The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL (original) (raw)
Some nuclear bodies are formed with an architectural RNA (arcRNA) as the structural core. Here, Mannen et al. screened for new nuclear bodies built on unidentified arcRNAs and found that the Sam68 nuclear body (SNB) is composed of two distinct RNase-sensitive substructures. HNRNPL acts as the adaptor to combine the two substructures, thus forming the full SNB.
Abstract
The mammalian cell nucleus contains membraneless suborganelles referred to as nuclear bodies (NBs). Some NBs are formed with an architectural RNA (arcRNA) as the structural core. Here, we searched for new NBs that are built on unidentified arcRNAs by screening for ribonuclease (RNase)-sensitive NBs using 32,651 fluorescently tagged human cDNA clones. We identified 32 tagged proteins that required RNA for their localization in distinct nuclear foci. Among them, seven RNA-binding proteins commonly localized in the Sam68 nuclear body (SNB), which was disrupted by RNase treatment. Knockdown of each SNB protein revealed that SNBs are composed of two distinct RNase-sensitive substructures. One substructure is present as a distinct NB, termed the DBC1 body, in certain conditions, and the more dynamic substructure including Sam68 joins to form the intact SNB. HNRNPL acts as the adaptor to combine the two substructures and form the intact SNB through the interaction of two sets of RNA recognition motifs with the putative arcRNAs in the respective substructures.
Introduction
The mammalian cell nucleus is highly organized and composed of multiple distinct structures called nuclear bodies (NBs). NBs are subnuclear membrane-less granular structures that contain various proteins and RNA factors, many of which function as the sites of the biogenesis, storage, and sequestration of specific RNAs, proteins, and ribonucleoprotein (RNP) complexes (Mao et al., 2011; Sleeman and Trinkle-Mulcahy, 2014). NBs can also serve as scaffolds to epigenetically regulate the activity of specific chromosome loci that attach to them (Wang et al., 2004; Dundr et al., 2007; McStay and Grummt, 2008; West et al., 2014). In general, NBs likely serve to concentrate proteins and RNAs involved in common processes to enhance reaction efficiency and facilitate regulation or sequestrate factors to regulate the nucleoplasmic concentration of the active factors.
The specificity of the proteins incorporated into NBs can be reflected by the affinity with the core molecules of the respective NBs. Recent studies revealed that some NBs are constructed using specific long noncoding RNAs (lncRNAs) as their scaffolds (Chujo et al., 2016). Therefore, lncRNA-dependent NBs are composed of numerous RNA-binding proteins. The most remarkable example is paraspeckles, which were initially defined as foci in close proximity to nuclear speckles and are enriched in characteristic RNA-binding proteins (Fox et al., 2002). Nuclear paraspeckle assembly transcript 1 (NEAT1), a lncRNA, localizes exclusively to paraspeckles and acts as an essential structural component of these massive RNP complexes (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). Paraspeckles regulate the expression of a number of genes via the sequestration of specific proteins and RNAs (Prasanth et al., 2005; Hirose et al., 2014; Imamura et al., 2014) and are physiologically involved in the development of the corpus luteum and the mammary gland in mice (Nakagawa et al., 2014; Standaert et al., 2014). Paraspeckle formation is initiated by NEAT1 transcription at the NEAT1 locus on human chromosome 11 and proceeds in conjunction with the biogenesis of NEAT1 lncRNA with >40 proteins. Among 40 paraspeckle proteins, only seven RNA-binding proteins are essential for paraspeckle formation (Naganuma et al., 2012). These essential paraspeckle proteins contain the characteristic low-complexity domain (or prion-like domain [PLD]) (Yamazaki and Hirose, 2015). Recently, the PLD was shown to accelerate formation of hydrogels and liquid-liquid phase separation (LLPS) in vitro (Kato et al., 2012; Burke et al., 2015; Molliex et al., 2015; Patel et al., 2015). We recently demonstrated that the PLD of at least two paraspeckle proteins is essential for formation of the paraspeckle structure in vivo (Hennig et al., 2015).
It has been proposed that the class of lncRNAs functioning as the scaffold of NBs be designated architectural RNAs (arcRNAs) (Clemson et al., 2009; Chujo et al., 2016). Presently, four lncRNAs are classified as arcRNAs in addition to NEAT1, namely, intergenic spacer lncRNAs in the nucleolar detention center (Audas et al., 2012), human satellite III lncRNA in the nuclear stress body (Biamonti and Vourc’h, 2010), Drosophila melanogaster heat shock RNA omega in the omega speckle (Prasanth et al., 2000), and fission yeast meiRNA in the Mei2 dot (Watanabe and Yamamoto, 1994). Thus, arcRNAs are widely used in eukaryotes, from mammals to insects and yeast. It is expected that more lncRNAs with an architectural function remain uncharacterized under various conditions.
Among the identified NBs, several contain unidentified RNAs. For example, the Sam68 nuclear body (SNB) was originally discovered as the NB that is usually observed in the perinucleolar region (Chen et al., 1999). The SNB contains Sam68 (Src associated in mitosis of 68-kD protein, or KHDRBS1), which is a member of the signal transduction and activation of RNA family of RNA-binding proteins characterized by an HNRNP K homology (KH) domain embedded in a highly conserved region called the GRP33/Sam68/GLD1 (GSG) domain (Jones and Schedl, 1995). Electron microscopy studies showed that SNBs contained nucleic acids, which are likely RNA, and targeting of Sam68 to SNBs involves the GSG domain (Chen et al., 1999). As additional SNB components, three RNA-binding proteins have been identified: two Sam68-like mammalian proteins, SLM1 and SLM2 with a KH domain, which are predominantly expressed in neuronal cells, and HNRNPL, with four RNA recognition motifs (RRMs; Chen et al., 1999; Rajan et al., 2009).
Here, we searched for new arcRNA-dependent NBs by screening nuclear foci whose structures are disintegrated by RNase treatment. In our previous study, we used a Venus-tagged human full-length cDNA library (32,651 clones) and obtained 571 cDNA clones whose products (463 proteins) localize in certain nuclear foci (Hirose and Goshima, 2015). In this study, we investigated which nuclear foci disappeared after RNase treatment to select candidates of RNase-sensitive nuclear foci that potentially contain arcRNAs. Here, we show that the SNB is an RNase-sensitive NB composed of two distinct RNase-sensitive substructures. We identified additional protein components of the SNB, and characterization of each protein revealed that Sam68 and HNRNPD are essential components of the SNB. Detailed domain dissection revealed that the RNA-binding domains of Sam68 and HNRNPD as well as the PLD of HNRNPD play significant roles in SNB formation and that HNRNPL acts as the adaptor to combine the two substructures to form the intact SNB through interacting with RNA molecules residing in each substructure.
Results
Identification of the SNB as an RNase-sensitive NB
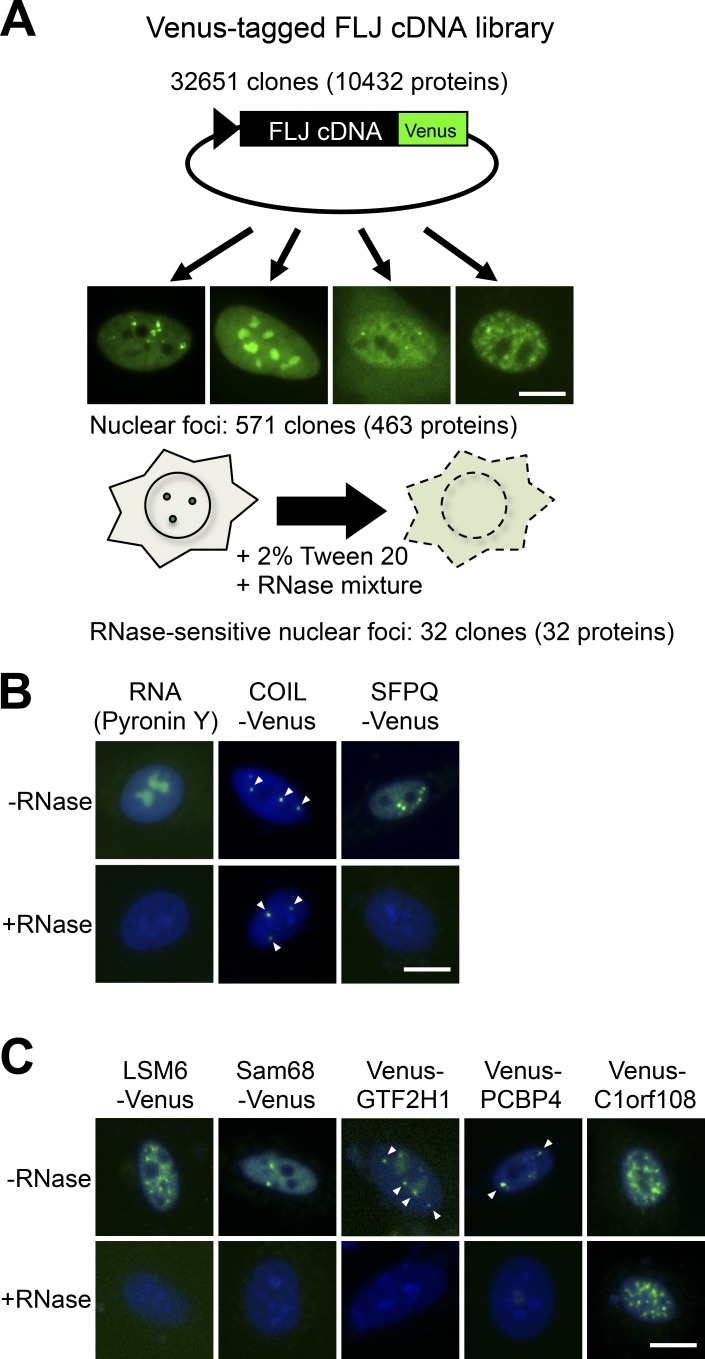
We attempted to newly identify NBs that are disrupted by RNase treatment. We used the fluorescently tagged human full-length cDNA library (32,651 clones encoding 10,432 proteins) that we previously used to identify novel paraspeckle proteins by colocalization screening (Hirose and Goshima, 2015). During the previous screening, we detected 571 human cDNA clones whose protein products (463 proteins) localize to some nuclear foci in HeLa cells (Naganuma et al., 2012). In this experiment, we investigated whether each of the nuclear foci marked by 463 cDNA products were disrupted by RNase treatment (Fig. 1 A). HeLa cells transfected with the cDNA plasmids were first permeabilized with Tween-20 followed by treatment with RNase mixtures (Fig. 1 A). If the nuclear foci visualized by Venus signals disappear after RNase treatment, it is possible that these nuclear foci are RNase-sensitive structures. RNase treatment sufficiently degraded cellular RNAs, as confirmed by staining with Pyronin Y, and it also disrupted paraspeckles marked by SFPQ-Venus, which are arcRNA-dependent NBs, but not Cajal bodies marked by COIL-Venus, which are RNase-resistant NBs (Fox et al., 2005; Sasaki et al., 2009; Naganuma et al., 2012; Fig. 1 B). The nuclear foci marked by 32 Venus-tagged proteins appeared to be disrupted by RNase treatment (Table 1).
Figure 1.
RNase sensitivity screening of NBs. (A) Brief procedure of the screening. Venus-tagged human FLJ cDNA clones were transfected into HeLa cells. cDNA clones whose products localized to certain nuclear foci were selected. Subsequently, the RNase sensitivity of the nuclear foci labeled by Venus was investigated. To this end, the cells were permeabilized with 2% Tween-20, followed by treatment with an RNase mixture. (B) Control experiment (Pyronin Y) and examples of RNase-resistant NBs (Cajal bodies marked by COIL-Venus) and RNase-sensitive NBs (paraspeckles marked by SFPQ-Venus). (C) Demonstration of the RNase-sensitive localization of four Venus-tagged proteins in nuclear foci selected from Table 1. Venus-C1orf108 is an example of RNase-resistant localization in nuclear foci. Arrowheads indicate nuclear foci. Bars, 10 µm.
Table 1. Nuclear body proteins identified by RNase sensitivity screening.
| Nuclear body and protein | RNA-binding domains | Other domains or motifs | Colocalized markers | Novel | Low-complexity domains | Prion-like domains | Disassembly rate |
|---|---|---|---|---|---|---|---|
| % | |||||||
| Paraspeckle | |||||||
| CPSF7 | RRM | - | PSPC1 | No | Yes | No | 12 |
| ELAVL1 | 3 RRMs | - | PSPC1 | Yes | No | No | 7 |
| FUS | RRM | ZnF_RanBP2 | PSPC1 | No | Yes | Yes | 19 |
| HNRNPA1 | 2 RRMs | - | PSPC1 | No | Yes | Yes | 5 |
| HNRNPUL1 | - | SAP, SPRY | PSPC1 | No | Yes | Yes | 3 |
| PSPC1 | 2 RRMs | - | PSPC1 | No | Yes | Yes | 8 |
| RBFOX2 | RRM | - | PSPC1 | Yes | Yes | No | 16 |
| RBM7 | RRM | - | PSPC1 | No | Yes | No | 6 |
| RBM14 | 2 RRMs | - | PSPC1 | No | Yes | Yes | 2 |
| RRM | RS | PSPC1 | No | Yes | No | 11 | |
| TARDBP | 2 RRMs | - | PSPC1 | No | Yes | Yes | 4 |
| Nuclear speckle | |||||||
| FYTTD1 | - | UAP56-binding | SRSF2/SC35 | No | No | No | 4 |
| LSM6 | - | - | SRSF2/SC35 | Yes | No | No | 0 |
| RBM5 | 2 RRMs | G-patch, ZnF_RanBP2, ZnF_C2H2 | SRSF2/SC35 | No | Yes | Yes | 17 |
| SMNDC1 | - | Tudor | SRSF2/SC35 | No | Yes | No | 2 |
| SNRPE | - | - | SRSF2/SC35 | No | Yes | No | 18 |
| SRSF1 | 2 RRMs | RS | SRSF2/SC35 | No | Yes | No | 3 |
| SRSF7 | RRM | RS, ZnF_CCHC | SRSF2/SC35 | No | Yes | No | 5 |
| YTHDC1 | - | YTH, Coiled_coil | SRSF2/SC35 | No | Yes | No | 9 |
| ZC3H11A | - | Coiled_coil, 3 ZnF_C3H1 | SRSF2/SC35 | No | Yes | No | 18 |
| SNB | |||||||
| CCAR2/DBC1 | - | Coiled_coil | KHDRBS1/Sam68 | Yes | Yes | No | 17 |
| KHDRBS1/Sam68 | KH | - | KHDRBS1/Sam68 | No | Yes | No | 4 |
| HNRNPD | 2 RRMs | - | KHDRBS1/Sam68 | Yes | Yes | Yes | 15 |
| STRBP | 2 DSRM | DZF | KHDRBS1/Sam68 | Yes | Yes | No | 4 |
| ZMAT4 | - | 4 ZnF_C2H2 | KHDRBS1/Sam68 | Yes | No | No | 13 |
| ZNF346 | - | 4 ZnF_C2H2 | KHDRBS1/Sam68 | Yes | No | No | 11 |
| ZNF385B | - | 4 ZnF_C2H2 | KHDRBS1/Sam68 | Yes | Yes | No | 0 |
| Cajal body | |||||||
| GTF2H1 | - | 2 BSD | COIL | No | Yes | No | 14 |
| SNRPB2 | 2 RRMs | - | COIL | No | Yes | Yes | 10 |
| Centromere | |||||||
| ZNF549 | - | 15 ZnF_C2H2 | CENPB | Yes | No | No | 9 |
| Unknown | |||||||
| C1orf147 | - | - | - | Yes | No | No | 13 |
| PCBP4 | 3 KH | - | - | Yes | Yes | No | 5 |
Many of the selected RNase-sensitive nuclear foci overlapped with known NBs. In addition to nine known components, two proteins (ELAVL1 and RBFOX2) were selected as potentially localizing to paraspeckles (Table 1). The localization of nine proteins to nuclear speckles was RNase-sensitive (Table 1 and Fig. 1 C). The nuclear speckle itself marked by SC35 is not an RNA-dependent NB (Sytnikova et al., 2011), which suggests that some RNA molecules bridge these nine proteins with nuclear speckles. Two Cajal body–localized proteins as well as a centromere-localized protein also require RNAs for their localization in the respective structures, which are not RNase-sensitive (Table 1 and Fig. 1 C). We discovered seven proteins that localized to SNBs whose signals commonly disappeared after RNase treatment (Table 1 and Fig. 1 C). This strongly suggests that the SNB itself is an RNase-sensitive NB. Except for KHDRBS1/Sam68, the SNB-localized proteins found were all unidentified as SNB components before this work. The two remaining proteins (C1orf147 and PCBP4) localized in unidentified nuclear foci (Table 1 and Fig. 1 C); however, the localization of the endogenous proteins in the same foci remains to be investigated because antibodies against these proteins are not available. In the next part of this study, we focused on the SNB to shed light on its structure and functions.
SNB formation requires two RNA-binding proteins
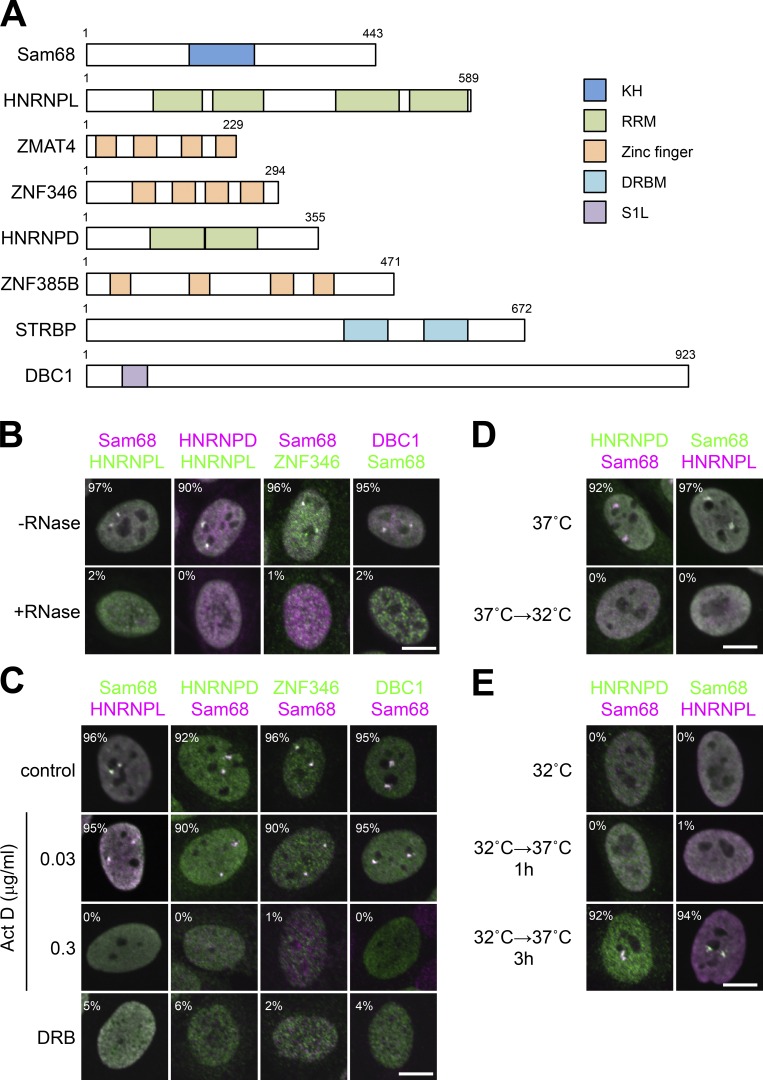
All the annotated SNB-localized proteins possess putative RNA-binding domains such as a KH domain, an RRM, a zinc finger, a double-stranded RNA-binding domain, and an S1-like domain (S1L; Fig. 2 A). We confirmed the colocalization of each of the SNB-localized proteins (HNRNPD, deleted in breast cancer 1 [DBC1], and ZNF346) and a previously identified SNB component, Sam68 or HNRNPL (Rajan et al., 2009; Fig. 2 B). For the remaining three SNB proteins (ZMAT4, ZNF385B, and STRBP), SNB localization was supported only by detection of Venus signals (Fig. S1 A) because they were poorly expressed and hardly detectable in HeLa cells by Western blotting (Fig. S1 B). The focal signals of all five SNB proteins synchronously disappeared upon RNase treatment (Fig. 2 B, bottom) or treatment with the transcription inhibitors actinomycin D (at a concentration of 0.3 µg/ml but not 0.03 µg/ml) or 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Fig. 2 C). These data strongly suggest that the SNB itself is an RNase-sensitive structure that is formed with RNA polymerase II (RNAPII) transcripts.
Figure 2.
Components and features of the SNB. (A) RNA-binding domains of SNB components. Numbers represent the amino acid counts of each protein. (B) Immunofluorescence analysis of SNB components and the effect of RNase treatment. Five SNB components colocalized in SNBs, and all protein signals dispersed upon RNase treatment (+RNase). (C) SNB formation requires RNAPII transcription. Immunofluorescence analysis of SNB components was performed in HeLa cells treated with actinomycin D (Act D; 0.03 and 0.3 µg/ml) or DRB (100 µM). The control cells were treated with DMSO. (D and E) A temperature shift reversibly regulates SNB formation. The temperature for cell culture was shifted from 37°C to 32°C, and cells were incubated for 24 h (D). The cells were incubated at 32°C for 24 h, shifted to 37°C, and incubated for 1 or 3 h (E). SNBs were detected by immunofluorescence labeling of Sam68, HNRNPD, and HNRNPL. The cell populations (%) in which each protein and Sam68 or HNRNPL signals overlap are shown in B, C, D, and E (>100 cells, n = 3). Bars, 10 µm.
To monitor SNBs, we performed temperature shifts that reportedly affect certain subnuclear structures (Liu and Dreyfuss, 1996; Biamonti, 2004). SNBs disappeared upon a temperature shift from 37 to 32°C for 24 h (Fig. 2 D). SNBs reformed when the temperature was returned to 37°C for 3 h (Fig. 2 E). The levels of all detected SNB proteins remained constant during these temperature shifts (Fig. S1 C). We confirmed that the disintegrated SNB components likely diffused to the nucleoplasm and did not relocalize to other NBs (Fig. S1 D).
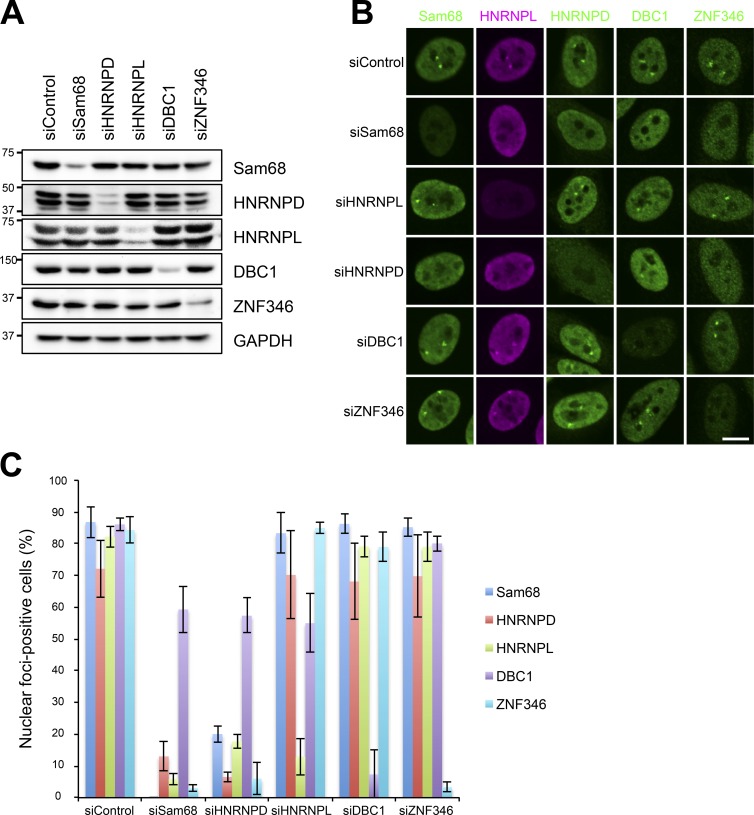
To investigate how the SNB forms with the identified components, we performed reciprocal depletion of each of the five SNB components (Sam68, HNRNPL, HNRNPD, DBC1, and ZNF346) by RNAi in HeLa cells (Fig. 3 A). Immunofluorescence analysis of the five SNB components in siRNA-treated cells revealed that depletion of Sam68 or HNRNPD resulted in the disappearance of SNBs (see siSam68 and siHNRNPD in Fig. 3, B and C), whereas depletion of the three other proteins (HNRNPL, DBC1, and ZNF346) hardly affected the integrity of SNBs (see siHNRNPL, siDBC1, and siZNF346 in Fig. 3, B and C). It should be noted that the focal signal of DBC1 was not dramatically reduced upon depletion of Sam68 or HNRNPD (discussed in the section SNB is composed of two distinct RNase-sensitive substructures). These results indicate that Sam68 and HNRNPD are both essential components for formation of the RNase-sensitive SNB structure in HeLa cells, although we cannot rule out the possibility that unidentified core factors are present in SNBs.
Figure 3.
Identification of the essential SNB components. (A and B) Five SNB components were reciprocally knocked down by RNAi. The efficient and specific knockdown of each protein by RNAi was detected by Western blotting (A). The molecular mass marker (kD) is shown on the left. Each protein was detected by immunofluorescence (B). (C) Quantitation of nuclear foci–positive cells in the experiments shown in B (>100 cells, ±SD, n = 3). Bar, 10 µm.
SNB formation requires both the RRMs and PLD of HNRNPD
The functional domains of Sam68 were dissected previously, and the GSG domain was identified to be responsible for its SNB localization (Chen et al., 1999). We constructed a series of deletion mutants of Sam68 fused with Venus and then investigated whether each Sam68 mutant was able to colocalize with endogenous HNRNPL in SNBs (Fig. S2 A). We confirmed that the GSG domain, particularly the N-terminal-to-KH-domain region (NK region) and the RNA-binding KH domain but not the C-terminal-to-KH-domain region (CK region) in this domain, was required for SNB localization (Fig. S2 B). Deletion of the C-terminal region (ΔCT) resulted in predominant mislocalization in the cytoplasm; therefore, the nuclear localization signal (NLS; Ishidate et al., 1997) was retained in ΔCT+NLS, which properly localized in SNBs (Fig. S2 B). We also investigated the rescue activities of a series of Sam68 mutants by transfecting them into HeLa cells in which endogenous Sam68 had been depleted by RNAi. Counting the numbers of rescued cells in which SNBs were detectable (using HNRNPL as a marker) revealed that the rescue activity of the mutants correlated with the ability to localize to SNBs, with ΔGSG, ΔNK, ΔKH, and ΔCT lacking the rescue ability (Fig. S2 C).
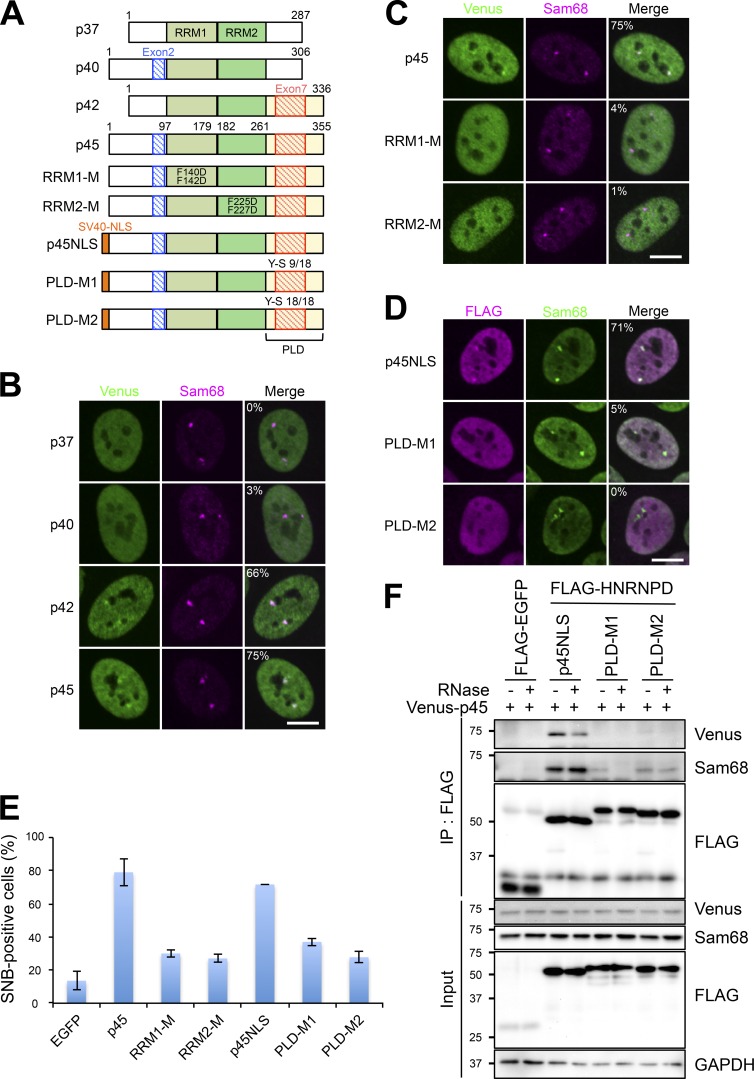
We next attempted to dissect the functional domains of another essential SNB factor, HNRNPD. There are four isoforms of HNRNPD (p37, p40, p42, and p45) synthesized by alternative pre-mRNA splicing of exon 2 and 7 (Fig. 4 A; Zucconi et al., 2010). Venus-tagged constructs of the four HNRNPD isoforms were transfected into HeLa cells. All four isoforms localized in the nucleoplasm and only p42 and p45 were prominently enriched in SNBs (Fig. 4 B). The siRNAs against the common exons (siExon 4 and siExon 5), one of which was used in Fig. 3, depleted all four isoforms (Fig. S3, A and B) and disrupted SNBs (Fig. 3). Even the siRNAs against exon 7 (siExon 7A and siExon 7B), which specifically depleted p42 and p45 but not p37 or p40 (Fig. S3, A and B), sufficiently disrupted SNBs (Fig. S3 C). Furthermore, p42 and p45 had more potent rescue abilities for SNB formation (Fig. S3 D). These results indicate that the isoforms containing exon 7 (p42 and p45) localize in SNBs and function in their formation. It should be noted that p37 and p40 also possessed weak rescue abilities (Fig. S3 D). Coimmunoprecipitation (coIP) with FLAG-tagged HNRNPD isoforms indicated that p42 and p45 were efficiently coimmunoprecipitated with Sam68, HNRNPL, and DBC1 (Fig. S3 E).
Figure 4.
The RNA-binding domain and PLD of HNRNPD are required for SNB formation. (A) Schematics of all the HNRNPD constructs used in the rest of the figure. Four isoforms (p37, p40, p42, and p45) are shown above. p45 is the standard isoform construct corresponding to HNRNPD shown in Fig. 2 A. The alternatively selected exons 2 and 7 are shown in blue and red, respectively. Schematics of two RRM mutants (RRM1-M and RRM2-M), in which two phenylalanine residues in each of RRM1 and RRM2 (pale green and green boxes, respectively) were substituted with aspartic acid (F140D and F142D for RRM1-M and F225D and F227D for RRM2-M), are shown in the middle. The schematics of PLD mutants are shown at the bottom. Nine or 18 of the 18 tyrosine residues present in the PLD (yellow box) were mutated to serine residues (Y-S 9/18 and Y-S 18/18) to generate PLD-M1 and PLD-M2, respectively. The PLD mutation resulted in the cytoplasmic mislocalization of the mutant HNRNPDs; therefore, a SV40-NLS was inserted at the N terminus (orange box). (B) Isoform-specific localization of HNRNPD in SNBs. The Venus-tagged HNRNPD isoforms shown on the left were transfected and their localizations were detected. Sam68 was detected as the marker of SNBs. (C) RNA binding of HNRNPD through RRM1 and RRM2 is required for its SNB localization. Venus-tagged p45 and two RRM mutants were transfected, and their localizations were detected as in B. (D) The PLD is required for the SNB localization of HNRNPD. The FLAG-tagged HNRNPD isoforms were transfected, and their localizations were detected as in B. The cell populations (%) in which Venus or FLAG and Sam68 signals overlap are shown in the merge panels in B, C, and D (>100 cells, n = 3). (E) The RRMs and PLD are required for SNB formation. Rescue of the defect in SNB formation by the HNRNPD mutant constructs shown in A. The HNRNPD constructs were transfected into HeLa cells in which endogenous HNRNPD had been depleted by RNAi, and then SNB-positive cells (Sam68 foci–positive cells) were counted (>100 cells, ±SD, n = 3). As a negative control, the FLAG-EGFP plasmid was transfected (EGFP). (F) The PLD is required for protein–protein interactions of HNRNPD. The FLAG-tagged PLD mutants used in D were immunoprecipitated to detect the interaction with Sam68 and cotransfected Venus-tagged HNRNPD (p45). GAPDH was used as the input control. The molecular mass marker (kD) is shown on the left. Bars, 10 µm.
HNRNPD possesses two RRMs; therefore, we asked whether the RNA-binding activity of HNRNPD is required for its SNB localization. Based on Venus-p45 as the wild-type (WT) Venus-HNRNPD construct, two phenylalanine residues essential for RNA binding in each of RRM1 and RRM2 were mutated to aspartic acid to create RRM1-M (F140D/F142D) and RRM2-M (F225D/F227D; Fig. 4 A), and their localizations were detected by Venus signals (Fig. 4 C). Both RRM point mutants failed to localize to SNBs, indicating that RNA binding of HNRNPD through the two RRMs simultaneously is essential for its SNB localization. We also investigated the rescue activity of the RRM mutants for SNB formation in HNRNPD-depleted cells. The Flag-tagged HNRNPD constructs, including p45 (WT), RRM1-M, and RRM2-M, and an EGFP plasmid (as a control) were transfected into HeLa cells in which endogenous HNRNPD had been depleted by RNAi. Counting the numbers of SNB-positive cells revealed that both RRM mutants almost completely lacked the rescue activity for SNB formation (RRM1-M and RRM2-M in Fig. 4 E), indicating that the RNA binding of HNRNPD is required not only for SNB localization but also for SNB formation.
Exon 7, which is present in p42 and p45, is embedded in the PLD (Fig. 4 A). The PLD is glycine-, glutamine-, and tyrosine-rich and contributes to protein aggregation by forming hydrogel-like structures, which is critical for the formation of cellular bodies (Hennig et al., 2015). To examine whether the prion-like property of this domain is required for SNB formation, the tyrosine residues in the PLD of p45 were mutated to serine to create the partial Y-S mutant (PLD-M1: nine tyrosine residues were altered) and full Y-S mutant (PLD-M2: all 18 tyrosine residues were altered; Figs. 4 A and S3 F). Because PLD-M2 failed to localize in the nucleus, SV40-NLS was attached at the N terminus of all the constructs (Fig. 4 A) to examine the nuclear role of the PLD. As expected, both PLD mutants failed to localize to SNBs (Fig. 4 D). A plasmid rescue experiment showed that the PLD mutants had a significantly reduced ability to rescue the defect in SNB formation in HNRNPD-depleted cells (Fig. 4 E). CoIP with FLAG-tagged PLD mutants revealed that these mutants neither interacted with Sam68 nor cotransfected Venus-tagged p45 itself (Fig. 4 F). This indicates that the HNRNPD PLD is required for the interaction with Sam68 and for the homodimeric interaction of HNRNPD in SNBs.
SNB is composed of two distinct RNase-sensitive substructures
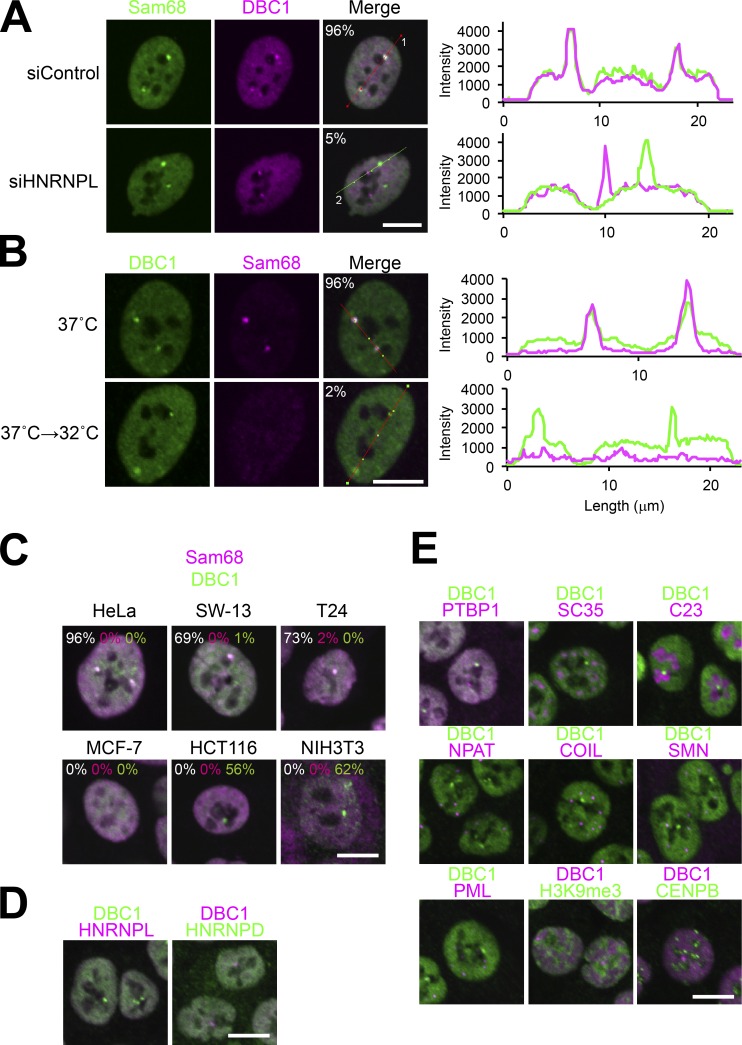
Unlike the immunostaining signals of the other SNB components, a substantial DBC1 signal was detected when an essential SNB component, either Sam68 or HNRNPD, was knocked down (Fig. 3). Furthermore, when HNRNPL was knocked down, we detected the DBC1 signal in nuclear foci distinct from those labeled with Sam68 and HNRNPD (Figs. 5 A and S4 A). Both nuclear foci (the Sam68 substructure and the DBC1 substructure) were sensitive to RNase treatment (Fig. S4 A), indicating that both require RNA molecules to maintain their structures.
Figure 5.
DBC1 is a component of a SNB substructure and a novel RNase-sensitive NB. (A) The SNB is separated into two distinct substructures upon HNRNPL depletion. Immunofluorescence analysis of Sam68 and DBC1 in control and HNRNPL-depleted (siHNRNPL) cells is shown on the left. Quantification of fluorescence by the line scan marked in the merge panel (marked as 1 and 2) is shown on the right. The green and pink lines correspond to the signals of Sam68 and DBC1, respectively. The cell populations (%) in which Sam68 and DBC1 signals overlap are shown in the merge panels (>100 cells, n = 3). (B) DBC1 foci are detectable under the cold shock conditions shown on the left. Immunofluorescence analysis was performed as in A. (C) Distinct DBC1 foci (DBC1 bodies) are detectable in other cell lines. Immunofluorescence analysis was performed to detect Sam68 and DBC1 in five human cell lines (HeLa, SW-13, T24, MCF-7, and HCT116) and a mouse cell line (NIH3T3). In the merge panels, the cell populations (%) in which Sam68 and DBC1 signals overlap are shown by white numbers, and those with nonoverlapping Sam68 and DBC1 signals are shown by magenta and green numbers, respectively (>100 cells, n = 3). (D) No other SNB components localize to the DBC1 body in HCT116 cells. Immunofluorescence analysis of HNRNPL and HNRNPD with DBC1 was performed in HCT116 cells. (E) The DBC1 body does not overlap with other known NBs in HCT116 cells. Immunofluorescence analysis of various NB markers with DBC1 was performed in HCT116 cells. Bars, 10 µm.
The DBC1 substructure was still detectable under the cold shock condition at 32°C, in which the Sam68 and HNRNPD focal signals disappeared (Fig. 5 B). This indicates that the Sam68 substructure within the SNB was selectively depleted and that the DBC1 substructure remained under the cold shock condition. It should be noted that the remaining DBC1 substructure was RNase sensitive (Fig. S4 B).
Through the detection of SNBs in various cultured cell lines, we found that SNBs in which Sam68 and DBC1 overlapped, as observed in HeLa cells, were detected in two additional cell lines (SW-13 and T24), and DBC1 foci, but not Sam68 foci, were detectable in specific cell lines such as human HCT116 and mouse NIH3T3 (Fig. 5 C). We confirmed that no other SNB components (HNRNPD and HNRNPL) localized in DBC1 foci (Fig. 5 D), which were RNase sensitive (Fig. S4 C). Our observations indicate that DBC1 foci are distinct NBs in these cell lines and also in certain physiological conditions, such as cold shock. To determine whether the distinct DBC1 foci in HCT116 cells overlapped with some of the known NBs, coimmunostaining was performed using antibodies against the marker proteins of known NBs. No known NB markers examined overlapped with DBC1 foci (Fig. 5 E), indicating that DBC1 foci are a novel RNase-sensitive NB, which we therefore termed the DBC1 body.
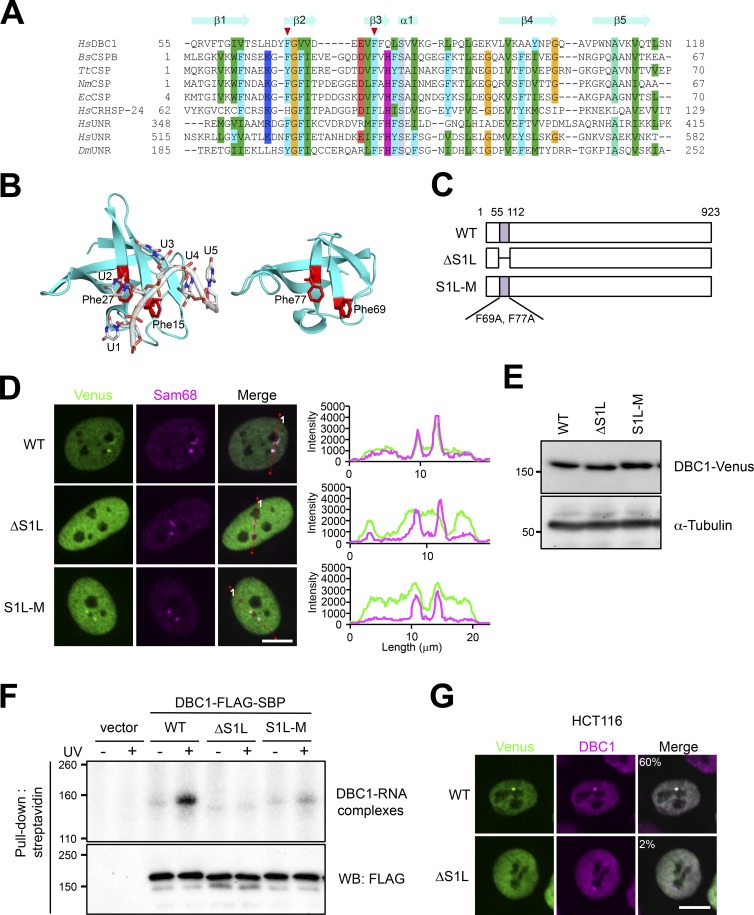
Previously, mRNA-bound proteome analysis showed the possible mRNA-binding ability of DBC1 (KIAA1967; Baltz et al., 2012). DBC1 possesses an S1L domain in the N-terminal region, which potentially acts as the RNA-binding domain (Anantharaman and Aravind, 2008). We asked whether SNB localization of DBC1 requires its RNA-binding activity. Using the structure of Bacillus subtilis CSPB (BsCSPB) complexed with oligo U RNA (Sachs et al., 2012) as the template, a homology model of the S1L domain of human DBC1 was constructed. Based on the sequence alignment and complex structure of BsCSPB, the highly conserved phenylalanines of DBC1 (Phe 69 and Phe 77) were chosen as putative RNA-interacting residues (Fig. 6, A and B). We constructed Venus-tagged mutant DBC1 lacking the S1L domain (ΔS1L) and in which the two phenylalanines were substituted with alanine (S1L-M; Fig. 6 C) and investigated the contribution of the S1L domain to its localization and RNA binding. Both ΔS1L and S1L-M were diffusely localized in the nucleoplasm, and a small fraction of S1L-M was still detectable in the SNB, but ΔS1L was undetectable in the SNB (Fig. 6 D). We confirmed that S1L mutations did not affect the expression level of DBC1 (Fig. 6 E). UV cross-linking pull-down of SBP-tagged DBC1 followed by the detection of bound RNAs with 32P labeling revealed that WT DBC1 was able to directly interact with RNA; however, the RNA-binding ability was almost completely abolished by the ΔS1L mutation and was markedly reduced by the S1L-M mutation (Fig. 6 F). Furthermore, the ΔS1L mutation abolished localization in the distinct DBC1 body in HCT116 cells (Fig. 6 G). These data suggest that the RNA binding of DBC1 is required for localization to the SNB in HeLa cells as well as the DBC1 body in HCT116 cells.
Figure 6.
DBC1 is a component of a SNB substructure and a novel RNase-sensitive NB. (A) Structure-based sequence alignment of the S1 domain. The mutated residues are indicated by red arrowheads above the alignment. HsDBC1, Homo sapiens DBC1; BsCSPB, B. subtilis cold shock protein (CSP) B; TtCDP, Thermus thermophilus CSP; NmCPS, Neisseria meningitidis CSP; Ec CSP, Escherichia coli CSP; HsCRHSP-24, H. sapiens CRHSP-24; HsUNR, H. sapiens UNR; DmUNR, D. melanogaster UNR. (B) Structure of the BsCSP-RNA complex (PDB ID: 3pf5) is shown on the left, and the BsCPSB-based homology model of the S1L domain of HsDBC1 is shown on the right. Positions of mutated Phe 69 and Phe 77 are shown. (C) Schematics of WT protein and mutants with the S1L domain deletion (ΔS1L) and the S1L domain point mutations (S1L-M). Amino acid numbers are shown above. (D) The S1L of DBC1 is required for localization to the SNB in HeLa cells. Venus-tagged WT, ΔS1L, and S1L-M were transfected into HeLa cells. Endogenous Sam68 is a marker of the SNB in HeLa cells. The line scan data are shown on the right. (E) The mutation in the S1L domain does not affect expression. Expression of the three DBC1 proteins shown in C was measured by Western blotting. The molecular mass marker (kD) is shown on the left. α-Tubulin is a control. (F) The S1L domain is required for RNA binding of DBC1. UV cross-linking and streptavidin pull-down of FLAG-SBP-tagged WT, ΔS1L, and S1L-M was performed. After RNase digestion, the cross-linked RNA fragments were 32P-labeled with polynucleotide kinase. The radiolabeled DBC1-RNA complexes were separated by SDS-PAGE. IP was confirmed by Western blotting (WB) with an anti-FLAG antibody. The molecular mass marker (kD) is shown on the left. (G) The S1L domain of DBC1 is required for localization to the DBC1 body in HCT116 cells. Venus-tagged WT and ΔS1L were transfected into HCT116 cells. Endogenous DBC1 is a marker of the DBC1 body. The cell populations (%) in which DBC1-Venus and the endogenous DBC1 signals overlap are shown in the merge panels (>100 cells, n = 3). Bars, 10 µm.
HNRNPL acts as the adaptor to combine the two substructures
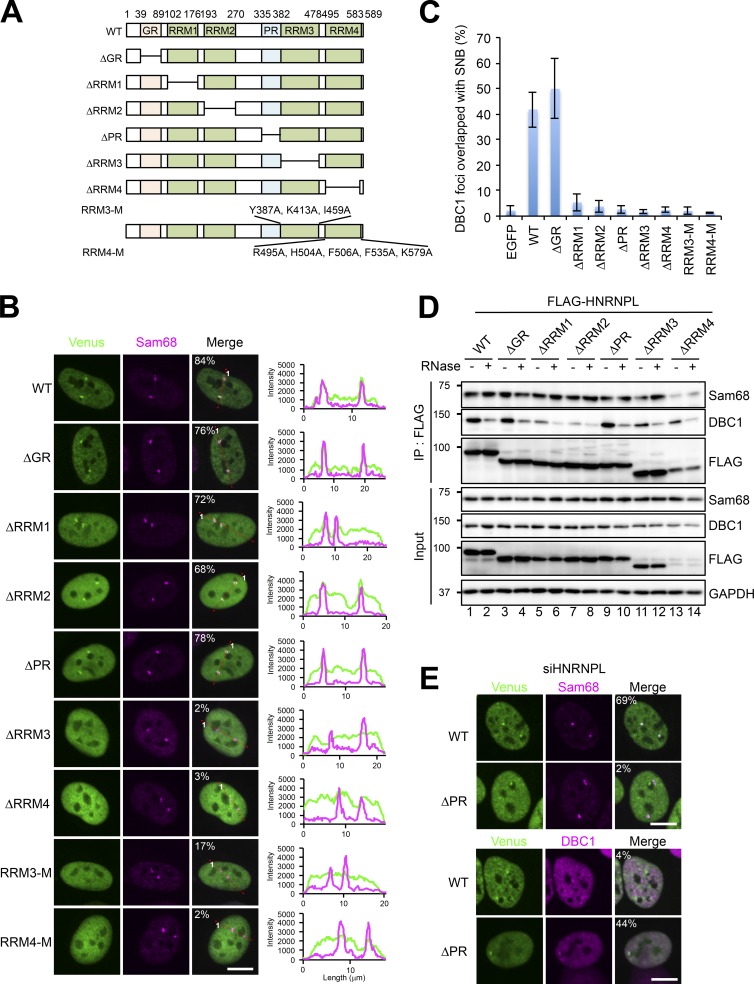
HNRNPL knockdown resulted in the clear separation of SNBs into the Sam68 substructure and DBC1 substructure, suggesting that HNRNPL functions as the adaptor of the two substructures. The annotated domains in HNRNPL were deleted to construct a series of deletion mutants fused with Venus (Fig. 7 A). The localization of each HNRNPL deletion mutant, which was monitored by Venus signals, showed that ΔRRM3 and ΔRRM4 failed to localize to SNBs labeled with endogenous Sam68 (Fig. 7 B). We further investigated whether the RNA-binding ability of RRM3 and RRM4 is responsible for the SNB localization of HNRNPL. The RRMs of HNRNPL exhibit noncanonical features that lack the conserved aromatic residues found in canonical RRMs. We referred to the crystal structure of RRM3 and RRM4 (Zhang et al., 2013) and mutated a number of residues critical for RNA binding in each (RRM3-M and RRM4-M in Fig. 7 A). Both of these RRM mutations abolished the localization to SNBs (Fig. 7 B). UV cross-linking immunoprecipitation (CLIP) revealed that simultaneous mutations of RRM3 and RRM4 (R3/4-M in Fig. S5 A) abolished RNA-binding activity in vivo (Fig. S5 C). These data strongly suggest that RNA binding via RRM3 and RRM4 is required for SNB localization of HNRNPL. Deletion mutants of RRM1 and RRM2 (ΔRRM1 and ΔRRM2) still partially retained the ability to localize in SNBs, with increased nucleoplasmic signals (see line scan data in Fig. 7 B), suggesting that RRM1 and RRM2 of HNRNPL have a moderate effect on its SNB localization (see Discussion).
Figure 7.
HNRNPL is the adaptor to form the intact SNB composed of two distinct RNase-sensitive substructures. (A) Schematics of the deletion mutants and point mutants of HNRNPL. For two point mutants (RRM3-M and RRM4-M), the mutated residues are shown. (B) Identification of the domains required for the SNB localization of HNRNPL. The Venus-tagged deletion mutants and point mutants of HNRNPL were transfected, and their localizations were monitored. Sam68 is the marker of endogenous SNBs. The cell populations (%) in which Venus and Sam68 signals overlap are shown in the merge panels (>100 cells, n = 3). The line scan data are shown on the right. (C) Identification of the domains of HNRNPL required for formation of the SNB by connecting two distinct substructures. A series of FLAG-tagged HNRNPL deletion mutants were transfected into HeLa cells in which HNRNPL had been depleted by RNAi before the plasmid transfection, and then the numbers of foci in which Sam68 and DBC1 signals overlapped were counted. The ratio of the overlapped foci relative to the total number of DBC1 foci counted is plotted in the graph (>100 cells, ±SD, n = 3). (D) Identification of the HNRNPL domains required for the interaction with Sam68 and DBC1. A series of FLAG-tagged HNRNPL deletion mutants were immunoprecipitated in the presence and absence of RNase treatment, and coprecipitated Sam68 and DBC1 were detected by Western blotting. GAPDH is the input control. The molecular mass marker (kD) is shown on the left. (E) The ΔPR mutant colocalizes with the DBC1 substructure in the absence of endogenous HNRNPL. Venus-tagged ΔPR was transfected into HeLa cells in which endogenous HNRNPL had been depleted by RNAi before the plasmid transfection. Sam68 and DBC1 were detected by immunofluorescence analysis as the markers of each substructure. The cell populations (%) in which Venus and Sam68 signals (top) or Venus and DBC1 signals (bottom) overlap are shown in the merge panels (>100 cells, n = 3). Bars, 10 µm.
Next, the ability to form the intact SNB by combining the separated DBC1 substructure and Sam68 substructure in HNRNPL-depleted cells was monitored using the HNRNPL mutant series. WT and ΔGR had combination activity leading to the formation of intact SNBs in ∼50% of cells; however, deletion of any of the four RRMs (ΔRRM1–4) abolished this combination activity (Fig. 7 C). Consistently, both RRM3-M and RRM4-M did not have combination activity (Fig. 7 C). These data indicate that mutations in either RRM3 or RRM4 abolish the SNB localization of HNRNPL, which results in the loss of combination activity. Notably, ΔRRM1 and ΔRRM2 lacked combination activity, despite localizing in the SNB, suggesting that RRM1 and RRM2 are simultaneously required to combine the DBC1 substructure with the Sam68 substructure when HNRNPL itself is attached.
CoIP with the FLAG-HNRNPL mutants revealed that the domains responsible for the combination activity described earlier are involved in the interaction with DBC1 or Sam68. WT HNRNPL interacted with Sam68 and DBC1 in RNase-resistant and RNase-sensitive manners, respectively (Fig. 7 D, lanes 1 and 2). Both ΔRRM1 and ΔRRM2 abolished the RNase-sensitive interaction with DBC1, but the interaction with Sam68 was retained (Fig. 7 D, lanes 5–8). These results strongly suggest that the HNRNPL–DBC1 interaction is bridged by the RNA molecule, which binds to RRM1 and RRM2 of HNRNPL. This is consistent with the result of the rescue experiment described earlier (Fig. 7 C) and further supports that the RNA-mediated interaction between DBC1 and HNRNPL via RRM1 and RRM2 sustains the construction of the intact SNB by recruiting the DBC1 substructure. CLIP revealed that simultaneous deletion of RRM1 and RRM2 (ΔR1/2) reduced RNA binding (∼55%); however, that of RRM3 and RRM4 abolished RNA binding, even though RRM1 and RRM2 remained intact (Fig. S5 C). Meanwhile, deletion of RRM3 or RRM4, although ΔRRM4 expression was markedly low, affected neither the RNase-resistant interaction with Sam68 nor the RNase-sensitive interaction with DBC1 (Fig. 7 D, lanes 11–14). This result suggests that the RNA–protein interaction via RRM3 and RRM4, rather than the protein–protein interaction between HNRNPL and Sam68, sustains the ability of HNRNPL to localize to the SNB and function in SNB formation.
In addition to the four ΔRRM mutants, ΔPR (proline-rich) almost completely abolished the rescue ability (Fig. 7 C). In contrast, ΔPR localized to the SNB (Fig. 7 B) and interacted with both Sam68 and DBC1 (Fig. 7 D). Quantitation of the level of WT HNRNPL and ΔPR in SNBs (which was normalized by the level of Sam68) revealed that the level of ΔPR localized in SNBs was diminished to ∼60% relative to WT HNRNPL (Fig. S5 D). Depletion of DBC1 resulted in ∼50% reduction in the localization of both WT HNRNPL and ΔPR in the Sam68 substructure, leading to a marked reduction (<30%) in the localization of ΔPR relative to that of WT HNRNPL in SNBs (Fig. S5 D). These data suggest that the PR is not essential but substantially facilitates the SNB localization of HNRNPL. Furthermore, the localization pattern was altered when endogenous HNRNPL was depleted with siRNA (siHNRNPL) before expression of ΔPR protein at a moderate level (see Materials and methods). In this condition, ΔPR did not localize in the Sam68 substructure but instead localized in the DBC1 substructure, which were separated as a consequence of HNRNPL depletion (Fig. 7 E). This changeable localization of ΔPR may be caused by its diminished ability to associate with SNBs, and endogenous HNRNPL may partially compensate for the defect in the SNB localization of ΔPR. These results are consistent with our aforementioned observation that ΔPR lacks rescue ability.
Discussion
In this article, we showed that the SNB is an RNase-sensitive NB built with multiple RNA-binding proteins. We identified Sam68 and HNRNPD as the essential proteins and further showed that their RNA-binding abilities are required for SNB formation. As in the case of paraspeckles, it was suggested that RNA–protein interactions between essential RNA-binding proteins and an unidentified RNA molecule are a prerequisite for SNB formation. Furthermore, as in the case of paraspeckles, we demonstrated that SNB formation also requires the PLD in HNRNPD through an interaction with Sam68 and HNRNPD itself. RNA binding of PLD-containing proteins results in an increase in the local concentration, triggering PLD-mediated LLPS (Molliex et al., 2015). This mechanism may be applicable to the arcRNA-dependent formation of NBs, including paraspeckle and SNB.
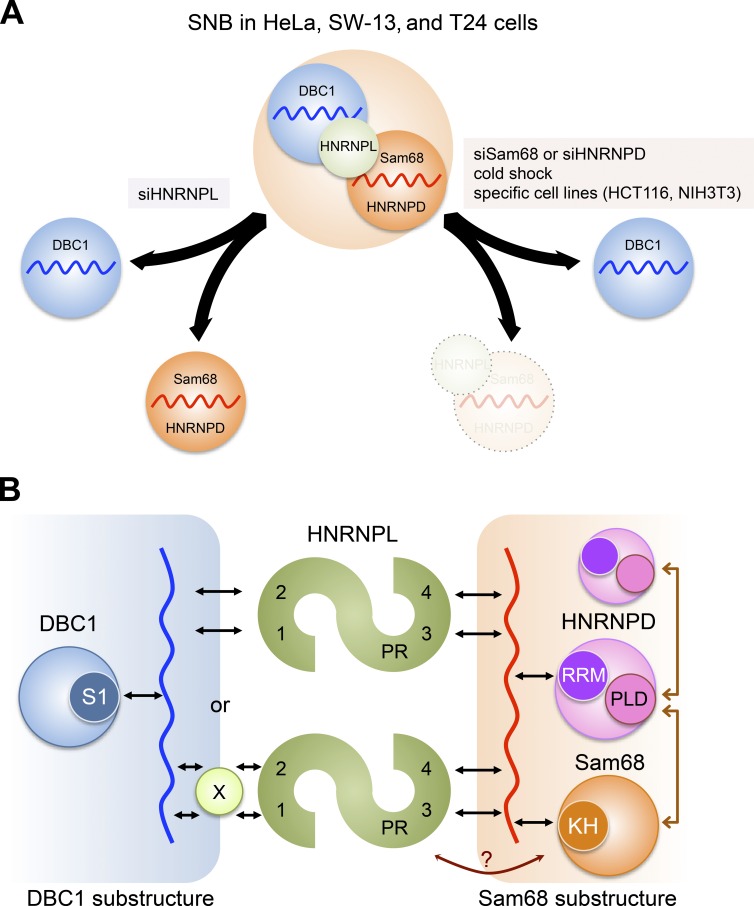
We also revealed that the typical SNB observed in HeLa cells is formed by a combination of two distinct substructures, which are both RNase sensitive (Fig. 8 A). The presence of the Sam68 substructure and DBC1 substructure in SNBs was revealed by four independent observations: (a) the DBC1 focal signals remained detectable even when Sam68 or HNRNPD was depleted; (b) the SNB was separated into two distinct foci when HNRNPL was depleted; (c) the Sam68 substructure, but not the DBC1 substructure, was reversibly disintegrated upon cold shock; and (d) specific cell lines contained only DBC1 foci. Collectively, these data indicate that the typical SNB in HeLa cells is composed of two substructures, which are fused to form the SNB but can be separated under certain conditions (Fig. 8 A). Importantly, the separated substructures as well as SNBs are both RNase sensitive, suggesting that each substructure contains its own distinct arcRNA. Under the cold stress condition, the overlapped Cajal bodies and gems are separated into distinct nuclear bodies (Liu and Dreyfuss, 1996), suggesting that subnuclear reorganization occurs. We found that the Sam68 substructure disintegrated and the DBC1 substructure remained detectable upon cold stress. All the identified SNB proteins were constantly expressed during the temperature shift, which raises the intriguing possibility that the putative arcRNA for the Sam68 substructure is specifically down-regulated in this cold shock condition. From this point of view, the arcRNA for the Sam68 substructure, rather than ubiquitous HNRNPL and other SNB proteins, may be silenced in cell lines in which the DBC1 substructure is solely detectable as the DBC1 body, although other possibilities such as protein modifications cannot be ruled out. Both of the putative arcRNAs are likely RNAPII transcripts, because RNAPII inhibitors obliterated the focal signals of Sam68 and DBC1 in HeLa cells. We confirmed that none of the abundant nuclear RNAPII transcripts (e.g., U-snRNAs, box C/D and H/ACA small nucleolar RNAs, and MALAT1 and NEAT1 lncRNAs) were detectable in SNBs, and the mixture of poly(A)+ RNAs, which are detectable in nuclear speckles and stress-induced cytoplasmic stress granules, were not detected in SNBs (Fig. S1 E), suggesting that the SNB arcRNA is a specific noncanonical RNAPII transcript that might lack a poly(A) tail.
Figure 8.
Model of the SNB architecture in HeLa cells. (A) The SNB is formed by a combination of two distinct substructures, each of which is built on distinct arcRNAs (red and blue wavy lines). HNRNPL acts as the adaptor of the two substructures, namely, the Sam68 substructure (orange circle) and the DBC1 substructure (blue circle), to form the intact SNB in HeLa, SW-13, and T24 cells. The SNB can be separated into two substructures upon knockdown of HNRNPL (siHNRNPL). The Sam68 substructure disappears and the DBC1 substructure remains under certain conditions, including knockdown of either Sam68 or HNRNPD (siSam68 or siHNRNPD) and cold shock, and in specific cell lines such as HCT116 and NIH3T3. (B) Molecular interactions within SNB. In the Sam68 substructure (orange rectangle on the right), the RRM and PLD of HNRNPD (pink circle), which are essential to form the SNB, likely interact with the putative arcRNA (red wavy line) and Sam68 (orange circle)/HNRNPD, respectively. Sam68 likely interacts with the putative arcRNA through the KH domain (KH). In the DBC1 substructure (blue rectangle on the left), DBC1 interacts with the putative arcRNA (blue wavy line) through the N-terminal S1L domain. HNRNPL acts as the adaptor (green circle) of the two substructures. RRM1 and RRM2 (shown as 1 and 2) of HNRNPL may directly or indirectly interact with the putative arcRNA for the DBC1 substructure. X is a hypothetical protein. RRM3 and RRM4 (shown as 3 and 4) likely interact with the putative arcRNA for the Sam68 substructure. PR acts to facilitate the interaction of HNRNPL with the Sam68 substructure. At least two HNRNPL molecules cooperatively function as the adaptor to combine the two substructures. RNA–protein interactions and protein–protein interactions are represented by black and brown arrows, respectively.
The DBC1 body is a novel NB that is RNase sensitive and detectable in both human and mouse. The DBC1 body is suggested to contain its own arcRNA, and we experimentally showed that DBC1 directly binds to RNA through the N-terminal S1L domain. DBC1 mutated in the S1L domain failed to localize to the SNB in HeLa cells and the DBC1 body in HCT116 cells, strongly suggesting that RNA–protein interaction through the S1L domain contributes to the localization of DBC1 and likely the formation of the DBC1 body. It should be noted that we cannot rule out the possibility that the DBC1 substructure in the SNB of HeLa cells is distinct from the DBC1 body in HCT116 cells. In this case, different arcRNAs may capture DBC1 to form distinct NBs.
HNRNPL acts as the adaptor of the two distinct substructures described earlier to form the intact SNB. HNRNPL associates preferentially with the Sam68 substructure because the HNRNPL focal signal concomitantly disappeared when the Sam68 substructure, but not the DBC1 substructure, was disintegrated by depletion of Sam68 or HNRNPD and in the cold stress condition (Fig. 8 A). RNA binding through RRM3 and RRM4 of HNRNPL was required for its SNB localization and also the ability to combine the two substructures. Mutations in residues critical for RNA binding in each of RRM3 and RRM4 abolished the localization of HNRNPL to the Sam68 substructure, suggesting that RRM3 and RRM4 cooperatively and directly bind to the putative arcRNA of the Sam68 substructure (Fig. 8 B). HNRNPL associated with the Sam68 substructure can capture the DBC1 substructure to form the intact SNB. RRM1 and RRM2 of HNRNPL are also suggested to act for the capture of the DBC1 substructure through an interaction with RNA in the DBC1 substructure. Because it remains ambiguous whether RRM1 and RRM2 associate with certain RNAs in vivo, we cannot rule out the possibility that an unidentified protein mediates the interaction between HNRNPL and the DBC1 substructure (Fig. 8 B). In addition to the four RRMs, we showed that PR contributes to the combination of the two substructures. The PR mutation resulted in mislocalization of HNRNPL in the DBC1 substructure only when endogenous HNRNPL was absent, suggesting that at least two HNRNPL molecules function as the adaptor, meaning that WT HNRNPL can partially compensate for the defect in the adaptor function of ΔPR (Fig. 8 B).
SNB components such as Sam68 and HNRNPL are regulators of alternative splicing (Hung et al., 2008; Chawla et al., 2009). DBC1 is the component of the DBIRD complex that integrates alternative splicing with RNAPII transcript elongation (Close et al., 2012). Considering that the DBC1 body combines with the Sam68 substructure to form SNBs, the SNB may act as the regulatory factory of coupled transcription-splicing events in which Sam68, HNRNPL, and DBC1 regulate the optimal splicing patterns coupled with RNAPII transcription of specific chromosomal loci located near SNBs. To further understand the function of the SNB, it is crucial to identify their arcRNAs. Functional analyses of arcRNAs will elucidate the mechanism underlying SNB formation and dynamics as well as the biological functions of SNBs. Detailed analyses of SNBs will reveal the commonality of the mechanism underlying arcRNA actions in the formation of arcRNA-dependent NBs.
Materials and methods
Plasmid construction
The PCR-amplified Venus sequence was inserted into the pcDNA3 vector (Thermo Fisher Scientific) between the HindIII and BamHI sites, generating the 5′-Venus plasmid. The PCR-amplified Venus sequence was inserted into the pcDNA3 vector between the EcoRV and NotI sites, generating the 3′-Venus plasmid. The PCR-amplified 3xFLAG sequence was inserted into the pcDNA5 vector (Thermo Fisher Scientific) at HindIII, generating the 5′-FLAG plasmid. A SV40-NLS fragment with BglII and BamHI sites was produced by annealing a set of DNA oligonucleotides (5′-GATCTGATCCAAAAAAGAAGAGAAAGGTAGATACGGCCG-3′ and 5′-GATCCGGCCGTATCTACCTTTCTCTTCTTTTTTGGATCA-3′) and was inserted into the 5′-FLAG plasmid at the BamHI site, generating the 5′-FLAG-NLS plasmid. The PCR-amplified 3xFLAG-SBP fragment was inserted into the pcDNA3 vector between the EcoRI and NotI sites, generating the 3′-FLAG-SBP plasmid. The PCR-amplified HNRNPD (p45) cDNA fragment was inserted into the pBluescript II SK(+) vector (Agilent Technologies) between the BamHI and XhoI sites, followed by construction of the siRNA-resistant HNRNPD, HNRNPD RRM mutants (F140D/F142D and F225D/F227D), and HNRNPD isoforms (p37, p40, and p42) by site-directed mutagenesis using PCR. The HNRNPD fragments constructed as detailed earlier were cloned into the 5′-Venus and 5′-FLAG plasmids between BamHI and XhoI. The HNRNPD p45 PLD mutants (residues 262–355) were generated by artificial gene synthesis (GenScript). The mutated residues of HNRNPD PLD-M1 were Y294S, Y302S, Y305S, Y307S, Y312S, Y315S, Y323S, Y329S, and Y339S, and those of HNRNPD PLD-M2 were Y263S, Y294S, Y297S, Y302S, Y305S, Y307S, Y312S, Y315S, Y318S, Y320S, Y323S, Y326S, Y327S, Y329S, Y332S, Y339S, Y352S, and Y355S. The HNRNPD PLD mutant fragments were PCR-amplified using the synthesized DNAs as templates, followed by cloning into the 5′-FLAG-NLS plasmid using the Gibson assembly system (New England Biolabs, Inc.). The PCR-amplified DBC1 fragment was inserted into the 3′-Venus plasmid between HindIII and EcoRV, followed by construction of the DBC1 ΔS1L and S1L-M mutants by site-directed mutagenesis using PCR. The mutated residues in DBC1 were F69A and F77A. The DBC1 fragments constructed as described earlier were cloned into the 3′-FLAG-SBP plasmid between HindIII and EcoRV. The PCR-amplified HNRNPL fragment was inserted into pBluescript II SK(+) between BamHI and XhoI, followed by construction of the siRNA-resistant HNRNPL, HNRNPL ΔGR, ΔRRM1, ΔRRM2, ΔPR, ΔRRM3, ΔRRM4, and ΔR1/R2 mutants by site-directed mutagenesis using PCR. These HNRNPL constructs were cloned into the 5′-Venus and 5′-FLAG plasmids. The HNRNPL RRM3 and RRM4 mutants were generated by artificial gene synthesis (GenScript). The mutated residues in HNRNPL were Y387A, K413A, and I459A for RRM3-M; R495A, H504A, F506A, F535A, and K579A for RRM4-M; and Y387A, K413A, I459A, R495A, H504A, F506A, F535A, and K579A for R3/4-M. The HNRNPL RRM mutant fragments were PCR-amplified using the synthesized DNAs as templates, followed by cloning into the 5′-Venus and 5′-FLAG plasmids using the Gibson assembly system. The siRNA-resistant Sam68, ΔNT, ΔNK, ΔKH, ΔCK, ΔGSG ΔCT, and ΔCT+NLS Sam68 mutants were generated by site-directed mutagenesis using PCR with the Sam68 cDNA cloned into pENTR (Thermo Fisher Scientific). The Sam68 fragments constructed as described earlier were cloned into the pDESTMN-Venus and pcDNAnFLAG-DEST plasmids using the Gateway cloning system (Thermo Fisher Scientific).
Quantitative RT-PCR
Total RNA (1 µg) was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) and purchased from Sigma-Genosys or Thermo Fisher Scientific. Aliquots of cDNA were amplified by qPCR using LightCycler 480 SYBR Green I Master reagent (Roche), according to the manufacturer’s protocol. The primers used were as follows: HNRNPD Exon 4–5 forward, 5′-AAGGGCCAAAGCCATGAAAA-3′; HNRNPD Exon 4–5 reverse, 5′-CCACCTCACCAAAACCACCA-3′; HNRNPD Exon 7 forward, 5′-CCCCAGTCAAAACTGGAACC-3′; and HNRNPD Exon 7 reverse, 5′-CCATAACCACCGTAACCTTGGC-3′.
Cell culture
HeLa and MCF-7 cells were grown and maintained in MEM (Gibco), T24 and HCT116 cells were grown and maintained in McCoy’s 5A medium (Gibco), and SW-13 and NIH3T3 cells were grown and maintained in DMEM (Nacalai Tesque). All media were supplemented with 10% FBS (Gibco). The cells were grown at 37°C in a humidified incubator with 5% CO2. For cold shock experiments, cells were incubated for 24 h at 32°C and then allowed to recover for 1 or 3 h at 37°C. Some cells were treated with actinomycin D (0.03 or 0.3 µg/ml) for 4 h or DRB (100 µM) for 6 h.
RNase treatment of cells
For RNase sensitivity screening of nuclear foci, HeLa cells (6.0 × 103) were grown on 96-well glass-bottom plates (Iwaki) and transfected with the Venus-tagged cDNA clones using TransIT LT1 reagent (Takara Bio Inc.), according to the manufacturer’s instructions. After 24 h, the cells were rinsed briefly in PBS and then rinsed in permeabilization buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.5 mM EGTA, and complete EDTA-free protease inhibitor [Roche]). Subsequently, cells were permeabilized for 10 min at RT in permeabilization buffer containing 2% Tween-20 and then rinsed once with permeabilization buffer. The permeabilized cells were incubated with Riboshredder RNase Blend (Epicentre; 50 U/ml prepared in PBS) for 20 min at RT. After RNase treatment, cells were rinsed with PBS and fixed with 4% paraformaldehyde prepared in PBS at RT for 10 min. The fixed cells were stained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) and Pyronin Y (Sigma-Aldrich) to monitor RNase digestion. Fluorescence images were acquired using IN Cell Analyzer 1000 (GE Healthcare).
Plasmid transfection
For microscopy observation, cells were seeded onto a multichamber culture slide (BD Falcon). The plasmids were introduced into the cells using TransIT LT1, according to the manufacturer’s instructions. The cells were usually fixed 24 h after transfection. For siRNA transfection and plasmid rescue experiments, cells were grown in six-well plates and transfected with siRNA using Lipofectamine RNAi MAX (Thermo Fisher Scientific), according to the manufacturer’s instructions. After 24 h, the cells were trypsinized and seeded into multichamber culture slides. At 48 h after siRNA transfection, cells were transfected with plasmids using Lipofectamine 2000 (Thermo Fisher Scientific), according to the manufacturer’s instructions. The cells were usually fixed 24 h after transfection.
RNAi
Stealth siRNAs were purchased from Thermo Fisher Scientific. The siRNAs against exon 7 of HNRNPD (siExon 7A or 7B) were purchased from Integrated DNA Technologies. HeLa cells (5.0 × 105) were grown in six-well tissue culture dishes and transfected with siRNAs (final concentration, 33 nM) using Lipofectamine RNAi MAX reagent, according to the manufacturer’s instructions. After 24 h, the cells were trypsinized and seeded into six-well tissue culture dishes for the preparation of RNA and protein or onto a multichamber culture slide for immunofluorescence. The cells were then cultured for a further 24 h before harvesting. The siRNAs used were as follows (asterisks represent deoxynucleotides): siSam68 sense, 5′-UUUCUGAAUCUUCUCAAUUUCUGCC-3′; siSam68 antisense, 5′-GGCAGAAAUUGAGAAGAUUCAGAAA-3′; siHNRNPL sense, 5′-UCAAAUUCCACCAUCGCCUGAACUC-3′; siHNRNPL antisense, 5′-GAGUUCAGGCGAUGGUGGAAUUUGA-3′; siExon 4 sense, 5′-UCACCUUCCCAUUCAAUUUAUGUUC-3′; siExon 4 antisense, 5′-GAACAUAAAUUGAAUGGGAAGGUGA-3′; siExon 5 (used as siHNRNPD in Fig. 3) sense, 5′-AAGGUAAUAAAGCAGAACCCACGCC-3′; siExon 5 (used as siHNRNPD in Fig. 3) antisense, 5′-GGCGUGGGUUCUGCUUUAUUACCUU-3′; siExon 7A sense, 5′-ACCAGGGAUAUAGUAACUAUUGGA*A*-3′; siExon 7A antisense, 5′-UUCCAAUAGUUACUAUAUCCCUGGUUC-3′; siExon 7B sense, 5′-UGACUACACUGGUUACAACAACUA*C*-3′; siExon 7B antisense, 5′-GUAGUUGUUGUAACCAGUGUAGUCAUA-3′; siDBC1 sense, 5′-CCAUCUGUGACUUCCUAGAACUCCA-3′; siDBC1 antisense, 5′-UGGAGUUCUAGGAAGUCACAGAUGG-3′; siZNF346 sense, 5′-UUCAGCUUUAAGUUCUUUGCGUGGG-3′; and siZNF346 antisense, 5′-CCCACGCAAAGAACUUAAAGCUGAA-3′. siControls were purchased from Thermo Fisher Scientific (12935300) and Integrated DNA Technologies (51-01-14-03; used in Fig. S3).
RNA-FISH
The RNA probes were synthesized using SP6 RNA polymerase and a digoxigenin (DIG) RNA labeling kit (Roche). Linearized plasmids (1 µg) containing a NEAT1 fragment (+1 to +1,000) and MALAT1 (+5,114 to +5,712) were used as templates for transcription. The oligo(dT)50 probes were synthesized using a DIG oligonucleotide 3′-end labeling kit (Roche). RNA-FISH was performed as described previously (Naganuma et al., 2012). Briefly, the cells were seeded onto a multichamber culture slide, washed with PBS, and fixed with 4% paraformaldehyde prepared in PBS at RT for 10 min. The fixed cells were permeabilized with 0.5% Triton X-100 prepared in PBS for 20 min and then rinsed with PBS. The slides were incubated with prehybridization solution (2× SSC, 1× Denhardt’s solution, 50% formamide, 10 mM EDTA, pH 8.0, 100 µg/ml yeast tRNA, and 0.01% Tween-20) at 55°C (for RNA probes) or 37°C [for oligo(dT)50 probes] for 2 h. The prehybridized slides were then incubated with hybridization solution (prehybridization solution containing 5% dextran sulfate and 2 µg/ml DIG-labeled RNA probe) at 55°C (for RNA probes) or 37°C [for oligo(dT)50 probes] for 16 h. After hybridization, the slides were washed twice with prewarmed wash buffer (2× SSC, 50% formamide, and 0.01% Tween-20) at 55°C for 30 min, and excess RNA probes were digested by incubation with 10 µg/ml RNase A prepared in NTET buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 500 mM NaCl, and 0.1% Tween-20) at 37°C for 1 h. For oligo(dT)50 probes, this RNase treatment can be omitted. For RNA probes, the slides were then washed once with buffer A (2× SSC and 0.01% Tween-20) at 55°C for 30 min and twice with buffer B (0.1× SSC and 0.01% Tween-20) at 55°C for 30 min. For oligo(dT)50 probes, the slides were washed three times with prewarmed wash buffer C (2× SSC) at 37°C for 10 min and once with buffer D (1× SSC) at 37°C for 10 min. For detection, the slides were washed with Tris-buffered saline containing 0.1% Tween-20 (TBST), incubated with blocking solution (3% BSA prepared in TBST) at RT for 1 h, and incubated with anti-DIG antibodies diluted in blocking solution at 4°C overnight. Unbound antibodies were removed by three 15-min washes in TBST. The slides were then incubated with a fluorophore-conjugated secondary antibody diluted in blocking solution for 1 h at RT. After washing, the slides were mounted with Vectashield (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole. Fluorescence images were visualized at RT on a microscope (FluoView FV1000D IX81; Olympus) equipped with U-Plan Apochromat 40×/0.95 objective lenses (Olympus). FluoView FV10-ASW1.7 software (Olympus) was used for image acquisition and processing. All overlaid images were transferred as high-resolution TIFF files. Figures were compiled using Photoshop (Adobe Systems).
Immunofluorescence
Immunofluorescence analyses were performed as described previously (Naganuma et al., 2012). In brief, cells were seeded onto a multichamber culture slide and fixed with 4% paraformaldehyde prepared in PBS at RT for 10 min. The fixed cells were permeabilized with 0.2% Triton X-100 prepared in PBS for 15 min, rinsed, and blocked with 3% BSA prepared in PBS containing 0.1% Tween-20 (PBST) for 1 h. The slides were incubated at 4°C overnight with primary antibodies (diluted in PBST containing 3% BSA) against specific proteins. Unbound antibodies were removed by three 10-min washes with PBST. The slides were then incubated with Alexa-conjugated secondary antibodies (Molecular Probes) for 1 h at RT, washed, and mounted with Vectashield. Immunostained cells were examined using a confocal laser scanning microscope (FV1000D; Olympus). Each data series (samples on a single 8-chamber culture slide probed with the same antibodies) was processed with fixed parameters to enable comparison of the signal intensities. The antibodies used are listed in Table S1.
Immunoblotting
Cells were lysed in immunoprecipitation (IP) lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, and complete EDTA-free protease inhibitor) and then disrupted by three pulses of sonication for 5 s. The cell extracts were cleared by centrifugation, and the protein concentration was determined using the Bradford method. An equal volume of 2× SDS sample buffer was added, and the samples were heated before separation by SDS-PAGE. After fractionation, the proteins were transferred to FluoroTrans W membranes (Pall Corporation) by electroblotting. The antibodies used are listed in Table S1.
IP
HeLa cell pellets (5 × 106 cells) were suspended in IP lysis buffer (1 ml) and sonicated with three 5-s pulses. The resultant cell extracts were treated with or without RNase A (1 µg/ml) at 4°C for 1 h, and then cleared by centrifugation at 10,000 g for 10 min. The antibodies were mixed with Dynabeads Protein-G (Thermo Fisher Scientific) at 4°C for 1 h and then washed five times with IP lysis buffer. The supernatant of HeLa cell extracts was mixed with the antibody–Dynabead conjugates and rotated at 4°C overnight. The beads were finally washed five times with IP lysis buffer. The IP samples were recovered by direct addition of SDS-PAGE loading buffer. The antibodies used are listed in Table S1.
UV cross-linking and pull-down of RNP complexes
UV cross-linking was basically performed as described previously (Hafner et al., 2010). After transfection for 4–6 h, 4-thiouridine (Sigma-Aldrich) was added to the cell culture medium at a final concentration of 100 µM, and the cells were further incubated for 16 h. The cells were washed with PBS and cross-linked twice by irradiation with 365-nm UV light at 200 mJ/cm2. HeLa cells were suspended in lysis buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, and complete EDTA-free protease inhibitor) and sonicated with three 5-s pulses. The cell extracts were incubated with RNase T1 (0.05 U/µl; Roche) at RT for 15 min and then cleared by centrifugation at 10,000 g for 10 min. For precipitation of SBP-tagged DBC1, the supernatants were mixed with Dynabeads MyOne Streptavidin T1 (Thermo Fisher Scientific) and rotated for 1 h at 4°C. For IP of Flag-tagged HNRNPL, the supernatants were mixed with the antibody–Dynabead conjugates and rotated for 1 h at 4°C. The beads were washed three times with IP wash buffer (20 mM Hepes, pH 7.4, 300 mM NaCl, 1 mM DTT, 0.05% NP-40, and complete EDTA-free protease inhibitor). The beads were incubated with RNase T1 (10 U/µl) at RT for 15 min and washed three times with high-salt wash buffer (20 mM Hepes, pH 7.4, 500 mM NaCl, 1 mM DTT, 0.05% NP-40, and complete EDTA-free protease inhibitor). Subsequently, beads were incubated with calf intestinal alkaline phosphatase (0.1 U/µl; Toyobo) for 10 min at 37°C to dephosphorylate the RNAs. The beads were washed twice with PNK buffer lacking DTT (50 mM Tris-HCl, pH 7.4, 50 mM NaCl, and 10 mM MgCl2) and incubated for 30 min at 37°C with T4 polynucleotide kinase (0.1 U/µl, Takara Bio Inc.) and 10 µCi γ-[32P]ATP (3,000 Ci/mmol and 10 mCi/ml; PerkinElmer) to radiolabel the cross-linked RNAs. The RNA–protein complexes were eluted with 1× LDS loading buffer (Thermo Fisher Scientific) and separated by SDS-PAGE (NuPAGE Novex 4–12% Bis-Tris gel; Thermo Fisher Scientific) in MOPS buffer.
Construction of the homology model of the S1L domain of human DBC1
The homology model of the S1L domain of human DBC1 was constructed using the SWISS-MODEL server (Biasini et al., 2014). BsCSPB complexed with oligo U RNA (PDB ID: 3PF5) was used as the template of the homology model. Based on the sequence alignment and complex structure of BsCSPB, the highly conserved phenylalanine residues of human DBC1 (Phe 69 and Phe 77) were chosen as the putative RNA-interacting residues and were mutated to alanine in the cell-based assay.
Online supplemental material
Fig. S1 shows the localization and expression of other SNB components, features of SNBs under cold shock conditions, and data excluding the possibility that well-characterized nuclear RNAPII transcripts are the SNB arcRNA. Fig. S2 shows that RNA binding is required for the function of Sam68 in SNB formation. Fig. S3 shows the significance of specific isoforms of HNRNPD that possess the PLD. Fig. S4 shows that the DBC1 substructure is sensitive to RNase treatment and the effect of DBC1 knockdown on HNRNPL localization. Fig. S5 shows the in vivo RNA-binding ability of HNRNPL RRMs and the effect of DBC1 depletion on localization of the ΔPR HNRNPL mutant. Table S1 lists the antibodies used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201601024/DC1.
Supplementary Material
Supplemental Materials (PDF)
Acknowledgments
The authors thank Y. Kawamura and H. Mochizuki for their support in the preparation of the Venus-tagged cDNA library, S. Nakanishi and A. Kubota for technical assistance, and the members of the Hirose laboratory for useful discussions.
This research was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (26113002 and 26291001).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
arcRNA
architectural RNA
coIP
coimmunoprecipitation
DIG
digoxigenin
DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
GSG
GRP33/Sam68/GLD1
IP
immunoprecipitation
KH
K homology
lncRNA
long noncoding RNA
NB
nuclear body
PLD
prion-like domain
RNAPII
RNA polymerase II
RRM
RNA recognition motif
S1L
S1-like domain
SNB
Sam68 nuclear body
WT
wild type
References
- Anantharaman V., and Aravind L.. 2008. Analysis of DBC1 and its homologs suggests a potential mechanism for regulation of sirtuin domain deacetylases by NAD metabolites. Cell Cycle. 7:1467–1472. 10.4161/cc.7.10.5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas T.E., Jacob M.D., and Lee S.. 2012. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell. 45:147–157. 10.1016/j.molcel.2011.12.012 [DOI] [PubMed] [Google Scholar]
- Baltz A.G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., et al. . 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 46:674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Biamonti G. 2004. Nuclear stress bodies: A heterochromatin affair? Nat. Rev. Mol. Cell Biol. 5:493–498. 10.1038/nrm1405 [DOI] [PubMed] [Google Scholar]
- Biamonti G., and Vourc’h C.. 2010. Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2:a000695 10.1101/cshperspect.a000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T.. 2014. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42(W1):W252–W258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke K.A., Janke A.M., Rhine C.L., and Fawzi N.L.. 2015. Residue-by-residue view of in vitro FUS granules that bind the c-terminal domain of RNA polymerase II. Mol. Cell. 60:231–241. 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G., Lin C.H., Han A., Shiue L., Ares M. Jr., and Black D.L.. 2009. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol. Cell. Biol. 29:201–213. 10.1128/MCB.01349-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., and Carmichael G.G.. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 35:467–478. 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Boisvert F.M., Bazett-Jones D.P., and Richard S.. 1999. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell. 10:3015–3033. 10.1091/mbc.10.9.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T., Yamazaki T., and Hirose T.. 2016. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim. Biophys. Acta. 1859:139–146. 10.1016/j.bbagrm.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., and Lawrence J.B.. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 33:717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P., East P., Dirac-Svejstrup A.B., Hartmann H., Heron M., Maslen S., Chariot A., Söding J., Skehel M., and Svejstrup J.Q.. 2012. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 484:386–389. 10.1038/nature10925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Ospina J.K., Sung M.H., John S., Upender M., Ried T., Hager G.L., and Matera A.G.. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179:1095–1103. 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., and Lamond A.I.. 2002. Paraspeckles: A novel nuclear domain. Curr. Biol. 12:13–25. 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- Fox A.H., Bond C.S., and Lamond A.I.. 2005. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 16:5304–5315. 10.1091/mbc.E05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M., et al. . 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A., et al. . 2015. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 210:529–539. 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., and Goshima N.. 2015. Genome-wide co-localization screening of nuclear body components using a fluorescently tagged FLJ cDNA clone library. Methods Mol. Biol. 1262:155–163. 10.1007/978-1-4939-2253-6_9 [DOI] [PubMed] [Google Scholar]
- Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Bénard M., Fox A.H., and Pierron G.. 2014. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 25:169–183. 10.1091/mbc.E13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.H., Heiner M., Hui J., Schreiner S., Benes V., and Bindereif A.. 2008. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 14:284–296. 10.1261/rna.725208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. . 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 53:393–406. 10.1016/j.molcel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Ishidate T., Yoshihara S., Kawasaki Y., Roy B.C., Toyoshima K., and Akiyama T.. 1997. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 409:237–241. 10.1016/S0014-5793(97)00455-9 [DOI] [PubMed] [Google Scholar]
- Jones A.R., and Schedl T.. 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9:1491–1504. 10.1101/gad.9.12.1491 [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. . 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., and Dreyfuss G.. 1996. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Zhang B., and Spector D.L.. 2011. Biogenesis and function of nuclear bodies. Trends Genet. 27:295–306. 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., and Grummt I.. 2008. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24:131–157. 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., and Hirose T.. 2012. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31:4020–4034. 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., Fujita Y., Fujimori T., Standaert L., Marine J.C., and Hirose T.. 2014. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 141:4618–4627. 10.1242/dev.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. . 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Rajendra T.K., Lal A.K., and Lakhotia S.C.. 2000. Omega speckles: A novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 113:3485–3497. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., and Spector D.L.. 2005. Regulating gene expression through RNA nuclear retention. Cell. 123:249–263. 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- Rajan P., Dalgliesh C., Bourgeois C.F., Heiner M., Emami K., Clark E.L., Bindereif A., Stevenin J., Robson C.N., Leung H.Y., and Elliott D.J.. 2009. Proteomic identification of heterogeneous nuclear ribonucleoprotein L as a novel component of SLM/Sam68 nuclear bodies. BMC Cell Biol. 10:82 10.1186/1471-2121-10-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs R., Max K.E., Heinemann U., and Balbach J.. 2012. RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. RNA. 18:65–76. 10.1261/rna.02809212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y.T., Ideue T., Sano M., Mituyama T., and Hirose T.. 2009. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA. 106:2525–2530. 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E., and Trinkle-Mulcahy L.. 2014. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr. Opin. Cell Biol. 28:76–83. 10.1016/j.ceb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- Standaert L., Adriaens C., Radaelli E., Van Keymeulen A., Blanpain C., Hirose T., Nakagawa S., and Marine J.C.. 2014. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 20:1844–1849. 10.1261/rna.047332.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., and Spector D.L.. 2009. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19:347–359. 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnikova Y.A., Kubarenko A.V., Schäfer A., Weber A.N., and Niehrs C.. 2011. Gadd45a is an RNA binding protein and is localized in nuclear speckles. PLoS One. 6:e14500 10.1371/journal.pone.0014500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P.J., Islam S.A., Freemont P.S., and Sheer D.. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164:515–526. 10.1083/jcb.200305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., and Yamamoto M.. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 78:487–498. 10.1016/0092-8674(94)90426-X [DOI] [PubMed] [Google Scholar]
- West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., and Kingston R.E.. 2014. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 55:791–802. 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., and Hirose T.. 2015. The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci. (Elite Ed.). 7:1–41. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zeng F., Liu Y., Zhao Y., Lv H., Niu L., Teng M., and Li X.. 2013. Crystal structures and RNA-binding properties of the RNA recognition motifs of heterogeneous nuclear ribonucleoprotein L: Insights into its roles in alternative splicing regulation. J. Biol. Chem. 288:22636–22649. 10.1074/jbc.M113.463901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi B.E., Ballin J.D., Brewer B.Y., Ross C.R., Huang J., Toth E.A., and Wilson G.M.. 2010. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J. Biol. Chem. 285:39127–39139. 10.1074/jbc.M110.180182 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials (PDF)