An oxygen-regulated switch in the protein synthesis machinery (original) (raw)
. Author manuscript; available in PMC: 2016 Aug 4.
Published in final edited form as: Nature. 2012 May 6;486(7401):126–129. doi: 10.1038/nature11055
SUMMARY
Protein synthesis involves the translation of ribonucleic acid information into proteins, the building blocks of life. The initial step of protein synthesis consists of the eukaryotic translation initiation factor 4E (eIF4E) binding to the 7-methylguanosine (m7-GpppG) 5′cap of mRNAs1,2. Low oxygen tension (hypoxia) represses cap-mediated translation by sequestering eIF4E through mammalian target of rapamycin (mTOR)-dependent mechanisms3–6. While the internal ribosome entry site is an alternative translation initiation mechanism, this pathway alone cannot account for the translational capacity of hypoxic cells7,8. This raises a fundamental question in biology as to how proteins are synthesized in periods of oxygen scarcity and eIF4E inhibition9. Here, we uncover an oxygen-regulated translation initiation complex that mediates selective cap-dependent protein synthesis. Hypoxia stimulates the formation of a complex that includes the oxygen-regulated hypoxia-inducible factor 2α (HIF-2α), the RNA binding protein RBM4 and the cap-binding eIF4E2, an eIF4E homologue. PAR-CLIP10 analysis identified an RNA hypoxia response element (rHRE) that recruits this complex to a wide array mRNAs, including the epidermal growth factor receptor (EGFR). Once assembled at the rHRE, HIF-2α/RBM4/eIF4E2 captures the 5′cap and targets mRNAs to polysomes for active translation thereby evading hypoxia-induced repression of protein synthesis. These findings demonstrate that cells have evolved a program whereby oxygen tension switches the basic translation initiation machinery.
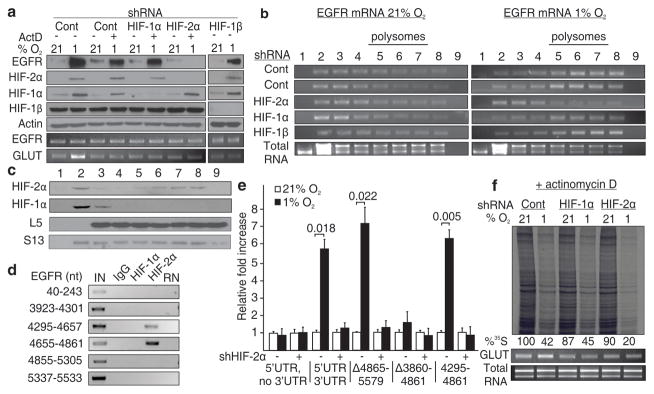
Here, we describe an oxygen-regulated mechanism that mediates selective cap-dependent translation during hypoxia and eIF4E inactivation (Supplementary Fig. 1). We began our investigation into this alternative protein synthesis mechanism by examining the EGFR, a receptor tyrosine kinase that plays a critical role in cell proliferation, tissue development and cancer11. Hypoxia activates translation of EGFR mRNA through HIF-2α12, a member of the HIF family of transcription factors involved in maintaining cellular oxygen homeostasis13–17. We hypothesized that HIF-2α orchestrates a gene program that enables translation of EGFR mRNA during periods of eIF4E inactivation. Cells were treated with transcription inhibitors to preclude activation of HIF-2α target genes during hypoxia. To our surprise, hypoxia caused the accumulation of EGFR protein in a HIF-2α-dependent manner even in transcription-incompetent glioma or primary cultures of renal epithelial cells (Fig. 1a and Supplementary Fig. 2). The EGFR mRNA was captured by polysomes and de novo translated in hypoxic cells treated with actinomycin D (Fig. 1b and Supplementary Figs. 3–4). Silencing of HIF-2α abolished the association of EGFR mRNA with polysomes and prevented its de novo synthesis (Fig. 1b and Supplementary Figs. 3–4). In contrast, silencing of HIF-1α, a paralog of HIF-2α, did not prevent hypoxic induction of EGFR translation and capture by polysomes (Fig. 1a–b and Supplementary Figs. 3–4). In addition, ablation of HIF-1β, a protein required for HIF transcriptional activity, had no effect on hypoxia-inducible translation of the EGFR mRNA (Fig. 1a–b and Supplementary Fig. 3). HIF-2α, but not HIF-1α, was observed in polysome fractions of hypoxic cells suggesting its direct involvement in the translational machinery (Fig. 1c and Supplementary Fig. 5). RNA immunoprecipitation revealed that HIF-2α associates with the EGFR mRNA 3′UTR between nucleotides 4295-4861 (Fig. 1d, Supplementary Fig. 6 and Supplementary Table 1). This segment was both necessary and sufficient to enhance translation of a luciferase reporter during hypoxia in a HIF-2α-dependent manner even in the absence of transcription (Fig. 1e and Supplementary Fig. 7). Silencing HIF-2α, but not HIF-1α, considerably reduced the rate of global hypoxic translation highlighting its participation in hypoxic protein synthesis beyond EGFR translation (Fig. 1f and Supplementary Fig. 8).
Figure 1. HIF-2α activates EGFR mRNA translation by interacting with its 3′UTR.
a–b, Western blot and polysomal distribution of EGFR protein and mRNA in HIF-2α, HIF-1α or HIF-1β knockdown cells in the presence of actinomycin D. c, Polysomal distribution of HIF-2α and HIF-1α in hypoxia. d, RNA immunoprecipitation of HIF-1α and HIF-2α. IN, input; RN, RNase-treated. e, Dual luciferase assays in cells transfected with EGFR 3′UTR reporter constructs. Significance of fold change (Student’s t test) is shown. Columns, mean (n = 3); error bars, s.e.m. f, Global translation rates of transcription-incompetent cells expressing shRNA targeting HIF-2α or HIF-1α. Experiments performed in U87MG glioblastoma.
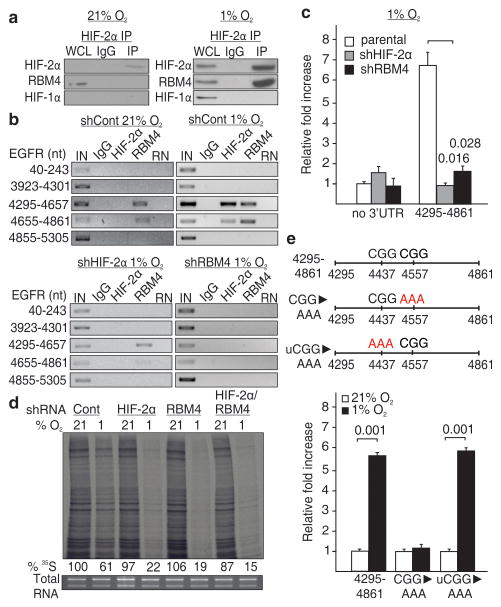
HIF-2α does not contain a classical RNA recognition motif therefore we searched for potential interacting partners that could bind the EGFR 3′UTR. Immunoprecipitation revealed a band of 40 kDa specifically associated with HIF-2α that was identified as RNA-binding motif protein 4 (RBM4) (Supplementary Fig. 9), a protein involved in translation control18,19. Co-immunoprecipitation revealed that endogenous RBM4 interacted with the N-terminal region of HIF-2α but not with HIF-1α during hypoxia (Fig. 2a and Supplementary Fig. 10). Furthermore, RBM4 assembled with the EGFR 3′UTR in vivo and in vitro independently of oxygen tension (Fig. 2b and Supplementary Fig. 11). Silencing experiments revealed that RBM4 is essential for HIF-2α recruitment to the EGFR 3′UTR (Fig. 2b and Supplementary Fig. 11a), the hypoxic induction of EGFR protein (Supplementary Fig. 12), and the ability of the 4295-4861 EGFR 3′UTR segment to induce hypoxia-dependent translation of a reporter construct (Fig. 2c). In addition, depletion of RBM4 caused reduced hypoxic cellular translation to levels similar to those observed in HIF-2α-incompetent cells (Fig. 2d and Supplementary Fig. 13). Consistent with the RNA immunoprecipitation, multiple PAR-CLIP sequenced reads for HIF-2α/RBM4 and RBM4 concentrated at the same site within the EGFR 3′UTR 4295-4861 fragment that confers hypoxic translation to a reporter protein but not elsewhere on the transcript (Supplementary Figs. 14–15). The crosslink sites were near a CGG trinucleotide, a feature of RBM4 binding motifs20,21. Mutation of this CGG motif, but not another CGG sequence, was sufficient to disrupt the secondary structure and to abolish hypoxia-inducible translation of a reporter construct (Fig. 2e and Supplementary Figs. 16–17). In addition, shorter segments of the EGFR 3′UTR that altered the secondary structure were unresponsive to hypoxia in luciferase assays (Supplementary Figs. 18–19). Consistent with the 35S-labeling experiments, a wide array of mRNAs interacted with the HIF-2α/RBM4 complex (Supplementary Figure 20a, Supplementary Data 1–2 and Supplementary Table 2). Similar to EGFR, multiple sequenced reads for HIF-2α/RBM4 and RBM4 concentrated at the same site near CG(G) nucleotides in the majority of candidates (Supplementary Fig. 20b–c). Several PAR-CLIP candidates were validated for HIF-2α-dependent hypoxic induction (Supplementary Figs. 20c–21). We suggest that RBM4 binds to specific regions in the 3′UTR of mRNAs to recruit HIF-2α and induce hypoxic translation. These RNA sequences, which share commonality with RBM4 binding sites, are referred to as rHRE.
Figure 2. RBM4 recruits HIF-2α to the 3′UTR for hypoxic translation.
a, Co-immunoprecipitation of HIF-2α in normoxia and hypoxia. WCL, whole cell lysate. b, RNA immunoprecipitation of HIF-2α and RBM4 in HIF-2α or RBM4 knockdown cells. IN, input; RN, RNase-treated. c, Effect of silencing HIF-2α or RBM4 on the hypoxic expression of a luciferase reporter fused to the 4295-4861 segment of the EGFR 3′UTR. d, Global translation rates in normoxic or hypoxic HIF-2α and/or RBM4 knockdown cells. e, Expression of a luciferase reporter containing a CGG to AAA mutation near RBM4 crosslinking sites (red arrows), or in an unrelated upstream region (uCGG). c and e, Columns, mean (n = 3); error bars, s.e.m. Significance of fold change (Student’s t test) is shown. Experiments performed in U87MG glioblastoma.
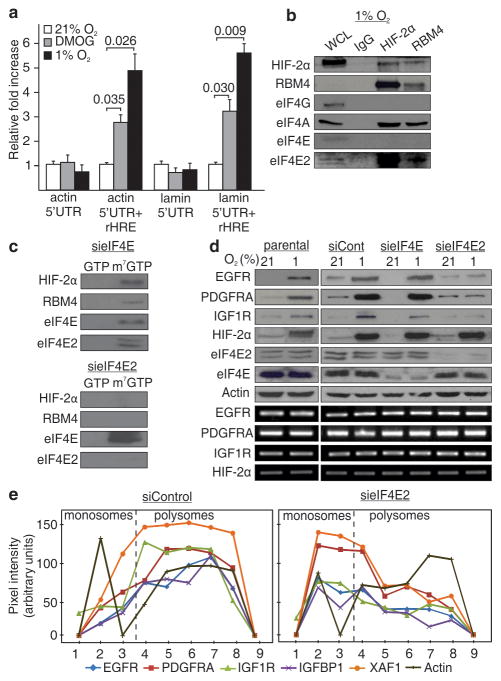
A key characteristic of the EGFR rHRE is that it confers hypoxia-inducible translation to several unrelated 5′UTRs that are otherwise unable to initiate translation during hypoxia (Fig. 3a). We thus suspected that the rHRE might exploit the cap as it is a common feature used by the 5′UTRs of mRNAs to initiate protein synthesis. The RBM4/HIF-2α complex of hypoxic cells was captured by m7-GTP beads (Supplementary Fig. 22). Interestingly, immunoprecipitation experiments demonstrated that HIF-2α/RBM4 specifically assembles with the cap-binding protein eIF4E2, a homologue of eIF4E (Fig. 3b and Supplementary Fig. 23a–b). We thus reasoned that eIF4E2 is recruited by HIF-2α/RBM4 to activate selective cap-dependent translation of rHRE-containing mRNAs during hypoxia and inhibition of eIF4E by 4EBP6,22,23. Immunoprecipitation revealed that 4EBP displays more affinity to eIF4E than eIF4E2, consistent with previously published data (Supplementary Fig. 23c)24,25. Silencing of eIF4E2, but not eIF4E, prevented binding of HIF-2α/RBM4 from hypoxic cells to m7-GTP beads (Fig. 3c). In addition, ablation of eIF4E2 prevented hypoxic induction of multiple proteins identified by PAR-CLIP, including EGFR, whereas silencing of eIF4E had no discernable effect (Fig. 3d and Supplementary Figs. 20b and 24). Ablation of eIF4E2 prevented the capture of rHRE-containing mRNAs by polysomes (Fig. 3e and Supplementary Figs. 25–26). The HIF-2α/RBM4/eIF4E2 complex also recruited the RNA helicase eIF4A, a fundamental component of translation initiation26 (Fig. 3b and Supplementary Fig 23b). Taken together, these data demonstrate that eIF4E2 is a member of a hypoxic translation initiation complex that mediates selective cap-dependent protein synthesis independently of eIF4E.
Figure 3. HIF-2α/RBM4 recruits the m7-GTP cap via an interaction with eIF4E2.
a, Dual luciferase assays in cells transfected with reporter constructs containing the 5′UTR of actin or lamin a/c with or without a 3′ rHRE. Significance of fold change (Student’s t test) is shown. Columns, mean (n = 3); error bars, s.e.m. b, Co-immunoprecipitation of HIF-2α and RBM4 in hypoxia. WCL, whole cell lysate. c, Capture assays using m7-GTP beads in hypoxic cell lysates depleted in eIF4E or eIF4E2. GTP, proteins dislodged from the beads by GTP. m7-GTP, proteins bound to m7-GTP beads after GTP wash. d, Western blot of total EGFR, PDGFRA, IGF1R, HIF-2α, eIF4E and eIF4E2 levels in eIF4E or eIF4E2 knockdown cells. e, Polysomal distribution of HIF-2α/RBM4 mRNA targets in hypoxic eIF4E2 knockdown cells. Experiments performed in U87MG glioblastoma.
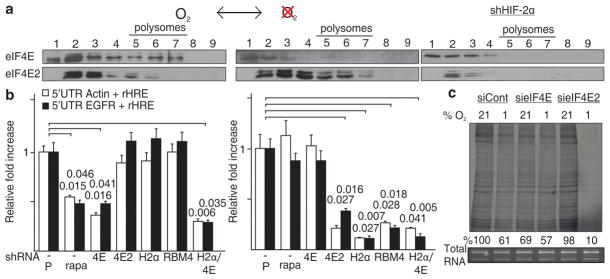
Figure 3d revealed that eIF4E might be involved in the translation of EGFR, PDGFRA and IGF1R mRNAs during normoxia. This raises the intriguing possibility that the cap-dependent translational machinery switches from eIF4E to eIF4E2 as a function of oxygen tension. This oxygen-dependent switch was clearly observed between eIF4E and eIF4E2 in polysomes whereby eIF4E participation in the translational machinery was essentially limited to normoxia and eIF4E2 to hypoxia (Fig. 4a and Supplementary Figs. 27–28). Ablation of HIF-2α abolished the hypoxic shift of eIF4E2 to polysomes attesting to the role of HIF-2α as the oxygen-regulated subunit of the eIF4E2/RBM4/HIF-2α complex (Fig. 4a and Supplementary Fig. 27c). In contrast, treatment with rapamycin, an inhbitior of mTOR and eIF4E, prevented the accumulation of eIF4E in normoxic polysomes but had no effect on eIF4E2 (Supplementary Fig. 27e). Treatment with rapamycin, or silencing of eIF4E, significantly reduced the expression of rHRE-containing luciferase mRNAs in normoxia but had no effect in hypoxia (Fig. 4b and Supplementary Fig. 29). Silencing any of the participants of the HIF-2α/RBM4/eIF4E2 cap-binding complex prevented translation of rHRE-containing mRNAs in hypoxia but not in normoxia. Silencing of eIF4E reduced the rate of cellular protein synthesis in normoxia but had no effect in hypoxia (Fig. 4c and Supplementary Fig. 30). In stark contrast, depletion of eIF4E2 considerably limited the global rate of hypoxic translation without affecting protein synthesis of cells maintained in normoxia (Fig. 4c and Supplementary Fig. 30). These results demonstrate that oxygen tension regulates the cap-dependent protein synthesis machinery by switching from eIF4E to eIF4E2-dependent translation in a HIF-2α-dependent manner (Supplementary Fig. 1).
Figure 4. An oxygen-regulated switch from eIF4E- to eIF4E2-dependent protein synthesis.
a, eIF4E and eIF4E2 polysome association in normoxia and hypoxia. b, Dual luciferase assays in normoxic (left) and hypoxic (right) cells transfected with constructs containing actin or EGFR 5′UTRs and EGFR rHREs. Assays were performed on cells treated with rapamycin, an inhibitor of mTOR and eIF4E, and in knockdown cells of eIF4E, eIF4E2, HIF-2α or RBM4 and in a co-eIF4E/HIF-2α knockdown. Significance of fold change (Student’s t test) is shown. P, parental; rapa, rapamycin. Columns, mean (n = 3); error bars, s.e.m. c, Global translation rates in normoxic or hypoxic eIF4E or eIF4E2 knockdown cells. Experiments performed in U87MG glioblastoma.
In summary, this report identifies a selective cap-dependent translation initiation mechanism that operates independently of eIF4E and that targets mRNAs for protein synthesis during hypoxia. The data suggest that the HIF-2α/RBM4/eIF4E2 complex is extensively involved in coordinating the translation response to low oxygen availability and thus is essential in cellular oxygen homeostasis. This complex likely recruits functional homologs of the canonical eIF4E-dependent pathway, as well as distinct components, to initiate hypoxic protein synthesis. This process is regulated by the oxygen sensing machinery first identified as the main regulator of the transcriptional response to hypoxia13–16. A human population that recently migrated to the Tibetan highlands contains a point mutation in the HIF-2α gene (EPAS1) further emphasizing the evolutionarily role HIF-2α plays in the adaptation to high altitude and low oxygen tension27. The target mRNAs code for proteins including the EGFR, PDGFRA and IGF1R that are implicated in the adaptive response to hypoxia as well as a wide variety of biological processes including development and cancer. The role of these receptor tyrosine kinases in human malignancy is particularly well-documented and they are at the center of targeted therapy11,28. EGFR is often overproduced by tumors that harbor a wild-type EGFR gene suggesting that cancer cells hijack the eIF4E2 pathway for their proliferative advantage29,30. The data shown here provide the foundation to further investigate the adaptive properties of the basic protein synthesis machinery in response to environmental conditions.
METHODS
Cell culture and reagents
Human renal proximal tubular epithelial cells (HRPTEC) were maintained in epithelial cell medium (ScienCell). All other cell lines were obtained from the American Type Culture Collection and propagated as suggested. Cells were incubated at 37 °C in a 5% CO2 environment. Hypoxia was induced by incubating at 37 °C in a 1% O2, 5% CO2 and N2-balanced atmosphere for 24 h unless otherwise indicated. Heat shock was induced by incubating cells at 42 °C for 30 min. Dimethyl oxallylglycine (DMOG; Cayman Chemical) was used at a concentration of 60 μg/mL to stabilize HIF-2α. Transcription inhibitors actinomycinD (EMD Biosciences), 5,6-dichloro-1-β-D-ribobenzimidazole (DRB; EMD Biosciences), and α-amanitin (Sigma) were added 30 min prior to hypoxic exposure and used at concentrations of 10 μg/mL, 78 μM and 10 μg/mL, respectively. Cells were treated with 10 nM rapamycin (Sigma) for 1 h prior to hypoxia.
Western Blot analysis
Western blot was performed using standard techniques. Monoclonal antibodies were used to detect EGFR (Ab-12; LabVision), GFP (Roche), HIF-1α (Novus), and HIF-1β (Novus). Polyclonal antibodies were used to detect HIF-2α (Novus), actin (Sigma), L26 (abcam), L5 (abcam), S13 (abcam), PDGFRA (Assay Biotech), IGF1R (Cell Signaling), RBM4 (ProteinTech Group), 4EBP1 (Cell Signaling), 4EBP1-P (Cell Signaling), AKT (Cell Signaling), AKT-P (Cell Signaling), EGFR-P (Cell Signaling), S6-P (Cell Signaling), eIF4E (Santa Cruz), eIF4E2 (Genetex), and eIF4G1 (Novus). Primary antibody against eIF4A1 was created in the lab of Dr. Nahum Sonenberg. Secondary antibodies were HRP-conjugated anti-mouse (Amersham Biosciences) or anti-rabbit (Jackson ImmunoResearch Inc.). Bands were detected by enhanced chemiluminescence (Pierce). Whole cell lysate (WCL) is defined as 5% of the input used for immunoprecipitations.
Protein synthesis by 35S-Met incorporation
Cells were grown in 10-cm plates for 48 h. Serum-free conditions supplemented with 1% insulin-transferrin-selenium (Invitrogen) were used when cells were incubated in hypoxia to detect stronger de novo EGFR accumulation. Actinomycin D was added to fresh media at a concentration of 10 μg/mL for 30 min prior to the addition of DMOG for the indicated times. GLUT mRNA was used as a control for actinomycin D activity. One hour before the end point, media was changed to glutamine-, methionine- and cysteine-free DMEM and labeled 30 min later with [35S]Met [33 μCi/mL] for 30 min. Cells were lysed in 1 mL modified RIPA (50 mM Tris-HCl, 1% Igepal, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4 and 1 mM NaF, 1 μg/mL aprotinin, leupeptin and pepstatin) for 30 min at 4 °C. Immunoprecipitation of EGFR was performed using 50 μL of agarose-conjugated EGFR antibody (Santa Cruz). Samples were run on a 6% SDS-PAGE gel, dried at 80 °C for 90 min and exposed to X-ray film overnight at −80 °C. For total cellular protein synthesis rates, samples were loaded on a per cell basis (500,000 cells). Cell viability assays were performed by incubating cells for 5 min in 3 μg/mL propidium iodide (Sigma; to stain dead cells), 3 μg/mL Hoescht (Invitrogen; to stain nuclei), and 3 μM fluorescein diacetate (Sigma; to stain live cells). % live cells over dead cells were plotted. Pixel intensity was measured by densitometry using Adobe Photoshop CS5.1.
RNA Immunoprecipitation
1% formaldehyde was added to cells for 30 min at RT. 200 mM glycine was added for 5 min to stop the reaction followed by two washes with cold PBS. Cells were lysed in 1 mL modified RIPA. RNase-inhibitor (40 U/mL; Ambion) was added to modified RIPA just before use. Samples were sonicated at 50% amplitude for two cycles of 30 sec (2 sec on/2 sec off) with a 1 min pause in between cycles. DNase-treatment (12 μL of 20 mg/mL DNaseI, 25 mM MgCl2 and 5 mM CaCl2) was carried out for 30 min at 37 °C. The reaction was stopped by adding 20 mM EDTA. For RNase-treated control samples, 5 μL of a 10 mg/mL RNAse A solution was added for 30 min at 37 °C (RNase A; Fermentas). Samples were pre-cleared using 10 μL Dynabeads (Invitrogen) for 15 min at 4 °C. Beads were removed using a magnetic stand (Promega). Immunoprecipitation was carried out at 2 μg/mL of primary antibody overnight at 4 °C. Samples were centrifuged at 12,000 g for 15 min at 4 °C. The supernatant was incubated with 20 μL Dynabeads equilibrated in 2% BSA for 1 h at 4 °C. Beads were recovered and washed five times with modified RIPA and eluted with 20 μL 0.1 M glycine (pH 3.0). Bound proteins were removed by adding 200 mM NaCl and 20 μg Proteinase K to the supernatant and incubating for 1 h at 42 °C. Crosslinking was reversed by incubating overnight at 65 °C. RNA extraction and RT-PCR analysis was performed to identify interacting RNA segments. Inputs are 2% of the sample. Primers listed in Supplementary Table 1.
Adenoviral infections
Adenoviruses encoding GFP, HIF-1α and HIF-2α were generated and used as previously described31,32.
Analysis of cap-binding proteins
Cells on two 150 mm plate were washed with PBS and lysed in 1 mL of Nature Lysis Buffer (NLB; 20 mM Tris-HCl, 100 mM NaCl, 25 mM MgCl2, 0.5% NP40 + standard protease and phosphatase inhibitors). Extracts were clarified by centrifugation at 10,000×g for 10 min at 4 °C. Supernatants were pre-cleared with 30 μL of Sepharose 4B beads (Sigma) for 10 min at 4 °C. Beads were removed by centrifugation at 500×g for 30 sec, and supernatants were incubated with 50 μL 7-methyl GTP-Sepharose 4B beads (GE Healthcare) for 1 h at 4 °C. Pelleted beads were washed 4 times with 0.5 mL NLB and resuspended in 0.6 mL NLB + 1 mM GTP for 1 h at 4 °C. Following four final washes with NLB, the beads were re-suspended in sample buffer and boiled for 1 min. Concentrated GTP wash, m7-GTP-bound proteins, as well as 5% input taken just before m7-GTP beads were added were run on an SDS-PAGE.
Statistical analysis
P values associated with all comparisons were based on paired two-tailed Student’s t tests. Results are mean (n = 3) ± standard error of the mean (s.e.m.).
Supplementary Material
1
2
3
4
Acknowledgments
This work was funded by the Canadian Institutes of Health Research (S.L. and M.H.). J.U. is a Research Fellow of The Terry Fox Foundation (Canadian Cancer Society Award #700014). A.F. was supported by a Terry Fox Foundation Studentship from the Canadian Cancer Society. We thank Jocelyn Côté, William Kaelin, Christopher Kennedy and Thomas Tuschl for reagents and technical advice.
Footnotes
AUTHOR CONTRIBUTIONS
J.U. performed most experiments and made most plasmid constructs with assistance from C.E.H. (identified RBM4 interaction with HIF-2α, participated in PAR-CLIP experiments, and made some luciferase contructs), G.L. (performed experiments with HRPTEC), A.F. (performed actinomycin D experiments on total hypoxic EGFR levels, created stable shHIF-2α and shHIF-1α cell lines and some plasmid constructs), M.D.J. (created luciferase constructs for CGG mutagenesis and rHRE mapping and created HIF-2α truncation mutants), M.R.F. (performed eIF4E2 co-IP assays), and J.P. (created some luciferase constructs). J.U., C.E.H., G.L., A.F., M.R.F., M.H., A.P. and S.L. conceived experiments and analyzed data. J.U. and S.L. wrote the paper.
The authors declare no competing financial interests.
References
- 1.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunstein S, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–12. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koritzinsky M, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. Embo J. 2006;25:1114–25. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L, et al. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–31. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–27. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 8.Young RM, et al. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J Biol Chem. 2008;283:16309–19. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrick WC. Eukaryotic protein synthesis: still a mystery. J Biol Chem. 285:21197–201. doi: 10.1074/jbc.R110.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 12.Franovic A, et al. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc Natl Acad Sci U S A. 2007;104:13092–7. doi: 10.1073/pnas.0702387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 14.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–78. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 17.Wiesener MS, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. Faseb J. 2003;17:271–3. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 18.Lin JC, Hsu M, Tarn WY. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci U S A. 2007;104:2235–40. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JC, Tarn WY. RNA-binding motif protein 4 translocates to cytoplasmic granules and suppresses translation via argonaute2 during muscle cell differentiation. J Biol Chem. 2009;284:34658–65. doi: 10.1074/jbc.M109.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput Biol. 6:e1000832. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray D, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–70. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 22.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 23.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 24.Rom E, et al. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem. 1998;273:13104–9. doi: 10.1074/jbc.273.21.13104. [DOI] [PubMed] [Google Scholar]
- 25.Tee AR, Tee JA, Blenis J. Characterizing the interaction of the mammalian eIF4E-related protein 4EHP with 4E-BP1. FEBS Lett. 2004;564:58–62. doi: 10.1016/S0014-5793(04)00313-8. [DOI] [PubMed] [Google Scholar]
- 26.Parsyan A, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 12:235–45. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 27.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 329:75–8. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehn BM. Genomics illuminates a deadly brain cancer. Jama. 303:925–7. doi: 10.1001/jama.2010.236. [DOI] [PubMed] [Google Scholar]
- 29.Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2alpha oncogenic axis. Proc Natl Acad Sci U S A. 2009;106:21306–11. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giatromanolaki A, et al. Expression of hypoxia-inducible carbonic anhydrase-9 relates to angiogenic pathways and independently to poor outcome in non-small cell lung cancer. Cancer Res. 2001;61:7992–8. [PubMed] [Google Scholar]
- 31.Gunaratnam L, et al. Hypoxia inducible factor activates the transforming growth factor-alpha/epidermal growth factor receptor growth stimulatory pathway in VHL(−/−) renal cell carcinoma cells. J Biol Chem. 2003;278:44966–74. doi: 10.1074/jbc.M305502200. [DOI] [PubMed] [Google Scholar]
- 32.Smith K, et al. Silencing of epidermal growth factor receptor suppresses hypoxia-inducible factor-2-driven VHL−/− renal cancer. Cancer Res. 2005;65:5221–30. doi: 10.1158/0008-5472.CAN-05-0169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4