Role of the Pilot Protein YscW in the Biogenesis of the YscC Secretin in Yersinia enterocolitica (original) (raw)
Abstract
The YscC secretin is a major component of the type III protein secretion system of Yersinia enterocolitica and forms an oligomeric structure in the outer membrane. In a mutant lacking the outer membrane lipoprotein YscW, secretion is strongly reduced, and it has been proposed that YscW plays a role in the biogenesis of the secretin. To study the interaction between the secretin and this putative pilot protein, YscC and YscW were produced in trans in a Y. enterocolitica strain lacking all other components of the secretion machinery. YscW expression increased the yield of oligomeric YscC and was required for its outer membrane localization, confirming the function of YscW as a pilot protein. Whereas the pilot-binding site of other members of the secretin family has been identified in the C terminus, a truncated YscC derivative lacking the C-terminal 96 amino acid residues was functional and stabilized by YscW. Pulse-chase experiments revealed that ∼30 min were required before YscC oligomerization was completed. In the absence of YscW, oligomerization was delayed and the yield of YscC oligomers was strongly reduced. An unlipidated form of the YscW protein was not functional, although it still interacted with the secretin and caused mislocalization of YscC even in the presence of wild-type YscW. Hence, YscW interacts with the unassembled YscC protein and facilitates efficient oligomerization, likely at the outer membrane.
Type III protein secretion systems are found in many pathogenic gram-negative bacteria and mediate the introduction of proteins from the bacterial cytoplasm directly into eukaryotic target cells (20). The type III secretion system of Yersinia enterocolitica consists of ∼25 Ysc proteins and is assembled in the cell envelope when the bacteria are grown at 37°C. However, only upon contact with eukaryotic cells (31) or when the bacteria are grown in a medium depleted of calcium is the system activated, resulting in the secretion of 11 Yop proteins. The ysc and yop genes are located on a large virulence plasmid, pYV (27, 43).
YscC is the only integral outer membrane constituent of the Y. enterocolitica type III protein secretion system, and it belongs to the family of secretins. Secretins of gram-negative bacteria form a distinct class of outer membrane proteins (OMPs) that participate in various systems for the transport of macromolecules, such as type II and III protein secretion, type IV pilus assembly, and filamentous phage release (15). They form stable oligomeric structures consisting of 12 to 14 subunits with a ring-like appearance, as shown by electron microscopy (2). The homology within the secretin family is especially pronounced in their C-terminal halves. This part of the protein is involved in oligomerization and is thought to form, in analogy to other OMPs, a β-barrel structure based on secondary structure predictions and circular dichroism analysis performed on isolated oligomers (4). Secretins are synthesized in the cytoplasm as precursors with an N-terminal signal sequence, which targets them to the Sec machinery in the inner membrane (32). After transport, the signal sequence is cleaved off by a signal peptidase, and the processed intermediate has to be transported across the peptidoglycan-containing periplasm in order to reach the outer membrane. During this process, the protein has to fold and to oligomerize. The pIV secretin of filamentous phage f1 has been shown to exist for several minutes as a periplasmic intermediate before it becomes associated with membranes (3), but more information on the biogenesis of secretins is not available.
Secretins appear to follow to some extent the same sorting pathway as other OMPs, since oligomerization and outer membrane insertion of the PilQ secretin of Neisseria meningitidis were dependent on the Omp85 protein (42). Omp85 was shown to play a crucial role in OMP assembly, since depletion of this protein in a conditional mutant of N. meningitidis caused the accumulation of unassembled forms of all OMPs tested. In other aspects, secretins are very different from other OMPs. First, they lack the C-terminal consensus motif that is present in most other OMPs and that is thought to be important for their assembly into the outer membrane (39). Second, as described above, they form large oligomeric structures, a property that is expected to demand additional adjustments to facilitate their passage through the periplasm. Indeed, several secretins require a specific pilot protein for outer membrane localization and for protection against proteolytic degradation. These pilot proteins are small outer-membrane-located lipoproteins, which share no or very limited sequence homology. Examples of these pilot proteins include PulS and InvH, which promote the localization and stability of the secretins PulD of the type II secretion system of Klebsiella oxytoca (18) and InvG of the type III secretion system of Salmonella enterica serovar Typhimurium (11, 13), respectively. In general, pilot proteins interact with a domain at the extreme C terminus of their cognate secretin (12, 13, 35, 36, 37) and, in the case of the Pul system, PulS remains associated with PulD after its assembly into the outer membrane (29).
Previously, it was shown that in a mutant of Y. enterocolitica lacking the lipoprotein YscW (formerly designated VirG) the total amount of YscC oligomers was reduced and that the secretin did not appear to be properly localized in the outer membrane (23). Therefore, YscW was proposed to represent the pilot protein of YscC, despite the lack of any sequence homology with the known pilot proteins. Furthermore, YscC has a C-terminal extension beyond the conserved homology domain, which could possibly function as the pilot-binding site. However, the putative YscC-YscW interaction was not further investigated. Here, the role of YscW in the biogenesis of the YscC secretin was further explored by expressing the two proteins in a strain lacking all other Ysc components and by studying the kinetics of YscC oligomerization in pulse-chase experiments in the presence or absence of the YscW protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used are listed in Table 1. Escherichia coli strains DH5α, S17-1, and CJ236 were used for routine gene cloning, conjugational transfer of plasmids to Y. enterocolitica, and site-directed mutagenesis, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80d_lacZ_ΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK−) phoA supE44 λ−thi-1 gyrA96_Δ_relA | 17 |
| S17-1 | F−thi pro hsdR recA; RP4-2 (Tc::Mu Km::Tn_7_) | 38 |
| CJ236 | F′ cat(=pCJ105; M135 Cmr)/dut ung-1 thi-1 relA1 spoT1 mcrA | 25 |
| Y. enterocolitica | ||
| W22703 | (Res−, Mod+) of the serotype O:9 strain W227 | 9 |
| KNG22703 | W22703 blaA replaced by the luxAB genes | 22 |
| KNG22703(pRS227) | KNG22703 carrying the aphA-3 cassette in the yscW gene | 1 |
| KNG22703(pAA203) | KNG22703 carrying the aphA-3 cassette in the yscC gene | 23 |
| CE1525 | KNG22703 without virulence plasmid pYV22703 | 5 |
| Plasmids | ||
| pBBR1MCS-5 | Gmr; cloning vector | 24 |
| pBC18R | Apr; cloning vector | 7 |
| pSM3 | Apr; pMMBH67EH; _virF_′ _yscB yscC yscD_′ | 23 |
| pSM3km | Kmr; pUR6500; _virF_′ _yscB yscC yscD_′ | 5 |
| pSM9 | Apr; pMMBH67EH; yscC | 23 |
| pCK4 | Apr; pBC18R; 3′ end of yscC, _yscD_′ | This study |
| pCK44 | Apr; pBC18R; 3′ end of yscC with BglII at 287 bp before stop codon, _yscD_′ | This study |
| pSM9.4 | Apr; pMMBH67EH; 3′ end of yscC with BglII at 287 bp before stop codon, _yscD_′ | This study |
| pSM9.4T | Apr; pMMBH67EH; 3′ end of _yscC_ΔC-term | This study |
| pSM3Δ7 | Kmr; pUR6500; _virF_′ _yscB yscC_ΔC-term | This study |
| pBBR1MCS-5-YscAC | Gmr; pBBR1MSC-5; _virF_′ _yscB yscC_ΔC-term | This study |
| pSM3kmΔ7 | Kmr; pUR6500; _virF_′ _yscB yscC_ΔC-term | This study |
| pCK8 | Apr; pBC18R; 5′ end of yscC | This study |
| pCK8.5 | Apr; pBC18R; 5′ end of yscC with BglII site at bp 85 | This study |
| pRS6 | Tcr; pTM100; yscW | 1 |
| pEW3 | Apr; pET16b; (His6)-_yscW_Δsig | 5 |
| pEW4 | Apr; pBC18R: _yscC_sig-_yscW_Δsig (yscW_Δ_lip) | This study |
Routinely, the E. coli strains were grown at 37°C in a modified Luria-Bertani broth (LB) (40), and the Y. enterocolitica strains were grown in LB supplemented with 0.4% glucose at room temperature. The M9 minimal medium that was used in the pulse-chase experiments was composed of M9 salts (34), 0.2% glucose, 0.0001% thiamine, 246 mg of MgSO4 · 0.7H2O/ml, and 1 mg of FeSO4 · 0.7H2O/ml. To induce for Yop secretion, Y. enterocolitica strains were inoculated at an optical density at 600 nm (OD600) of 0.1 in brain heart infusion broth supplemented with 0.4% glucose, 20 mM MgCl2, and 20 mM sodium oxalate (BHI-OX). The cultures were grown for 2 h at room temperature, followed by 4 h of growth at 37°C. Similar growth conditions were applied in the experiments in which the production of YscC oligomers was studied, except that the strains were grown for 2 h instead of 4 h at 37°C. To induce the tac or lac promoter, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM. Antibiotics were used at the following concentrations: for Y. enterocolitica, ampicillin at 1 mg/ml, kanamycin at 50 μg/ml, nalidixic acid at 25 μg/ml, and tetracycline at 10 μg/ml in LB and at 20 μg/ml in BHI-OX and for E. coli, ampicillin at 100 μg/ml, gentamicin at 10 μg/ml, kanamycin at 25 μg/ml, and tetracycline at 10 μg/ml in LB and at 20 μg/ml in BHI-OX.
Cloning of the yscC and yscW genes.
Recombinant DNA methods were performed essentially as described previously (34). The plasmids used in the present study are listed in Table 1. They were introduced in E. coli by transformation by using the CaCl2 procedure (34) and into Y. enterocolitica by electroporation (8) or by conjugation on LB-agar plates overnight at room temperature.
To create a truncated YscC derivative lacking the C-terminal 96 amino acid residues, the BamHI-PstI fragment of pSM3 carrying the last 1,077 bp of yscC and the first 98 bp of yscD was cloned into vector pBC18R, resulting in pCK4. By site-directed mutagenesis (25), a BglII site was introduced at 287 bp before the stop codon of yscC on pCK4 by using primer MIPA264 (5′-CGGATTATTGACGAAAGATCTGCGCATCATTTAGCGTTAG-3′). The BamHI-PstI fragment of this pCK44 plasmid was introduced into pSM9, generating pSM9.4. From this plasmid the last 287 bp of yscC and the first 98 bp of yscD were excised with BglII and PstI. The 5′ overhang of the BglII site and the 3′ overhang of the PstI site of the remaining plasmid were filled in and removed, respectively, by treatment with T4 DNA polymerase (Pharmacia), and the plasmid was circularized with T4 ligase (Fermentas), creating pSM9.4T. Plasmid pSM3Δ7 was obtained by cloning the EcoRI-BamHI fragment of pSM3 into pSM9.4T. The EcoRI-HindIII fragment of pSM3Δ7 was cloned into vector pBBR1MCS-5, creating pBBR1MCS-5-YscAC. The EcoRI-SalI fragment of this plasmid was cloned into pUR6500, generating pSM3kmΔ7. To create a plasmid encoding the unlipidated YscW derivative, the HpaI_-_BamHI fragment of pSM3, carrying the first 747 bp of the yscC gene, was cloned in between the SmaI and BamHI sites of pBC18R, thereby creating pCK8. By site-directed mutagenesis, a BglII site was introduced at the end of the signal sequence-encoding part of yscC, i.e., 86 bp from the start codon, by using primer MIPA265 (5′-GGGCGCAAGAACTAGATCTGTTGCCTATACCTT-3′), resulting in pCK8.5. The yscW gene was excised from pEW3 with BglII and HindIII and cloned into pCK8.5, resulting in pEW4 encoding mature YscW without its N-terminal cysteine residue behind the signal sequence of YscC.
SDS-PAGE and immunoblotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26), with 0.2% SDS in the running gel. The proteins were stained in the gel with Coomassie brilliant blue or silver (28). Alternatively, they were transferred onto nitrocellulose membranes by semidry electroblotting. To improve the immunodetection of the YscC oligomers, the gels were, prior to the blotting procedure, incubated for at least 1 h in 10% trichloroacetic acid (TCA), washed four times in distilled water, and washed two times in SDS-PAGE electrode buffer. After incubation with the primary antibodies and with horseradish peroxidase-coupled anti-rabbit immunoglobulin G (1:3,000; Biosource), the immunoblots were developed by enhanced chemiluminescence (Pierce). As primary antibodies, 1:1,000 dilutions of polyclonal rabbit antisera directed against a synthetic YscC peptide (23) or against YscW (5) were used. In the case of the pulse-chase experiments, the polyacrylamide gels were, after electrophoresis, treated with Amplify (Amersham), vacuum dried, and exposed to X-ray films at −80°C. Quantification of protein bands was performed by using the program ImageQuant. after the dried gels were exposed in a PhosphorImager (Molecular Dynamics).
Pulse-chase experiments.
The pulse-labeling experiments were performed as described previously (6) with minor modifications. Briefly, cells of overnight cultures, which were grown in LB at room temperature, were pelleted and used to inoculate M9 medium, which was supplemented with all amino acids except for methionine and cysteine (i.e., 0.12% threonine, serine, alanine, proline, valine, leucine, isoleucine, phenylalanine, asparagine, glutamine, lysine, arginine, histidine, and glycine and 0.1% aspartic acid, glutamic acid, tyrosine, and tryptophan), to an OD600 of 0.2. The cultures were grown for 3 h at 37°C, after which IPTG was added to a final concentration of 0.1 mM to induce the tac or lac promoter, and growth was continued for another 30 min. The cells were then pelleted and resuspended in 1/10 of the original volume of the growth medium. Cells were pulse-labeled for 1 min with 100 μCi of Redivue l-[35S]methionine (Amersham)/ml at 37°C. The chase was initiated by the addition of 9 volumes of prewarmed M9 minimal medium with 5% Casamino Acids and 25 μg of chloramphenicol/ml and then continued for 3 h at 37°C. Samples of 500 μl of cell suspension were quickly frozen in acetone-CO2 solid). After thawing, cells were pelleted, and YscC was isolated by immunoprecipitation.
Immunoprecipitation.
The immunoprecipitation experiments were performed as described previously (6) with only minor modifications. Cells or proteins were resuspended in 50 μl of 4% SDS-0.5 M Tris-HCl (pH 8.0) for 20 min and incubated at 100°C for 10 min. Insoluble material was removed by centrifugation at 20,000 × g for 5 min, and 40 μl of supernatant was added to 1 ml of radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl [pH 8.0]) containing a 1:1,000 dilution of a rabbit polyclonal antiserum (Eurogentec) raised against purified YscC oligomers (5). After vortexing and incubation for 1 h at room temperature, 50 μl of protein A-CL-4B Sepharose (Amersham; 50% [vol/vol] in 150 mM NaCl-10 mM EDTA-50 mM Tris-HCl [pH 7.5]) was added. The mixtures were gently rocked for 1 h, and the immunocomplexes were collected by centrifugation at 8,000 × g for 1 min and washed three times in radioimmunoprecipitation assay buffer. YscC was eluted by boiling in 20 μl of SDS-PAGE sample buffer for 10 min.
Dissociation of the YscC oligomer.
The YscC oligomer was dissociated by treatment with TCA and trifluoroacetic acid (TFA). Then, 1 volume of cell culture was mixed with 1 volume of 10% TCA, and the mixture was incubated for 20 min on ice. Insoluble material was collected by centrifugation at 20,000 × g for 20 min at 4°C, washed twice with 2 volumes of ice-cold acetone, and dried. To increase the dissociation efficiency, 50 μl of TFA was added to the pellet, which was then immediately evaporated by a stream of air.
Cell fractionation and extraction.
Secreted proteins were precipitated from culture supernatants with 35% ammonium sulfate (10). To separate the soluble and membrane fractions, whole cells were washed in 0.9% NaCl, concentrated to an OD600 of 10 in 2 mM EDTA-50 mM Tris-HCl (pH 8.5), and frozen at −20°C. After they thawed, the cells were disrupted by sonication and centrifuged for 1 h at 150,000 × g at 4°C. Pellets containing the cell envelopes were suspended in 2 mM Tris-HCl (pH 7.8).
For separation of inner and outer membranes, cells were washed in 0.9% NaCl and then suspended in 7 ml of ice-cold 0.75 M sucrose with 10 mM Tris-HCl (pH 7.8). After the addition of 0.35 ml of 2 mg of egg white lysozyme (Merck)/ml, the cells were incubated for 1 min on ice. With gentle shaking, 14.7 ml of 1.5 mM EDTA was slowly added to the suspension. After 1.5 h of incubation on ice, cells were inactivated for 30 min with 0.5 mg of streptomycin/ml. Subsequently, dithiothreitol and phenylmethylsulfonyl fluoride were added to a final concentration of 1 mM, and cells were broken with a French pressure cell at 8,000 lb/in2. Unbroken cells were removed by centrifugation for 30 min at 750 × g at 4°C. Cell envelopes were collected by centrifugation for 1 h at 150,000 × g at 4°C and layered on top of a discontinuous sucrose gradient spanning 30 to 55% (wt/wt) sucrose in 5 mM EDTA-0.2 mM dithiothreitol. The gradients were centrifuged at 38,000 rpm in an SW41 rotor (Beckman) for 20 h at 4°C and then fractionated. NADH-oxidase activity was determined as described previously (30) and always peaked at 37 to 41% sucrose. The presence of porins in the different fractions was evaluated by SDS-PAGE. The porins always peaked at 47 to 52% sucrose. All of the localization studies were performed at least twice.
RESULTS
YscW is required for localization and stabilization of the YscC oligomer.
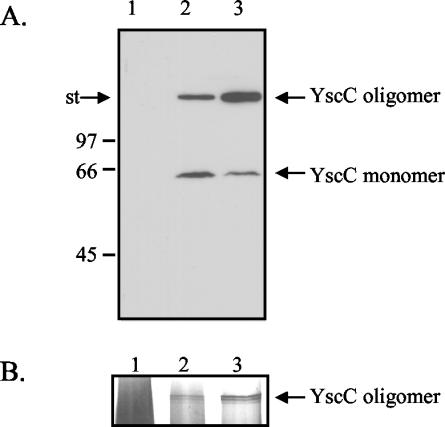
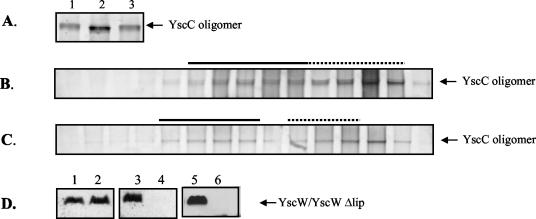
In a Y. enterocolitica mutant lacking the YscW lipoprotein, the amount of YscC oligomers is reduced and, when YscC is overproduced in this strain, the secretin does not correctly localize to the outer membrane (23). However, it is possible that the yscW mutation has a primary effect on another Ysc component of the machinery and that the effect on YscC is only indirect. In order to determine whether yscW expression directly affects the yield and localization of YscC oligomers, the yscC and yscW genes were cloned on two separate plasmids, pSM3km and pRS6, respectively, and introduced into the pYV-cured Y. enterocolitica strain CE1525. Both the oligomeric and the monomeric forms of YscC were detectable on immunoblots of the membrane proteins of the strain harboring only yscC on a plasmid (Fig. 1A, lane 2). Coexpression of yscW caused a significant increase in the yield of YscC oligomer, which was accompanied by a decrease in the monomeric form (Fig. 1A, lane 3). However, the amounts of YscC oligomer identified by immunodetection varied between experiments, probably due to inefficient transfer to the nitrocellulose membrane. Therefore, the relative amounts of YscC oligomers produced by the strains were also estimated from silver-stained SDS-PAGE gels. This experiment confirmed that the yield of oligomeric YscC was significantly increased by the presence of YscW (Fig. 1B). YscC oligomers often appeared as multiple bands, probably due to limited proteolysis. YscC was never detected in the soluble fraction (data no shown).
FIG. 1.
Analysis of the cell envelope proteins of the pYV-cured strain CE1525 with either plasmid pRS6 (yscW) (lane 1), plasmid pSM3km (yscC) (lane 2), or both plasmids (lane 3). (A) Cell envelopes were analyzed by SDS-PAGE on an 11% polyacrylamide gel, and YscC was detected by immunoblotting. The positions of the molecular mass standard proteins (in kilodaltons) and the boundary between the stacking and running gel (st) are indicated at the left. (B) Cell envelopes were analyzed by SDS-PAGE on a 3 to 9% polyacrylamide gradient gel, and proteins were stained with silver. Only the relevant part of the gel is shown. Strains were grown at 37°C in BHI-OX with IPTG to induce yscC expression. Cell envelopes from an equal amount of cells were loaded in each lane.
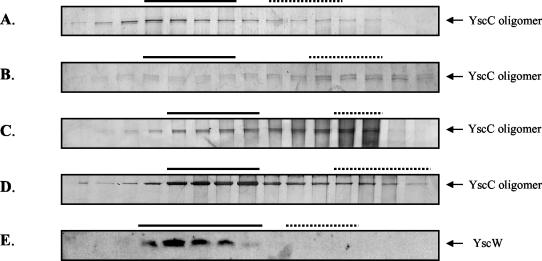
To study the role of YscW in the localization of YscC oligomers, inner and outer membranes were separated by isopycnic sucrose density gradient centrifugation. NADH-oxidase activity and the porins were used as markers for the inner and outer membranes, respectively. In the Y. enterocolitica wild-type strain KNG22703, oligomers were almost exclusively found in the outer membrane, as expected (Fig. 2A). In the yscW mutant KNG22703(pRS227), the majority of the YscC oligomers colocalized with the NADH-oxidase activity (Fig. 2B). Also in the pYV-cured strain CE1525 containing pSM3km, the oligomers were mostly found in the inner membrane fractions (Fig. 2C). However, when yscW was coexpressed in this strain from pRS6, the oligomers were associated with the outer membrane (Fig. 2D). It can be concluded that YscW is directly required for the correct localization of the secretin in the outer membrane and for optimal yield of YscC oligomers and that no other Ysc component is required for this process.
FIG. 2.
Subcellular localization of YscC in the presence or absence of YscW and of YscW itself. Membrane preparations of French press lysates of the Y. enterocolitica wild-type strain KNG22703 (A), the yscW mutant KNG22703(pRS227) (B), and the pYV-cured strain CE1525 with either plasmid pSM3km (yscC) (C), plasmids pSM3km (yscC) and pRS6 (yscW) (D), or plasmid pRS6 (yscW) (E) were applied onto 30 to 55% sucrose gradients, which were centrifuged and fractionated. Aliquots of the fractions were analyzed by SDS-PAGE on 3 to 9% polyacrylamide gradient gels (A to D) or on a 14% polyacrylamide gel (E). For the detection of YscC oligomers, protein bands were stained with silver (A to D), and the YscW protein was immunodetected on a Western blot (E). The fractions that represent the inner (dotted line) and outer membranes (continuous line) were identified based on NADH-oxidase activity and the presence of the porins, respectively. The strains were grown at 37°C in BHI-OX with IPTG to induce yscC expression.
The subcellular localization of YscW was determined as well. In the pYV-cured strain CE1525 bearing yscW on pRS6, YscW localized to the outer membranes (Fig. 2E). Similarly, in the Y. enterocolitica wild-type strain and the yscC mutant KNG22703(pAA203), YscW cofractionated with the porins (data not shown). Thus, the pilot protein is associated with the outer membrane, and its localization is not dependent on other Ysc proteins.
C terminally truncated YscC interacts with YscW.
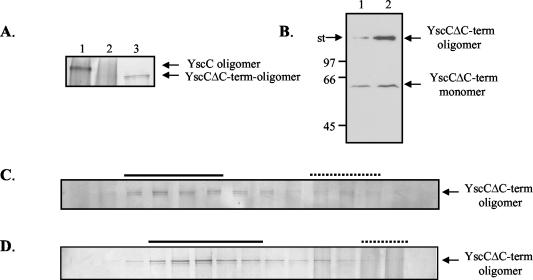
In all cases studied thus far, pilot proteins were found to interact with the C terminus of their cognate secretin (12, 13, 35, 36, 37). To determine the importance of the C-terminal part of the YscC secretin for the YscC-YscW interaction, we constructed plasmid pSM3kmΔ7, encoding a truncated derivative of YscC lacking the last 96 amino acid residues (YscCΔC-term). When YscCΔC-term was produced in the pYV-cured strain CE1525, a high-molecular-weight complex was formed that was hardly detectable (Fig. 3A, lane 2) unless the gel was stained for a prolonged time (data not shown) or immunodetection was used (Fig. 3B, lane 1). Also, the amount of monomeric YscCΔC-term was reduced, suggesting that deletion of the C terminus destabilizes the protein. Interestingly, in the presence of YscW, the amount of YscCΔC-term oligomers strongly increased, demonstrating that YscW is able to interact with and to stabilize the C-terminally truncated YscC derivative (Fig. 3A, lane 3, and B, lane 2). Determination of the subcellular localization of YscCΔC-term oligomers in the pYV-cured Y. enterocolitica strain CE1525 revealed that it localized to the outer membrane in the presence (Fig. 3D) but also in the absence of YscW (Fig. 3C). Thus, the C terminus of YscC is not required for outer membrane association, but it renders the protein dependent on YscW for proper localization.
FIG. 3.
Stability and localization of YscCΔC-term. (A) Cell envelopes of the pYV-cured Y. enterocolitica strain CE1525 with either plasmid pSM3km (yscC) (lane 1), plasmid pSM3kmΔ7 (yscCΔC-term) (lane 2), or plasmids pSM3kmΔ7 (yscCΔC-term) and pRS6 (yscW) (lane 3) were analyzed by SDS-PAGE on a 3 to 9% polyacrylamide gradient gel, and YscC was detected by silver staining. (B) Cell envelopes of the pYV-cured Y. enterocolitica strain CE1525 with either plasmid pSM3kmΔ7 (yscCΔC-term) (lane 1) or plasmids pSM3kmΔ7 (yscCΔC-term) and pRS6 (yscW) (lane 2) were analyzed by SDS-PAGE on an 11% polyacrylamide gel, and YscC was detected by immunoblotting. The positions of the molecular mass standard proteins (in kilodaltons) and the boundary between the stacking and running gel (st) are indicated on the left. Cell envelopes from an equal amount of cells were loaded in each lane. (C and D) The cell envelope fraction of the French press lysates of the pYV-cured strain CE1525 with either plasmid pSM3kmΔ7 (yscCΔC-term) (C) or plasmid pSM3kmΔ7 and plasmid pRS6 (yscW) (D) were applied on a 30 to 55% sucrose gradient, which was centrifuged and fractionated. The samples were analyzed by SDS-PAGE on 3 to 9% polyacrylamide gradient gels, and YscC oligomers were detected by silver staining. The fractions that represent the inner (dotted line) and outer membranes (continuous line) were identified based on NADH-oxidase activity and the presence of the porins, respectively. The strains were grown at 37°C in BHI-OX with IPTG to induce yscC expression.
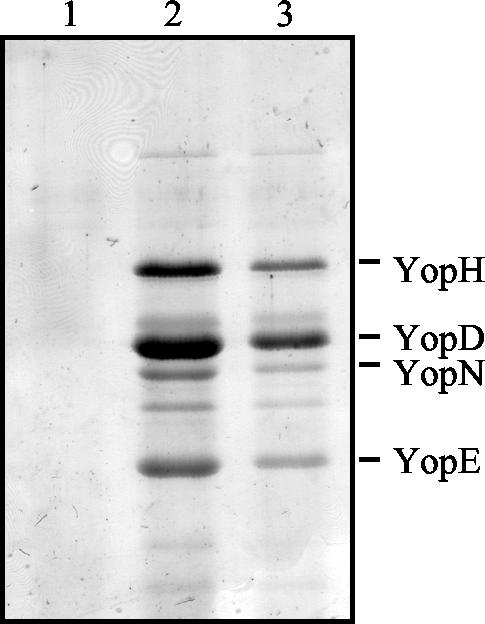
The effect of the C-terminal deletion on the functionality of YscC was determined by expressing yscCΔC-term in the Y. enterocolitica yscC mutant. The production of C terminally truncated YscC restored almost completely the secretion of Yops in the yscC mutant strain KNG22703(pAA203) (Fig. 4). Apparently, the C-terminal 96 amino acids of YscC are not essential for the interaction of the protein with YscW or for its function.
FIG. 4.
Complementation of an yscC mutation by YscCΔC-term. Secreted protein patterns are shown of the Y. enterocolitica yscC mutant KNG22703(pAA203) (lane 1) with pSM3 (yscC) (lane 2) or pSM3Δ7 (yscCΔC-term) (lane 3), which were grown at 37°C in BHI-OX. The expression of yscC or yscCΔC-term was induced by the addition of IPTG to a final concentration of 0.1 mM. Proteins from the extracellular medium were analyzed by SDS-PAGE on an 11% polyacrylamide gel and stained with Coomassie brilliant blue. The positions of the most abundant Yop proteins are indicated on the right.
Role of YscW in YscC oligomerization.
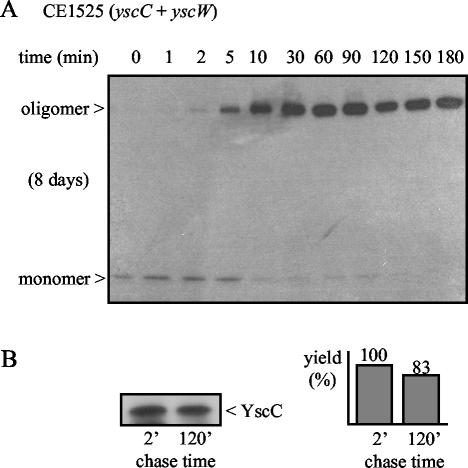
Several roles can be envisaged for the pilot protein in secretin assembly. Among other possibilities, the pilot protein could bind to the unstable monomer, thereby protecting it against proteases until a stable oligomer is formed. Alternatively, the YscW protein could interact with the oligomer and prevent its proteolytic degradation before its insertion into the outer membrane. The pilot protein could also affect oligomerization, either by stimulating it or, alternatively, by delaying it to allow for the transport of YscC in a monomeric form across the peptidoglycan-containing periplasm. To gain insight in these different possibilities, the kinetics of YscC oligomerization was studied in the presence or absence of YscW. Therefore, pulse-chase experiments were performed with the pYV-cured Y. enterocolitica strain CE1525, which expresses yscC and yscW from the plasmids pSM3km and pRS6, respectively, followed by immunoprecipitation with antibodies directed against the secretin. Already directly after the pulse, a small amount of labeled oligomer could be detected (data not shown). However, the yield increased strongly after a 5- to 10-min chase period and reached its maximum after ∼30 min (Fig. 5A). Concomitantly, the amount of YscC monomer decreased, showing that the monomer on the autoradiogram represents an assembly intermediate. However, we cannot exclude the possibility that this monomer represents an unstable oligomeric intermediate that dissociates during the procedure. No stable oligomeric intermediates could be detected.
FIG. 5.
Time course of the formation of stable oligomers of the YscC secretin. Cells of the strain CE1525 with pSM3km (yscC) and pRS6 (yscW) were induced for the production of YscC and pulse-labeled for 1 min with [35S]methionine. (A) Samples were taken after the chase periods indicated, and YscC was immunoprecipitated and analyzed by SDS-PAGE and autoradiography. The exposure time and the positions of the monomer and the oligomer are indicated on the left. The data are representative of three independent experiments. (B) Samples obtained after 2 and 120 min chase were treated with TCA and TFA to dissociate YscC oligomers, and YscC was immunoprecipitated and analyzed by SDS-PAGE and autoradiography (left). For quantification of the YscC levels, the autoradiogram was analyzed on a PhosphorImager (right), and the value obtained for the 2-min chase time was set at 100%.
The yield of oligomers appeared much higher than the initial amount of monomer directly after the pulse (Fig. 5A). This result could be explained by assuming that the oligomer is more efficiently immunoprecipitated than the monomer. To investigate this possibility, the samples taken after 2 and 120 min of chase were treated with TCA and TFA to dissociate the YscC oligomers prior to the immunoprecipitation procedure. In this case, there was no strong difference in the yield of labeled YscC between the samples (Fig. 5B). Hence, the apparent increase in the amount of labeled YscC during the pulse-chase experiment is indeed due to more efficient immunoprecipitation of YscC oligomers.
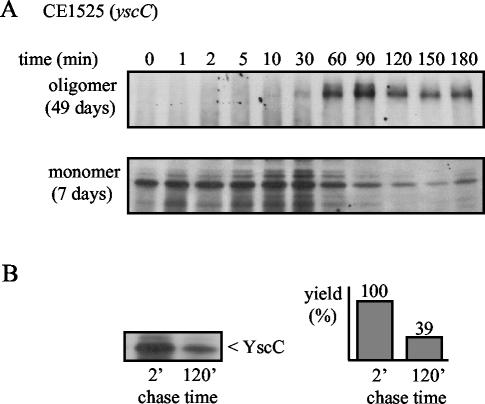
In the absence of the pilot protein, the yield of YscC oligomers was much lower. Only after a very long exposure time, they were detectable on the autoradiogram (Fig. 6A). In addition, the formation of oligomers was considerably delayed, since its yield reached its maximum only after a chase period of ∼90 min and the monomer remained visible during the entire experiment (Fig. 6A; note the difference in exposure time of the autoradiograms). The total amount of labeled YscC detected after dissociation of the oligomer decreased during the chase (Fig. 6B), a finding that is in line with the observation that the pilot protein increases the yield of YscC oligomers. These experiments demonstrate that the YscW lipoprotein is important for efficient oligomerization of YscC.
FIG. 6.
Time course of YscC oligomerization in the absence of YscW. Cells of strain CE1525 with pSM3km (yscC) were induced for the production of YscC and pulse-labeled for 1 min with [35S]methionine. (A) Samples were taken after the chase periods indicated, and YscC was immunoprecipitated and analyzed by SDS-PAGE and autoradiography. Different exposure times were used for the detection of the monomer (lower panel) and the oligomer (upper panel), as indicated in parentheses on the left. The data are representative of three independent experiments. (B) Samples obtained after 2 and 120 min of chase were treated with TCA and TFA to dissociate YscC oligomers, and YscC was immunoprecipitated and analyzed by SDS-PAGE and autoradiography (left). For quantification of the YscC levels, the gel was exposed in a PhosphorImager (right), and the value obtained for the 2-min chase time was set at 100%.
Outer membrane anchoring of YscW is essential for its function.
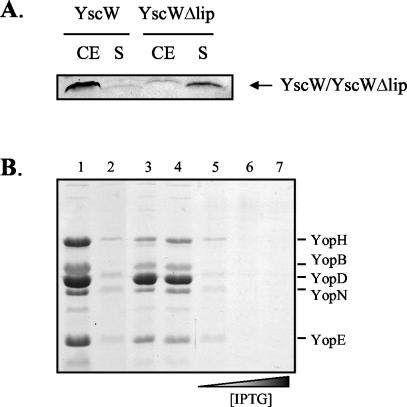
The YscW protein is associated with the outer membrane likely via the lipid moiety attached to its N-terminal cysteine. To determine whether targeting of the pilot protein to the outer membrane is essential for its function, we constructed plasmid pEW4 in which a DNA fragment encoding the YscC signal sequence replaces the fragment encoding the signal sequence and the N-terminal cysteine residue of YscW. Cell fractionation studies showed that the unlipidated form of YscW (YscWΔlip) was predominantly present in the soluble fraction, whereas native YscW was only found in association with the membranes (Fig. 7A). The amounts of YscWΔlip present in the membrane fraction varied between different experiments from <5 to 50%, indicating that the protein is loosely associated with the membranes. Separation of the membranes by sucrose density gradient centrifugation showed that membrane-associated YscWΔlip cofractionated with the inner membrane (data not shown).
FIG. 7.
Localization of unlipidated YscW and its dominant-negative effect on Yop secretion. (A) Cell envelopes (CE) and soluble fractions (S) of the Y. enterocolitica pYV-cured strain CE1525 with either pRS6 (yscW) or pEW4 (yscWΔlip) were analyzed by SDS-PAGE on a 14% polyacrylamide gel. YscW was detected by immunoblotting. (B) The Y. enterocolitica yscW mutant KNG22703(pRS227) with either pRS6 (yscW) (lane 1) or pEW4 (yscWΔlip) (lane 2), and the wild-type strain KNG22703 (lane 3) with either pRS6 (yscW) (lane 4) or pEW4 (yscWΔlip) (lanes 5 to 7) were grown at 37°C in BHI-OX. The expression of YscWΔlip was induced by the addition 0.1 mM (lanes 2 and 5), 0.5 mM (lane 6), or 1.0 mM (lane 7) IPTG. Yops from the extracellular medium were precipitated and analyzed by SDS-PAGE on an 11% polyacrylamide gel. Proteins were stained with Coomassie brilliant blue. The positions of the most abundant Yop proteins are indicated on the right.
The functionality of YscWΔlip was subsequently studied. Production of unlipidated YscW did not complement the yscW mutation in strain KNG22703(pRS227) for Yop secretion (Fig. 7B, lane 2). Moreover, production of YscWΔlip in the wild-type strain blocked secretion (Fig. 7B, lanes 5 to 7), demonstrating a dominant-negative effect of unlipidated YscW. To study whether YscWΔlip could stabilize and localize YscC, the relevant plasmids were introduced into the pYV-cured strain CE1525. In contrast to native YscW, YscWΔlip did not increase the yield of YscC oligomers (Fig. 8A) and was not able to promote their outer membrane localization (Fig. 8B). In the wild-type strain, which contains a wild-type copy of the yscW gene on the pYV plasmid, the coproduction of unlipidated YscW caused mislocalization of YscC oligomers to the inner membrane (Fig. 8C), a result consistent with the dominant-negative effect of yscWΔlip expression on Yop secretion. Surprisingly, whereas YscWΔlip could be detected in strain CE1525 carrying pEW4 (Fig. 8D, lane 3), it could not be detected anymore if coexpressed with the secretin (Fig. 8D, lane 4). In contrast, the level of native YscW was unaffected by the presence of YscC (Fig. 8D, lanes 1 and 2). A similar phenomenon was observed in the mutant KNG22703(pRS227), which lacks an intact copy of the yscW gene but has all of the other ysc genes, including yscC. After introduction of plasmid pRS6 into this strain, the native YscW could readily be detected (Fig. 8D, lane 5), whereas the introduction of pEW4 did not lead to detectable levels of YscWΔlip (Fig. 8D, lane 6). This indicates that unlipidated YscW is degraded by proteases in the presence of YscC. The dominant-negative effect of YscWΔlip on secretion in the wild-type strain suggests that this degradation occurs only after the protein has mislocalized YscC.
FIG. 8.
Stability and localization of YscC in the presence of YscWΔlip. (A) Cell envelope proteins of the pYV-cured Y. enterocolitica strain CE1525 with either plasmid pSM3km (yscC) (lane 1), plasmids pSM3km (yscC) and pRS6 (yscW) (lane 2), or plasmids pSM3km (yscC) and pEW4 (yscWΔlip) (lane 3) were separated by SDS-PAGE on a 3 to 9% polyacrylamide gradient gel. YscC oligomers were detected with Coomassie brilliant blue. Cell envelopes from an equal amount of cells were loaded in each lane. (B and C) The cell envelope fractions of French press lysates of the pYV-cured strain CE1525 with pEW4 (yscWΔlip) and pSM3km (yscC) (B) and that of the wild-type strain KNG22703 with pEW4 (yscWΔlip) (C) were applied onto 30 to 55% sucrose gradients, which were centrifuged and fractionated. Samples from all fractions were analyzed by SDS-PAGE on 3 to 9% polyacrylamide gradient gels. YscC oligomers were detected by silver staining. The fractions that represent the inner (dotted line) and outer membranes (continuous line) were identified based on NADH-oxidase activity and the presence of the porins, respectively. (D) Proteins from whole cells of the pYV-cured strain CE1525 bearing either plasmid pRS6 (yscW) (lane 1), plasmids pRS6 (yscW) and pSM3km (yscC) (lane 2), plasmid pEW4 (yscWΔlip) (lane 3), or plasmids pEW4 (yscWΔlip) and pSM3km (yscC) (lane 4) and the Y. enterocolitica yscW mutant KNG22703(pRS227) with either plasmid pRS6 (yscW) (lane 5) or plasmid pEW4 (yscWΔlip) (lane 6) were separated on a 14% polyacrylamide gel, and YscW was immunodetected on Western blots. The same amount of total cell extracts was loaded in each lane. The strains were grown at 37°C in BHI-OX with IPTG to induce yscC and/or yscWΔlip expression.
DISCUSSION
The biogenesis of several secretins is assisted by small outer-membrane-associated lipoproteins, which are termed pilot proteins. A mutant of Y. enterocolitica lacking the outer membrane lipoprotein YscW, showed reduced protein secretion via the type III pathway and had a lower yield of the YscC secretin. Therefore, it was proposed that YscW might belong to the family of pilot proteins (23), even though the protein does not show any sequence homology with known pilot proteins. However, the sequence homology within this group of proteins is in general very low. We show here that coexpression of yscW increased the yield of YscC oligomers. Furthermore, the YscW protein was required for outer membrane localization of YscC. Since these results were obtained in a strain expressing no other components of the Ysc apparatus, YscW appears to exert its effect directly on the secretin and, therefore, the results confirm the proposed role of YscW as a pilot protein for YscC.
The nonconserved C terminus of various secretins has been identified as the pilot-binding site. Fusing this part of PulD (12) or InvG (13) to the pilot-independent pIV secretin of filamentous phage rendered the resulting hybrid proteins dependent on the cognate pilot proteins in order to function in phage assembly. We show here that the reverse is also true. By deleting the C-terminal 96 amino acids of YscC, the secretin became independent of YscW for targeting to the outer membrane. Thus, YscC intrinsically contains the information that is required for its outer membrane localization. Since YscCΔC-term restored Yop secretion in a Y. enterocolitica yscC mutant to almost wild-type levels, it appears that the extreme C terminus is also not essential for the function of the secretin in type III secretion. Similarly, a C terminally truncated OutD derivative partially restored type II secretion in an Erwinia chrysanthemi outD mutant (36). Deletion of the C terminus did result in a highly unstable protein, which, surprisingly, was still stabilized by YscW. This result shows that the C-terminal domain of YscC is not essential for the interaction with its pilot protein. Altogether, these findings imply that an important function of the C terminus is to render YscC dependent on YscW for correct localization. Mechanistically, the C terminus might shield a domain that facilitates transfer of YscC to the outer membrane. An interaction with YscW may induce a conformational change in YscC, thereby relieving the adverse effect of the C terminus. The biological advantage of the YscW-dependent localization of the YscC protein remains speculative, but it is possible that the pilot protein improves the efficiency of YscC biogenesis or facilitates interactions with other Ysc components. Noteworthy in this respect is the observation that some secretins appear to be pilot-independent. Moreover, although the PscC secretin of the type III system in Pseudomonas aeruginosa and YscC are very homologous, the putative yscW homolog in this system, exsB, is very likely unexpressed (16). In this system, a different protein might have taken over the function of YscW or, alternatively, PscC does not require a pilot protein to be functional.
To obtain more insight into the role of the YscW protein in the assembly of the secretin, we also investigated the kinetics of YscC oligomer formation in pulse-chase experiments. The time course of oligomerization of secretins has to our knowledge not been described before and was found to be a relatively slow process. Although some oligomers were already detected directly after the pulse, it took ∼30 min before all pulse-labeled monomers were converted into oligomers. In comparison, studies on porins describe the conversion into stable trimers already within a 30-s pulse period, although a slower pathway, requiring several minutes for completion, has also been observed (21, 33, 41). We did not identify assembly intermediates, such as dimers and trimers. Possibly, oligomeric intermediates dissociate into monomers during the procedure or they only exist for a very short time span. Alternatively, the formation of the oligomer is a coordinated process, requiring all subunits simultaneously. When the pulse-chase experiments were performed in the absence of the pilot protein YscW, the time course of oligomer formation was delayed, which demonstrates a direct or indirect role for the pilot protein in stimulating the oligomerization of the secretin. This result was consistent with the data obtained from the steady-state experiments, which showed an increase in the amount of monomeric YscC in the absence of the pilot protein. We can therefore conclude that YscW interacts with the YscC protein prior to oligomerization and that its function is likely not to delay oligomerization to enable the protein to reach its destination as a monomer. In the absence of YscW, the final yield of YscC oligomers formed during the chase was also strongly reduced, a finding consistent with its function in stabilization. However, the monomer was not very unstable in the absence of YscW and remained detectable for several hours after the pulse. This observation is not in agreement with the possibility that the pilot functions to protect the monomer from proteolytic degradation prior to its assembly into the oligomer. Also, the oligomers formed in the absence of YscW were not particularly unstable. We assume therefore that the reduction in the total yield of YscC during the chase is due to a slow decay of the unassembled protein. Thus, the stabilizing role of YscW is probably an indirect consequence of the more efficient oligomerization of YscC in the presence of YscW.
By constructing a nonlipidated YscW variant, we established that outer membrane targeting of the pilot protein is essential for its function. It has been reported in other studies that similar variants of other pilot proteins retain their ability to stabilize their cognate secretin, although they can no longer localize it to the outer membrane (19, 37), thereby showing an uncoupling of the two functions. In contrast, unlipidated YscW did not increase the amount of YscC oligomer. However, YscWΔlip could not be detected in the cell when coexpressed with YscC, which might explain the inability of the pilot protein to increase the yield of oligomeric YscC. We have considered the possibility that mislocalization of YscC induces a periplasmic stress response via the σE or Cpx pathway, leading to an increased expression of proteases. However, the production of YscC in combination with YscWΔlip did not lead to an increased production of the periplasmic chaperone Skp (unpublished results), which is known to be a component of both pathways (14). Possibly, an interaction between YscC and YscW induces a conformational change in the pilot protein, leading to increased sensitivity of the YscWΔlip to proteases. Despite the fact that YscWΔlip was proteolytically degraded in the presence of YscC, it was still capable of interacting with and mislocalizing YscC even in the presence of lipidated YscW. This result suggests that the mutant YscW, because of its periplasmic location, could interact earlier with the YscC secretin than the native YscW protein, which localizes to the outer membrane. Consequently, outer membrane localization of the pilot protein and the secretin may be successive instead of simultaneous events.
Although the precise function of the YscW protein in secretin assembly is still not understood, a number of important conclusions can be drawn from the experiments. First, the pilot protein does not delay oligomerization to increase the time span for the YscC monomer to reach the outer membrane. In contrast, YscW promotes formation of the oligomeric form of YscC and thus interacts with the YscC protein prior to its oligomerization. Second, the pilot protein does not act by preventing proteolytic degradation of unassembled monomers prior to their oligomerization, but the stabilizing function of YscW appears to be the consequence of efficient oligomerization. Third, association of YscW with the outer membrane is a prerequisite for its function in the biogenesis of YscC. It is thus possible that it only indirectly promotes oligomerization, e.g., the secretin may remain associated with the inner membrane until it interacts with YscW in the outer membrane, where its oligomerization can be induced by an interaction with another outer membrane component, such as lipopolysaccharide or Omp85. The role of the C terminus of the secretin might be to prevent detachment from the inner membrane until another part of the protein interacts with the pilot protein.
Acknowledgments
This research was supported by grant 700-97-012 from the Council for Chemical Sciences of The Netherlands Organization of Scientific Research and by grant HPRN-CT-2000-00075 from the European Community.
REFERENCES
- 1.Allaoui, A., R. Scheen, C. Lambert de Rouvroit, and G. R. Cornelis. 1995. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to exsB of Pseudomonas aeruginosa. J. Bacteriol. 177**:**4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitter, W. 2003. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch. Microbiol. 179**:**307-314. [DOI] [PubMed] [Google Scholar]
- 3.Brissette, J. L., and M. Russel. 1990. Secretion and membrane integration of a filamentous phage-encoded morphogenetic protein. J. Mol. Biol. 211**:**565-580. [DOI] [PubMed] [Google Scholar]
- 4.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294**:**1169-1179. [DOI] [PubMed] [Google Scholar]
- 5.Burghout, P., R. van Boxtel, P. Van Gelder, S. A. Müller, J. Tommassen, and M. Koster. 2004. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 186**:**4645-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24**:**757-765. [DOI] [PubMed] [Google Scholar]
- 7.China, B., T. Michiels, and G. R. Cornelis. 1990. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol. Microbiol. 4**:**1585-1593. [DOI] [PubMed] [Google Scholar]
- 8.Conchas, R. F., and E. Carniel. 1990. A highly efficient electroporation system for transformation of Yersinia. Gene 87**:**133-137. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87**:**285-291. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G., J. C. Vanootegem, and C. Sluiters. 1987. Transcription of the yop regulon from Y. enterocolitica requires _trans_-acting pYV and chromosomal genes. Microb. Pathog. 2**:**367-379. [DOI] [PubMed] [Google Scholar]
- 11.Crago, A. M., and V. Koronakis. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30**:**47-56. [DOI] [PubMed] [Google Scholar]
- 12.Daefler, S., I. Guilvout, K. R. Hardie, A. P. Pugsley, and M. Russel. 1997. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol. Microbiol. 24**:**465-475. [DOI] [PubMed] [Google Scholar]
- 13.Daefler, S., and M. Russel. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28**:**1367-1380. [DOI] [PubMed] [Google Scholar]
- 14.DiGiuseppe, P. A., and T. J. Silhavy. 2004. Pushing the envelope: lessons learned from stressing bacteria. ASM News 70**:**71-79. [Google Scholar]
- 15.Genin, S., and C. A. Boucher. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243**:**112-118. [DOI] [PubMed] [Google Scholar]
- 16.Goranson, J., and D. W. Frank. 1996. Genetic analysis of exoenzyme S expression by Pseudomonas aeruginosa. FEMS Microbiol. Lett. 135**:**149-155. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166**:**557-580. [DOI] [PubMed] [Google Scholar]
- 18.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15**:**978-988. [PMC free article] [PubMed] [Google Scholar]
- 19.Hardie, K. R., A. Seydel, I. Guilvout, and A. P. Pugsley. 1996. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol. 22**:**967-976. [DOI] [PubMed] [Google Scholar]
- 20.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62**:**379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen, C., M. Heutink, J. Tommassen, and H. de Cock. 2000. The assembly pathway of outer membrane protein PhoE of Escherichia coli. Eur. J. Biochem. 267**:**3792-3800. [DOI] [PubMed] [Google Scholar]
- 22.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109**:**137-141. [DOI] [PubMed] [Google Scholar]
- 23.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26**:**789-797. [DOI] [PubMed] [Google Scholar]
- 24.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166**:**175-176. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154**:**367-382. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227**:**680-685. [DOI] [PubMed] [Google Scholar]
- 27.Laroche, Y., M. van Bouchaute, and G. Cornelis. 1984. A restriction map of virulence plasmid pVYE439-80 from a serogroup 9 Yersinia enterocolitica strain. Plasmid 12**:**67-70. [DOI] [PubMed] [Google Scholar]
- 28.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117**:**307-310. [DOI] [PubMed] [Google Scholar]
- 29.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96**:**8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium: isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247**:**3962-3972. [PubMed] [Google Scholar]
- 31.Pettersson, J., R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson, and H. Wolf Watz. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273**:**1231-1233. [DOI] [PubMed] [Google Scholar]
- 32.Rapoza, M. P., and R. E. Webster. 1993. The filamentous bacteriophage assembly proteins require the bacterial SecA protein for correct localization to the membrane. J. Bacteriol. 175**:**1856-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid, J., H. Fung, K. Gehring, P. E. Klebba, and H. Nikaido. 1988. Targeting of porin to the outer membrane of Escherichia coli: rate of trimer assembly and identification of a dimer intermediate. J. Biol. Chem. 263**:**7753-7759. [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schuch, R., and A. T. Maurelli. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 183**:**6991-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevchik, V. E., J. Robert-Baudouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16**:**3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shevchik, V. E., and G. Condemine. 1998. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology 144**:**3219-3228. [DOI] [PubMed] [Google Scholar]
- 38.Simon, R., V. Priefer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1**:**784-791. [Google Scholar]
- 39.Struyvé, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218**:**141-148. [DOI] [PubMed] [Google Scholar]
- 40.Tommassen, J., H. van Tol, and B. Lugtenberg. 1983. The ultimate localization of an outer membrane protein of Escherichia coli K-12 is not determined by the signal sequence. EMBO J. 2**:**1275-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vos-Scheperkeuter, G. H., and B. Witholt. 1984. Assembly pathway of newly synthesized LamB protein an outer membrane protein of Escherichia coli K-12. J. Mol. Biol. 175**:**511-528. [DOI] [PubMed] [Google Scholar]
- 42.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299**:**262-265. [DOI] [PubMed] [Google Scholar]
- 43.Zink, D. L., J. C. Feeley, J. G. Wells, C. Vanderzant, J. C. Vickery, W. D. Roof, and G. A. O'Donovan. 1980. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature 283**:**224-226. [DOI] [PubMed] [Google Scholar]