Resolving Early Mesoderm Diversification through Single Cell Expression Profiling (original) (raw)
. Author manuscript; available in PMC: 2017 Jan 14.
Published in final edited form as: Nature. 2016 Jul 14;535(7611):289–293. doi: 10.1038/nature18633
Summary
In mammals, specification of the three major germ layers occurs during gastrulation, when cells ingressing through the primitive streak differentiate into the precursor cells of major organ systems. However, the molecular mechanisms underlying this process remain unclear, as numbers of gastrulating cells are very limited. In the E6.5 mouse embryo, cells located at the junction between the extra-embryonic region and the epiblast on the posterior side of the embryo undergo an epithelial-to-mesenchymal transition (EMT) and ingress through the primitive streak (PS). Subsequently, cells migrate, either surrounding the prospective ectoderm contributing to the embryo proper, or into the extra-embryonic region to form the yolk sac (YS), umbilical cord and placenta. Fate mapping has shown that mature tissues such as blood and heart originate from specific regions of the pre-gastrula epiblast1 but the plasticity of cells within the embryo and the function of key cell type-specific transcription factors remain unclear. Here we analyse 1,205 cells from the epiblast and nascent Flk1+ mesoderm of gastrulating mouse embryos using single cell RNA-sequencing, representing the first transcriptome-wide in vivo view of early mesoderm formation during mammalian gastrulation. Additionally, using knock-out mice, we study the function of Tal1, a key hematopoietic transcription factor (TF), and demonstrate, contrary to previous studies performed using retrospective assays2,3, that_Tal1_ knock out does not immediately bias precursor cells towards a cardiac fate.
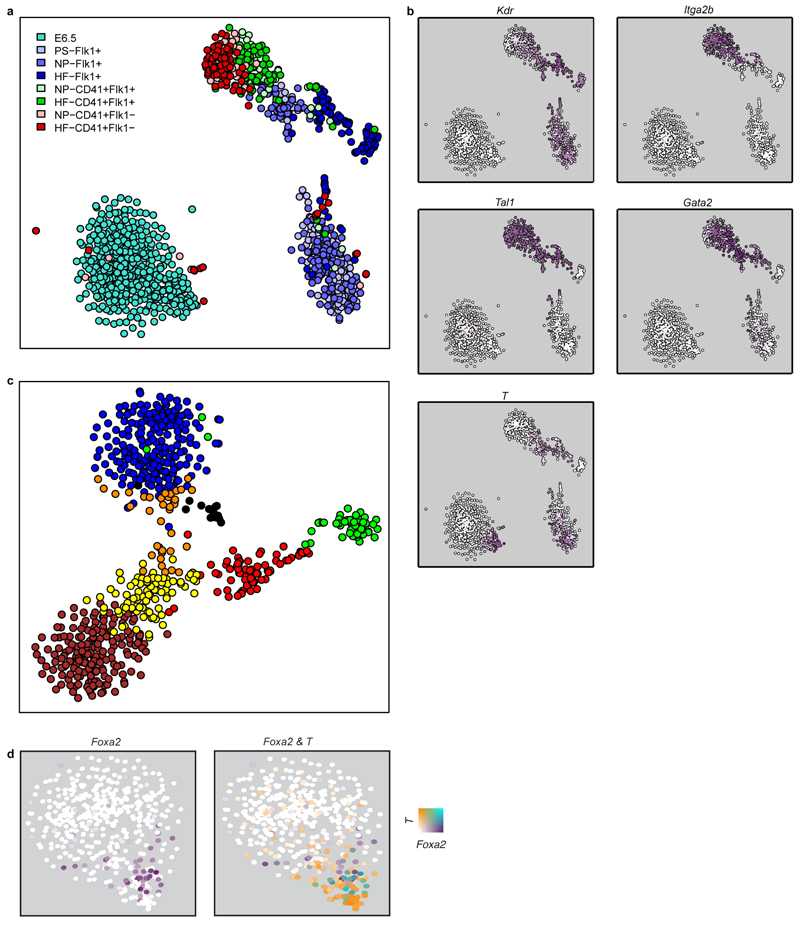
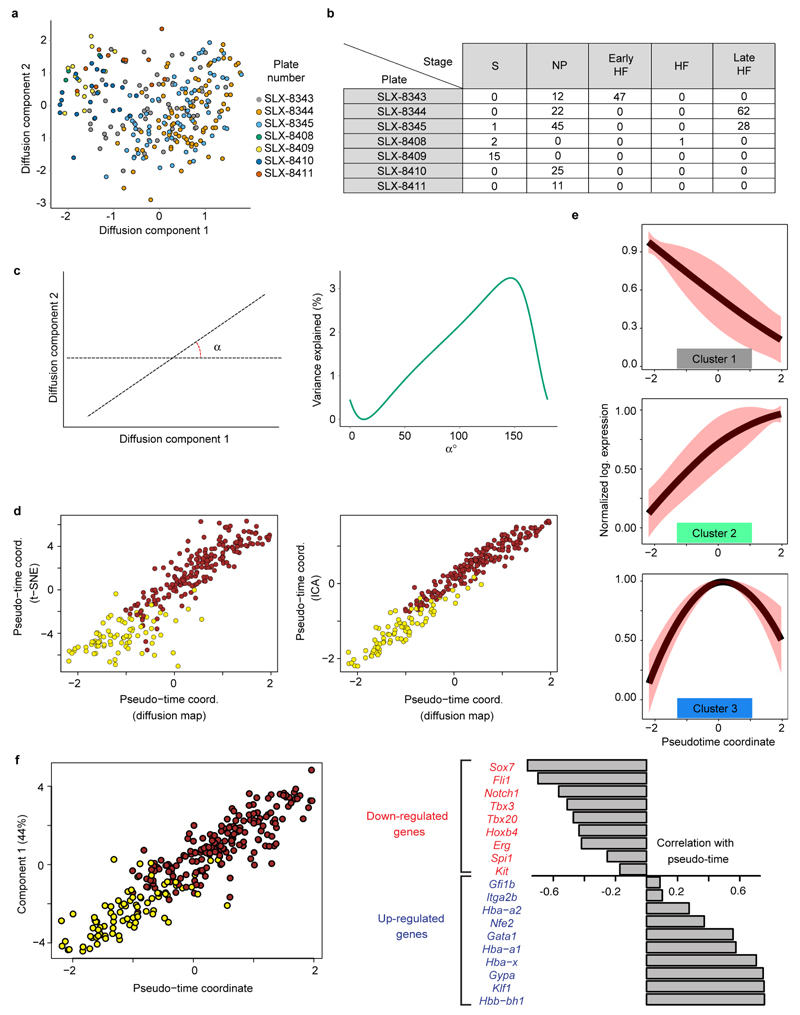
Traditional experimental approaches for genome-scale analysis rely on large numbers of input cells and therefore cannot be applied to study early lineage diversification directly in the embryo. To address this, we used single cell transcriptomics to investigate mesodermal lineage diversification towards the haematopoietic system in 1,205 single cells covering a timecourse from early gastrulation at embryonic day E6.5 to the generation of primitive red blood cells at E7.75 (Figure 1a, Extended Data Fig. 1a,2a). Using previously published metrics (Methods), we observed that the data were of high quality. 501 single cell transcriptomes were obtained from dissected distal halves of E6.5 embryos sorted for viability only, which contain all of the epiblast cells, including the developing PS, and a limited number of visceral endoderm and extra-embryonic ectoderm cells. From E7.0, embryos were staged according to anatomical features (Methods) as primitive streak (S), neural plate (NP) and head fold (HF). The VEGF receptor Flk1 (Kdr) was used to capture cells as it marks much of the developing mesoderm4. During subsequent blood development, Flk1 is down-regulated and CD41 (Itga2b) is up-regulated5. We therefore also sampled cells expressing both markers and CD41 alone at the NP and HF stages (Figure 1a; Extended Data Fig. 1b, 2a), giving a total of 138 cells from E7.0 (S), 259 from E7.5 (NP) and 307 from E7.75 (HF).
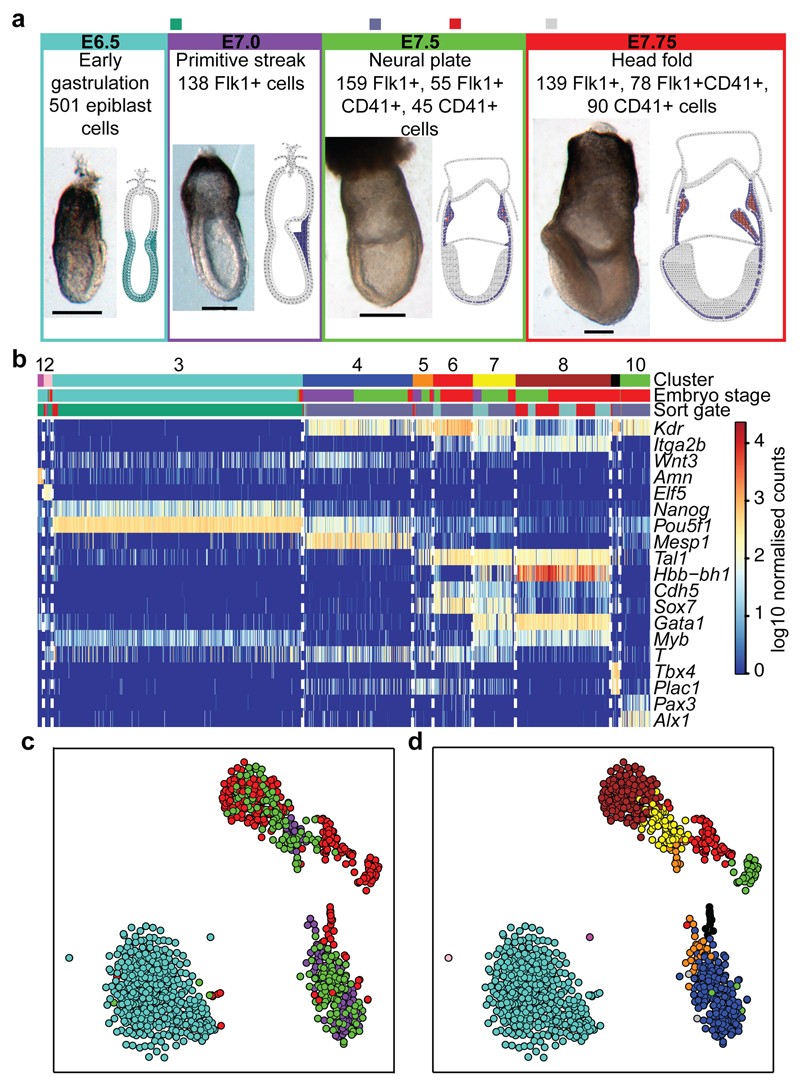
Figure 1. Single cell transcriptomics identifies 10 populations relevant to early mesodermal development.
a) Whole mount images and schematics of E6.5 to E7.75 embryo sections. Colors indicate approximate locations of sorted cells. Anterior, left; posterior, right. Scale bars: 200µm. b) Heatmap showing key genes distinguishing 10 clusters. Colored bars indicate assigned cluster (top), stage (middle: turquoise, E6.5; purple, primitive streak (S, E7.0); green, neural plate (NP, E7.5); red, headfold (HF, E7.75)) and the sorted population (bottom: green, E6.5 epiblast; blue, Flk1+; turquoise, Flk1+CD41+; red, Flk1-CD41+). c) t-SNE of all 1205 cells colored by embryonic stage, and d) according to clusters in (b).
Following rigorous quality control, 2,085 genes were identified as having significantly more heterogeneous expression across the 1,205 cells than expected by chance (Extended Data Fig. 2b-d). Unsupervised hierarchical clustering in conjunction with a dynamic hybrid cut (Methods) yielded 10 robust clusters with varying contributions from the different embryonic stages (Figure 1b, Extended Data Fig. 3, Methods, cell numbers in Extended Data Fig. 3h). Using t-Stochastic Neighbor Embedding (t-SNE) dimensionality reduction to visualise the data, three major groups were observed, one comprising almost all E6.5 cells, another mainly consisting of earlier S and NP stage cells, and a third containing predominantly later HF stage cells (Figure 1c). Importantly, clusters were coherent with the t-SNE visualization except for the small cluster 5 (Figure 1d).
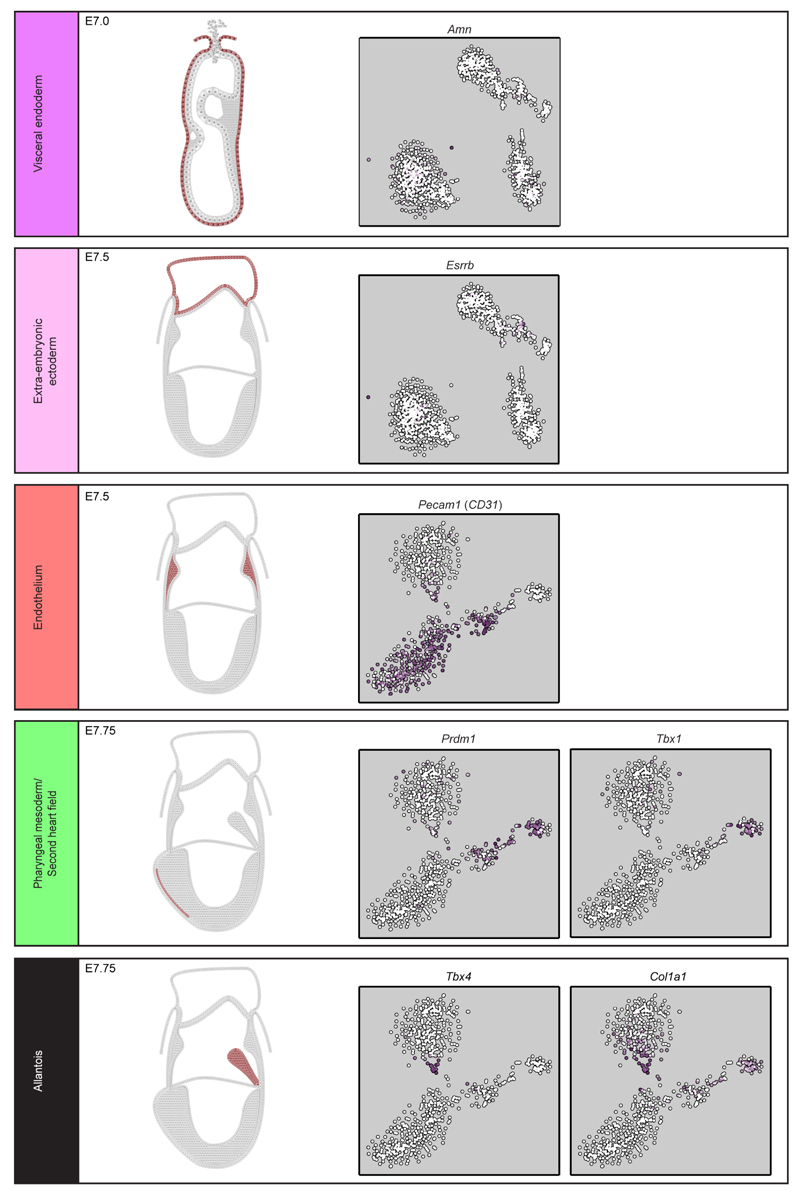
The expression of key marker genes allowed us to assign identities to each cluster: visceral endoderm, extra-embryonic ectoderm, epiblast, early mesodermal progenitors, posterior mesoderm, endothelium, blood progenitors, primitive erythrocytes, allantoic mesoderm, pharyngeal mesoderm (Figure 1b,Extended Data Fig. 4 and Extended Data Fig. 3h). Because of the limited cell numbers and lack of markers for their prospective isolation, conventional bulk transcriptome analysis of these key populations has never before been attempted.
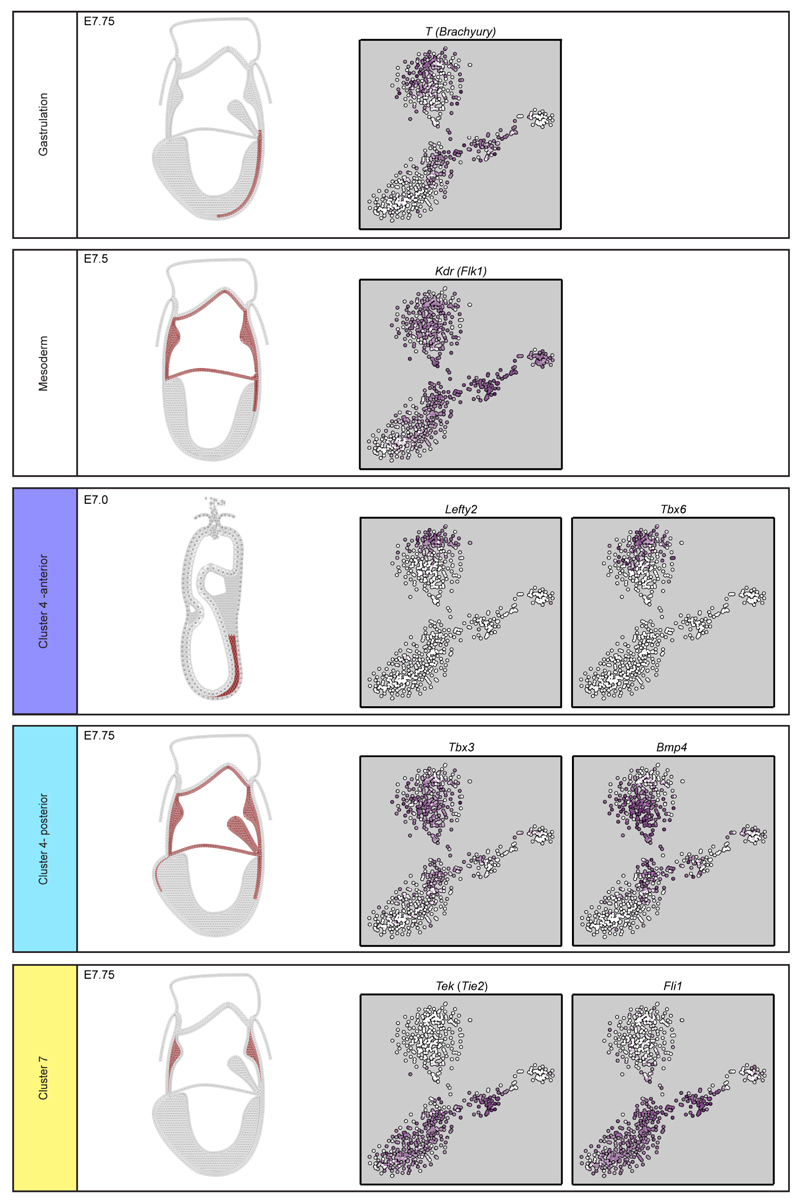
Since the T-box transcription factor Brachyury - encoded by the_T_ gene – marks the nascent PS6, we investigated the gene expression programs associated with_T_ induction in the E6.5 cells (cluster 3). T_expression was restricted to a distinct subset of epiblast cells found closest to cluster 4 (Figure 1d, Extended Data Fig. 5b), with rare isolated cells within the bulk of the epiblast population also expressing moderate levels, consistent with priming events for single gastrulation-associated genes. T expression correlated with other gastrulation-associated genes including Mixl1 and_Mesp1 (Figure 2a), with_Mesp1_ highly expressed only in the small subset of cells situated at the pole of the E6.5 epiblast cluster (association of T and_Mesp1_ expression: p-value 3x10-15, Fisher’s exact test). We also observed a subset of cells distinct from the_T_+/Mesp1+ population, which expressed Foxa2, suggestive of endodermal priming7 (Extended Data Fig. 5d).
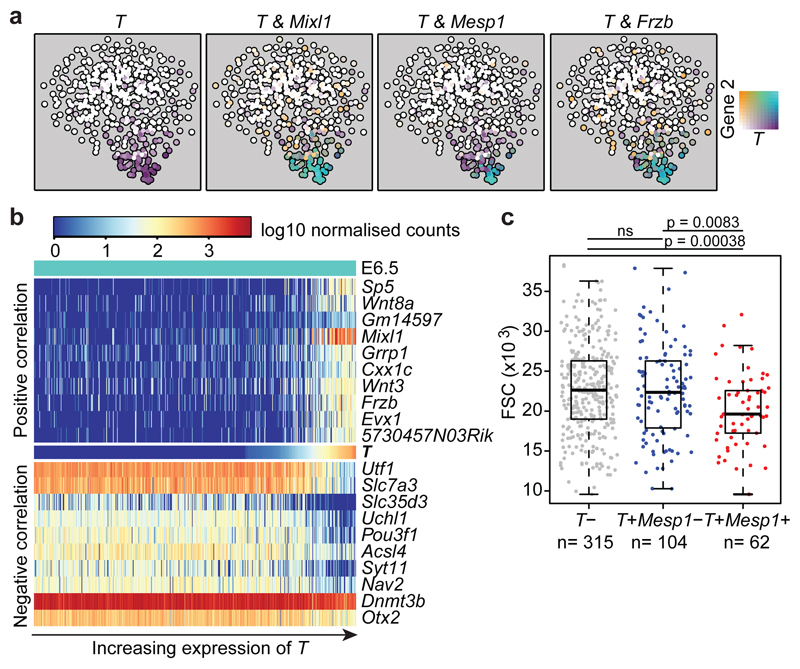
Figure 2. Transcriptional program associated with T induction in E6.5 epiblast cells.
a) t-SNE of the 481 E6.5 cells in cluster 3. Points are colored by expression of T (Brachyury) and Mixl1, Mesp1 and Frzb. b) Heatmap showing the 10 genes most highly positively and negatively correlated with_T_ (Supplementary Information Table 1). c) Forward scatter (FSC) for the 481 E6.5 epiblast cells in cluster 3, with cells grouped according to T/Mesp1 expression. Boxplots indicate the median and interquartile range. P-values were calculated using a two-sided Welch’s t-test for samples with unequal variance, with FDR correction for multiple testing.
We next identified genes displaying correlated expression with_T_, which identified known markers and regulators such as_Mixl1_, and also genes not previously implicated in mammalian gastrulation, such as Slc35d3, an orphan member of a nucleotide sugar transporter family8 and the retrotransposon-derived transcript Cxx1c9 (Figure 2b and Supplementary Information Table 1). Genes negatively correlated with T were consistently expressed across the majority of epiblast cells, suggesting that cells outside the PS have not yet committed to a particular fate, consistent with the known plasticity of epiblast cells in transplant experiments10. Ingressing epiblast cells undergo an EMT, turning from pseudo-stratified epithelial cells into individual motile cells, a conformational change associated with alterations in cell size and shape11. Our E6.5 epiblast cells were isolated using index sorting thus providing a forward scatter (FSC) value for each cell. As shown in Figure 2c,T+/Mesp1+ co-expressing cells showed a significant reduction in FSC values compared to_T_+/Mesp1_- and_T- cells. Since FSC correlates positively with cell size, this observation provides a direct link between specific transcriptional programs and characteristic physical changes associated with gastrulation. As_T_+/Mesp1+ cells also express_Mesp2_, this observation was consistent with the known EMT defect in_Mesp1_/Mesp2 double knock-out embryos12. Index sorting therefore linked expression changes with dynamic physical changes similar to those recognised to occur during chicken gastrulation13.
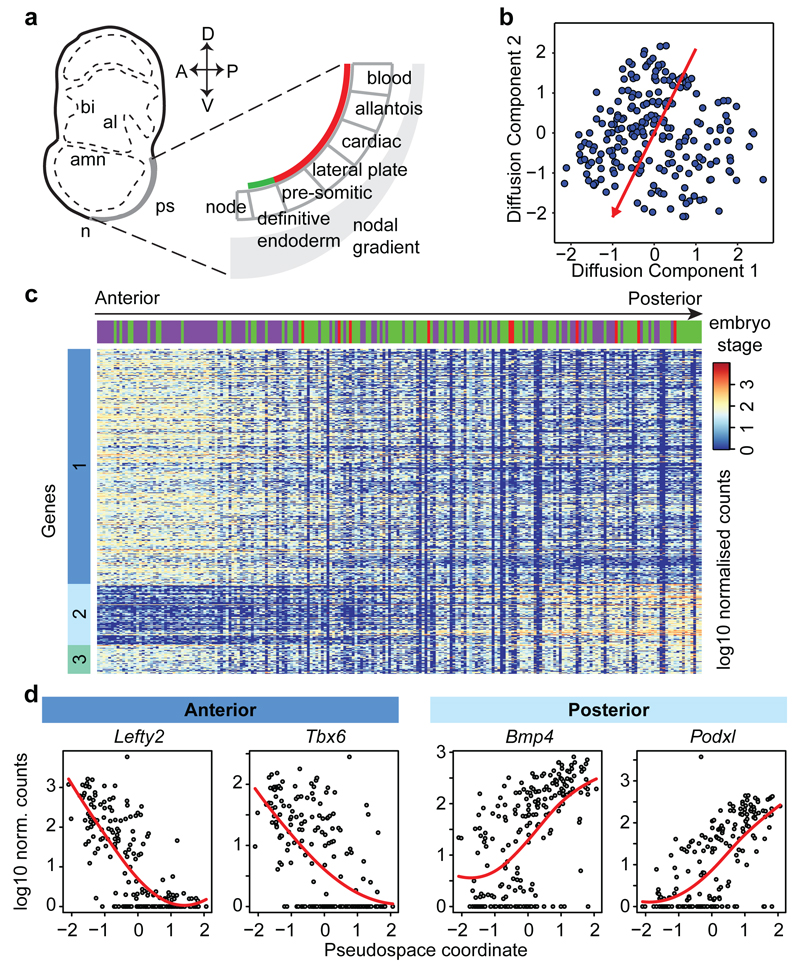
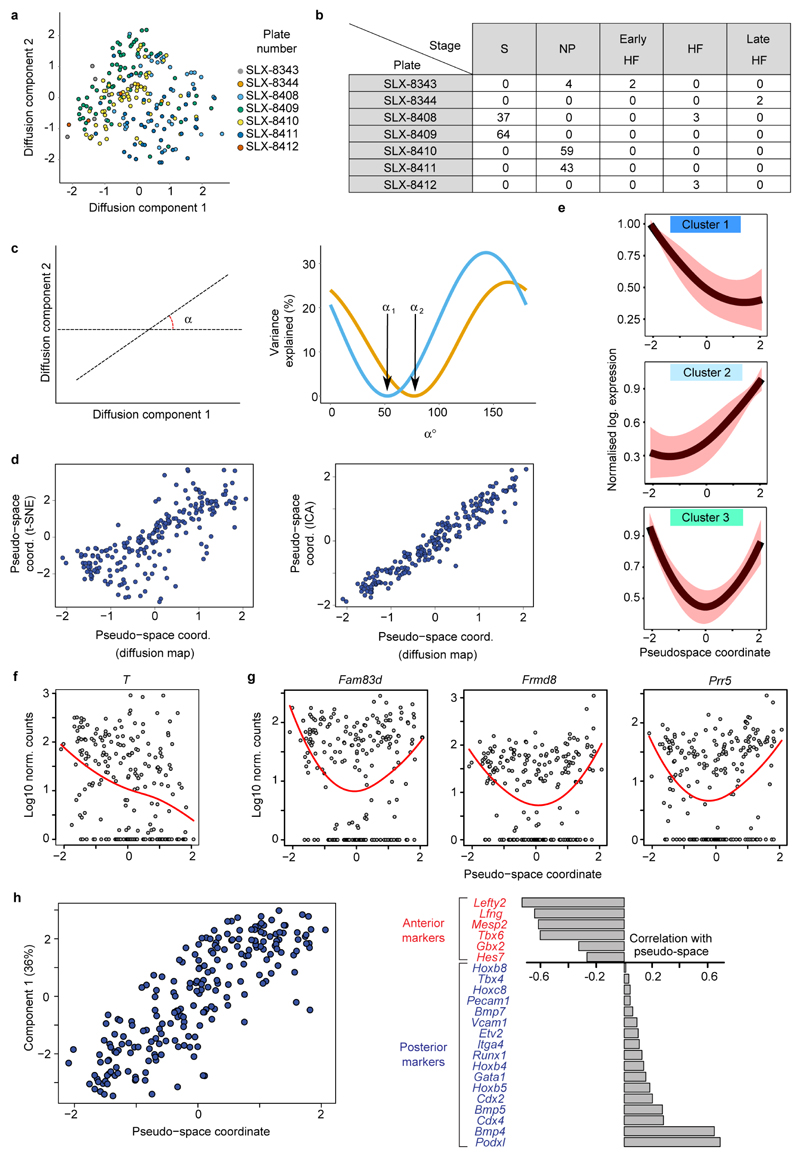
We next focused on mesodermal lineage divergence during and immediately after gastrulation. We reasoned that approaches analogous to those used to order single cells in developmental pseudotime could be used to infer the location of cells in pseudo_space_, specifically with respect to the anterior-posterior axis of the primitive streak (Figure 3a). To this end, we used diffusion maps14, a dimensionality reduction technique particularly suitable for reconstructing developmental trajectories15. We identified the diffusion-space direction that most likely represents true biological effects (see Methods), which we interpreted as the pseudospace coordinate (red line in Figure 3b, Extended Data Fig. 6a-d). Hierarchical clustering revealed three groups of genes (Figure 3c, Extended Data Fig. 6e, Supplementary Information Table 4) showing a gradient of expression along the pseudospace axis. These were assigned as anterior (darker blue, 334 genes) and posterior (lighter blue, 87 genes) due to the enrichment of genes with known differential expression along the anterior-posterior axis of the PS (Figure 3d, Extended Data Fig. 6f-h, 7). A third cluster was expressed highly at either end of the pseudospace axis (turquoise, 41 genes). Interestingly, the more posterior Flk1+ mesodermal cells are associated with the allantois, blood and endothelial clusters (Figure 1d, Extended Data Fig. 5c), which are known to arise from the posterior primitive streak. Gene ontology analysis revealed that the putative anterior genes were associated with terms relating to somite development, endoderm development and Notch signaling, consistent with a more anterior mesoderm identity16 (Supplementary Information Table 2a; Extended Data Fig. 6h). Conversely, the putative posterior mesoderm cluster was associated with BMP signaling, hindlimb development and endothelial cell differentiation, which consistent with the posterior portion of the streak17.
Figure 3. Dimensionality reduction reveals transcriptional profiles associated with cell location in the embryo.
a) Schematic of tissue emergence along the anterior-posterior primitive streak, derived from 29. Mesodermally- and endodermally-derived tissues are marked by a red and green line, respectively. b) Diffusion map of 216 cells in cluster 4 with pseudospace axis in red. Projections onto this axis represent pseudospace coordinates. c) Heatmap for differentially expressed genes along the pseudospace axis, showing genes more highly expressed in the anterior (dark blue) and posterior region (light blue), or highly expressed at either end (aquamarine). d) Expression profiles for example genes.
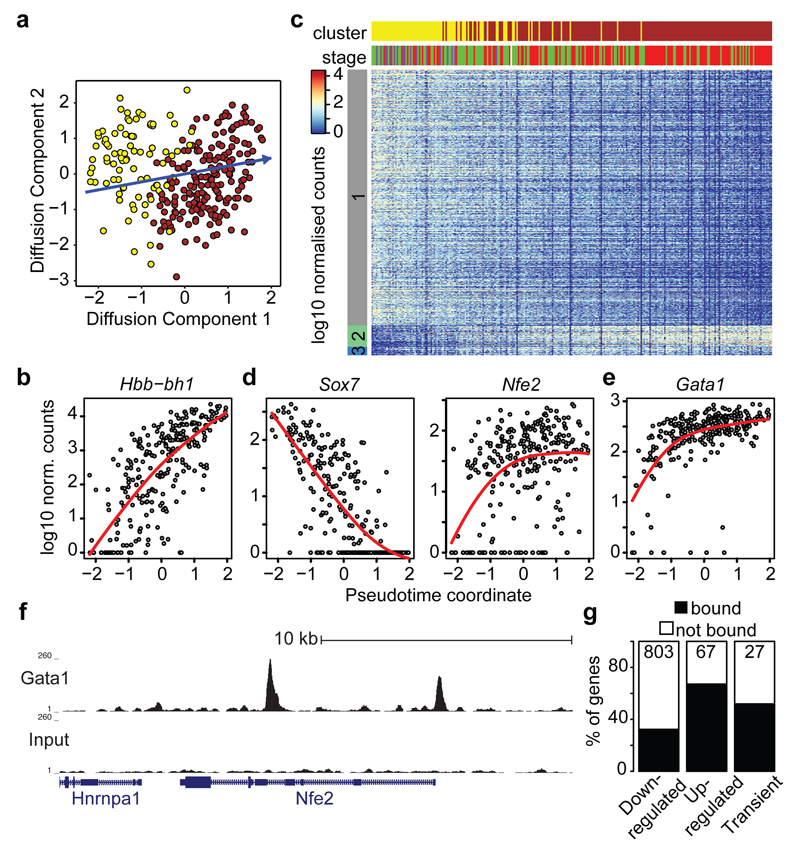
Although derived from the same embryonic stages as the mesodermal progenitor cells, cluster 7 lacks expression of genes such as Mesp1, yet expresses_Tal1, Sox7, Tek_ (Tie2) and Fli1 which are vital for extra-embryonic mesoderm formation (Figure 1b,Extended Data Fig. 5, 7). Expression of Kdr and Itga2b(Extended Data Fig. 5b) further highlights clusters 7 and 8 (brown) as corresponding to the developmental journey towards blood (Figure 4a), with a transition to mostly HF stage cells in cluster 8 and increasing expression of embryonic haemoglobin_Hbb-bh1_ (Figure 4b). Given the apparent trajectory of blood development from cluster 7 to 8, we used an analogous approach to that described above to recover a pseudotemporal ordering of cells (Figure 4a, Extended Data Fig. 8a-d and Methods). 803 genes were down-regulated, including the haematovascular TF Sox7 which is known to be down-regulated during blood commitment15 (Figure 4c,d, Extended Data Fig. 8e,f). 67 genes were up-regulated including the erythroid-specific TFs Gata1 and Nfe2, and embryonic globin Hbb-bh1 (Figure 4b, d, e,Extended Data Fig. 8). 27 genes were transiently expressed, including the known erythroid regulator Gfi1b. Significant GO terms associated with the up-regulated genes were indicative of erythroid development, while down-regulated genes were associated with other mesodermal processes including vasculogenesis and osteoblast differentiation (Supplementary Information Table 2b).
Figure 4. Inferring the transcriptional program underlying primitive erythropoiesis.
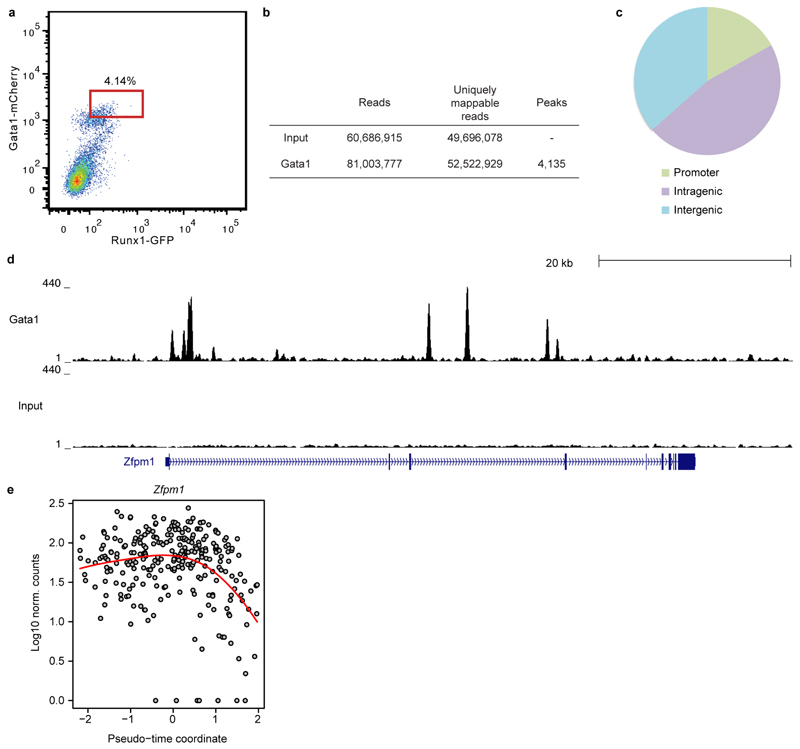
a) Diffusion map of 271 cells in clusters 7 and 8 displaying the inferred pseudotime axis (blue). b) Expression of_Hbb-bh1_ ordered by pseudotime (local polynomial fit).c) Heatmap ordered along the pseudo-time axis. Horizontal bars indicate cluster and developmental stage. Genes shown were repressed (grey), activated (green) or transiently expressed (blue). d) Examples of activated and repressed genes and e) Gata1 as in (b). f) UCSC Browser tracks for Gata1 ChIP-seq and input in Runx1+Gata1+ cells; the_Nfe2_ locus is shown. g) Percentage of genes in each group identified in C overlapping Gata1 targets.
Gata1 null embryos die at around E10.5 due to the arrest of yolk sac erythropoiesis18. We generated genome-wide ChIP-seq data for Gata1 in haematopoietic cells derived following 5 days of ESC in vitro differentiation (Extended Data Fig. 9a-c). The group of up-regulated genes from the pseudotime analysis showed a pronounced overlap with Gata1 targets (p < 2.2x10-16, Fisher’s test) including known targets such as Nfe2 and Zfpm1 (Figure 4f,g, Extended Data Fig. 9d,e, Supplementary Information Table 6). Integration of single cell transcriptomics with complementary TF binding data therefore predicts likely in vivo targets of developmental regulators such as Gata1.
Two contrasting mechanisms are commonly invoked to explain how drivers of cell fate determination regulate cell type diversification. The first involves fate restriction through a step-wise sequence of binary fate choices and is supported by mechanistic investigations using embryonic stem cell (ESC) differentiation2,19. The alternative invokes acquisition of diverse fates from independent precursor cells and is commonly supported by cell transplantation and lineage tracing analysis (Figure 5a)1,10,20,21. In contrast to the retrospective nature of transplantation and lineage tracing experiments where measurements are typically obtained a day or more after cell fate decisions are made, single cell transcriptomics allows cellular states to be determined at the moment when fate decisions are executed since low cell numbers are not a limiting factor.
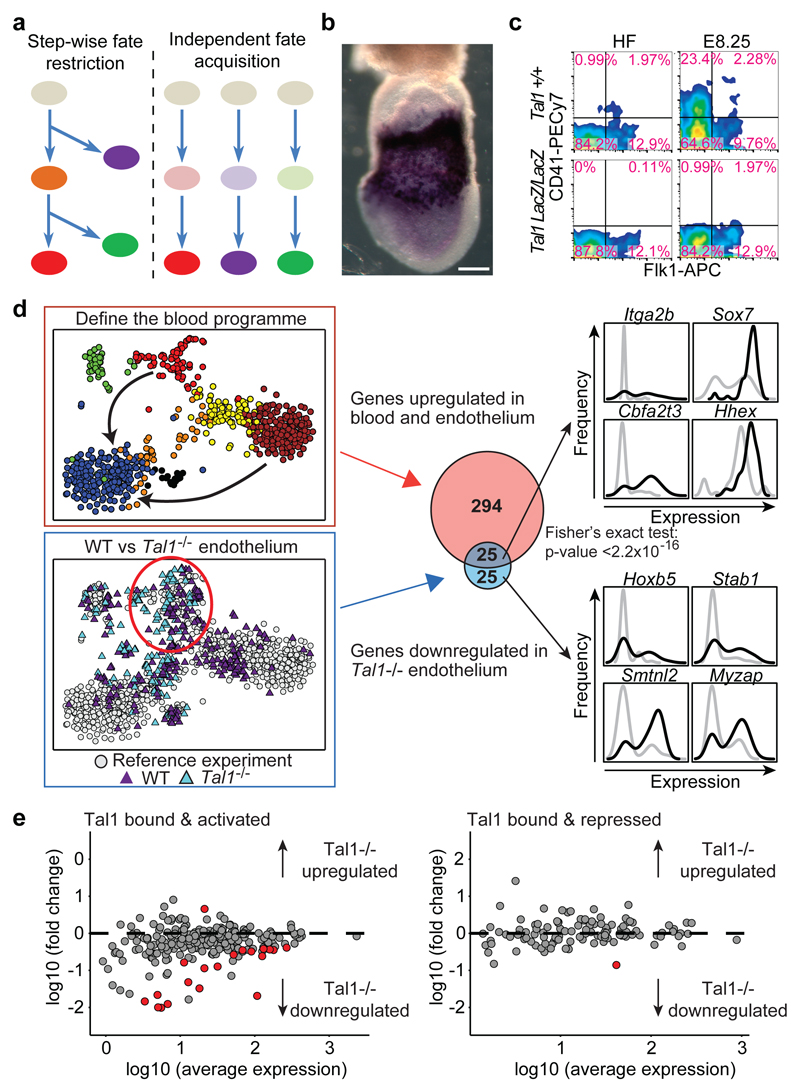
Figure 5. Analysis of _Tal1_-/- embryos suggests independent fate acquisition.
a) Schematic of two cell fate diversification models.b) Tal1 in situ hybridization at HF stage. Scale bar: 200µm**.** c) Flow cytometry of WT and _Tal1_-/- mice at HF and E8.25. d) Blood program genes are differentially expressed between nascent mesoderm (blue) and endothelial (red) and blood cells (brown). Differential expression between 45 _Tal1_-/- and 59 WT endothelial cells (lower left t-SNE) identified 50 down-regulated genes. Gene set overlap (central panel) indicates failure to induce the blood program in_Tal1_-/- endothelium (p < 2.2 x 10-16, Fisher's test). On the right are expression distributions for selected genes in wild type (black) or _Tal1_-/- (grey) endothelial cells.**e)** For genes previously reported3 to be bound and activated (left panel) or bound and repressed (right panel) by Tal1, fold change between _Tal1_-/-and WT endothelium (defined in D) is plotted against average expression. Red circles: genes with a fold change > 1.5 and an FDR < 0.05.
The bHLH TF Tal1 (also known as Scl) is essential for the development of all blood cells22,23 with strong expression in posterior mesodermal derivatives (Figure 5b). _Tal1_-/- bipotential blood/endothelial progenitors cannot progress to a haemogenic endothelial state19, Tal1 overexpression drives transdifferentiation of fibroblasts into blood progenitors24 and _Tal1_-/- mesodermal progenitors from the yolk sac give rise to aberrant cardiomyocyte progenitors when cultured in vitro2. However, the precise nature of the molecular defect within _Tal1_-/- mesodermal progenitors within the embryo has remained obscure, because cell numbers are too small for conventional analysis.
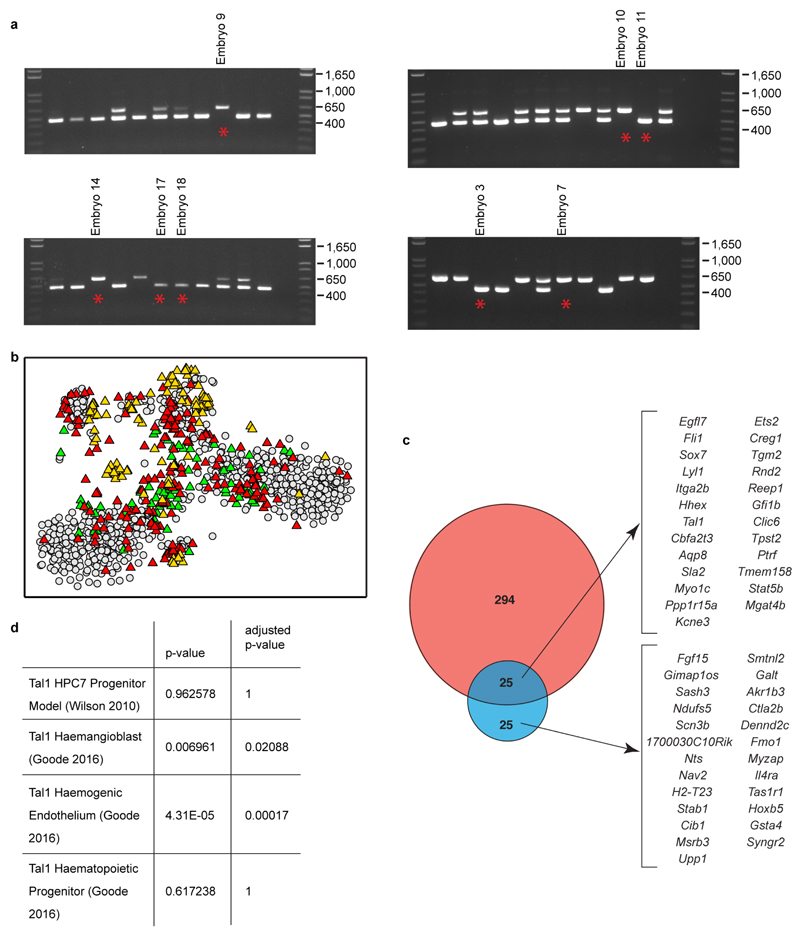
We profiled single Flk1+ cells from 4 wild type and 4_Tal1_-/- embryos obtained from E7.5 (NP) to E8.25 (4 somite stage) (256 wild type and 121 _Tal1_-/- cells; Figure 5c, Extended Data Fig. 10), and computationally assigned cells to the previously defined 10 clusters (Methods). Cells from wild type embryos contributed to all clusters, while _Tal1_-/- embryos did not contain any cells corresponding to the blood progenitor and primitive erythroid clusters (yellow and brown, Figure 5d) consistent with the known failure of primitive erythropoiesis in _Tal1_-/- embryos23 and their lack of CD41 expression (Figure 5c).
Fifty-one _Tal1_-/- cells were mapped to the endothelial (red) cluster, which therefore allowed us to investigate the early consequences of Tal1 deletion in this key population for definitive haematopoietic development (Figure 5d, Supplementary Information Table 7, 8). Fifty genes were down-regulated in Tal1_-/- endothelial cells (fold change < 0.67, 5% FDR). These include known regulators of early blood development (Itga2b, Lyl1, Cbfa2t3, Hhex, Fli1, Ets2, Egfl7, Sox7, Hoxb5), consistent with Tal1 specifying a haematopoietic fate in embryonic endothelial progenitor cells19, and in particular_Hoxb5, which has recently emerged as a powerful marker for definitive blood stem cells25. Single cell profiling also identified genes with altered distributions of expression. For example,Sox7 changed from a largely unimodal pattern in wild-type cells to a bimodal on/off pattern in Tal1_-/- endothelial cells, while_Cbfa2t3 showed the opposite pattern (Figure 5d).
However, we did not observe up-regulation of cardiac markers in_Tal1_-/- endothelial cells (Figure 5e, Supplementary Information Table 8, 9). Previously, this up-regulation had been observed in YS endothelial cells collected 1-1.5 days later than our data2, and had been taken as evidence that Tal1 acts as a gatekeeper controlling the balance between alternative cardiac and blood/endothelial fates within single multipotent mesodermal progenitors3. Our results however suggest that the primary role of Tal1 is induction of a blood program, and the subsequent ectopic expression of cardiac genes may be the result of secondary induction events acting on a still relatively plastic mesodermal cell blocked from executing its natural developmental program.
Here we have used single cell transcriptomics to obtain a comprehensive view of the transcriptional programs associated with mammalian gastrulation and early mesodermal lineage diversification. Further technological advances to resolve epigenetic processes at single cell resolution26 and match single cell expression profiles with spatial resolution27,28 are likely key drivers of future progress in this field. Finally, our analysis of_Tal1_-/- embryos illustrates how the phenotypes of key regulators can be re-evaluated at single cell resolution to advance our understanding of early mammalian development.
Methods
Timed matings and embryo collection
All procedures were performed in strict adherence to United Kingdom Home Office regulations. Timed matings were set up between CD1 mice (which produce large litters). Embryos were staged according to the morphologic criteria of Downs and Davies30, and classified broadly as primitive streak (S), neural plate (NP) or head fold (HF) stage. Suspensions of cells from individual embryos were prepared by incubating with TrypLE Express dissociation reagent (Life Technologies) at 37°C for 10 minutes and quenching with heat inactivated serum. All cells were stained with DAPI for viability. At E6.5, the distal half of the embryo was dissected and dissociated into a single cell suspension, and live cells were sorted. For E7.0 and older, suspensions consisted of the whole embryo and were also stained with Flk1-APC (AVAS12 at 1:400 dilution; BD Bioscience) and only Flk1+cells were collected. For cell sorting of CD41+Flk1- and CD41+Flk1+ cells from NP stage and HF stages, suspensions were stained with Flk1-APC, PDGFRa-PE (APA5 at 1:200 dilution; Biolegend) and CD41-PEcy7 (MWReg30 at 1:400 dilution; Biolegend) for 20 minutes at 4°C as described31. Cells were sorted from 7 E6.5 embryos. Flk1+ cells were sorted from 3 S stage, 4 NP stage and 3 HF stage embryos (Extended Data Fig. 1a). CD41+Flk1- and CD41+Flk1+ cells were sorted from the same embryos, an additional 8 each at NP and HF stages (Extended Data Fig. 1b). Cell sorting was performed with a BD Influx cell sorter in single cell sort mode with index sorting to confirm the presence of a single event in each well. Additional cells were sorted into tissue culture plates to visually confirm the presence of single events.
To obtain Tal1_-/- cells, timed matings were set up between Tal1LacZ/+ mice32. Flk1+ cells were sorted as above from 4 embryos for each genotype: from 1 embryo for each genotype at NP and 4 somite (4S) stages, from 2 HF stage embryos for Tal1LacZ/LacZ(designated Tal1-/-),_ 1 HF stage WT embryo and 1 WT embryo intermediate between NP and HF stages. Genotyping PCR using 1/20 suspension cells was performed as described previously32.
Single-cell RNA sequencing
Library preparation and mapping of reads
scRNA-seq analysis was performed using the Smart-seq2 protocol as previously described33. Single cells were sorted by FACS into individual wells of a 96-well plate containing lysis buffer (0.2 % (v/v) Triton X-100 and 2 U/μl RNase inhibitor (Clontech)) and stored at -80°C. Libraries were prepared using the Illumina Nextera XT DNA preparation kit and pooled libraries of 96 cells were sequenced on the Illumina Hi-Seq 2500. Reads were mapped simultaneously to the M. musculus genome (Ensembl version 38.77) and the ERCC sequences using GSNAP (version 2014-10-07) with default parameters. HTseq-count34 was used to count the number of reads mapped to each gene (default options).
Identification of poor quality cells
To assess data quality35, five metrics were used: (i) total number of mapped reads, (ii) fraction of total reads mapped to endogenous genes, (iii) fraction of reads mapped to endogenous genes that are allocated to mitochondrial genes, (iv) fraction of total reads mapped to ERCC spike-ins and (v) level of sequence duplication (as estimated by FastQC, version 0.11.4, http://www.bioinformatics.babraham.ac.uk/projects/fastqc).
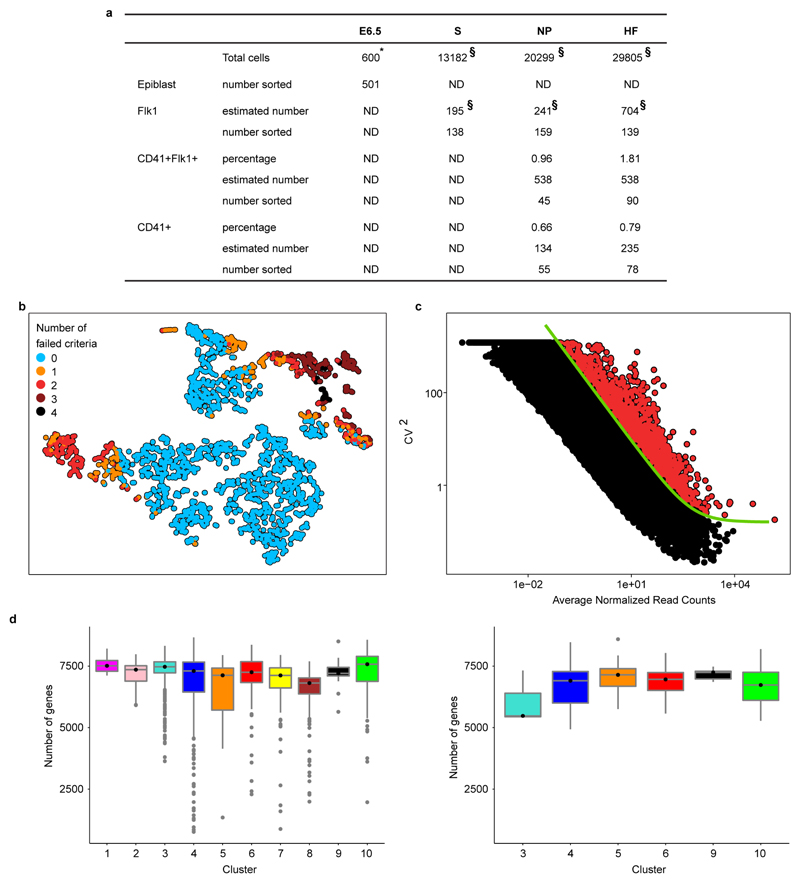
For all downstream analyses, we only retained samples that had (i) more than 200,000 reads mapped (either to ERCC spike-ins or endogenous mRNA), (ii) more than 20% of total reads mapped to mRNA, (iii) less than 20% of mapped reads allocated to mitochondrial genes, (iv) less than 20% of reads mapped to ERCC spike-ins and (v) less than 80% of duplicated sequences. Out of the 2,208 cells that were captured across the two experiments, 1,582 (i.e., ~72% of the total) passed our quality check. A t-SNE projection36 of the values of these five metrics (Extended Data Fig. 2b) shows that most of discarded cells tend to cluster together and fail at least two criteria. All metrics were standardised before applying t-SNE with the “RtSNE” function (default parameters) from the R package “RtSNE” (version 0.1) 37.
Normalization of read counts
The data was normalised for sequencing depth using size factors38 calculated on endogenous genes. By doing so, we also normalise for the amount of RNA obtained from each cell39, which is itself highly correlated with cell cycle stage40.
Highly variable genes and GO enrichment analysis
Highly variable genes were identified by using the method described in Brennecke et al.39. In brief, we fitted the squared coefficient of variation as function of the mean normalised counts39. In the fitting procedure, in order to minimise the skewing effect due to the lowly expressed genes39, only genes with a mean normalised count greater than 10 were used. Genes with an adjusted p-value (Benjamini-Hochberg method) less than 0.1 were considered significant (red circles in Extended Data Fig. 2c). This set of highly variable genes was used for the clustering analysis discussed below. The GO enrichment analysis was carried out using TopGO in its “elimination mode” with Fisher’s exact test; we considered as significant GO categories with an unadjusted p-value below 10-4 .
Differentially expressed genes
In order to find genes differentially expressed between two groups of cells we used edgeR41 (version 3.12). Before running edgeR, we excluded genes annotated as pseudogenes in Ensembl, sex-related genes (Xist and genes on the Y chromosome) and genes that were not detected or were expressed at very low levels (we considered only genes that had more than 10 reads per million in a number of cells greater than 10% of the cells in the smaller group being compared). The function “glmTreat” was then used to identify the genes having a fold change significantly greater than 1.5 at a FDR threshold equal to 0.05.
Clustering analysis
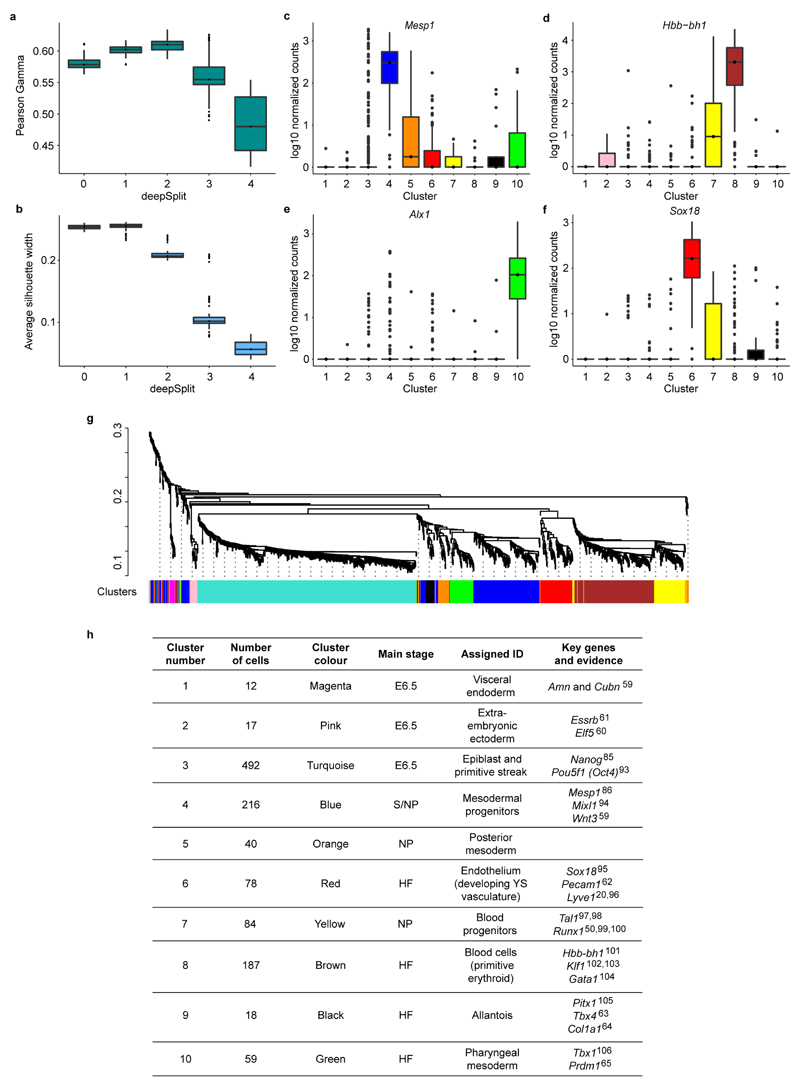
Clustering analysis was performed on the 1,205 WT cells from the first experiment that passed the QC. The Spearman correlation coefficient, ρ, was computed between the transcriptome of each pair of cells, which was then used to build a dissimilarity matrix defined as (1-ρ)/2. Hierarchical clustering was carried out (“hclust” R function with the “average” method) on the dissimilarity matrix and clusters were identified by means of the Dynamic Hybrid Cut algorithm42. The R function “cutreeDynamic” with the “hybrid” method and a minimum cluster size equal to 10 cells was used (“dynamicTreeCut” package, version 1.62). This function allows the user to specify the “deepSplit” parameter that controls the sensitivity of the method: higher values of this parameter correspond to higher sensitivity and can result in more clusters being identified, but also entail an increased risk of overfitting the data. The optimal trade-off between robustness of clustering and sensitivity was found by analyzing the results of the algorithm with all possible values of the deepSplit parameter (i.e. integer values from 0 to 4) on 100 subsamples of our data. In particular, in each subsample, we removed 10% of genes randomly selected before computing the dissimilarity matrix and applying the clustering algorithm.
The statistics of the Pearson gamma and the average silhouette width (computed with the “cluster.stats” function included in the R package “fpc”, version 2.1-10)43,44 of the subsamples (see Extended Data Fig. 3a,b) suggest that with “deepSplit=2” a good compromise is reached between robustness and sensitivity for our data. We identified 10 different clusters as well as two outlier cells that, although similar in gene expression to the mesodermal progenitor cells (cluster 4), were not assigned to any cluster by the algorithm, probably due to their relatively poor quality.
We then evaluated how specifically each gene is expressed in any given cluster. First, we found the differentially expressed genes (as described above) between all pairs of clusters. Marker genes for cluster_i_ are expected to be significantly up-regulated in_i_ across all pairwise comparisons involving cluster_i_. The average rank of a marker gene across the pairwise comparisons provides a measure of how specifically the marker is expressed in the cluster. Extended Data Fig. 3c-f show the expression values of marker genes for four different clusters. We provide the full list of markers in Supplementary Information Table 3. The clusters were visualised by using t-SNE (as implemented in the “RtSNE” R package) on the dissimilarity matrix.
Single-cell trajectories in pseudo-space: the anterior/posterior axis of the primitive streak
As discussed in the main text, cells allocated to cluster 4 (Figure 1b-d) are cells that have likely exited the primitive streak only recently. We sought to align the cells along a pseudospatial trajectory representing the anterior-posterior axis of the primitive streak, which would allow us to identify the likely original locations of each cell along such an axis.
To do this we adopted an unsupervised approach: we did not use any prior information about marker genes, but selected the strongest signal present in this cluster of cells (controlling for potential batch effects) and later verified its biological meaning. We first used a diffusion map-based technique to reduce the dimensionality of the dataset. Diffusion maps have recently been successfully applied to identify developmental trajectories in single-cell qPCR and RNA-seq data14,15. We used the implementation of the “destiny” R package (“DiffusionMap” function) developed by Angerer et al.45. We restricted the analysis to genes that are highly variable among cells in the blue cluster and have an average expression above 10 normalised read counts. The centered cosine similarity was used (“cosine” option in the “DiffusionMap” function) and only the first two diffusion components (DC1 and DC2) were retained for downstream analysis.
In addition to biologically meaningful signals, batch effects (due to cells being sorted and processed on different plates) can also be present and induce structure within the data. While in our dataset the batch effect does not strongly influence the definition of different populations of cells, it might become relevant when finer structures within a single cluster of cells are considered (see Extended Data Fig. 6a). In order to tease apart the signals due to biological and batch effects, we computed the fraction of variance attributable to the batch effect along each direction in the diffusion space using a linear regression model. The direction “orthogonal” to the batch effect, i.e., the direction associated with the smallest fraction of variance explained by the batch effect, was considered as mostly driven by a biologically relevant signal. Hence, all cells were projected on this direction to obtain a “pseudo-coordinate” representing the state of a cell relative to the biological process captured by the diffusion map. The direction was identified by the angle α that it formed with the DC1 axis (Extended Data Fig. 6c).
Cells considered here are mostly from 2 batches including cells from the Primitive Streak stage (plate SLX-8408 and SLX-8409) and 2 batches including cells from the Neural Plate stage (plate SLX-8410 and SLX-8411;Extended Data Fig. 6b). For each of these two sets of batches, we computed the fraction of variance that can be explained by the batch covariate along any possible direction in the diffusion plot. The angles α1 and α2corresponding to the directions orthogonal to the two batch effects are very close to each other (Extended Data Fig. 6c); we took the average ᾱ between these two angles to approximate the direction orthogonal to both batch effects.
Cells’ coordinates in the diffusion space were projected along the direction identified by ᾱ, and this projection was interpreted as a “pseudospace” coordinate representing the position of cells along the primitive streak (see main text and Figure 3). We tested the robustness of such a pseudospace coordinate by repeating the same analysis with alternative dimensionality reduction techniques (t-SNE and Independent Component Analysis), which gave highly correlated coordinates (see Extended Data Fig. 6d). A Principal Component Analysis carried out with a set of previously known markers for the anterior and posterior regions of the primitive streak also yielded a first component highly correlated with the pseudospace coordinate (see Extended Data Fig. 6d left panel). Moreover, the pseudospace coordinate had a positive (negative) correlation with the posterior (anterior) markers used (see Extended Data Fig. 6d right panel). These results strongly support the robustness of the signal we identified as well as its biological interpretation.
Once the pseudospace trajectory was defined, we selected genes that were differentially expressed along the trajectory. First, we removed all genes that were not detected in any cell. Then, for each gene, we fitted the log10-expression levels (adding a pseudocount of 1) by using two local polynomial models: one with degree 0 and another with degree 2 (“locfit” function in “locfit” R package, nearest neighbor component parameter equal to 1). The first, simpler model is better suited for genes that do not change their expression level along the trajectory. The second model has a greater number of parameters and is able to reproduce the more complex dynamics of genes that are differentially expressed.
We evaluated these two models by using the Akaike information criterion (AIC), a score that measures how well the data are reproduced by the model and includes a penalization for more complex models46. Better models according to this criterion correspond to smaller AIC scores.
To compute the AIC scores for the two models, we used the “aic” function available in the “locfit” R package, and then calculated the difference:
ΔAIC = AIC(degree = 2) – AIC(degree = 0)
Negative values indicate that the more complex model with degree 2 local polynomials perform better, and therefore correspond to genes that are more likely to be differentially expressed. Genes having a ΔAIC < -2 were considered to be significantly differentially expressed along the trajectory46.
A hierarchical tree was built with the normalised expression patterns of the 462 differentially expressed genes (function “hclust” with average linkage method and dissimilarity based on Spearman correlation) and a dynamic hybrid cut algorithm (“cutreeDynamic” function, minimum cluster size equal to 5) split this set of genes in 3 clusters according to the type of dynamics they have (see Figure 3, Extended Data Fig. 6e and Supplementary Information Table 4).
Single-cell trajectories in pseudo-time: the blood developmental trajectory
As discussed in the main text, clusters 7 and 8 (yellow and brown clusters in Figure 1b and 1d) include blood progenitors at different stages of differentiation. By using a procedure analogous to the one described above, we aligned these cells along a trajectory representing embryonic blood development.
Extended Data Fig. 8a shows the diffusion plot with cells from the yellow and the brown clusters. Most of these cells come from plates SLX-8344 and SLX-8345 that were collected from embryos at Neural Plate and late Head Fold stages (see Extended Data Fig. 8b). With a linear regression model, where we controlled for biological parameters like stage and sorting, we found the direction that correlates the least with the batch effect associated to these two plates and projected all cells onto it (Extended Data Fig. 8c). Note that the minimum correlation with the batch effect is achieved at a very small value of α (~10°, see Extended Data Fig. 8c), suggesting that the first diffusion component is mainly driven by a biologically meaningful signal and the batch effect plays a minor role here even at this more detailed scale of analysis. The new cell coordinate obtained from the projection was interpreted as a “pseudotime” coordinate, which represents the differentiation stage of each cell along their journey towards erythroid fate. As expected, cells in the yellow cluster have a smaller pseudotime coordinate compared to the brown cluster that is mainly composed of more differentiated primitive erythroid cells. An analysis with alternative dimensionality reduction techniques yielded highly correlated pseudotime coordinates, suggesting the robustness of the signal (Extended Data Fig. 8d). Furthermore, our biological interpretation of the pseudotime coordinate is supported by the expression pattern of genes that are known to be up-regulated or down-regulated along the blood developmental trajectory, as is clear via Principal Component Analysis (see Extended Data Fig. 8f).
By using the filtering and clustering procedure described in the previous section, we were able to detect 897 genes that were differentially expressed along the trajectory, which were divided in 3 clusters, each displaying a different type of dynamics (see Extended Data Fig. 8e and Supplementary Information Table 5).
Random Forest to allocate cells to previously identified clusters
Cells captured in the Tal1 experiment (testing dataset) were allocated to the clusters we previously identified by using a Random Forest algorithm47 (R package “randomForest”, version 4.6-12)48 trained on the cells captured in the first experiment (training dataset). The rank-normalised expression levels of all highly variable genes in the training dataset were used as variables (the R function “rank” was used for normalization, ties were averaged). The random forest algorithm was first used on the training data to assess variable importance with 1,000 classification trees. The 25% most important variables were selected to grow another set of 1,000 trees that were then used for the classification of the testing dataset. With this filtered set of variables the out-of-bag error estimate was ~4.8%.
The quality of allocation of each cell in the testing dataset was verified by computing the median of pairwise dissimilarities (defined as (1-ρ)/2, with ρ being the Spearman correlation) of that cell to all other cells in the training data allocated to the same cluster. Cells in the testing dataset having a median pairwise dissimilarity larger than the maximum of the medians of pairwise dissimilarities of cells in the training data were considered to be “unclassified” (~1.8% of all cells from the testing dataset). For the identification of differentially expressed genes between clusters in the testing data, only cells that were confidently allocated to the clusters (i.e., cells with a minimum difference of 10% probability between the best and the second best cluster allocation) were used.
Generation, maintenance and haematopoietic differentiation of Runx1-GFP/Gata1-mCherry ESCs
Runx1GFP/+:Gata1mCherry/Y ESCs were generated from morulae as described previously49,50. ESCs were grown on gelatinised plates (0.1 % gelatin in water) at 37°C and 5 % CO2 in ESC media (Knockout DMEM (Life Technologies) with 15 % FCS (batch-tested for ESC culture; Life Technologies), 2mM L-glutamine (PAA Laboratories), 0.5 % P/S, 0.1mM β-mercaptoethanol (Life Technologies) and 103 U/ml recombinant LIF (ORF Genetics)). Cells were passaged with TrypLE Express dissociation reagent (Life Technologies) every 1-3 days. ESCs were differentiated as EBs as previously described31,51. EBs were harvested into Falcon tubes after 5 days of culture and dissociated with TrypLE Express dissociation reagent and prepared for FACS.
ChIP-seq
ChIP was performed as described52with modifications for low cell numbers53. Approximately 7x106 FACS-sorted day 5 EB cells (Runx1-ires-GFP+/Gata1-mCherry+; Extended Data Fig. 9a) per ChIP were cross-linked using formaldehyde to a final concentration of 1%. As samples were pooled from several sorts, isolated nuclei were frozen on dry ice-cold isopropanol and stored at -80°C. During the immunoprecipitation step, 4 μl recombinant histone 2B (New England Biolabs) and 1 μl of mouse RNA (Qiagen; diluted 1/5 in IP dilution buffer) were added as carriers, followed by 7 μg of primary antibody (rabbit anti-Gata1, Abcam ab11963). Sequencing libraries were prepared using the TruSeq Kit (Illumina) for high throughput sequencing on an Illumina HiSeq 2500, according to manufacturer’s instructions, with size selection for fragments of 150-400 bp.
ChIP-seq mapping and analysis
Alignment of the ChIP-seq reads to the mouse mm10 genome, quality control and peak calling were performed according to the data pipeline set out by Sanchez-Castillo et al54. Peak calling was performed using MACS255 with p=1e-6. Post-processing using in-house scripts converted the peak coordinates to 400 bp based on peak summits given in the MACS output. Coordinates of genomic regions that lie at the end of chromosomes and/or in repeat regions were discarded from the final high-confidence peak lists. PolyAPeak56 was run in R to remove abnormally shaped peaks. Peaks were assigned to genes using an in-house script according to whether they overlapped with a known TSS or fell within 50 kb each side of a gene.
In situ hybridization
Whole-mount in situ hybridization for Tal1 was performed as described previously57. In situ hybridization probe for Tal1 was synthesised using published sequence (Tal1 860-1428, accession number M59764) with the DIG RNA labeling kit (Roche).
Code availability
All data were analysed with standard programs and packages, as detailed above. Code is available on request.
Extended Data
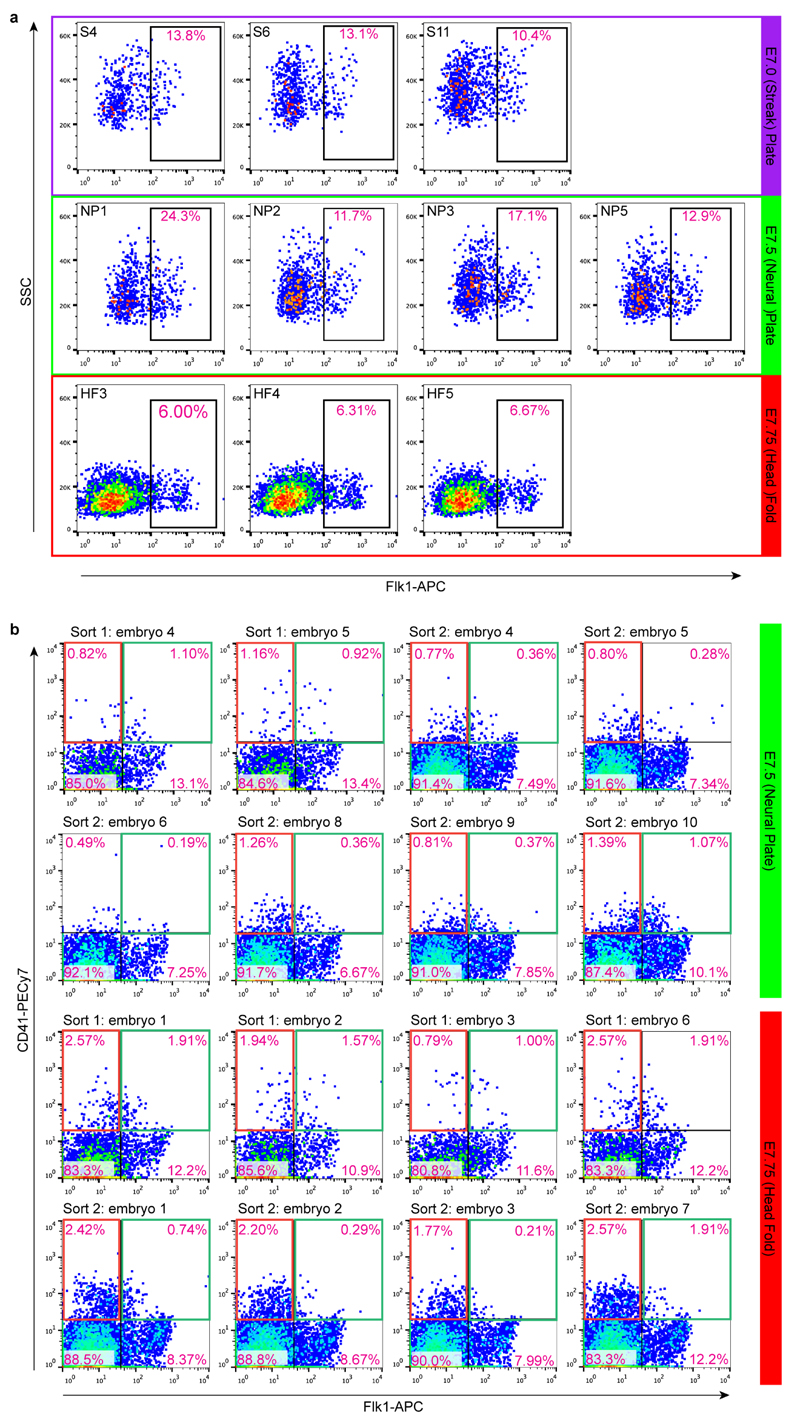
Extended Data Figure 1. Fluorescence activated cell sorting of single cells.
a) Flk1+ cells were sorted from 3 embryos at S and HF stages and 4 embryos at NP stage. Labels such at ‘S4’ refer to the embryo number in the metadata available online at http://gastrulation.stemcells.cam.ac.uk/scialdone2016.b) CD41+Flk1- cells (red gate) and CD41+Flk1+ (green gate) cells were sorted from 8 embryos each at NP and HF stages. Each stage was sorted on two occasions. Labels above FACS plots refer to the sort and embryo number in the metadata available online, as above. In all plots, pink text indicates the percentage of cells in that gate.
Extended Data Figure 2. Quality control of single cell RNA-seq data.
a) Table showing numbers and estimates of numbers of cells of different phenotypes present in embryos between E6.5 and E7.75 (HF stage) and numbers sorted for this study. (*) Total cell numbers for E6.5 are from Beddington and Robertson (1999)58 and (§) total numbers and numbers of Flk1+ cells are from Moignard et al., (2015)15. Percentages of cells expressing Flk1 and/or CD41 at NP and HF stages are the average values from the embryos used in this study and were used to calculate the estimated numbers present in embryos from the total cell numbers. ND, not done. b) t-SNE representation of the five metrics used to assess the quality of the samples. Only cells that passed all criteria (blue circles) were used for downstream analysis.c) Squared coefficient of variation (CV2) as a function of the mean normalised counts (µ) for genes across all cells. The green line shows the fit CV2 = a1/μ + α0. All highly variable genes (with an adjusted p-value < 0.1) are marked by red circles. d) Number of genes detected (i.e., with more than 10 normalised read counts) in cells across the different clusters in the WT (left panel) and the Tal1-/-(right panel) mice.
Extended Data Figure 3. Identifying cell clusters.
The Dynamic Hybrid Cut algorithm was used with all the possible values of the “deepSplit” parameter on 100 bootstrapped subsamples. To assess the quality of the clustering, the Pearson gamma**(a)** and the average silhouette width (b) were calculated. Higher values of these parameters correspond to better clustering. The Pearson gamma represents the correlation between the dissimilarity of samples and a binary variable that equals 1 for pairs of samples in the same cluster and 0 for samples in different clusters. The average silhouette width measures the average separation between neighboring clusters43,44. At “deepSplit”=2 the Pearson gamma is highest whereas the average silhouette width begins to decrease. This suggests that at such a value of the “deepSplit” parameter a good compromise between robustness and sensitivity is achieved. The Pearson gamma and the average silhouette width were computed with the R function “cluster.stats” in the “fpc” package (version 2.1-9). (c-f) Examples of marker genes for four clusters:Mesp1 for cluster 4 (top-ranked marker)(c), Sox18 for cluster 6 (second-ranked)(d), Hbb-bh1 for cluster 8 (fourth-ranked)(e) and Alx1 for cluster 10 (top-ranked)**(f).**The y-axis shows the log10 normalised expression of the genes. g) Dendrogram showing the clustering of the cells in the first experiment. The colors at the bottom indicate the cluster each cell was assigned to by the dynamic hybrid cut algorithm. Cluster assignment was used to sort cells in Figure 1b. h) Identities were assigned to the 10 clusters in Figure 1c based on the expression of key genes associated with various mesodermal lineages or spatial locations within the embryo.
Extended Data Figure 4. Expression of key marker genes in E7.0-7.75 embryos.
Schematic representations of expression patterns were generated from published in situ hybridization data (see citations) for key markers of clusters 1 (magenta, visceral endoderm59), 2 (pink, extra-embryonic ectoderm61), 6 (red, YS endothelium62), 9 (black, allantois63,64) and 10 (green, second heart field65,66). Anterior is shown on the left and posterior on the right. Also shown is the t-SNE for all cells or cells from E7.0 onwards (S, NP and HF stages) indicating expression of each gene (white, low; purple, high).
Extended Data Figure 5. Expression of key genes used for sorting single cells.
**a)**t-SNE as in Figure 1 showing the sorting strategy for each of the 1,205 cells.**b)**Expression of Flk1 (Kdr), CD41 (Itga2b), Scl (Tal1),Gata2 and T(Brachyury) superimposed onto the t-SNE.c) t-SNE showing only the cells from S, NP and HF stages, colored according to cluster as in Figure 1c and E. d) t-SNE for the 481 E6.5 cells in cluster 3, as in Figure 2a. Each point is colored by expression of T and Foxa2.
Extended Data Figure 6. Pseudospace analysis of cluster 4 correlates with anterior-posterior position along the primitive streak.
a) Diffusion plot of cells in cluster 4. Different colors correspond to different plates and different lanes of flow cells.b) Table showing the number of cells in each stage analysed on the different lanes of flow cells (S, Primitive streak; NP, Neural plate; HF, Head fold). c) A direction in the diffusion space can be identified by the angle α that it forms with the first diffusion component (left panel). For each value of α the right panel shows the percentage of variance explained by the batch effect associated to plates SLX-8408 and SLX-8409 (orange line) and plates SLX-8410 and SLX-8411. α1 and α2 are the angles corresponding to directions that correlate the least with the batch effect (i.e., variance explained by the batch effect is minimum). **d)**The use of alternative dimensionality reduction techniques results in the identification of highly correlated pseudo-space coordinates. A t-SNE projection of the dissimilarity matrix was carried out (perplexity set to 50), and the direction corresponding to the pseudo-space coordinate was estimated by minimizing the correlation with the batch effect (left panel; Spearman correlation between the two pseudo-space coordinates 0.79, p-value < 2.2x10-16). Independent Component Analysis was performed on the dissimilarity matrix with the “fastICA” R function, and 3 independent components (corresponding to the 2 batch effects and the biological effect) were estimated. The presumptive pseudo-space coordinate is the component having the smallest correlation with the batch effects (right panel; Spearman correlation coefficient is 0.97, p-value < 2.2x10-16). e) Plots showing the average expression of genes in clusters 1-3 of Figure 3c along the pseudospace axis. Gene expression levels are normalised between 0 and 1. Black lines indicate the normalised mean expression levels of genes in each cluster as obtained from the fitting procedure and red shaded area indicates standard deviation. **f)**Expression of T as function of the pseudospace coordinate.g) Gene expression levels for example genes showing high-low-high expression pattern across the blue cluster. In B and C, putative anterior cells are to the left and posterior to the right as in C. Each dot represents a cell and red lines indicate fits based on local polynomial functions (see Methods).h) We performed Principal Component Analysis (PCA) on the cells in cluster 4 by using markers of presomitic mesoderm as anterior mesoderm markers and genes expressed in haemato-vascular and allantoic mesoderm as posterior markers67–80, as well as_Podxl_ which was shown to separate distinct Flk1+ mesodermal lineages81. The first component explained 36% of the total variance and was highly correlated with the pseudo-space coordinate (left panel; Spearman rank correlation 0.84, p-value < 2.2x10-16). All the anterior markers were negatively correlated with the pseudo-space coordinate, whereas all posterior markers had a positive correlation (right panel).
Extended Data Figure 7. Expression of key genes along the anterior-posterior axis of the primitive streak in E7.0-7.75 embryos.
Schematic representations of gene expression were generated from published in situ hybridization data (see citations) for key markers of clusters 4 (blue, mesoderm) and 7 (yellow, posterior mesoderm/blood progenitors). Expression of T(Brachyury)82and Flk1 (Kdr – from in house data) are shown to illustrate the extent of the primitive streak at E7.5.Lefty2 and Tbx659 are expressed in the putative anterior portion of cluster 4 and in more anterior regions of the primitive streak in in situ analysis. Tbx383 and_Bmp4_84 are expressed in the more posterior portion of cluster 4 and in the embryo are expressed in the more posterior region of the primitive streak around the amnion and into the extra-embryonic mesoderm. Tek and_Fli1_ (from in-house data) are expressed in cluster 7 and in the embryo are found exclusively in the extra-embryonic portion. Also shown is the t-SNE for the cells from E7.0 onwards (S, NP and HF stages) indicating expression of each gene (white, low; purple, high).
Extended Data Figure 8. Pseudotime analysis of primitive erythroid development.
a) Diffusion plot of cells in cluster 7 and 8. Different colors correspond to different plates and lanes of flow cells.b) Table showing the number of cells in each stage collected on the different plates (S, primitive streak; NP, Neural plate; HF, Head fold). c) Analogously to Extended Data Fig. 6, the angle α identifies a direction in the diffusion space (left panel). The percentage of variance explained by the batch effect associated to plates SLX-8344 and SLX-8345 is plotted as function of α in the right panel. d) The pseudo-time coordinate is robust to the use of different dimensionality reduction techniques, as shown in the left panel with t-SNE (Spearman correlation 0.92, p-value < 2.2x10-16) and in the right panel with Independent Component Analysis (Spearman correlation 0.97, p-value < 2.2x10-16; same procedure described in Extended Data Fig. 6d). **e)**Plots showing the average expression of genes in clusters 1-3 of Figure 4c along the pseudotime axis. Gene expression levels are normalised between 0 and 1. Black lines are the average expression levels of genes in each cluster as obtained from the fitting procedure, after normalization. Red shaded areas indicate standard deviation. f) Principal Component Analysis was carried out on the expression pattern of genes known from previous studies to be up-regulated or down-regulated along the blood developmental trajectory15,85–90. The first principal component (explaining 44% of total variance) showed a very strong correlation with the pseudo-time coordinate (left panel; Spearman correlation coefficient 0.91, p-value < 2.2x10-16). All up-regulated (down-regulated) genes positively (negatively) correlate with the pseudotime coordinate (right panel).
Extended Data Figure 9. ChIP-seq for Gata1 in ESC-derived haematopoietic cells.
a) Flow cytometry for Gata1-mCherry and Runx1-IRES-GFP knock-in reporter genes in embryoid body cells after 5 days of haematopoietic differentiation. Cells were sorted for the expression of both Runx1-IRES-GFP and Gata1-mCherry knock-in reporter genes to provide in vitro equivalents of the developing primitive erythrocytes assayed by RNA-seq. The gate used for sorting is shown in red. b) Numbers of reads and peaks identified for Gata1 and an input sample after mapping and peak calling. 4,135 Gata1 peaks were identified. c) Distribution of Gata1 peaks between promoter, intragenic and intergenic sequences.d) UCSC genome browser tracks for Gata1 and input sample at the Zfpm1 (Fog1) locus known to be a target of Gata1, indicating the quality of the ChIP-seq data. e) Expression of Gata1 target Zfpm1 during the pseudotimecourse for erythroid development, as in Figure 4.
Extended Data Figure 10. Collection of embryos from Tal1 LacZ/+ crosses.
a) Genotyping PCR for embryos from Tal1 LacZ/+ crosses. Lower band is the wild type allele and upper band is the mutant allele carrying a neomycin knock in. Presence of both bands indicates heterozygosity. Embryos from which sequencing data were obtained are indicated with a red star and the number given corresponds to embryo identity in the metadata available online with the sequencing data.b) t-SNE as in Figure 5d showing Tal1 data (triangles) and original wild type data (grey circles). Tal1 data are colored according to the embryo stage from which they were collected: green, NP; red, HF; orange, 4SP. c) As in Figure 5d, showing the complete list of genes. **d)**Gene Set Control Analysis91 (GSCA) was used to identify statistically significant overlaps between genes significantly down-regulated in _Tal1_-/- compared with WT cells in the endothelial cluster (see Figure 5) and Tal1 targets identified by ChIP-seq. GSCA identified an enrichment of our gene set with Tal1 ChIP-seq in ESC-derived haemangioblasts and haemogenic endothelium92, but not ESC-derived haematopoietic progenitors92 or a haematopoietic progenitor cell line52.
Supplementary Material
supp_guide
supp_table1
supp_table2
supp_table3
supp_table4
supp_table5
supp_table6
supp_table7
supp_table8
supp_table9
Acknowledgements
The authors would like to thank Marella de Bruijn, Alfonso Martinez-Arias, Jenny Nichols and Carla Mulas for discussion, the Cambridge Institute for Medical Research Flow Cytometry facility for their expertise in single cell index sorting, and Stephan Lorenz from the Sanger Single Cell Genomics Core for supervising purification of _Tal1_-/- sequencing libraries. ChIP-seq reads were processed by Rebecca Hannah. Research in the authors' laboratories is supported by the MRC, CRUK, BBSRC, Bloodwise, the Leukemia and Lymphoma Society, the Sanger-EBI Single Cell Centre and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust - MRC Cambridge Stem Cell Institute and by core funding from Cancer Research UK and the European Molecular Biology Laboratory. YT was supported by a fellowship from the Japan Society for the Promotion of Science. WJ is a Wellcome Trust Clinical Research Fellow. AS is supported by the Sanger-EBI Single Cell Centre. This work was funded as part of Wellcome Trust Strategic Award 105031/D/14/Z "Tracing early mammalian lineage decisions by single cell genomics" awarded to Drs Wolf Reik, Sarah Teichmann, Jenny Nichols, Ben Simons, Thierry Voet, Shankar Srinivas, Ludovic Vallier, Bertie Göttgens and John Marioni.
Footnotes
Author Contributions
AS and WJ processed and analysed single cell RNA-Seq data. AS and VM generated figures. YT and WJ performed embryo dissection. NKW, VM and ICM carried out single cell RNA-Seq experiments. YT carried out flow cytometry, embryonic stem cell differentiation and in situ hybridization. VM performed ChIP-Seq assays. AS, WJ, YT, VM, BG and JCM interpreted results and wrote the paper. BG and JCM supervised and conceived the study.
Competing financial interests statement
The authors declare no competing financial interests.
References
- 1.Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 2.Van Handel B, et al. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell. 2012;150:590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Org T, et al. Scl binds to primed enhancers in mesoderm to regulate hematopoietic and cardiac fate divergence. EMBO J. 2015;34:759–77. doi: 10.15252/embj.201490542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ema M, et al. Primitive erythropoiesis from mesodermal precursors expressing VE-cadherin, PECAM-1, Tie2, endoglin, and CD34 in the mouse embryo. Blood. 2006;108:4018–24. doi: 10.1182/blood-2006-03-012872. [DOI] [PubMed] [Google Scholar]
- 5.Mikkola HKA, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–16. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 7.Burtscher I, Lickert H. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development. 2009;136:1029–38. doi: 10.1242/dev.028415. [DOI] [PubMed] [Google Scholar]
- 8.Chintala S, et al. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet-dense granules. Blood. 2007;109:1533–40. doi: 10.1182/blood-2006-08-040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henke C, et al. Selective expression of sense and antisense transcripts of the sushi-ichi-related retrotransposon--derived family during mouse placentogenesis. Retrovirology. 2015;12:9. doi: 10.1186/s12977-015-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tam PPL, Zhou SX. The Allocation of Epiblast Cells to Ectodermal and Germ-Line Lineages Is Influenced by the Position of the Cells in the Gastrulating Mouse Embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 11.Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annu Rev Cell Dev Biol. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- 12.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–26. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 13.Rozbicki E, et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat Cell Biol. 2015;17:397–408. doi: 10.1038/ncb3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–98. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 15.Moignard V, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015 doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saga Y. Segmental border is defined by the key transcription factor Mesp2, by means of the suppression of Notch activity. Dev Dyn. 2007;236:1450–5. doi: 10.1002/dvdy.21143. [DOI] [PubMed] [Google Scholar]
- 17.Lawson KA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–36. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–8. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancrin C, et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–5. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padrón-Barthe L, et al. Clonal analysis identifies hemogenic endothelium and not hemangioblasts as the source of the blood-endothelial common lineage in the mouse embryo. Blood. 2014 doi: 10.1182/blood-2013-12-545939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tam PP, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–42. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- 22.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 23.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–4. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 24.Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014;9:1871–84. doi: 10.1016/j.celrep.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JY, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530:223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bheda P, Schneider R. Epigenetics reloaded: the single-cell revolution. Trends Cell Biol. 2014;24:712–723. doi: 10.1016/j.tcb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Achim K, et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33:503–9. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- 28.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson EJ. Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin Cell Dev Biol. 2014;32:73–9. doi: 10.1016/j.semcdb.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–66. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson AC, et al. Single site-specific integration targeting coupled with embryonic stem cell differentiation provides a high-throughput alternative to in vivo enhancer analyses. Biol Open. 2013;2:1229–38. doi: 10.1242/bio.20136296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elefanty AG, et al. Characterization of hematopoietic progenitor cells that express the transcription factor SCL, using a lacZ ‘knock-in’ strategy. Proc Natl Acad Sci U S A. 1998;95:11897–902. doi: 10.1073/pnas.95.20.11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–45. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 36.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 37.van der Maaten L. Barnes-Hut-SNE. Int Conf Learn Represent. 2013 at < https://goo.gl/m7gER3>. [Google Scholar]
- 38.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–5. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 40.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–60. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–20. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 43.Halkidi M, Batistakis Y, Vazirgiannis M. On Clustering Validation Techniques. J Intell Inf Syst. 2001;17:107–145. [Google Scholar]
- 44.Rousseeuw Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 45.Angerer P, et al. bioRxiv. Cold Spring Harbor Labs Journals; 2015. destiny – diffusion maps for large-scale single-cell data in R. [DOI] [PubMed] [Google Scholar]
- 46.Burnham KP, Anderson DR. Model Sel. Multimodel Inference - A Pract. Springer; 2002. at < http://www.springer.com/gb/book/9780387953649>. [Google Scholar]
- 47.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 48.Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 49.Bryja V, Bonilla S, Arenas E. Derivation of mouse embryonic stem cells. Nat Protoc. 2006;1:2082–7. doi: 10.1038/nprot.2006.355. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka Y, et al. Circulation-Independent Differentiation Pathway from Extraembryonic Mesoderm toward Hematopoietic Stem Cells via Hemogenic Angioblasts. Cell Rep. 2014;8:31–9. doi: 10.1016/j.celrep.2014.05.055. [DOI] [PubMed] [Google Scholar]
- 51.Sroczynska P, Lancrin C, Pearson S, Kouskoff V, Lacaud G. In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol Biol. 2009;538:317–34. doi: 10.1007/978-1-59745-418-6_16. [DOI] [PubMed] [Google Scholar]
- 52.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–44. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 53.Zwart W, et al. A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics. 2013;14:232. doi: 10.1186/1471-2164-14-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez-Castillo M, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, Ji H. PolyaPeak: detecting transcription factor binding sites from ChIP-seq using peak shape information. PLoS One. 2014;9:e89694. doi: 10.1371/journal.pone.0089694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilkinson DG. In Situ Hybridization. Oxford University Press; 1999. at < https://global.oup.com/academic/product/in-situ-hybridization-9780199636587?cc=gb&lang=en&>. [Google Scholar]
- 58.Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 59.Du J, et al. O-fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev Biol. 2010;346:25–38. doi: 10.1016/j.ydbio.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donnison M, et al. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- 61.Mitsunaga K, et al. Loss of PGC-specific expression of the orphan nuclear receptor ERR-beta results in reduction of germ cell number in mouse embryos. Mech Dev. 2004;121:237–46. doi: 10.1016/j.mod.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Baldwin HS, et al. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–53. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- 63.Naiche LA, Arora R, Kania A, Lewandoski M, Papaioannou VE. Identity and fate of Tbx4-expressing cells reveal developmental cell fate decisions in the allantois, limb, and external genitalia. Dev Dyn. 2011;240:2290–300. doi: 10.1002/dvdy.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamplin OJ, et al. Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics. 2008;9:511. doi: 10.1186/1471-2164-9-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent SD, et al. Prdm1 functions in the mesoderm of the second heart field, where it interacts genetically with Tbx1, during outflow tract morphogenesis in the mouse embryo. Hum Mol Genet. 2014;23:5087–101. doi: 10.1093/hmg/ddu232. [DOI] [PubMed] [Google Scholar]
- 66.Brown CB, et al. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev Biol. 2004;267:190–202. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 67.Brennan J, et al. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–9. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- 68.Meno C, et al. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–98. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- 69.Bessho Y, et al. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001;15:2642–7. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oginuma M, Niwa Y, Chapman DL, Saga Y. Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development. 2008;135:2555–62. doi: 10.1242/dev.019877. [DOI] [PubMed] [Google Scholar]
- 71.Forlani S, Lawson KA, Deschamps J. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development. 2003;130:3807–19. doi: 10.1242/dev.00573. [DOI] [PubMed] [Google Scholar]
- 72.Zeigler BM, et al. The allantois and chorion, when isolated before circulation or chorioallantoic fusion, have hematopoietic potential. Development. 2006;133:4183–4192. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- 73.Downs KM, Hellman ER, McHugh J, Barrickman K, Inman KE. Investigation into a role for the primitive streak in development of the murine allantois. Development. 2004;131:37–55. doi: 10.1242/dev.00906. [DOI] [PubMed] [Google Scholar]
- 74.Caprioli A, Jaffredo T, Gautier R, Dubourg C, Dieterlen-Lièvre F. Blood-borne seeding by hematopoietic and endothelial precursors from the allantois. Proc Natl Acad Sci U S A. 1998;95:1641–6. doi: 10.1073/pnas.95.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Nes J, et al. The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development. 2006;133:419–28. doi: 10.1242/dev.02216. [DOI] [PubMed] [Google Scholar]
- 76.Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development. 1995;121:549–60. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- 77.Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–68. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 78.Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–9. [PubMed] [Google Scholar]
- 79.Lee D, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carapuco M. Hox genes specify vertebral types in the presomitic mesoderm. Genes Dev. 2005;19:2116–2121. doi: 10.1101/gad.338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang H, et al. Expression of podocalyxin separates the hematopoietic and vascular potentials of mouse embryonic stem cell-derived mesoderm. Stem Cells. 2014;32:191–203. doi: 10.1002/stem.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herrmann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991;113:913–7. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- 83.Weidgang CE, et al. TBX3 Directs Cell-Fate Decision toward Mesendoderm. Stem Cell Reports. 2013;1:248–265. doi: 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perea-Gómez A, Shawlot W, Sasaki H, Behringer RR, Ang S. HNF3beta and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo. Development. 1999;126:4499–511. doi: 10.1242/dev.126.20.4499. [DOI] [PubMed] [Google Scholar]
- 85.Morkel M, et al. Beta-catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development. 2003;130:6283–94. doi: 10.1242/dev.00859. [DOI] [PubMed] [Google Scholar]
- 86.Saga Y, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–47. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 87.Trimborn T, Gribnau J, Grosveld F, Fraser P. Mechanisms of developmental control of transcription in the murine alpha- and beta-globin loci. Genes Dev. 1999;13:112–24. doi: 10.1101/gad.13.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kingsley PD, Malik J, Fantauzzo KA, Palis J. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood. 2004;104:19–25. doi: 10.1182/blood-2003-12-4162. [DOI] [PubMed] [Google Scholar]
- 89.Hodge D, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–70. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Isern J, et al. Single-lineage transcriptome analysis reveals key regulatory pathways in primitive erythroid progenitors in the mouse embryo. Blood. 2011;117:4924–34. doi: 10.1182/blood-2010-10-313676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joshi A, Hannah R, Diamanti E, Göttgens B. Gene set control analysis predicts hematopoietic control mechanisms from genome-wide transcription factor binding data. Exp Hematol. 2013;41:354–66.e14. doi: 10.1016/j.exphem.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goode DK, et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev Cell. 2016;36:572–87. doi: 10.1016/j.devcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 94.Pearce JJH, Evans MJ. Mml, a mouse Mix-like gene expressed in the primitive streak. Mech Dev. 1999;87:189–192. doi: 10.1016/s0925-4773(99)00135-5. [DOI] [PubMed] [Google Scholar]
- 95.Pennisi D, et al. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet. 2000;24:434–7. doi: 10.1038/74301. [DOI] [PubMed] [Google Scholar]
- 96.Gordon EJ, Gale NW, Harvey NL. Expression of the hyaluronan receptor LYVE-1 is not restricted to the lymphatic vasculature; LYVE-1 is also expressed on embryonic blood vessels. Dev Dyn. 2008;237:1901–9. doi: 10.1002/dvdy.21605. [DOI] [PubMed] [Google Scholar]
- 97.Kallianpur AR, Jordan JE, Brandt SJ. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood. 1994;83:1200–8. [PubMed] [Google Scholar]
- 98.Robb L, et al. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci U S A. 1995;92:7075–9. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanaka Y, et al. The transcriptional programme controlled by Runx1 during early embryonic blood development. Dev Biol. 2012;366:404–19. doi: 10.1016/j.ydbio.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.North T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–75. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 101.Palis J, McGrath KE, Kingsley PD. Initiation of hematopoiesis and vasculogenesis in murine yolk sac explants. Blood. 1995;86:156–63. [PubMed] [Google Scholar]
- 102.Drissen R, et al. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol Cell Biol. 2005;25:5205–14. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Southwood CM, Downs KM, Bieker JJ. Erythroid Krüppel-like factor exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev Dyn. 1996;206:248–59. doi: 10.1002/(SICI)1097-0177(199607)206:3<248::AID-AJA3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 104.Silver L, Palis J. Initiation of murine embryonic erythropoiesis: a spatial analysis. Blood. 1997;89:1154–64. [PubMed] [Google Scholar]
- 105.Lanctôt C, Lamolet B, Drouin J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–17. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 106.Lania G, Ferrentino R, Baldini A. TBX1 Represses Vegfr2 Gene Expression and Enhances the Cardiac Fate of VEGFR2+ Cells. PLoS One. 2015;10:e0138525. doi: 10.1371/journal.pone.0138525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supp_guide
supp_table1
supp_table2
supp_table3
supp_table4
supp_table5
supp_table6
supp_table7
supp_table8
supp_table9