Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma–like and Non–Small Cell Carcinoma–like Subsets (original) (raw)
. Author manuscript; available in PMC: 2016 Aug 24.
Abstract
Purpose
Pulmonary large cell neuroendocrine carcinoma (LCNEC) is a highly aggressive neoplasm, whose biologic relationship to small cell lung carcinoma (SCLC) versus non-SCLC (NSCLC) remains unclear, contributing to uncertainty regarding optimal clinical management. To clarify these relationships, we analyzed genomic alterations in LCNEC compared with other major lung carcinoma types.
Experimental Design
LCNEC (n = 45) tumor/normal pairs underwent targeted next-generation sequencing of 241 cancer genes by Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) platform and comprehensive histologic, immunohistochemical, and clinical analysis. Genomic data were compared with MSK-IMPACT analysis of other lung carcinoma histologies (n = 242).
Results
Commonly altered genes in LCNEC included TP53 (78%), RB1 (38%), STK11 (33%), KEAP1 (31%), and KRAS (22%). Genomic profiles segregated LCNEC into 2 major and 1 minor subsets: SCLC-like (n = 18), characterized by TP53+RB1 co-mutation/loss and other SCLC-type alterations, including MYCL amplification; NSCLC-like (n = 25), characterized by the lack of coaltered TP53+RB1 and nearly universal occurrence of NSCLC-type mutations (STK11, KRAS, and KEAP1); and carcinoid-like (n = 2), characterized by MEN1 mutations and low mutation burden. SCLC-like and NSCLC-like subsets revealed several clinicopathologic differences, including higher proliferative activity in SCLC-like tumors (P < 0.0001) and exclusive adenocarcinoma-type differentiation marker expression in NSCLC-like tumors (P = 0.005). While exhibiting predominant similarity with lung adenocarcinoma, NSCLC-like LCNEC harbored several distinctive genomic alterations, including more frequent mutations in NOTCH family genes (28%), implicated as key regulators of neuroendocrine differentiation.
Conclusions
LCNEC is a biologically heterogeneous group of tumors, comprising distinct subsets with genomic signatures of SCLC, NSCLC (predominantly adenocarcinoma), and rarely, highly proliferative carcinoids. Recognition of these subsets may inform the classification and management of LCNEC patients.
Introduction
Pulmonary large cell neuroendocrine carcinoma (LCNEC) is defined pathologically as a neoplasm that shares with small cell lung carcinoma (SCLC) a neuroendocrine (NE) phenotype and an extremely high proliferation rate but which lacks the classic cytomorphology of SCLC. Diagnostic criteria for LCNEC were first introduced in 1991 by Travis and colleagues (1), and this category became adopted as a distinct entity by the World Health Organization (WHO) classification of lung tumors in 1999 and subsequent editions (2). Prior to this, these unconventional neuroendocrine tumors were classified under a variety of diagnostic terms and criteria (3). While the LCNEC category captured in a single group highly proliferative non–small cell neuroendocrine neoplasms, it has long been recognized that tumors falling under this umbrella term are histologically heterogeneous (4, 5). In particular, although the prototypical LCNECs are morphologically sharply distinct from SCLC, some LCNECs enter in a close morphologic differential diagnosis with SCLC (6). Clinically, LCNEC are generally associated with high rate of metastases and poor patient survival, but remarkably broad survival ranges have been reported (7). In addition, highly variable results have been reported regarding the chemosensitivity of LCNEC to platinum/etoposide–based regimens utilized for SCLC (8–10), resulting in the lack of consensus on whether LCNEC should be clinically managed as SCLC versus non-SCLC (NSCLC).
At that crux of the problem is the unresolved biologic relationship between LCNEC and SCLC, which is compounded by the paucity of clinical studies and relative rarity of these tumors (~3% of all lung carcinomas; ref. 11). Because LCNEC shares several fundamental clinicopathologic features with SCLC, including aggressive clinical behavior, strong link to smoking, exceptionally high proliferation rates, neuroendocrine gene expression, and some morphologic features (in at least a subset of cases), combined with the evidence from experimental models of a close relationship between SCLC and LCNEC (12), it has been postulated that LCNECs may represent a variant of SCLC. To date, gene expression and limited genomic analyses have yielded conflicting results regarding the biologic relationship between LCNEC and SCLC. Both entirely overlapping (13) and distinct (14) gene expression profiles have been reported. Genomically, LCNEC is known to share with SCLC frequent alterations in RB1 and TP53, albeit with variable reported frequencies (15–17). Currently, only limited published data are available on comprehensive genomic profiles of LCNEC by next-generation sequencing (NGS) methods. Notably, in a study that included whole-exome sequencing of 15 LCNECs, it was concluded that LCNEC was overall similar to SCLC but harbored occasional alterations characteristic of other tumor types (17). On the other hand, there have been several reports of molecular alterations typical of adenocarcinoma in histologically pure LCNEC with no overt adenocarcinoma component, including EGFR mutations (18, 19), ALK rearrangements (20), and KRAS mutations (21), drawing a sharp contradistinction with classic SCLC, which in the pure de novo form is consistently devoid of adenocarcinoma-type driver mutations (22–24).
The goal of this study was therefore to perform detailed genomic characterization of LCNEC to clarify its biologic relationship with SCLC and other major lung cancer types, as well as to describe the landscape of potentially targetable molecular alterations in these tumors. Here, we report the results of targeted NGS of 45 LCNECs, in conjunction with detailed morphologic, immunohistochemical, and clinicopathologic analysis. Furthermore, we compared mutation profiles in LCNEC with those of other lung cancer histologies analyzed by the same NGS platform.
Materials and Methods
Patients and samples
The study was performed with the approval of the Institutional Review Board of the Memorial Sloan Kettering Cancer Center (MSKCC, New York, NY). Surgically resected LCNECs were included in the study after central pathology review by 3 thoracic pathologists (N. Rekhtman, A.L. Moreira, and W.D. Travis). Only histologically pure LCNECs were included [i.e., tumors lacking morphologically identifiable adenocarcinoma, squamous cell carcinoma (SqCC), or SCLC component]. All cases met the WHO criteria for LCNEC, including (i) neuroendocrine morphology, including organoid nesting, palisading, trabeculae, and/or rosettes; (ii) high grade, defined as >10 mitoses per 10 high-power fields (HPF) and typically necrosis; and (iii) IHC expression of at least one neuroendocrine marker (synaptophysin, chromogranin-A, CD56/NCAM) in ≥10% of tumor cells (2). LCNEC was distinguished from SCLC based on a constellation of cytomorphologic features, including the presence of nucleoli (contrasting with the sine qua non nuclear appearance of SCLC characterized by dispersed granular chromatin lacking prominent nucleoli) and/or abundant cytoplasm, and typically but not invariably larger nuclear size (2).
Molecular analysis
DNA was extracted from approximately 80 μm of formalin-fixed, paraffin-embedded tumor sections and matched benign tissue. Each tumor sample was macrodissected to enrich for tumor cells. Tissue was deparaffinized and DNA extracted using DNeasy Blood & Tissue Kit (Qiagen).
NGS of paired tumor/normal tissue samples was performed using Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) platform, a custom hybrid capture-based assay for targeted deep sequencing of all exons and selected introns of key cancer genes. The selected genes (n = 241) encompass the majority of established oncogenes and tumor suppressors, including actionable targets of approved therapies and agents under clinical investigation at MSKCC (full list in Supplementary Table S1). Briefly, barcoded DNA libraries from tumor and normal samples were captured using custom oligonucleotide probes, massively parallel sequenced on an Illumina HiSeq 2500 platform, and subjected to a custom analysis pipeline to identify single-nucleotide variants, small indels (<30 bp), copy number alterations (CNA), and selected structural rearrangements (including ALK fusions), as described in detail elsewhere (25). All candidate mutations and indels were manually reviewed using Integrative Genomics Viewer.
Mutations predicted to be deleterious, defined as being reported in the Catalogue of Somatic Mutation in Cancer or having medium/high functional impact score by Mutation Assessor [accessed via cBioPortal for Cancer Genomics (26)], were examined and manually curated for potential actionability using approved or investigational compounds. For comparison with other lung tumor histologies (N = 242), MSK-IMPACT results were reviewed for lung adenocarcinomas (n = 151), SqCC (n = 36), SCLC (n = 42), and carcinoids (n = 13) that were consecutively sequenced as part of routine clinical care under an institutional initiative (27), or as part of clinical protocols or retrospective studies, with detailed results reported in part elsewhere (24, 28).
Histologic, immunohistochemical, and clinical analysis
Hematoxylin and eosin–stained tissue sections were reviewed to quantify the mitotic rate by manually counting the mitotic figures in 10 HPFs, calculated as an average of at least 5 sets of 10 HPF counts. Prominence of nucleoli and abundance of cytoplasm, the key histologic features distinguishing LCNEC from SCLC (2), were recorded. Tumors featuring prominent nucleoli (defined as nucleoli that were readily identifiable using a 10× microscope objective) and/or abundant cytoplasm (defined as cytoplasm of sufficient volume to render cytoplasmic membranes readily visible) were classified as “NSCLC-spectrum” morphology, whereas tumors with identifiable yet not prominent nucleoli and cytoplasm were classified as “SCLC-spectrum” morphology (i.e., the morphology that enters in the differential diagnosis with SCLC, yet is sufficiently distinct to qualify for the diagnosis of LCNEC). Tumors with intermediate or geographically mixed morphologic features were classified as “mixed.” IHC was performed utilizing the following primary antibodies: synaptophysin (clone Snp88), chromogranin-A (LK2H10), CD56 (MRQ-42), Ki67 (MIB1), ASCL1 (24B72D11.1), pRb (13A100), STK11 (Ley37D/G6), Napsin-A (IP64), and p40 (BC28; see Supplementary Table S2 for detailed protocols and scoring criteria).
Electronic medical records were reviewed to collect demographic and clinical information. Staging was performed according to the American Joint Committee on Cancer 7th Edition. Overall and recurrence-free survival were estimated using the Kaplan–Meier method, with the mean follow-up period of 4.1 years (range 0.02–14.5 years). For patients with localized disease treated with curative intent, recurrence-free survival was determined as time to recurrence or death, whichever came first. To assess treatment response to first-line chemotherapy, CT scans were reviewed up to 7 months (mean 3 months) from a baseline scan using the standard RECIST guidelines (version 1.1) if scans were available for review.
Statistical analysis
Statistical analysis was performed using R software (R version 3.2.2). P values were computed using Fisher exact test and non-parametric Mann–Whitney U test for categorical and continuous variables, respectively. Survival analysis was performed using survival package in CRAN, version 2.38.
Results
Clinicopathologic characteristics
Tumor and patient characteristics of 45 LCNECs in this series are summarized in Supplementary Table S3. All but one patient were current or former smokers, with a mean smoking history of 50 pack-years (range 0–150). The mean age at diagnosis was 64 (range 45–79) and 51% were male. Stage distribution was as follows: stage I, 40%; stage II–IIIA, 44%; and stage IIIB/IV, 15%. The mean mitotic rate was 68/10 HPF (range 14–110), and the mean Ki67 proliferation index was 75% (range 30%–100%). All tumors expressed at least one conventional neuroendocrine marker (synaptophysin, chromogranin-A, CD56), 80% of tumors expressed 2 to 3 conventional markers, and 71% expressed the investigational neuroendocrine marker ASCL1.
Overall landscape of somatic mutation
Targeted NGS of 45 LCNEC tumor/normal tissue pairs by MSK-IMPACT platform identified a total of 535 nonsynonymous somatic mutations (392 missense, 66 nonsense, 37 splice site, 34 frameshift, 4 deletion, 2 insertion), 61 gains, and 15 losses (Supplementary Table S4). Given the total protein-coding territory of MSK-IMPACTof 1.2 Mb, we estimate that the mean rate of nonsynonymous mutations in LCNEC is 10.5/Mb. Of the single-nucleotide variants, 47.5% were G>T (C>A) transversions, typical of tobacco-induced carcinogenesis. The mean number of nonsynonymous mutations, gains, and losses per case was 11.8 (range 1–30), 1.3 (range 0–7), and 0.3 (0–2), respectively. The mean tumor coverage depth was 486-fold (range 60–1,086 fold).
Recurrently altered genes
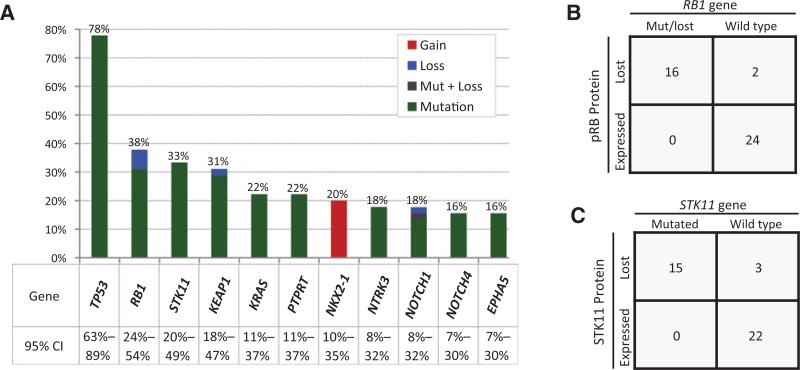
Commonly altered genes in LCNEC included TP53 (n = 35; 78%), RB1 (n = 17; 38%), STK11 (n = 15; 33%), KEAP1 (n = 14; 31%), and KRAS (n = 10; 22%; Fig. 1A). Analogous to lung adenocarcinoma, KRAS, STK11, and KEAP1 mutations were not mutually exclusive (30, 31). Ninety-five percent of TP53 mutations affected the functionally critical DNA-binding domain. The majority (81%) of RB1 mutations and many STK11 and KEAP1 mutations were protein truncating (frameshift and nonsense), consistent with their tumor suppressor role. KRAS mutations comprised the canonical missense mutations: G12C (n = 6), G12D (n = 1), G13D (n = 2), and Q61L (n = 1; example of _KRAS_-mutant LCNEC shown in Supplementary Fig. S1).
Figure 1.
Commonly altered genes in LCNEC. A, histogram of commonly altered genes detected by NGS (MSK-IMPACT) in LCNEC. CI, confidence intervals. B, RB1 gene alterations versus pRb expression by IHC. C, STK11 mutations versus STK11 expression by IHC. IHC not available for pRB and STK11 IHC in 3 and 5 cases, respectively. Mut, mutated.
Given the prior observations that the loss of pRB (29) and STK11 (30, 31) expression may occur by nonmutational mechanisms in lung carcinomas, LCNECs with sufficient tissue were further evaluated by anti-pRb (n = 42) and anti-STK11 (n = 40) IHC. As shown in Fig. 1B and C, all tested LCNECs with RB1 (n = 16) and STK11 (n = 15) gene alterations showed complete loss of the respective protein expression. In addition, complete loss of protein expression was identified in 2 of 26 cases lacking RB1 and 3 of 25 cases lacking STK11 gene alterations.
Molecular subsets
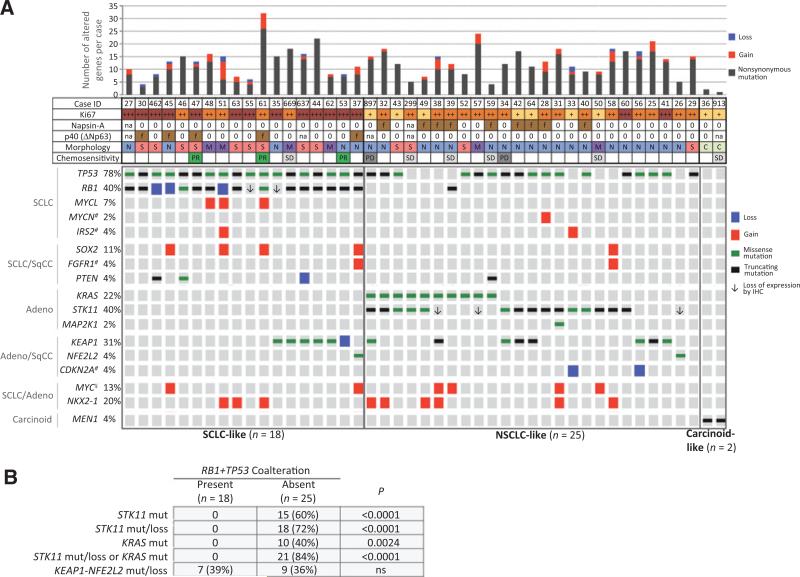
On the basis of the recent NGS studies showing that joint inactivation of RB1+TP53 is a virtually invariable signature event in SCLC (22), we classified LCNEC with coaltered RB1+TP53 as “SCLC-like” (n = 18; 40%), whereas tumors lacking this coalteration were classified as “NSCLC-like” (n = 25; 68%) after exclusion of 2 outlier cases with carcinoid-like molecular and cytomorphologic features (see below).
Cooccurring genomic alterations in these 3 subgroups are shown in Fig. 2A. The most striking finding was that nearly all LCNECs lacking RB1+TP53 alterations (NSCLC-like subset) harbored additional NSCLC-type mutations: STK11 and KRAS mutations, typical of adenocarcinoma, occurred in 15 (60%) and 10 (40%) cases, respectively, with a combined rate of STK11 mutations/protein loss in NSCLC-like tumors of 72% (18/25 cases). Either STK11 mutation/protein loss or KRAS mutation were present in 21 of 25 (84%) cases. KEAP1-NFE2L2 mutations, typical of either adenocarcinoma or SqCC, were present in 9 (36%) of cases. Overall, STK11, KRAS, or KEAP1-NFE2L2 mutations were present in 24 of 25 NSCLC-like LCNECs. Notably, all STK11 and KRAS alterations were entirely restricted to this subset, and none occurred in tumors bearing RB1+TP53 coalterations (P < 0.0001 and P = 0.0024, respectively; Fig. 2B). Conversely, KEAP1–NFE2L2 mutations were not mutually exclusive with RB1+TP53 (see below). Only a single NSCLC-like tumor (KRAS mutated) harbored RB1 mutation/protein loss, but this case lacked TP53 mutations. Several other alterations characteristic of NSCLC were identified in isolated cases this subset, including CDKN2A losses (n = 2), MAP2K1 (n = 1), and PIK3CA (n = 1) mutations. NKX2.1 amplification, an alteration characteristic of both lung adenocarcinoma and SCLC, was identified in both NSCLC-like (24%) and SCLC-like (17%) subsets. Interestingly, NSCLC-like tumors occasionally harbored alterations characteristic of SCLC and other neuroendocrine/neuronal tumors, including isolated cases with high MYCN amplification (8.5-fold) and IRS2 amplification.
Figure 2.
Molecular subsets of LCNEC. A, OncoPrint depicting coalterations in selected genes in LCNEC with presence or absence of RB1+TP53 coalteration defining the major SCLC-like and NSCLC-like subsets, respectively, and MEN1 mutations (mut) and low total mutation burden defining carcinoid-like subset. Left, conventional lung cancer types characteristically associated with indicated gene alterations are shown. ¥, MYC amplifications also occur in SqCC. #, genes for which only CNAs are shown. Loss of pRB and STK11 expression by IHC (↓) is only shown for cases without gene mutations/losses. For all other cases, loss of expression and molecular results were concordant. No pRB IHC available for cases 27, 897, 299; and no STK11 IHC available for cases 27, 55, 637, 299, and 913. Selected clinicopathologic features are designated as follows: Ki67: +++ >80%; ++ 60%–80%; + 40%–50%; Napsin-A and p40: f, focally expressed by IHC; 0, no expression; na, not available; morphology: S, SCLC spectrum; N, NSCLC spectrum; M, mixed; C, carcinoid-like; chemosensitivity: PR, partial response; SD, stable disease; PD, progressive disease. B, mutual exclusivity of RB1+TP53 and STK11/KRAS alterations. ns, not significant.
Conversely, tumor with RB1+TP53 comutation/loss (SCLC-like subset) featured exclusive or preferential coalterations of several genes typical of SCLC, including MYCL amplification (17%), SOX2 amplification (22%), PTEN mutation/loss (17%), and FGFR1 amplification (5%). In contrast, STK11 and KRAS mutations were entirely absent, whereas KEAP1–NFE2L2 alterations occurred in 39% of these cases (see Discussion).
The third, minor, subset of LCNEC, carcinoid-like, included 2 outlier cases, characterized by a low total number of detected alterations (two and one per case) compared with the mean of 13 (range 4–32) genomic alterations per case for other LCNECs (P = 0.007; Supplementary Fig. S2). In addition, in contrast to all other LCNEC in our dataset, both of these tumors harbored MEN1 mutations - the hallmark of carcinoid tumors. Both MEN1 mutations were protein truncating and were associated with high mutant allele frequencies (79% and 89%), suggesting associated loss-of-heterozygosity.
Other frequently altered genes and functional groups, including NOTCH family genes
We identified the following 4 gene families/functional groups to be frequently altered in LCNEC overall, with no predilection for either the SCLC-like or NSCLC-like subset. This included (i) chromatin modifiers (78% of LCNEC), with particularly high mutation frequency in MLL histone methyltransferases (33%) and SWI/SNF chromatin–remodeling genes (40%); (ii) genes implicated in neurogenesis, including NOTCH1–4 (33%) and NTRK2/3 (19%); (iii) DNA replication/repair genes (52%); and (iv) PI3K–AKT–mTOR pathway genes (49%; Supplementary Fig. S3). The majority (17/19) of NOTCH family mutations were overt loss-of-function (protein truncating) mutations or were predicted to be functionally deleterious by PROVEAN or SIFT (Supplementary Table S5). Many mutations were located in the extracellular EGF-like domain (Supplementary Fig. S4), analogous to the loss-of-function NOTCH mutations in SCLC (22) and SqCC (32).
Molecular alterations in LCNEC subsets versus conventional lung carcinomas
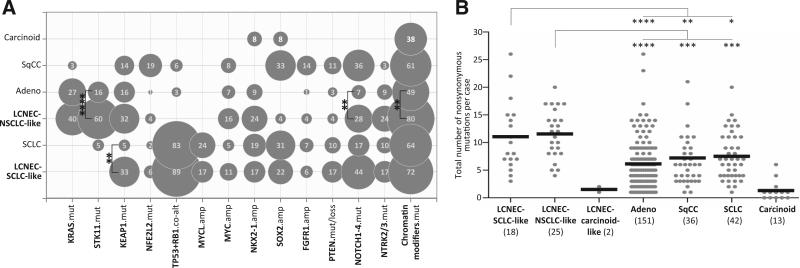
Figure 3A illustrates mutation frequencies in LCNEC versus other major lung cancer histologies analyzed by the same NGS platform (MSK-IMPACT), which highlighted the overall similarity between the SCLC-like subset and SCLC and the NSCLC-like subset and adenocarcinoma. This comparison also revealed that despite the overall similarity, there were several distinctive alterations in LCNEC subsets compared with their corresponding conventional carcinoma counterparts. Significant differences between NSCLC-like LCNEC and conventional adenocarcinoma included more frequent mutations in STK11 (60% vs. 16%; P = 0.0001), NOTCH1–4 (28% vs. 7%; P = 0.005), chromatin modifiers (80% vs. 49%, P = 0.005), fewer sensitizing EGFR mutations (0% vs. 25%, P = 0.003), as well as significantly elevated comutation rate in STK11+TP53 (40% vs. 8%; P = 0.0002), respectively. Conversely, SCLC-like LCNEC differed from SCLC by significantly higher rate of KEAP1 alterations (33% vs. 5%, respectively; P = 0.008).
Figure 3.
Genomic alteration in LCNEC versus other lung cancer types analyzed by MSK-IMPACT. A, bubble graph illustrating frequency of selected alterations in SCLC-like and NSCLC-like LCNEC subsets versus conventional SCLC (n = 42), adenocarcinoma (n = 151), SqCC (n = 36), and carcinoids (n = 13), with bubble area indicating frequency of an alteration. Significant differences (P < 0.01; Fisher exact test) are shown for SCLC-like LCNEC versus SCLC and NSCLC-like LCNEC versus adenocarcinoma. EGFR and ALK alterations (not shown) were absent in LCNEC, compared with 25% and 7% rate, respectively, in adenocarcinoma. See Supplementary Fig. S3 for individual genes in “chromatin modifiers” group. B, dot plot graph illustrating total number of nonsynonymous mutations (mut) per case in LCNEC versus other lung tumors. Lines indicate the mean. This comparison was performed for 222 genes shared across several MSK-IMPACT gene panel versions used to sequence different tumor types. Three data points (2 adenocarcinomas, 1 SqCC) are outside the axis limit. amp, amplification; co-alt, coalteration. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
We further noted that the profile of major mutations in NSCLC-like LCNEC was reminiscent of the “proximal-proliferative” transcriptional subset of lung adenocarcinoma in The Cancer Genome Atlas (TCGA) series (33), which is similarly characterized by frequent mutations in STK11, KEAP1, and KRAS. To test the hypothesis that NSCLC-like LCNEC may be related specifically to this subset of lung adenocarcinoma, we analyzed publicly available TCGA data-set for the expression of neuroendocrine genes, ASCL1 and dopa decarboxylase, previously found to be overexpressed in a fraction of lung adenocarcinomas (34–36). This analysis revealed that, indeed, proximal-proliferative subset was significantly enriched in tumors with high expression of ASCL1 (P = 6.92e–10) and dopa decarboxylase (P = 3.86e–14; Supplementary Fig. S5).
Finally, we compared overall mutation burden detected by MSK-IMPACT across LCNEC and other lung cancer types. This analysis revealed that both NSCLC-like and, to a lesser extent, SCLC-like LCNEC subsets harbored a higher mutation load than their conventional counterparts: NSCLC-like subset exhibited greater number of mutations than both lung adenocarcinoma (P < 0.0001) and SqCC (P = 0.0002), and the SCLC-like subset had marginally elevated mutation burden compared with conventional SCLC (P = 0.03; Fig. 3B).
Potentially targetable genomic alterations
No sensitizing EGFR mutations or ALK rearrangements were identified. Nevertheless, at least one alteration potentially targetable by investigational agents was present in 30 of 45 (67%) LCNEC, more commonly in NSCLC-like than SCLC-like subset (84% vs. 50%, respectively; P = 0.02). Individual drug targets are listed in Supplementary Table S6.
Molecular subsets of LCNEC and histomorphologic features
We next asked whether SCLC-like and NSCLC-like molecular subsets of LCNEC were distinguishable morphologically. Indeed, we found that at least focal SCLC-spectrum morphology (see Materials and Methods) was over-represented in SCLC-like molecular subset (72%), whereas NSCLC-spectrum morphology predominated in tumors with NSCLC-like molecular features (76%), P = 0.006 (Table 1; Fig. 4A and B). Despite this association, there was a substantial overlap in morphologic features between these subsets, with a total of 35% of cases exhibiting discordant or mixed morphology, particularly in SCLC-like molecular group.
Table 1.
Clinicopathologic features of SCLC-like versus NSCLC-like molecular subsets of LCNEC
| SCLC-like (n = 18) | NSCLC-like (n = 25) | P | |
|---|---|---|---|
| Age - mean (range) | 63 (50-78) | 64 (45-79) | 0.803 |
| Gender (female) | 6 (33%) | 16 (64%) | 0.067 |
| Smoking (pack-years) - mean (range) | 50 (25-80) | 51 (0-150) | 0.830 |
| Never smoker | 0 | 1 | |
| Current/former smoker | 18 | 24 | |
| Stage | |||
| I | 7 (39%) | 11 (44%) | 0.922 |
| II-IIIA | 8 (44%) | 10 (40%) | |
| IIIB/IV | 3 (17%) | 4 (16%) | |
| Syn, Chr, CD56 expressiona | |||
| ++ | 14 (78%) | 21 (84%) | 0.701 |
| + | 4 (22%) | 4 (16%) | |
| ASCL1 Expression | 10/16 (62%) | 19/24 (79%) | 0.295 |
| Ki67 (MIB1) Proliferation rateb - mean (range) | 88 (60-100) | 68 (40-90) | <0.0001 |
| >80% | 12 (67%) | 2 (8%) | |
| 60-80% | 6 (33%) | 17 (68%) | |
| 40-50% | 0 | 6 (24%) | |
| Mitotic count per 10 HPFb - mean (range) | 83 (40-110) | 60 (14-110) | 0.017 |
| Napsin-A expression | 0/17 | 9/24 (38%)c | 0.005 |
| p40 (ΔNp63) expression | 5/16 (31%)d | 0/23 | 0.006 |
| Cytomorphology | |||
| LCNEC, SCLC-spectrum | 9 (50%) | 4 (16%) | 0.006 |
| LCNEC, NSCLC-spectrum | 5 (28%) | 19 (76%) | |
| LCNEC, mixed | 4 (22%) | 2 (8%) | |
| Survival (log-rank test)e | |||
| Overall survival: 5-yr (median) | 59.5% (5.1 y) | 53.8% (5.3 y) | 0.883 |
| Recurrence-free survival: 5-yr (median) | 27.5% (0.9 y) | 51.4% (5.3 y) | 0.475 |
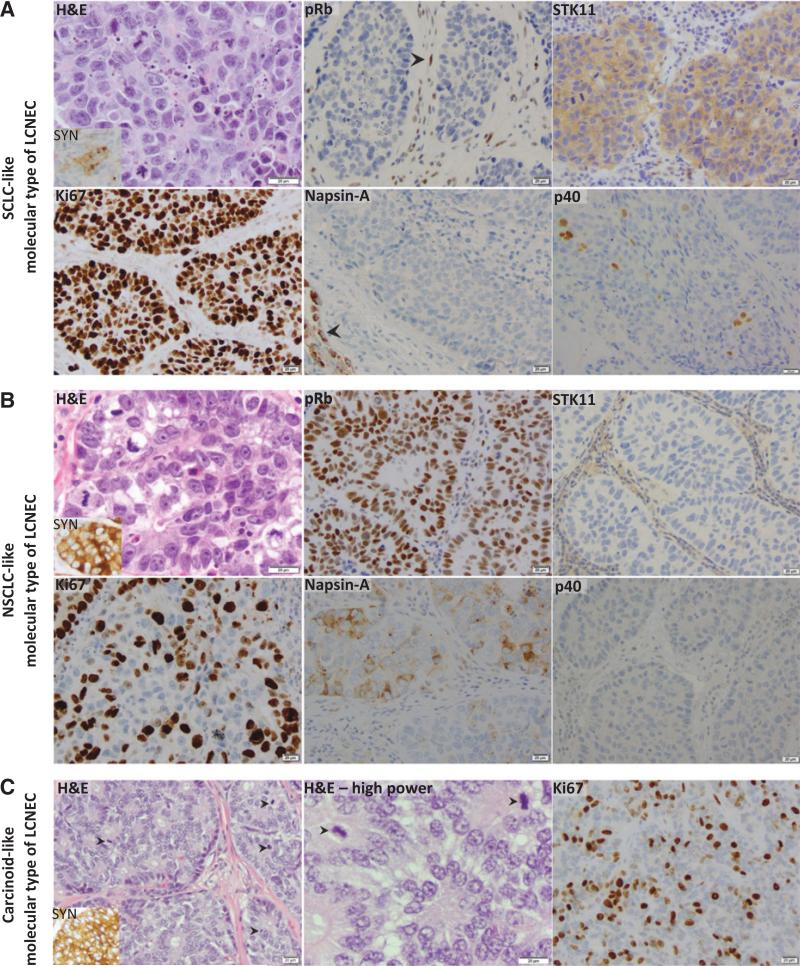
Figure 4.
Morphologic and immunohistochemical features of genomically defined LCNEC subsets. Representative examples from each molecular subset are illustrated. A, LCNEC with SCLC-like molecular profile (case ID LCNEC-47) and SCLC-spectrum morphology [identifiable but not prominent nucleoli and cytoplasm on hematoxylin and eosin (H&E)]. IHC illustrates complete loss of pRb in tumor cells (arrowhead indicates intact pRB expression in stromal cells, serving as internal positive control), intact STK11 expression, high Ki67 index (100%), negative Napsin-A (arrowhead indicates Napsin-A–positive peritumoral pneumocytes, serving as internal positive control), and focal (<5%) tumor cell labeling for p40. B, LCNEC with NSCLC-like molecular profile (case ID LCNEC-40, _STK11_-mutated) and NSCLC-spectrum morphology (prominent nucleoli and abundant cytoplasm on H&E). IHC illustrates intact pRB expression, complete loss of STK11 expression, Ki67 proliferation index of 70%, weak/focal Napsin-A expression, and negative p40. C, LCNEC with carcinoid-like molecular profile (case ID LCNEC-36). H&E illustrates morphology characteristic of carcinoid tumors (bland uniform nuclei, abundant rosettes), but with unusually-high mitotic rate (arrowheads) and elevated Ki67 rate (40%). SYN synaptophysin.
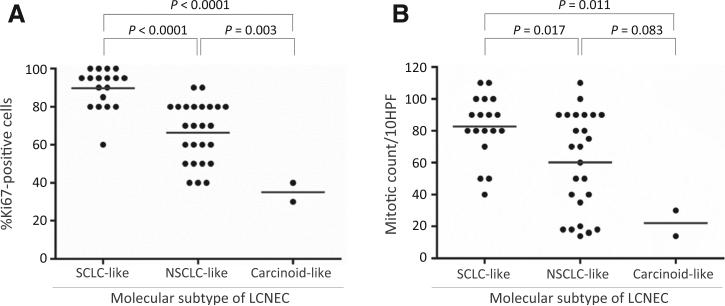
Two other histologic/immunophenotypic features differed in SCLC-like and NSCLC-like molecular subsets (Table 1). First, as a group, tumors with SCLC-like molecular features displayed significantly higher proliferative activity than NSCLC-like tumors, as revealed by Ki67 proliferation marker expression (P < 0.0001) and mitotic figure count (P = 0.017), although both subsets showed a wide range of proliferation rates with substantial overlap in individual cases (Fig. 5). Second, 9 (37%) NSCLC-like LCNECs exhibited low-level (weak and/or focal, 5%–30% tumor cells) reactivity for Napsin-A, a well-established and highly specific marker of exocrine differentiation, whereas none of the SCLC-like tumors expressed this marker (P = 0.005). Conversely, a low-level expression (≤10% tumor cells) of squamous differentiation marker, p40, was identified only in SCLC-like LCNECs (5 cases). Napsin-A and p40 expression was seen in areas of classic LCNEC morphology, lacking overt glandular or squamous differentiation (Fig. 4A and B). Neuroendocrine marker expression (synaptophysin, chromogranin-A, CD56, ASCL1) was equivalent in the SCLC-like and NSCLC-like subsets.
Figure 5.
Scatter plots for Ki67 proliferation index (A) and mitotic figure count in LCNEC subsets (B). Lines indicate the mean.
The 2 tumors with carcinoid-like molecular features met WHO criteria for LCNEC based on mitotic rate exceeding 10 mitosis/10 HPFs (featuring 30 and 14 mitoses per 10 HPF and Ki67 rate of 40% and 30%, respectively), but these tumors did have distinct carcinoid-type cytomorphology (Fig. 4C). Notably, although higher than typical of carcinoid tumors, these proliferation rates fell in the lowest range of proliferative activity seen in other LCNECs (Fig. 5).
Molecular subsets of LCNEC and clinical features, including response to cytotoxic therapy
Comparison of patient characteristics in the SCLC-like and NSCLC-like molecular subsets did not reveal significant differences in the distribution of patient age, gender, stage, smoking history, or survival, although NSCLC-like subset was numerically enriched in females compared with SCLC-like subset and included the sole never-smoker (Table 1). In addition, SCLC-like subset exhibited a nonsignificant trend toward shorter recurrence-free survival (Supplementary Fig. S6).
In this series, only 11 patients received cytotoxic chemotherapy (almost all platinum based) for the de novo or recurrent stage IV LCNEC (Supplementary Table S7). Only 3 of 11 patients had radiologic response to treatment (all partial responses). Although the small number of evaluable patients precludes statistical analysis, we did note that all 3 responders were in the SCLC-like molecular subset (3/4 treated patients), whereas none of the treated patients in NSCLC-like subset showed objective responses (0/6; Fig. 2), although 4 of 6 NSCLC-like patients did show stable disease on therapy, suggesting some treatment benefit. One patient with carcinoid-like LCNEC received cytotoxic chemotherapy and showed stable disease.
Discussion
NSCLC-like subset
The first major finding in this study is that 56% of LCNEC displayed NSCLC-like molecular features, characterized by the lack of RB1+TP53 coalteration and virtually invariable presence of NSCLC-type mutations (STK11, KRAS, KEAP1, NFE2L2), with STK11/KRAS mutations occurring in 80% of these tumors. In conventional lung carcinomas, both STK11 and KRAS mutations occur almost exclusively in adenocarcinomas (33), and they are either entirely absent or exceptionally rare in SCLC, as previously reported (22–24) and as seen in this study. This finding draws a sharp contrast between NSCLC-like LCNEC and SCLC and highlights a predominant genomic similarity between these tumors and lung adenocarcinoma. A link to adenocarcinoma is further supported by our finding that 37% of NSCLC-like LCNEC exhibited low-level expression of pulmonary exocrine cell marker Napsin-A - an aspartic proteinase involved in surfactant protein B processing, which is highly specific for lung adenocarcinoma and is consistently negative in SCLC (37). We suggest that NSCLC-like subset of LCNEC may represent the extreme end of the spectrum with tumors designated “NSCLC with NE differentiation,” the tumors in which neuroendocrine gene expression occur in the absence of overt neuroendocrine morphology (34–36). The known phenomenon that neuroendocrine gene expression has a predilection for lung adenocarcinomas and is rare in SqCC (35, 36) parallels our finding of a predominant relationship with adenocarcinoma in NSCLC-like LCNEC. Our observation that the profile of major mutations in NSCLC-like LCNEC resembles the “proximal-proliferative” transcriptional subgroup of lung adenocarcinoma in TCGA series (33), which is similarly dominated by STK11, KRAS, and KEAP1 mutations, and which we found to be significantly enriched in tumors with high neuroendocrine gene expression, further suggests a potential link between NSCLC-like LCNEC and this specific subset of lung adenocarcinoma.
Despite the overall genomic similarity with adenocarcinoma, we found several molecular differences between NSCLC-like LCNEC and conventional lung adenocarcinoma, which supports the concept that these tumors represent related but separate entities. The most striking difference was an unusually high rate of STK11 mutations (60%) in NSCLC-like LCNEC (with a combined rate of STK11 mutations and protein loss of 72%), compared with 16% to 17% STK11 mutation rate in lung adenocarcinoma in prior (33) and current series, respectively. STK11 (also known as LKB1) is a tumor suppressor, which affects diverse cell pathways, including cellular metabolism, pluripotency and phenotypic plasticity (38), and whose inactivation in lung adenocarcinoma is associated with accelerated tumor growth and metastases, particularly in cooperation with KRAS mutations (39, 40). These functional roles of STK11 may be contributing to dysregulated differentiation and aggressive clinical phenotype of LCNEC.
Other notable differences in NSCLC-like LCNEC from lung adenocarcinoma included frequent alterations in key neuro-genesis regulators, NOTCH family and NTRK2/3 and isolated alterations typical of SCLC and other neuroendocrine/neuronal tumors, MYCN, and IRS2 amplifications. In agreement with our findings, frequent mutations in the NOTCH family genes (41) and NTRK2/3 (42) were identified in LCNEC by other investigators. NOTCH family alterations are of particular interest given the strong evidence for their crucial role in neuroendocrine fate specification in normal development (43) and in SCLC (22). Also of potential relevance is our finding of a remarkably high rate of mutations in chromatin modifiers in LCNEC, as this feature has recently emerged as a hallmark of various neuroendocrine tumors (22, 44). Collectively, we hypothesize that alterations in genes regulating neurogenesis and the epigenetic landscape may contribute to the genesis of neuroendocrine phenotype in NSCLC-like LCNEC.
SCLC-like subset
The second major finding in this study is that 40% of LCNEC exhibited SCLC-like genomic profile, characterized by RB1+TP53 coalteration, complete absence of STK11 and KRAS mutations, and exclusive or enriched occurrence of MYCL, SOX2, and FGFR1 amplifications and PTEN mutation/loss, analogous to conventional SCLC (22–24). Importantly, the SCLC-like LCNECs displayed several clinicopathologic features that paralleled those seen in conventional SCLC, including higher proliferative activity, a trend toward shorter recurrence-free survival, and, in a subset analysis, a suggestion of greater platinum-based chemosensitivity (see below) than NSCLC-like tumors. Furthermore, SCLC-like molecular subset was enriched in tumors with morphology that approached the morphologic spectrum of SCLC, yet met the criteria for LCNEC. Although these data support a close relationship of SCLC-like LCNEC and conventional SCLC, whether these tumors may be regarded as a single group for management purposes will require further clinical studies.
Despite the overall similarity, a notable difference in SCLC-like LCNEC from conventional SCLC identified in this study was an elevated rate of KEAP1–NFE2L2 alterations (39%). These alterations only rarely occur in conventional SCLC, and in the absence of concurrent STK11/KRAS mutations, they are a common feature of SqCC. In fact, a close histogenetic relationship of some conventional SCLC and SqCC has been previously suggested, given that these tumors share several key genomic aberrations, including SOX2 and FGFR1 amplification and PTEN inactivation (45). It is thus tempting to speculate that at least some SCLC-like LCNEC may share an even stronger histogenetic link with SqCC than conventional SCLC, which may account for their atypical morphology and higher rate of KEAP1–NFE2L2 alterations. This hypothesis is supported by our finding of focal expression of squamous marker p40 in this group of tumors.
Although the joint loss of RB1+TP53 has emerged as a virtually universal feature of SCLC (22), this coalteration is not entirely specific to SCLC, as it also occurs in a minority of conventional NSCLC (≤6% in our series). Nevertheless, the use of RB1+TP53 coalteration as a classifier for SCLC-like subtype of LCNEC is supported by the evidence from experimental models showing that neuroendocrine cells are selectively vulnerable to pRB loss, contrasting with the ability of non-neuroendocrine cells to compensate for pRb inactivation (46), which suggests that pRB inactivation may expert selective growth-promoting effects specifically in the context of neuroendocrine carcinomas.
Carcinoid-like subset
The finding that among LCNECs in this series 2 cases had carcinoid-like molecular profiles, characterized by inactivating MEN1 mutations and low overall mutation burden, analogous to conventional carcinoids (44), should not be interpreted as evidence that some bona fide LCNEC represent transformed carcinoid tumors. It is important to emphasize that both carcinoid-like LCNECs were clear morphologic outliers in that they displayed the overt carcinoid-type morphology, unlike the other LCNECs in this series. Their classification as LCNEC was based entirely on their proliferation rate exceeding the maximal proliferative activity currently accepted for lung carcinoids (i.e., 10 mitoses/10 HPF, with typical Ki67 rate under 20%), which qualifies them for the diagnosis of LCNEC under the WHO criteria (2). This special issue will require further study, but overall, our data support carcinoid family kinship of these unusual tumors and expand the spectrum of proliferative capacity currently recognized for lung carcinoids, analogous to the recent findings for gastroenteropancreatic neuroendocrine tumors (47).
Therapeutic implications
A possible implication of our findings is that biologic subgroups of LCNEC could underlie the reported heterogeneity of responses to platinum-based chemotherapy in patients with LCNEC. Given the small number of patients with evaluable treatment responses in this study, we could not definitely assess this hypothesis. However, in a small subgroup analysis, there was a suggestion that SCLC-like LCNECs exhibited greater platinum-based chemosensitivity than other LCNECs. Prospective genomic analysis of patients with advanced disease, which is currently ongoing at our institution, should enable further correlation of molecular profiles and clinical behavior in LCNEC patients.
With respect to targeted therapy implications, the relative paucity of potentially actionable alterations in SCLC-like subset, which included rare PTEN and FGFR1 alterations, parallels the observations in conventional SCLC (22–24). For NSCLC-like subset, the profile of potential targets resembles those seen in smoking-related lung adenocarcinomas. Although in this study, we did not identify sensitizing EGFR mutations or ALK rearrangements, which are inversely associated with smoking history, these alterations have been reported in isolated cases of LCNEC (18–20). Despite the apparent low incidence, it is important to include LCNEC in screening for EGFR and ALK alterations because impressive clinical responses to tyrosine kinase inhibitors have been reported in several _EGFR_-mutated LCNECs (18, 19).
Our findings suggest several other special therapeutic considerations for LCNEC. First, the finding that both NSCLC-like and, to a lesser extent, SCLC-like LCNEC harbor extremely high mutation burden, exceeding that of conventional NSCLC and SCLC, suggests that these tumors may be particularly sensitive to PD-1/PD-L1 immune checkpoint inhibitors, whose activity has been shown to correlate with high nonsynonymous mutational load (48). In addition, frequently altered chromatin modifiers in LCNEC may serve as potential targets for emerging epigenetic therapies (49).
Pathology
An important practical question raised by our findings is whether SCLC-like and NSCLC-like subsets of LCNEC can be identified by routine pathologic examination in the absence of molecular analysis. Although we did find that genomically defined subgroups were enriched in tumors with SCLC-like and NSCLC-like morphologic features, respectively, and that, as a group, tumors with SCLC-like molecular features were more proliferative, we also found a substantial histologic overlap, emphasizing that these molecular subtypes cannot be accurately predicted by routine morphologic examination in individual cases. On the other hand, we found that RB1 alteration, the key defining feature of SCLC-like group, can be accurately identified by IHC, which also detected few instances of pRB protein loss occurring in the absence of gene mutations/CNAs, which may be mediated by mechanisms like promoter hypermethylation, as described in SCLC (29). Thus, in a case with LCNEC morphology, intact pRb expression by IHC could serve as a marker of NSCLC-like subtype, even in the absence of molecular testing. Conversely, the loss of pRb expression, although favoring SCLC-like subset, would require molecular confirmation of concurrent TP53 mutation given the rare possibility of pRb loss in the absence of TP53 mutation, as illustrated in a single case of _KRAS_-mutated LCNEC with isolated RB1 mutation in this study. Unlike pRb, we found p53 IHC to be an imperfect surrogate for molecular testing in LCNEC (data not shown). In addition, our data suggest that the loss of STK11 expression and/or presence of focal Napsin-A expression may serve as specific albeit incompletely sensitive IHC-based markers of NSCLC-like subset. Overall, pending further validation, our data suggest that IHC could serve to predict molecular subtype of LCNEC in at least a subset of cases at the time of histopathologic evaluation.
Finally, it is worth commenting on a potential limitation of our targeted NGS platform compared with more comprehensive NGS methodologies. Although 241 genes represented in our gene panel encompass the majority of the known functionally important cancer genes (27), we cannot exclude the possibility that our analysis may have failed to identify relevant alterations in untested genes or non-exonic regions. Therefore, studies utilizing whole-genome/exome sequencing technologies will be needed for more comprehensive characterization of LCNEC, including more accurate determination of overall mutation burden in these tumors, which can only be estimated on the basis of targeted NGS methods. Finally, analysis of a larger cohort of cases will be needed to capture a full spectrum of genomic profiles in LCNEC.
In conclusion, by performing targeted NGS in conjunction with detailed histopathologic and clinical analysis, we gained insight that LCNEC is composed of biologically distinct subgroups. These findings have potential implications for classification, clinical management, and future investigations of LCNEC. Notably, these data parallel the recent general trend of genomics providing essential insights into the biologic relationships of tumors that have been notoriously difficult to categorize by traditional methods (17, 50).
Supplementary Material
Figuers 1-6
Table 1
Table 2
Table 3
Table 4
Table 5
Table 6
Table 7
Translational Relevance.
Currently, the data on comprehensive genomic profiles of pulmonary large cell neuroendocrine carcinoma (LCNEC) are limited, contributing to uncertainty regarding its biologic relationship with other major lung cancer types and optimal clinical management. Here, we report on targeted next-generation sequencing of 45 LCNECs with detailed clinicopatho-logic and immunohistochemical correlation. This analysis revealed distinct subgroups within LCNEC, exhibiting genomic signatures of SCLC, non-SCLC (NSCLC; predominantly adenocarcinoma), and, in a minority of cases, carcinoid tumors. These subsets were associated with several distinct clinicopathologic features, including higher proliferative activity of SCLC-like tumors and exclusive exocrine differentiation marker expression in NSCLC-like tumors. We also describe the landscape of potentially targetable genomic alterations in LCNEC. Our data yield novel insight into the biology of LCNEC and may provide a useful framework for future clinical investigations of cytotoxic and targeted therapies in patients with these tumors.
Acknowledgments
Grant Support
This study was supported in part by the grant from the Fiona and Stanley Druckenmiller Center for Lung Cancer Research (to N. Rekhtman), MSKCC Department of Pathology Research and Development grant (to N. Rekhtman), and P01CA129243 (to M. Ladanyi, N. Rekhtman, and M.F. Berger). The MSK-IMPACT program is supported in part by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology at Memorial Sloan Kettering Cancer Center and Cycle for Survival. This research was made possible by infrastructure support provided by the NIH/NCI Cancer Center Support Grant P30 CA008748.
Footnotes
Disclosure of Potential Conflicts of Interest
M.C. Pietanza is an employee of Merck. M. Hellmann reports receiving a commercial research grant from Bristol-Myers Squibb and is a consultant/advisory board member for AstraZeneca, Bristol-Meyers Squibb, Genentech, Inovio, Merck, and Neon. C.M. Rudin is a consultant/advisory board member for Bristol-Meyers Squibb, Celgene, ImaginAb, and Medivation. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: N. Rekhtman, M.C. Pietanza, J. Naidoo, A.L. Moreira, W.D. Travis, M. Ladanyi
Development of methodology: N. Rekhtman, J. Naidoo, S.K. Tian, M.F. Berger, A.L. Moreira, W.D. Travis
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): N. Rekhtman, M.C. Pietanza, M.D. Hellmann, J. Naidoo, D.F. Halpenny, S.K. Tian, A.M. Litvak, P.K. Paik, M.F. Berger, A.L. Moreira, W.D. Travis, C.M. Rudin
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): N. Rekhtman, M.C. Pietanza, M.D. Hellmann, J. Naidoo, A. Arora, H. Won, D.F. Halpenny, S.K. Tian, P.K. Paik, A.E. Drilon, N. Socci, J.T. Poirier, R. Shen, M.F. Berger, A.L. Moreira, W.D. Travis, M. Ladanyi
Writing, review, and/or revision of the manuscript: N. Rekhtman, M.C. Pietanza, M.D. Hellmann, J. Naidoo, H. Won, D.F. Halpenny, S.K. Tian, P.K. Paik, A.E. Drilon, J.T. Poirier, A.L. Moreira, W.D. Travis, C.M. Rudin, M. Ladanyi
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): N. Rekhtman, J. Naidoo, H. Wang, S.K. Tian, W.D. Travis
Study supervision: N. Rekhtman, J. Naidoo, M. Ladanyi
References
- 1.Travis WD, Linnoila RI, Tsokos MG, Hitchcock CL, Cutler GB, Jr, Nieman L, et al. Neuroendocrine tumors of the lung with proposed criteria for large-cell neuroendocrine carcinoma. An ultrastructural, immunohistochemical, and flow cytometric study of 35 cases. Am J Surg Pathol. 1991;15:529–53. doi: 10.1097/00000478-199106000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG, editors. WHO Classification of tumours of the lung, pleura, thymus and heart. 4th ed. IARC Press; Lyon, France: 2015. [DOI] [PubMed] [Google Scholar]
- 3.McDowell EM, Wilson TS, Trump BF. Atypical endocrine tumors of the lung. Arch Pathol Lab Med. 1981;105:20–8. [PubMed] [Google Scholar]
- 4.Rekhtman N. Neuroendocrine tumors of the lung: an update. Arch Pathol Lab Med. 2010;134:1628–38. doi: 10.5858/2009-0583-RAR.1. [DOI] [PubMed] [Google Scholar]
- 5.Pelosi G, Hiroshima K, Mino-Kenudson M. Controversial issues and new discoveries in lung neuroendocrine tumors. Diagn Histopathol. 2014;20:392–7. [Google Scholar]
- 6.Travis WD, Gal AA, Colby TV, Klimstra DS, Falk R, Koss MN. Reproducibility of neuroendocrine lung tumor classification. Human Pathol. 1998;29:272–9. doi: 10.1016/s0046-8177(98)90047-8. [DOI] [PubMed] [Google Scholar]
- 7.Iyoda A, Hiroshima K, Nakatani Y, Fujisawa T. Pulmonary large cell neuroendocrine carcinoma: its place in the spectrum of pulmonary carcinoma. Ann Thorac Surg. 2007;84:702–7. doi: 10.1016/j.athoracsur.2007.03.093. [DOI] [PubMed] [Google Scholar]
- 8.Varlotto JM, Medford-Davis LN, Recht A, Flickinger JC, Schaefer E, Zander DS, et al. Should large cell neuroendocrine lung carcinoma be classified and treated as a small cell lung cancer or with other large cell carcinomas? J Thorac Oncol. 2011;6:1050–8. doi: 10.1097/JTO.0b013e318217b6f8. [DOI] [PubMed] [Google Scholar]
- 9.Rossi G, Cavazza A, Marchioni A, Longo L, Migaldi M, Sartori G, et al. Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRalpha, PDGFRbeta, and Met in large-cell neuroendocrine carcinoma of the lung. J Clin Oncol. 2005;23:8774–85. doi: 10.1200/JCO.2005.02.8233. [DOI] [PubMed] [Google Scholar]
- 10.Naidoo J, Santos-Zabala ML, Iyriboz T, Woo KM, Sima CS, Fiore JJ, et al. Large cell neuroendocrine carcinoma of the lung: Clinico-pathologic features, treatment, and outcomes. Clin Lung Cancer. 2016 doi: 10.1016/j.cllc.2016.01.003. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkaria IS, Iyoda A, Roh MS, Sica G, Kuk D, Sima CS, et al. Neoadjuvant and adjuvant chemotherapy in resected pulmonary large cell neuroendocrine carcinomas: a single institution experience. Ann Thorac Surg. 2011;92:1180–6. doi: 10.1016/j.athoracsur.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol. 2015;10:553–64. doi: 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones MH, Virtanen C, Honjoh D, Miyoshi T, Satoh Y, Okumura S, et al. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363:775–81. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 14.Bari MF, Brown H, Nicholson AG, Kerr KM, Gosney JR, Wallace WA, et al. BAI3, CDX2 and VIL1: a panel of three antibodies to distinguish small cell from large cell neuroendocrine lung carcinomas. Histopathology. 2014;64:547–56. doi: 10.1111/his.12278. [DOI] [PubMed] [Google Scholar]
- 15.Przygodzki RM, Finkelstein SD, Langer JC, Swalsky PA, Fishback N, Bakker A, et al. Analysis of p53, K-ras-2, and C-raf-1 in pulmonary neuroendocrine tumors. Correlation with histological subtype and clinical outcome. Am J Pathol. 1996;148:1531–41. [PMC free article] [PubMed] [Google Scholar]
- 16.Rusch VW, Klimstra DS, Venkatraman ES. Molecular markers help characterize neuroendocrine lung tumors. Ann Thorac Surg. 1996;62:798–809. doi: 10.1016/s0003-4975(96)00435-3. [DOI] [PubMed] [Google Scholar]
- 17.Clinical Lung Cancer Genome P, Network Genomic M A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Pas TM, Giovannini M, Manzotti M, Trifiro G, Toffalorio F, Catania C, et al. Large-cell neuroendocrine carcinoma of the lung harboring EGFR mutation and responding to gefitinib. J Clin Oncol. 2011;29:e819–22. doi: 10.1200/JCO.2011.36.2251. [DOI] [PubMed] [Google Scholar]
- 19.Aroldi F, Bertocchi P, Meriggi F, Abeni C, Ogliosi C, Rota L, et al. Tyrosine kinase inhibitors in EGFR-mutated large-cell neuroendocrine carcinoma of the lung? A case report. Case Rep Oncol. 2014;7:478–83. doi: 10.1159/000365413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omachi N, Shimizu S, Kawaguchi T, Tezuka K, Kanazu M, Tamiya A, et al. A case of large-cell neuroendocrine carcinoma harboring an EML4-ALK rearrangement with resistance to the ALK inhibitor crizotinib. J Thorac Oncol. 2014;9:e40–2. doi: 10.1097/JTO.0000000000000103. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson A, Brunnstrom H, Lindquist KE, Jirstrom K, Jonsson M, Rosengren F, et al. Mutational and gene fusion analyses of primary large cell and large cell neuroendocrine lung cancer. Oncotarget. 2015;6:22028–37. doi: 10.18632/oncotarget.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietanza MC, Won HH, Rekhtman N, Wang L, Travis WD, Krug LM, et al. Prospective molecular analysis of small cell lung cancer (SCLC) using next generation sequencing (NGS). J Clin Oncol. 2015;33(suppl) abstr 7518. [Google Scholar]
- 25.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman DM, Solit DB, Arcila ME, Cheng DT, Sabbatini P, Baselga J, et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015;20:1422–8. doi: 10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paik PK, Shen R, Won H, Rekhtman N, Wang L, Sima CS, et al. Next-Generation sequencing of stage IV squamous cell lung cancers reveals an association of PI3K aberrations and evidence of clonal heterogeneity in patients with brain metastases. Cancer Discov. 2015;5:610–21. doi: 10.1158/2159-8290.CD-14-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869–78. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–77. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, et al. LKB1 Loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. J Thorac Oncol. 2014;9:794–804. doi: 10.1097/JTO.0000000000000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108:17761–6. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci U S A. 2014;111:14788–93. doi: 10.1073/pnas.1410419111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosari F, Ida CM, Aubry MC, Yang L, Kovtun IV, Klein JL, et al. ASCL1 and RET expression defines a clinically relevant subgroup of lung adenocarcinoma characterized by neuroendocrine differentiation. Oncogene. 2014;33:3776–83. doi: 10.1038/onc.2013.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joubert P, Wang H, Roh MS, Pietanza MC, Sarkaria I, Travis WD, et al. Napsin A expression in neuroendocrine tumors of the lung. Mod Pathol. 2013;26456A(suppl) abstr 7518. [Google Scholar]
- 38.Lai D, Chen Y, Wang F, Jiang L, Wei C. LKB1 controls the pluri-potent state of human embryonic stem cells. Cell Reprogram. 2012;14:164–70. doi: 10.1089/cell.2011.0068. [DOI] [PubMed] [Google Scholar]
- 39.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 40.Calles A, Sholl LM, Rodig SJ, Pelton AK, Hornick JL, Butaney M, et al. Immunohistochemical loss of LKB1 is a biomarker for more aggressive biology in KRAS-mutant lung adenocarcinoma. Clin Cancer Res. 2015;21:2851–60. doi: 10.1158/1078-0432.CCR-14-3112. [DOI] [PubMed] [Google Scholar]
- 41.Meder L, Konig K, Ozretic L, Schultheis AM, Ueckeroth F, Ade CP, et al. NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas. Int J Cancer. 2016;138:927–38. doi: 10.1002/ijc.29835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchetti A, Felicioni L, Pelosi G, Del Grammastro M, Fumagalli C, Sciarrotta M, et al. Frequent mutations in the neurotrophic tyrosine receptor kinase gene family in large cell neuroendocrine carcinoma of the lung. Hum Mut. 2008;29:609–16. doi: 10.1002/humu.20707. [DOI] [PubMed] [Google Scholar]
- 43.Ball DW. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett. 2004;204:159–69. doi: 10.1016/S0304-3835(03)00452-X. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Cuesta L, Peifer M, Lu X, Sun R, Ozretic L, Seidel D, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat Commun. 2014;5:3518. doi: 10.1038/ncomms4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietanza MC, Ladanyi M. Bringing the genomic landscape of small-cell lung cancer into focus. Nat Genet. 2012;44:1074–5. doi: 10.1038/ng.2415. [DOI] [PubMed] [Google Scholar]
- 46.Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development. 2004;131:4299–310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
- 47.Basturk O, Yang Z, Tang LH, Hruban RH, Adsay V, McCall CM, et al. The high-grade (WHO G3) pancreatic neuroendocrine tumor category is morphologically and biologically heterogenous and includes both well differentiated and poorly differentiated neoplasms. Am J Surg Pathol. 2015;39:683–90. doi: 10.1097/PAS.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forde PM, Brahmer JR, Kelly RJ. New strategies in lung cancer: epigenetic therapy for non-small cell lung cancer. Clin Cancer Res. 2014;20:2244–8. doi: 10.1158/1078-0432.CCR-13-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rekhtman N, Tafe LJ, Chaft JE, Wang L, Arcila ME, Colanta A, et al. Distinct profile of driver mutations and clinical features in immuno-marker-defined subsets of pulmonary large-cell carcinoma. Mod Pathol. 2013;26:511–22. doi: 10.1038/modpathol.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figuers 1-6
Table 1
Table 2
Table 3
Table 4
Table 5
Table 6
Table 7