phosphorylation-dependent posttranslational modification (22, 23), given the absence of any reduction in mobility in SDS/PAGE analysis (Fig. 1_D_ Bottom). Based on a deletion analysis of NFAT5 dimerization and DNA-binding function (23), the NFAT5Δ254–380 protein expressed in Nfat5+/Δ cells likely functions to dominantly inhibit NFAT5 function by forming dimers with wild-type protein that are incapable of binding DNA in a sequence-specific manner.
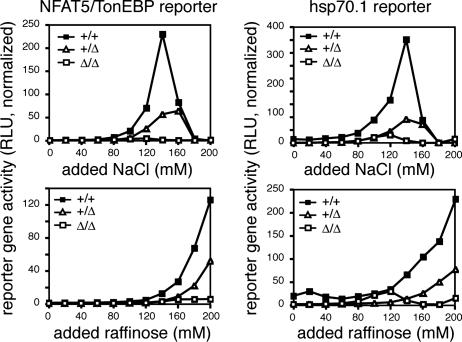
To verify that the _Nfat5_Δ allele conferred a loss of NFAT5 function in not only the homozygous but also the heterozygous states, the MEF cell lines were transfected with an NFAT5 reporter gene and subjected to culture under either isotonic or hypertonic conditions (Fig. 2). Whereas Nfat5+/+ cells exhibited marked induction of NFAT5-dependent reporter gene expression upon culture in either NaCl or raffinose, _Nfat5_Δ/Δcells showed no induction and Nfat5+/Δ cells exhibited significant but incomplete loss of NFAT5-dependent reporter activity. Loss of NFAT5 function was further demonstrated by measurement of transcription mediated by the hsp70.1 promoter, a previously defined NFAT5 target gene (12). Remarkably, hypertonicity-induced reporter gene expression mediated by the hsp70.1 promoter was completely eliminated in _Nfat5_Δ/Δ cells and markedly reduced in Nfat5+/Δ cells (Fig. 2). These results demonstrate not only complete loss of NFAT5 function resulting from homozygous deletion of exons 6 and 7, consistent with results from studies of NFAT5-null mice (14), but also partial loss of function in the heterozygous state, consistent with a dominant inhibitory function of the NFAT5Δ254–380 protein. In addition, these results indicate that NFAT5 is both necessary and sufficient for hypertonicity-dependent induction of the hsp70.1 promoter.
Fig. 2.
The _Nfat5_Δ allele confers either partial or complete loss of NFAT5 function. Immortalized MEF cell lines derived from Nfat5+/+, Nfat5+/Δ, and _Nfat5_Δ/Δ embryos were transfected with the indicated reporter gene. Approximately 24 h after transfection, the cells were cultured in complete medium with either NaCl or raffinose added to the indicated concentration. Cell extracts were prepared 16 h later for assay of reporter activity. The results represent luciferase reporter activity (relative light units, RLU) normalized to correct for variation in transfection efficiency and are representative of three independent experiments.
NFAT5 Is Essential for Viability. The genotype of litters obtained from matings of heterozygous Nfat5+/Δ mice demonstrated that complete loss of NFAT5 function due to homozygous deletion of exons 6 and 7 results in late gestational or perinatal lethality. No homozygous mice were present among 154 viable offspring from heterozygous mating. Moreover, the ratio of viable heterozygous to wild-type animals was 1.80, indicating that the _Nfat5_Δ allele in the heterozygous state does not significantly affect viability. Heterozygous matings of 129/SvJ × C57BL/6 hybrid mice that had been backcrossed less than two generations to C57BL/6 resulted in homozygous fetuses occurring at decreasing frequency from embryonic day 14.5 onward (data not shown), consistent with the similar timing of lethality observed in NFAT5-null animals (14). However, in matings of heterozygous animals backcrossed more than three generations to the C57BL/6 background, the homozygous genotype was represented at a frequency of 0.39 (n = 28) at embryonic days 18.5–20.5, indicating that death most likely occurred in the immediate perinatal period. These results indicate that the genetic background modifies the timing of death in homozygous _Nfat5_Δ/Δ animals.
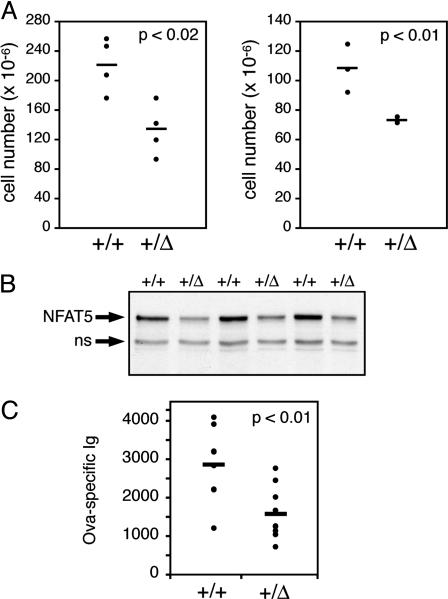
Impaired Immune Function in Nfat5+/Δ Mutant Mice. The lethal phenotype associated with complete loss of NFAT5 function limits analysis of the role of NFAT5 in regulating immune function. However, given that expression of the Nfat5_Δ allele confers partial loss of NFAT5-dependent transcriptional activity in the heterozygous state (Fig. 2), studies of immune function in heterozygous Nfat5+/Δ animals were pursued. Remarkably, the cellularity of the thymus and spleen from Nfat5+/_Δ animals was reduced by 40% and 32% relative to wild-type littermate controls (Fig. 3_A_). This phenotype is very similar to that of transgenic animals in which expression of a dominant negative form of NFAT5 was targeted to T lymphocytes by using the CD2 promoter (15), although the hypocellularity is greater in the Nfat5+/Δ mice. The observed reduction in cell number in thymuses from Nfat5+/Δ animals correlated with reduced expression of the NFAT5/TonEB protein (Fig. 3_B_). Composition of thymocyte subsets defined by the CD4 and CD8 markers of thymic development showed no significant differences (data not shown), again consistent with results obtained from the dominant negative NFAT5 transgenic mice (15). There also were no differences seen between the percentages of mature T and B cells within the spleens of wild-type and heterozygous animals (data not shown), indicating that the reduction in cell number was due to equivalent reductions in the absolute number of both T and B cells.
Fig. 3.
Partial loss of NFAT5 function results in impaired adaptive immunity. (A) The cellularity of the thymus (Left) and spleen (Right) from 5-week-old male Nfat5+/+ and Nfat5+/Δ littermates was determined by performing manual cell counts. The viable cells were distinguished by using trypan blue dye exclusion. (B) Whole-cell extracts of thymocytes from Nfat5+/Δ mice and Nfat5+/+ littermate controls were prepared and probed for NFAT5 expression by Western blot analysis with the C-terminal NFAT5 antisera. The indicated nonspecific (ns) band provides an internal control for equal protein loading and transfer. (C) Relative antigen-specific Ig levels were measured by ELISA of serum from Nfat5+/Δ and Nfat5+/+ mice collected 21 days after immunization with ovalbumin.
To determine whether partial loss of NFAT5 function resulted in impaired lymphocyte function in vivo, a T cell-dependent B cell immune response was induced by immunization with the protein antigen ovalbumin. Heterozygous Nfat5+/Δ animals were significantly impaired in their ability to mount an antigen-specific antibody response to this nominal protein antigen, exhibiting a 44% reduction in antigen-specific antibody, compared with wild-type controls (Fig. 3_C_). Similarly reduced antibody responses were observed upon secondary immunizations (data not shown). In contrast, there were no significant differences seen between Nfat5+/+ and Nfat5+/Δ animals in total serum IgM concentrations (10.3 ± 0.9 vs. 8.4 ± 0.7 μg/ml), indicating that the impaired antigen-specific response by Nfat5+/Δ animals does not simply reflect a difference in overall serum Ig concentration. Interestingly, total serum IgG levels were reduced in Nfat5+/Δ animals by 35%, compared with wild-type littermate controls (136 ± 17 vs. 88 ± 13 μg/ml; P < 0.04). This result is consistent with the observed reduction in antigen-specific antibody production upon immunization, given that total serum IgG levels reflect the transition from a naive IgM-positive B cell to an antigen-experienced, IgG-positive B cell, a process that requires T cell-dependent activation and proliferation within germinal centers. These results indicate that optimal T cell-dependent B cell responses in vivo are dependent on the function of NFAT5.
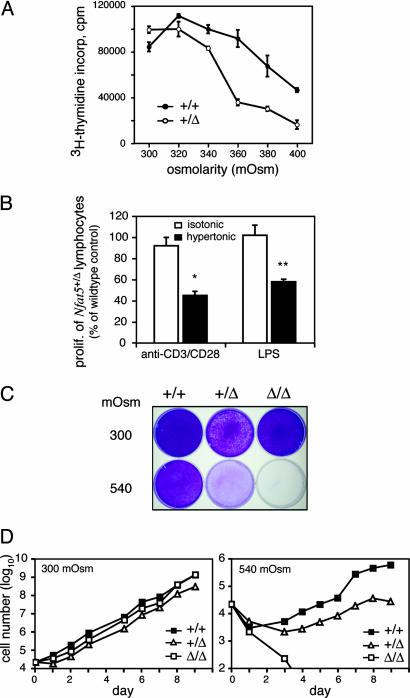
NFAT5 Is Necessary for Optimal Cell Growth in a Hyperosmotic Environment. To determine whether the impaired lymphocyte-dependent responses observed in vivo were due to an inability to compensate for osmotic stress, proliferation of splenocytes from Nfat5+/+ and Nfat5+/Δ animals was measured ex vivo under isotonic and hypertonic culture conditions. Whereas there was no difference between the proliferative responses of T cells from Nfat5+/+ and Nfat5+/Δ mice cultured under standard lymphocyte tissue culture conditions [RPMI 1640 complete medium, ≈300 milliosmoles per kg of water (mOsm)], the proliferation of Nfat5+/Δ T cells was increasingly compromised as the osmolality of the medium was increased by the addition of raffinose, which functions as a membrane-impermeant osmolyte (Fig. 4_A_). As shown, whereas the wild-type cells exhibited no impairment in proliferation when the osmolality of the medium was increased up to 360 mOsm, the proliferation of Nfat5+/Δ T cells was inhibited by >60%, demonstrating that NFAT5 function is essential for normal cell growth under conditions of hyperosmotic stress. The proliferation of wild-type cells was significantly enhanced upon culture in hypertonic medium (P < 0.01, 300 vs. 360 mOsm; 2.9-fold increase; _n_ = 5 independent experiments), consistent with the results previously reported by Junger _et al._ (24). An osmolality >360 mOsm consistently resulted in impaired proliferation, reflecting the enhanced sensitivity of lymphocytes to hyperosmotic stress, compared with nonlymphoid cells (25). The sensitivity of Nfat5+/Δ T cell proliferation to hyperosmotic stress was not due to impairment in IL2 production; exogenously added IL2 did not correct the defect (data not shown). Thus, NFAT5 function is essential for normal T cell growth under conditions of hyperosmotic stress that are significantly less than the extremes of hypertonicity present within the kidney in an antidiuretic state.
Fig. 4.
Normal cell proliferation under conditions of hyperosmotic stress requires NFAT5. (A) The splenocytes from Nfat5+/Δ and Nfat5+/+ littermates (5–8 weeks old) were cultured in standard complete medium (≈290 mOsm) or complete medium supplemented with raffinose to increase the osmolality of the culture conditions as indicated. The cells were stimulated with anti-CD3 plus anti-CD28 to induce T cell proliferation. (B) The splenocytes from Nfat5+/Δ mice and Nfat5+/+ littermate controls were cultured in standard complete medium (≈290 mOsm) or subjected to hypertonic stress (≈370 mOsm) through the addition of 80 mM raffinose. The cells were stimulated in the previously described method to induce T cell proliferation (n = 7 Nfat5+/+ and Nfat5+/Δ littermate pairs) or with LPS to induce B cell proliferation (n = 3 littermate pairs). Statistically significant differences are indicated (*, P < 0.01; **, P < 0.0001). (C) Immortalized MEF cell lines were cultured in complete medium adjusted to 300 mOsm in the absence and presence of additional NaCl (120 mM). The cells were seeded into the indicated medium and allowed to grow for 6 days, during which time the cells grown in standard medium were split and replated once. The plates were fixed and stained with crystal violet dye. (D) The MEF cell lines were seeded in 24-well tissue culture dishes and allowed to grow until ≈80% confluence, at which time the cultures were continued by replating. Manual cell counts were performed on the indicated days by using trypan blue exclusion to identify viable cells. The culture medium was replaced every 3 days. The data shown represent the mean of cell counts from triplicate wells. Standard errors, which were <5% of the mean, are not shown. These results are representative of at least four independent measurements of cell growth.
Splenocytes also were stimulated by using LPS under conditions of varying culture medium osmolality to determine whether B cell proliferation was similarly impaired by partial loss of NFAT5 function. Similar to T lymphocytes, proliferation of Nfat5+/Δ B cells was not impaired under isotonic culture conditions but was significantly inhibited upon culture in hypertonic medium (370 mOsm; Fig. 4_B_). Partial loss of NFAT5 function thus inhibits proliferation of both T and B cells when cultured under hypertonic, but not isotonic, conditions.
The effect of complete loss of NFAT5 function on cell growth was determined by comparing the growth of immortalized Nfat5+/+, Nfat5+/Δ, and _Nfat5_Δ/Δ MEF cell lines under normal vs. hyperosmotic culture conditions. Remarkably, whereas there were essentially no differences in cell growth under normal tissue culture conditions (≈300 mOsm), the growth of _Nfat5_Δ/Δ MEF cells under hyperosmotic conditions was markedly impaired (Fig. 4 C and D). The growth of heterozygous Nfat5+/Δ MEF cells exhibited partial impairment relative to wild-type cells, consistent with the partial loss of function demonstrated in NFAT5 reporter gene studies (Fig. 2) and with the partially compromised proliferative responses of Nfat5+/Δ lymphocytes (Fig. 4 A and B). These results clearly indicate that NFAT5 function is essential for normal cell proliferation under conditions of hyperosmotic stress. These results further suggest that, given the defect in lymphocyte proliferation observed upon partial loss of NFAT5 function (Fig. 4_A_), complete loss of function would likely result in an even more markedly impaired lymphocyte response.
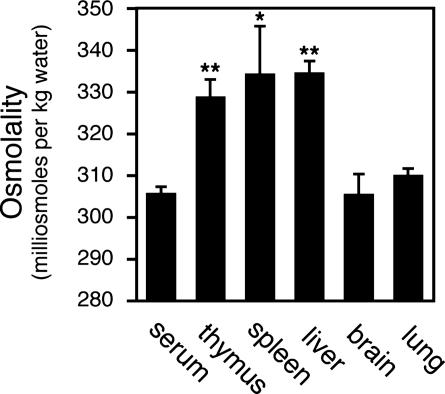
Lymphoid Tissue Hyperosmolality. The defective proliferative response of Nfat5+/Δ lymphocytes cultured ex vivo under conditions of hyperosmotic stress suggests that the impaired immune response observed in vivo is similarly due to an inability of lymphocytes to adapt to physiologic osmotic stress present within the lymphoid microenvironment. However, osmotic stress within the lymphoid microenvironment remains completely undefined. To specifically address this question, lymphoid tissue osmolality was measured directly by vapor pressure osmometry. Remarkably, in contrast to brain and lung, lymphoid tissues were significantly hyperosmolar relative to serum (Fig. 5). Liver tissue osmolality also was elevated. These results demonstrate that lymphocytes are exposed to physiologic hyperosmotic stress. Given that partial loss of NFAT5 function results in impaired lymphocyte function in vivo, this result suggests that NFAT5 functions to optimize lymphocyte function in vivo by regulating the transcriptional programs that enhance cellular adaptation to osmotic stress present within the lymphoid microenvironment.
Fig. 5.
Hyperosmolality of lymphoid tissues relative to blood. Tissue osmolality was determined by vapor pressure osmometry (see Materials and Methods). Statistically significant differences between blood and tissue osmolality are indicated (*, P < 0.02; **, P < 0.001).
Discussion
The relevance of osmotic stress to the function of T cells or peripheral blood mononuclear cells was first demonstrated in ex vivo studies showing that proliferation or cytokine production is enhanced upon culture in hyperosmotic medium (24, 26). However, the physiologic relevance of osmotic stress to lymphocyte function in vivo is unclear because it is generally assumed that the osmolality of tissues other than the kidney is similar to that of blood, which is maintained within a tightly defined and homeostatically regulated range (e.g., 285–295 mOsm in humans) (13). The observation that NFAT5, the only known mammalian osmosensitive transcription factor, is highly expressed in the thymus and is induced upon lymphocyte activation suggested that osmotic stress may be of physiologic relevance to lymphocyte function in vivo (7). The lymphoid tissue hypocellularity and impaired T cell-dependent antibody response observed in Nfat5+/Δ mice (Fig. 3) indicate that NFAT5 plays an important role in the function of the adaptive immune system and are consistent with previous functional studies employing transgenic mice in which expression of a dominant inhibitory form of NFAT5 was directed specifically to T cells using the CD2 transgene promoter (15). The observed correlation between the impaired lymphocyte responses in vivo resulting from partial loss of NFAT5 function and the impaired proliferative responses of Nfat5+/Δ lymphocytes ex vivo observed upon culture under conditions of hyperosmotic stress (Fig. 4) is consistent with the conclusion that NFAT5 functions as part of an osmotic stress response pathway that is active in lymphoid tissues. This conclusion is supported by the observation that lymphoid tissues are hyperosmotic relative to blood, based on direct measurements of tissue osmolality (Fig. 5).
A recent study by Zhang et al. (16) provides further support for this conclusion, demonstrating that the expression of NFAT5 protein in the thymus of rats rendered hypoosmotic was markedly reduced, compared with control animals. Given that the level of NFAT5 protein expression is regulated in response to changes in osmolality (27), this result indicates that within the thymus NFAT5 functions as part of an osmotic stress response. Although NFAT5 exhibited osmotic regulation in the thymus, mRNA levels of known NFAT5 target genes were apparently unaltered (16). However, this study described mRNA expression measured by real-time RT-PCR in relative and not absolute terms. Thus, it remains unclear to what extent these target genes are induced in the normal thymus at physiologic tissue osmolality. Known NFAT5 target genes have been identified primarily based on studies of hypertonic stress responses within the context of the kidney, which is exposed to extremes of hypertonic stress. As yet unidentified NFAT5-regulated genes may function to enable activated lymphocytes to compensate for osmotic stress present within the unique lymphoid microenvironment.
Several defining features of lymphocyte function likely confer upon lymphocytes a unique dependence on a functional osmotic stress response. First, extensive recombinational diversity of clonally expressed antigen receptors requires that lymphocytes be localized to nodal structures in which cells are present at high cell density. As suggested by measurements of tissue osmolality (Fig. 5), high cell density and/or the presence of metabolically highly active cells may give rise to a tissue microenvironment that is hyperosmotic relative to blood. Second, lymphocytes must undergo rapid cell proliferation in vivo (e.g., doubling times as short as ≈6 h) to ensure that the expansion of rare, antigen-specific lymphocytes is sufficient to mediate an effective immune response. Exposure of proliferating cells to hypertonic stress results in DNA damage, cell cycle arrest, and apotosis (28, 29). The results presented here clearly demonstrate that cell proliferation under conditions of hypertonic stress depends on the NFAT5 osmotic stress response (Fig. 4). Although hypertonic stress within the lymphoid microenvironment may not approach that within the kidney, environmental osmotic stress may be exacerbated by the depletion of intracellular osmolytes and reduction in available cell volume resulting from the massive induction of macromolecular biosynthesis that takes place during lymphocyte blastogenesis (30). Given the observed hyperosmolality of the liver, a nonproliferative but metabolically highly active tissue, osmotic stress in vivo may be more accurately considered a function of cell metabolism. And finally, a characteristic feature of adaptive immunity is abundant apoptosis, both in the thymus during T cell selection and in the periphery as a result of activation-induced cell death. Apoptosis, resulting in cell necrosis, may result in the generation of osmolytes within the tissue microenvironment that would further contribute to hyperosmotic stress. Considerations such as these suggest that the unique functions that define adaptive immunity in vivo may confer on lymphocytes a particular dependence on the NFAT5 osmotic stress response.
In the present study, the direct measurement of lymphoid tissue osmolality revealed values greater than serum osmolality but significantly less than the very high osmolality that can be reached within the renal medulla during antidiuresis (20). However, the values for tissue osmolality measured by vapor pressure osmometry likely represent an underestimate of actual tissue osmolality, because both intravascular as well as extravascular fluids contribute to the osmolality of the sample being measured. In addition, given the poor spatial resolution of the measurement, morphologically distinct regions within a tissue (e.g., germinal center) may exist in a microenvironment of much higher osmolality, compared with that of the surrounding tissue. Further elucidation of osmotic stress in vivo will require methods capable of measuring osmolality at single-cell resolution.
In summary, the results presented here demonstrate the critical importance of the NFAT5 transcription factor to the mammalian osmotic stress response. Moreover, these studies provide insight into the lymphoid microenvironment by revealing the relevance of the NFAT5 osmotic stress response to adaptive immunity. Further understanding of the unique features of the lymphoid microenvironment and the correspondingly unique mechanisms by which lymphocytes adapt to their environment may lead to new strategies to modulate lymphocyte function for clinically beneficial purposes.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. H. M. Kwon for providing Nfat5 genomic clones, TonEBP antisera, and HSP70-2 reporter construct; Dr. M. G. Rosenfeld for providing support in the generation of the knockout line; and Dr. G. Silverman and F. Sugiyami for their assistance with measurements of serum Ig levels. This work was supported by National Institutes of Health Grant GM59651 (to S.N.H.). W.Y.G. was supported in part by National Institutes of Health/National Institute of General Medical Sciences MSTP Training Grant PHSGM07198. Shared core facility resources were supported by National Cancer Institute Cancer Center Support Grant 2P30-CA23100-18.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NFAT, nuclear factor of activated T cells; TonEBP, tonicity enhancer binding protein; mOsm, milliosmoles per kg of water; DBD, DNA-binding domain; MEF, mouse embryonic fibroblast.
References
- 1.Wood, J. M., Bremer, E., Csonka, L. N., Kraemer, R., Poolman, B., van der Heide, T. & Smith, L. T. (2001) Comp. Biochem. Physiol. A Physiol. 130**,** 437–460. [DOI] [PubMed] [Google Scholar]
- 2.Hohmann, S. (2002) Microbiol. Mol. Biol. Rev. 66**,** 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D. & Somero, G. N. (1982) Science 217**,** 1214–1222. [DOI] [PubMed] [Google Scholar]
- 4.Burg, M. B. (1995) Am. J. Physiol. 268**,** F983–F996. [DOI] [PubMed] [Google Scholar]
- 5.Miyakawa, H., Woo, S. K., Dahl, S. C., Handler, J. S. & Kwon, H. M. (1999) Proc. Natl. Acad. Sci. USA 96**,** 2538–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Rodriguez, C., Aramburu, J., Rakeman, A. S. & Rao, A. (1999) Proc. Natl. Acad. Sci. USA 96**,** 7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trama, J., Lu, Q., Hawley, R. G. & Ho, S. N. (2000) J. Immunol. 165**,** 4884–4894. [DOI] [PubMed] [Google Scholar]
- 8.Ko, B. C., Turck, C. W., Lee, K. W., Yang, Y. & Chung, S. S. (2000) Biochem. Biophys. Res. Commun. 270**,** 52–61. [DOI] [PubMed] [Google Scholar]
- 9.Graef, I. A., Gastier, J. M., Francke, U. & Crabtree, G. R. (2001) Proc. Natl. Acad. Sci. USA 98**,** 5740–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo, K., Lee, D. & Kwon, H. M. (2002) Pflügers Arch. 444**,** 579–585. [DOI] [PubMed] [Google Scholar]
- 11.Burg, M. B., Kwon, E. D. & Kultz, D. (1997) Annu. Rev. Physiol. 59**,** 437–455. [DOI] [PubMed] [Google Scholar]
- 12.Woo, S. K., Lee, S. D., Na, K. Y., Park, W. K. & Kwon, H. M. (2002) Mol. Cell. Biol. 22**,** 5753–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verbalis, J. G. (2003) Best. Pract. Res. Clin. Endocrinol. Metab. 17**,** 471–503. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Rodriguez, C., Antos, C. L., Shelton, J. M., Richardson, J. A., Lin, F., Novobrantseva, T. I., Bronson, R. T., Igarashi, P., Rao, A. & Olson, E. N. (2004) Proc. Natl. Acad. Sci. USA 101**,** 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trama, J., Go, W. Y. & Ho, S. N. (2002) J. Immunol. 169**,** 5477–5488. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Z., Ferraris, J. D., Brooks, H. L., Brisc, I. & Burg, M. B. (2003) Am. J. Physiol. 285**,** F688–F693. [DOI] [PubMed] [Google Scholar]
- 17.Loyher, M. L., Mutin, M., Woo, S. K., Kwon, H. M. & Tappaz, M. L. (2004) Neuroscience 124**,** 89–104. [DOI] [PubMed] [Google Scholar]
- 18.Maouyo, D., Kim, J. Y., Lee, S. D., Wu, Y., Woo, S. K. & Kwon, H. M. (2002) Am. J. Physiol. 282**,** F802–F809. [DOI] [PubMed] [Google Scholar]
- 19.Tornheim, P. A. (1980) J. Neurosci. Methods 3**,** 21–35. [DOI] [PubMed] [Google Scholar]
- 20.Knepper, M. A. (1982) Kidney Int. 21**,** 653–655. [DOI] [PubMed] [Google Scholar]
- 21.Stroud, J. C., Lopez-Rodriguez, C., Rao, A. & Chen, L. (2002) Nat. Struct. Biol. 9**,** 90–94. [DOI] [PubMed] [Google Scholar]
- 22.Dahl, S. C., Handler, J. S. & Kwon, H. M. (2001) Am. J. Physiol. 280**,** C248–C253. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. D., Woo, S. K. & Kwon, H. M. (2002) Biochem. Biophys. Res. Commun. 294**,** 968–975. [DOI] [PubMed] [Google Scholar]
- 24.Junger, W. G., Liu, F. C., Loomis, W. H. & Hoyt, D. B. (1994) Circ. Shock 42**,** 190–196. [PubMed] [Google Scholar]
- 25.Bortner, C. D. & Cidlowski, J. A. (1996) Am. J. Physiol. 271**,** C950–C961. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro, L. & Dinarello, C. A. (1995) Proc. Natl. Acad. Sci. USA 92**,** 12230–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo, S. K., Dahl, S. C., Handler, J. S. & Kwon, H. M. (2000) Am. J. Physiol. 278**,** F1006–F1012. [DOI] [PubMed] [Google Scholar]
- 28.Kultz, D. & Chakravarty, D. (2001) Proc. Natl. Acad. Sci. USA 98**,** 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dmitrieva, N. I., Bulavin, D. V., Fornace, A. J., Jr., & Burg, M. B. (2002) Proc. Natl. Acad. Sci. USA 99**,** 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, S. N. (2003) Arch. Biochem. Biophys. 413**,** 151–157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information