Quantitative Acylcarnitine Determination by UHPLC-MS/MS – Going Beyond Tandem MS Acylcarnitine “Profiles” (original) (raw)
. Author manuscript; available in PMC: 2016 Dec 1.
Published in final edited form as: Mol Genet Metab. 2015 Oct 8;116(4):231–241. doi: 10.1016/j.ymgme.2015.10.002
Abstract
Tandem MS “profiling” of acylcarnitines and amino acids was conceived as a first-tier screening method, and its application to expanded newborn screening has been enormously successful. However, unlike amino acid screening (which uses amino acid analysis as its second-tier validation of screening results), acylcarnitine “profiling” also assumed the role of second-tier validation, due to the lack of a generally accepted second-tier acylcarnitine determination method. In this report, we present results from the application of our validated UHPLC-MS/MS second-tier method for the quantification of total carnitine, free carnitine, butyrobetaine, and acylcarnitines to patient samples with known diagnoses: malonic acidemia, short-chain acyl-CoA dehydrogenase deficiency (SCADD) or isobutyryl-CoA dehydrogenase deficiency (IBD), 3-methyl-crotonyl carboxylase deficiency (3-MCC) or β-ketothiolase deficiency (BKT), and methylmalonic acidemia (MMA). We demonstrate the assay’s ability to separate constitutional isomers and diastereomeric acylcarnitines and generate values with a high level of accuracy and precision. These capabilities are unavailable when using tandem MS “profiles”. We also show examples of research interest, where separation of acylcarnitine species and accurate and precise acylcarnitine quantification is necessary.
Keywords: Quantitative acylcarnitine analysis, second-tier analysis, newborn screening follow-up, carnitine analysis, metabolism research
1. Introduction
The widespread implementation of expanded newborn screening programs has been effective and beneficial [1]. These programs became possible because of advances in tandem mass spectrometric (MS) instrumentation and data analysis [2], allowing for “profiling” of amino acids and acylcarnitines using tandem MS [3,4]. Originally, “profiling” of acylcarnitines was conceived of as a qualitative screening method for detection of grossly elevated levels observed in rare, severe metabolic diseases [5]. The use of the jargon “profile” was intended to convey the inherent lack of analytical specificity and lack of quantitative accuracy that first-tier analyses sacrifice in exchange for rapidity, thus conceding the necessity of confirmatory second-tier analysis. The final validation of disease then often relied on enzyme activity measurements, while currently, a patient’s diagnosis is usually established using mutational analysis.
Today, the very different handling of the amino acid “profile” versus the acylcarnitine “profile” is a consequence of the historical development of their respective analytical technologies. Quantitative amino acid analysis has a long history, beginning with ion-exchange chromatography and ninhydrin derivatization in the 1950s [6], and continuing recently with HPLC-MS methods [7]. More importantly today, there are validated, commercially available amino acid analysis kits for selective, accurate, and fully quantitative amino acid analysis [8,9,10]. These kits provide both established chromatographic conditions for separation of isobaric compounds and standard compounds for quantitative accuracy. As a result, when first-tier tandem MS amino acid “profiles” were adopted in expanded newborn screening, the obvious second-tier analysis was already widely accepted. However, this was not the case with acylcarnitines, where qualitative soft ionization MS technologies were the dominant methodology [11,12]. With no generally accepted alternative second-tier methodology, the first-tier “profile” was simply repeated, coupled with semi-quantitative organic acid analysis of the patient’s urine [13]. This approach is and has been the state of the art for decades. However, today we can take advantage of more accurate and precise techniques due to advances in technology.
To move forward with acylcarnitine analysis, there are two features which any universally applicable, comprehensive, second-tier acylcarnitine method must provide:
- Discrimination of acylcarnitine constitutional isomers and diastereomers. Acylcarnitine diagnostic markers are often specific to a particular acylcarnitine isomer, reflecting the metabolic pathways that generated them. However, endogenous acylcarnitine species may be present in multiple isomeric forms, and since their isomer masses are identical, they are indistinguishable by tandem MS “profiling”. (U)HPLC separation of acylcarnitine constitutional isomers and diastereomers has been shown by several groups [14,15,16].
- Accurate Quantification. Acylcarnitine mass spectral responses are variable and compound specific. Therefore, accurate quantification requires standard compounds with accurately known concentrations to calibrate individual acylcarnitine compound responses. Tandem MS “profile” values are generated using single point calibration with labeled, standardized internal standards. The calculation of these values assumes that the relative responses of each acylcarnitine and its internal standard are proportional and linear throughout the entire detected range. This approach has been shown to be quantitatively inaccurate and imprecise [17,18]. However, protocols which comply with the FDA’s “Guidance for Industry: Analytical Procedures and Methods Validation for Drugs and Biologics” [19,20] can provide excellent accuracy and precision. These procedures use calibrated standards and internal standards to construct multiple-point calibration curves.
Although no commercial kit (analogous to those for amino acid analysis) exists for acylcarnitine analysis, many groups have published second-tier methods that are, to varying degrees, more selective and quantitatively rigorous than tandem MS “profiles” [21,22,23,24]. We developed second-tier methods for the quantification of carnitine beginning 40 years ago, using radioactively labeled acetyl-CoA and measuring the production of radiolabeled acetylcarnitine [25,26]. We next prepared a highly reactive, strongly UV absorbing derivatization reagent, 4′-bromophenacyl trifluoromethanesulfonate [27], and used it to develop HPLC-UV methods for the quantification of carnitine, butyrobetaine [28,29], and acylcarnitines [30,31]. These methods were further enhanced using the intensely fluorescent derivatization reagent 2-(2,3-naphthalimino)ethyl trifluoromethanesulfonate, and we quantified carnitine and acylcarnitines in plasma and tissues by HPLC-fluorescence [32,33]. These methods were selective (using HPLC), sensitive (spectrophotometric or fluorescence detection), and truly quantitative (with internal standards and calibration curves). With the advent of atmospheric pressure ionization (API) mass spectrometers, even more selectivity was available. We modified our procedure to use an ion trap mass spectrometer for detection following derivatization of carnitine and acylcarnitines with the MS specific reagent, pentafluorophenacyl trifluoromethanesulfonate [34,35]. Most recently, we incorporated greatly improved fast chromatography using fused-core C8 UHPLC columns, and used triple quadrupole mass spectrometers with multiple-reaction monitoring (MRM) for data collection [36]. Our improved, fast, validated method for quantification of total carnitine, free carnitine, butyrobetaine, and acylcarnitines fulfills both assay requirements for a second-tier acylcarnitine analysis method: 1) chromatographic separation of acylcarnitine constitutional isomers and diastereomers, and 2) accurate quantification using multiple point calibration curves.
For chromatographic separation of acylcarnitines, we developed a two-dimensional orthogonal chromatographic scheme in which cation-exchange is used to trap on-line all carnitine species (short-chain, medium-chain, long-chain, etc.) due to the positively charged trimethylammonium functional group of carnitine. This was followed by separation of carnitine and individual acylcarnitine constitutional isomers and diastereomers using reversed-phase UHPLC. We call this technique “sequential ion-exchange / reversed-phase chromatography” [28].
Few acylcarnitine standards are commercially available. Therefore, we used published methods for synthesis of acylcarnitines [37,38,39,40], and we prepared 66 different acylcarnitines, along with appropriate internal standards. Purification of synthesized acylcarnitines was accomplished using a two-step chromatographic process, combining cation-exchange solid-phase extraction (to remove unreacted acid) followed by preparative HPLC (to remove carnitine and synthetic impurities). We then standardized stock solutions of carnitine. This was necessary, since we have found errors in making standard solutions by weighing “pure” acylcarnitines because: 1) there is usually some free carnitine contamination in acylcarnitines that needs to be accounted for, and 2) synthesized acylcarnitines usually do not crystallize into readily transferable solids. They often are isolated as wet, viscous, hygroscopic materials that rapidly absorb additional atmospheric moisture. This prevents accurate weighing. Therefore, using standardized carnitine stock solutions [36], we standardized the acylcarnitine stock solutions with our method for free and total carnitine.
Based on literature reports, tandem MS acylcarnitine “profile” analysis has two basic problems: 1) Selectivity–isobaric contaminants and isomeric acylcarnitines have led to false negative [41] and false positive results [42,43,44], and 2) Quantification–Preparation of acylcarnitine butyl ester derivatives partially hydrolyzes some acylcarnitines, giving inaccurate free carnitine values [45]. Lacking rigorous quantitation (standardized calibrants, calibration curves, etc.), most reported “profile” values are pseudo-concentrations [17,18]. With our validated method, both of these problems are eliminated. This allows us to use our procedure for several distinct purposes:
- Follow-up to newborn screening – When confronted with ambiguous findings, we use our methodology to validate the indefinite screening results with a more rigorous technique, and thus resolve false-positive or false-negative screening results. We also use accurate quantification values for carnitine and butyrobetaine to characterize carnitine biosynthesis diseases.
- Follow-up to Patient Treatment – Patients receiving treatment are monitored, and require methodology capable of separating isomeric acylcarnitines with accurate quantification of carnitine, butyrobetaine, and acylcarnitines. From these determinations, appropriate modifications to the patient’s treatment regimen can be formulated.
- Metabolism Research – Acceptable research protocols require analytical methods that are selective, accurate, and precise. Selectivity is accomplished with UHPLC chromatography and mass spectrometry with multiple reaction monitoring (MRM). Accurate and precise values are generated using calibration standards and internal standards to construct multiple-point calibration curves. Our results are accurate (within ±20% of the true value at the lowest concentrations) with excellent precision [36].
This manuscript reports the application of this validated, second-tier method for quantification of total carnitine, free carnitine, butyrobetaine, and acylcarnitines in patient samples with known diagnoses, providing results that are not attainable when using tandem MS “profiles”. We also show examples of research interest, where accurate and precise quantification of acylcarnitines, as surrogates of their respective acyl-CoAs, is necessary.
2. Material and methods
2.1 Instrumentation
The instrumentation consisted of two Agilent UHPLC 1290 Infinity binary pumps, autosampler, and thermostated column compartment with a 6-port valve, and a 6460 QQQ triple quadrupole LC/MS purchased from Agilent Technologies (Santa Clara, CA). The chromatographic separation was accomplished with an SCX trap cartridge (2.1 mm × 5 mm) contained in an in-line holder purchased from Optimize Technologies (Oregon City, OR), connected in series (through the 6-port valve) with an Agilent Poroshell 120 EC-C8 column (3.0 × 100 mm, 2.7 μm). Configuration of the instrument and instrument parameters, construction of calibration curves, and generation of quantification values were as described [36].
2.2 Biological samples
Human plasma and urine from patients with known diagnoses were obtained from the College of American Pathologists (CAP; Northfield, IL). We participate in their Biochemical Genetic Survey program, and we analyzed these samples for proficiency testing purposes. Human cerebral spinal fluid was purchased from Golden West Biologicals, Inc. (Temecula, CA).
We also show results from analysis of perfused rat heart [46]. All procedures with animal materials were approved by the Case Western Reserve University Institutional Animal Care and Use Committee and performed in accordance with National Institutes of Health guidelines for care and use of animals in research. Six months old Fisher 344 rats were injected with 500 U of heparin (IP), anesthetized with sodium pentobarbital (100 mg/kg body weight), and the hearts were removed and canulated for perfusion. The perfusion protocol consisted of a 15 min non-recirculating perfusion in the Langendorff mode with Krebs–Henseleit buffer containing 5.5 mM glucose and 0.1 U/L insulin, followed by 60 min of perfusion in the working heart mode with 5.5 mM glucose/0.1 U/L insulin, and with either 1.2 mM palmitic acid or [1,2,3,4-13C4]-palmitic acid complexed to 3% BSA. At the end of perfusion, the heart was freeze-clamped, powdered under liquid nitrogen, and the powdered tissue was stored at −60 °C.
2.3 Sample preparation and analysis
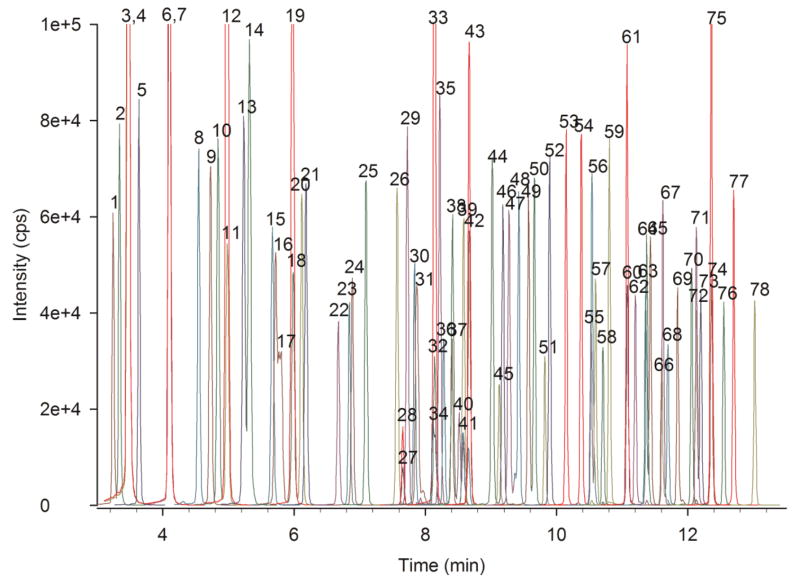
Samples were prepared and analyzed as described [36]. Briefly, to 10 μL of plasma, diluted urine, homogenized tissue, or cerebral spinal fluid (plus internal standards) was added organic solvents to precipitate salts and proteins. The resulting supernatant was then applied to a mixed-mode, reversed-phase/strong cation-exchange solid-phase extraction plate (Oasis MCX, purchased from Waters Corporation, Milford, MA). Carnitine and acylcarnitines were eluted, evaporated, and derivatized with pentafluorophenacyl trifluoromethanesulfonate [34], then injected into the UHPLC-MS/MS system. Carnitine and butyrobetaine were eluted in a 4 min chromatogram; optimized MRM transitions were collected for carnitine, d3-carnitine internal standard, butyrobetaine, and d3-butyrobetaine internal standard. Acylcarnitines were eluted in a 14 min chromatogram, and optimized MRM transitions were collected for acylcarnitines and their internal standards (see Fig. 1).
Fig. 1.
UHPLC-MS/MS chromatogram of an acylcarnitine calibration curve medium point. Overlaid extracted ion chromatograms of 78 transitions monitored for 66 compounds and 12 internal standards. Internal standards are colored red. Peak Identities: 1) _S_-3-hydroxy-butyryl-, 2) _R_-3-hydroxy-butyryl-, 3) acetyl-, 4) d6-acetyl-, 5) 3-hydroxy-isovaleryl-, 6) propionyl-, 7) d3-propionyl-, 8) _S_-3-hydroxy-hexanoyl-, 9) _R_-3-hydroxy-hexanoyl-, 10) isobutyryl-, 11) butyryl-,12) d3-butyryl-, 13) tigloyl-, 14) 3-methyl-crotonyl-, 15) benzoyl-, 16) pivaloyl- 17) 2-methyl-butyryl-, 18) isovaleryl-, 19) d9-isovaleryl-, 20) valeryl-, 21) phenylacetyl-, 22) _S_-3-hydroxy-octanoyl-, 23) _R_-3-hydroxy-octanoyl-, 24) phenylpropionyl-, 25) hexanoyl-, 26) 4-phenyl-butyryl-, 27) malonyl-, 28) d3-malonyl-, 29) 4-methyl-hexanoyl-, 30) succinyl-, 31) _cis_-3,4-methylene-heptanoyl-, 32) glutaroyl-, 33) d3-glutaroyl-, 34) methylmalonyl-, 35) valproyl-, 36) _S_-3-hydroxy-decanoyl-, 37) _R_-3-hydroxy-decanoyl-, 38) adipoyl-, 39) 5-decynoyl-, 40) 3- methyl-glutaroyl-, 41) ethylmalonyl-, 42) octanoyl-, 43) d3-octanoyl-, 44) 2,6-dimethyl-heptanoyl-, 45) suberoyl-, 46) 4-methyl-octanoyl-, 47) _cis_-3,4-methylene-nonanoyl-, 48) _cis_-4-decenoyl-, 49) _S_-3-hydroxy-lauroyl-, 50) _R_-3-hydroxy-lauroyl-, 51) sebacoyl-, 52) decanoyl-, 53) undecanedioyl-, 54) undecanoyl-, 55) _S_-3-hydroxy-myristoyl-, 56) _trans_-2-dodecenoyl-, 57) _R_-3-hydroxy-myristoyl-, 58) _cis,cis_-5,8-tetradecadienoyl-, 59) lauroylcarnitine, 60) myristoleoylcarnitine, 61) d3-myristoleoylcarnitine, 62) _cis_-5-tetradecenoylcarnitine, 63) _S_-3-hydroxy-palmitoyl-, 64) _R_-3-hydroxy-palmitoyl-, 65) _trans_-2-tetradecenoyl-, 66) α-linolenoyl-, 67) myristoyl-, 68) γ-linolenoyl-, 69) palmitoleoyl-, 70) linoleoyl-, 71) _S_-3-hydroxy-stearoyl-, 72) _R_-3-hydroxy-stearoyl-, 73) _trans_-2-hexadecenoyl-, 74) palmitoyl-, 75) d3-palmitoyl-, 76) oleoyl-, 77) heptadecanoyl-, 78) stearoylcarnitine.
3. Results and Discussion
3.1 Accurate Quantification: Malonylcarnitine in a CAP Plasma Sample from a patient with Malonic Acidemia
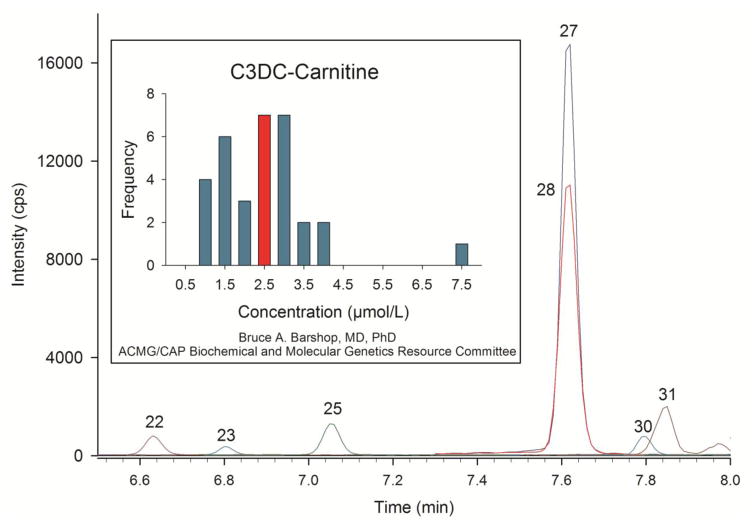
To comply with the requirements for our CLIA certification, we participate in the College of American Pathologists’ (CAP) proficiency testing for plasma acylcarnitine analysis. In one plasma sample we received, malonylcarnitine was highly elevated, consistent with a diagnosis of “malonic acidemia” (MA [47]). Fig. 2 shows part of the chromatogram from that analysis, highlighting malonylcarnitine (Peak 27) and its internal standard d3-malonylcarnitine (Peak 28). We quantified malonylcarnitine in this plasma (using a calibration curve constructed with standardized malonylcarnitine and with d3-malonylcarnitine as the internal standard) at 2.51 μmol/L. Normal values are less than 0.05 μmol/L. Later, we received our Participant Summary from CAP [48]. There were 46 total laboratories reporting, of which 44 used tandem MS; one lab used GC/MS and we were, presumably, the lab identified as “other”. From these participants, 39 (85%) reported the correct diagnosis, with 40 labs observing elevated malonylcarnitine. It is significant that 6 labs missed this large increase in malonylcarnitine. The Participant Summary also included a histogram showing the frequency distribution of the quantitative malonylcarnitine results from 32 participating labs, which we have reproduced in Fig. 2. We have highlighted the bar at 2.5 μmol/L, which was our result. CAP does not grade the quantitative value; the acceptable response for this specimen was detection of malonylcarnitine and reporting malonic acidemia as the correct diagnosis. However, our validation studies demonstrated accuracy and precision for malonylcarnitine quantification within ±15% of the correct value when using our method [36]. Therefore, we assert that the malonylcarnitine concentration value for the patient sample is indeed 2.5 μmol/L. We and 6 other labs measured the same quantitative value, but 26 other labs show widely variable results (from 1 to 7.5 μmol/L). Inconsistent quantitative results using tandem MS “profiles” is not unexpected. Chace et al. [17] reported a wide variation of results from 101 laboratories given identical bloodspots containing 0.58 μmol/L of malonylcarnitine. Using a variety of tandem MS methods, reported values varied from 0.2 to 2.0 μmol/L. The initial goal of their study was to determine what bias, if any, from the correct analytical result tandem MS “profile” quantification would yield. Assuming that the results would be precise and that any deviation from accuracy (bias) could be compensated for with correction factors, these authors instead found that “profile” results were neither accurate nor precise. The authors then abandoned the use of correction factors, and concluded that the numerical results from tandem MS analysis were pseudo-quantitative. This same group attempted a similar study with 3-hydroxyisovalerylcarnitine measurements by tandem MS, and they found again that tandem MS “profile” results were inconsistent and not quantitative [18].
Fig. 2.
UHPLC-MS/MS chromatogram of the acylcarnitine analysis of a plasma specimen provided by CAP with the expected diagnosis of malonic acidemia. Peak identities and colors are the same as in Fig. 1. Malonylcarnitine is Peak 27 and d3-malonylcarnitine internal standard is Peak 28. The inset graph is a histogram showing the frequency distribution of quantitative values determined by us and other labs participating in CAP testing. The highlighted bar at 2.5 μmol/L is the value we and six others determined from this analysis. Also present in this chromatogram are _S_-3-hydroxy-octanoyl- (Peak 22), _R_-3-hydroxy-octanoyl- (Peak 23), hexanoyl- (Peak 25), succinyl- (Peak 30) and _cis_-3,4-methylene-heptanoylcarnitine (Peak 31).
In Table 1, we report the absolute quantification of total carnitine, free carnitine, and the sum of all acylcarnitines quantified in this sample. We also calculated the concentration of the acylcarnitine pool by subtracting the free carnitine concentration from the total carnitine concentration (Total - Free). From these values, we then determined the percent of acylcarnitines accounted for by quantification of the individual acylcarnitines. For this sample the value is 93%. We call this calculation a “balance study”. Thus, we have demonstrated agreement (within 7%) between what the total and free carnitine concentrations tell us what the acylcarnitine concentration should be, and what we have actually identified and quantified. If the value for malonylcarnitine were 1.0 μmol/L (the low value on the histogram) or 7.5 μmol/L (the high value on the histogram), the “balance study” results would be 71% or 160%, respectively, and suggest quantitative inaccuracy. Instead, a value for malonylcarnitine of 2.51 μmol/L is consistent with the independently determined concentrations of free carnitine and total carnitine. The full analysis report is provided in the Supplemental Material.
Table 1.
Balance Studies: Demonstration of Quantitative Accuracy by Comparing Calculated (Total–Free) and Summed Acylcarnitine Values
| MA CAP plasma (μmol/L) | SCADD CAP plasma (μmol/L) | BKT CAP plasma (μmo/L)* | MMA CAP urine** (μmol/g creatinine) | Palmitate Perfused Rat Heart (nmol/g ww) | Cerebral Spinal Fluid (μmol/L)*** | |
|---|---|---|---|---|---|---|
| Total Carnitine | 40.8 | 47.1 | 80.7 | 24311 | 678.3 | 1.93 |
| Free Carnitine | 33.5 | 36.0 | 67.3 | 11049 | 284.1 | 1.41 |
| Total - Free | 7.27 | 11.1 | 13.4 | 13262 | 394.2 | 0.520 |
| Sum of Acylcarnitines | 6.74 | 9.92 | 10.3 | 10664 | 364.9 | 0.531 |
| % Acylcarnitines Accounted For | 93% | 90% | 77% | 80% | 93% | 102% |
3.2 Discrimination of Constitutional Isomers: Short-chain Acyl-CoA Dehydrogenase Deficiency or Isobutyryl-CoA Dehydrogenase Deficiency in a CAP Plasma Sample
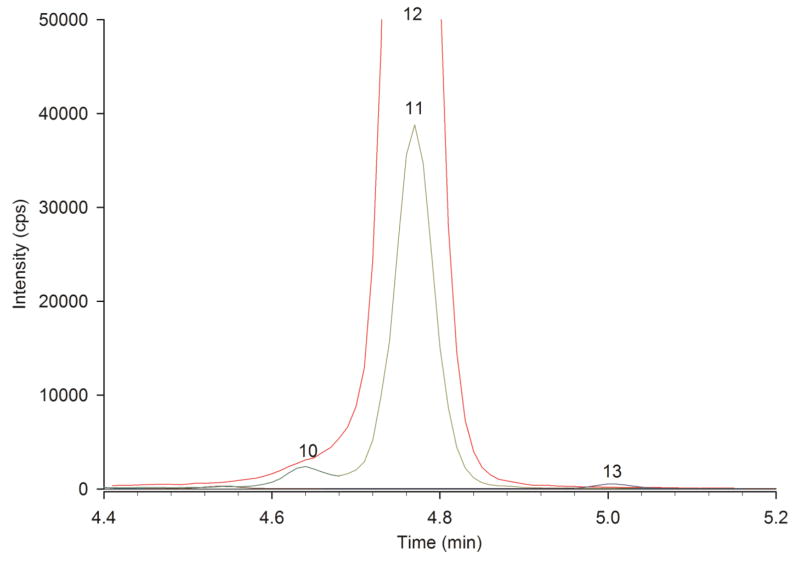
In another plasma sample we received from CAP, we determined that butyryl- but not isobutyrylcarnitine was elevated. The diagnosis we reported was short-chain acyl-CoA dehydrogenase deficiency (SCADD [49]). The expected diagnosis by CAP was either SCADD or isobutyryl-CoA dehydrogenase deficiency (IBD [50]). Because tandem MS “profiles” cannot distinguish between butyrylcarnitine (the marker for SCADD) and isobutyrylcarnitine (the marker for IBD), the diagnosis accepted by CAP was the less definite SCADD/IBD. Fig. 3 shows part of the chromatogram from our analysis, highlighting isobutyrylcarnitine (Peak 10), butyrylcarnitine (Peak 11) and their internal standard d3-butyrylcarnitine (Peak 12). We chromatographically resolved isobutyrylcarnitine from butyrylcarnitine, and determined their concentrations to be 0.05 μmol/L and 0.83 μmol/L, respectively. Normal values are <0.2 μmol/L. Since only butyrylcarnitine was elevated, the correct diagnosis was SCADD and not IBD. Table 1 includes the balance study for this sample (90% of acylcarnitines are accounted for) consistent with highly accurate quantification. The full analysis report is provided in the Supplemental Material. Later, we received our Participant Summary from CAP [51], and were informed that the sample had come from a patient with SCADD. There were 50 participants in the survey; we were the only lab not using tandem MS. Of the participants, 89% correctly identified an elevation in C4 acylcarnitine; however, because we separated butyryl- from isobutyrylcarnitine, we were the only lab that could make the correct diagnosis. The ability to resolve isomeric acylcarnitines is often underappreciated; we contributed to a report in which the inability of tandem MS “profiles” to resolve C8 acylcarnitine isomers resulted in misdiagnoses in newborn screens (see McCandless et al. [52], Supplemental Fig. 1).
Fig. 3.
UHPLC-MS/MS chromatogram of the acylcarnitine analysis of a plasma specimen provided by CAP with the expected diagnosis by tandem MS of either short-chain acyl-CoA dehydrogenase deficiency (SCADD) or isobutyryl-CoA dehydrogenase deficiency (IBD). Peak identities and colors are the same as in Fig. 1. This chromatogram illustrates the resolution between isobutyryl- (Peak 10) and butyrylcarnitine (Peak 11), leading us to the correct diagnosis of SCADD. Peak 12 is d3-butyrylcarnitine internal standard.
3.3 Discrimination of Diasteriomers: β-Ketothiolase Deficiency in a CAP Plasma Sample
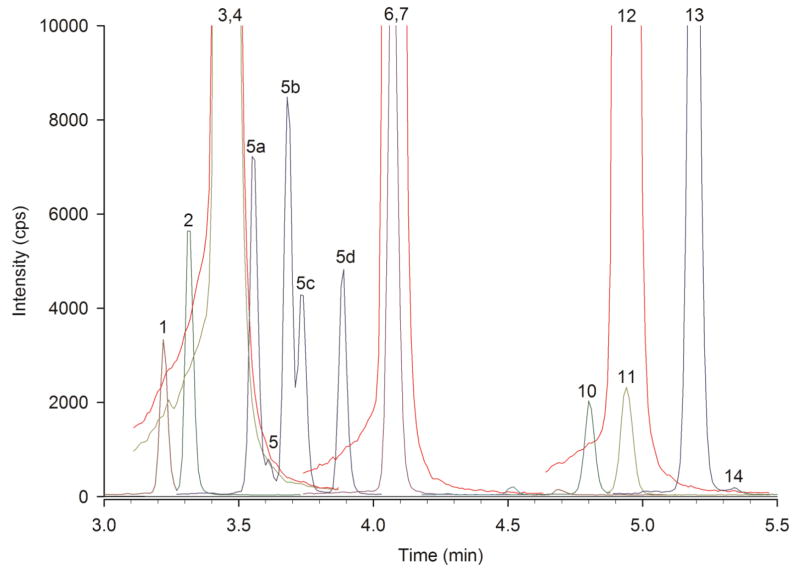
We analyzed a plasma specimen provided by CAP with the expected diagnosis of either 3-methyl-crotonyl carboxylase deficiency (3-MCC, [53]) or β-ketothiolase deficiency (BKT, [54]). The marker for these diseases is HO-C5 acylcarnitine, with 3-hydroxy-isovalerylcarnitine specific for 3-MCC and 3-hydroxy-2-methyl-butyrylcarnitines specific for BKT. Fig. 4 shows an acylcarnitine chromatogram from the sample with a tiny amount of 3-hydroxy-isovalerylcarnitine (Peak 5; 0.03 μmol/L). We identified this peak based on its identical retention time to that of the purified and standardized 3-hydroxy-isovalerylcarnitine, which was used to generate a calibration curve. However, there are 4 additional HO-C5 acyl-L-carnitine isomers (Peaks 5a–d), which by tandem MS “profile” analysis are indistinguishable from 3-hydroxy-isovalerylcarnitine. These are the four different diastereomers (_RR_-, _SS_-, _SR_-, and _RS_-) of 3-hydroxy-2-methyl-butyryl-L-carnitine, and they are chromatographically separable. Their concentrations, determined using the 3-hydroxy-isovalerylcarnitine calibration curve, were 0.09, 0.10, 0.06 and 0.07 μmol/L. The other marker for 3-MCC and BKT is C5:1 acylcarnitine, with 3-methyl-crotonylcarnitine present in 3-MCC, while tigloylcarnitine is present in BKT. The chromatogram in Fig. 4 shows an enormous peak of tigloylcarnitine (Peak 13) and essentially no 3-methyl-crotonylcarnitine (Peak 14). Table 1 includes the balance study for this sample, which accounts for 77% of the acylcarnitines. We included the 3-hydroxy-2-methyl-butyrylcarnitines in the calculation, with the remaining 23% consisting of small contributions from many other acylcarnitines. Because tandem MS acylcarnitine “profiles” cannot distinguish between 3-MCC and BKT, either diagnosis was acceptable to CAP. However, not only does our UHPLC-MS/MS procedure correctly identify an elevation in HO-C5 acylcarnitine, we can definitively identify the increase as 3-hydroxy-2-methyl-butyrylcarnitines and not 3-hydroxy-isovalerylcarnitine. Since we can also identify the elevated C5:1 acylcarnitine as tigloylcarnitine and not 3-methyl-crotonylcarnitine, we can unambiguously and correctly identify the patient’s condition as BKT and not 3-MCC. In our publication describing this procedure [36], we showed a 3-MCC sample in which we correctly identified the HO-C5 acylcarnitine as 3-hydroxy-isovalerylcarnitine and the C5:1 acylcarnitine as 3-methyl-crotonylcarnitine, providing in that case the correct and unambiguous diagnosis of 3-MCC.
Fig. 4.
UHPLC-MS/MS chromatogram of the acylcarnitine analysis of a plasma specimen provided by CAP with the diagnosis of either 3-methyl-crotonyl carboxylase deficency (3-MCC) or β-ketothiolase deficiency (BKT). Peak identities and colors are the same as in Fig. 1. Peaks 5a–d are the four stereoisomeric forms (RR-, SS-, SR-, and RS-) of 3-hydroxy-2-methyl-butyrylcarnitine. Tigloylcarnitine (Peak 13) and 3-methyl-crotonylcarnitine (Peak 14) are also chromatographically separated. This chromatogram illustrates the separation of HO-C5 and C5:1 acylcarnitines, leading us to the correct diagnosis of BKT.
3.4 Urine Acylcarnitines for Diagnosis and Treatment: Methylmalonic Acidemia in a CAP Urine Sample
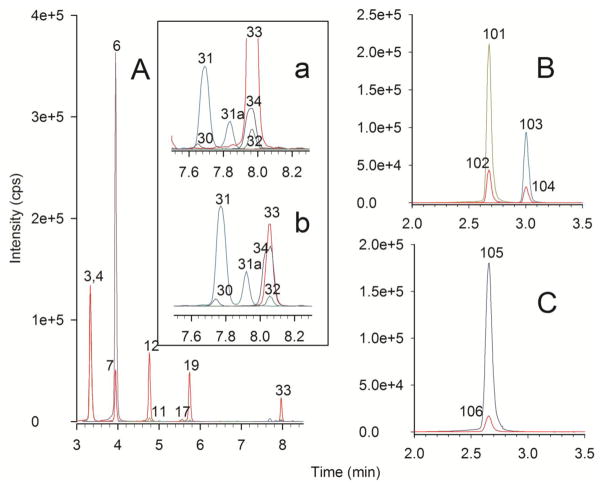
We also participated in CAP’s proficiency testing for urinary organic acid analysis. We analyzed a provided urine sample for urinary organic acids, made a diagnosis of methylmalonic acidemia (MMA), and this was the correct diagnosis. Concurrently, we analyzed this same sample for acylcarnitines. Tandem MS “profiles” of urine acylcarnitines are not commonly performed, and there is no proficiency testing for urine acylcarnitines from CAP. However, our sample preparation scheme is essentially sample matrix independent. We have analyzed a wide variety of specimen types without altering the basic protocol, including plasma, urine, tissue homogenates, cultured cells, blood spots, dietary supplements, and samples from a wide variety of research protocols. Fig. 5 shows the UHPLC-MS/MS chromatograms of the short- and medium-chain acylcarnitines, free carnitine, butyrobetaine, and total carnitine from analysis of the urine provided by CAP with the diagnosis of MMA, showing elevations in propionylcarnitine and methylmalonylcarnitine. Initially, the sample was prepared by first diluting by 1:5 with water, but the resulting chromatograms contained peaks that exceeded some of the calibration curves. The sample was then diluted 1:50 with water and reanalyzed, and this result is shown in Fig. 5A. Peak 6 is enormous, and it represents the excretion of propionylcarnitine (6950 μmol/g creatinine, normal values <2.8). Peak 7 is the internal standard d3-propionylcarnitine. The inset (Fig. 5Aa) is an expanded view highlighting methylmalonylcarnitine (Peak 34), along with the internal standard (Peak 33, d3-glutaroylcarnitine) used to construct the methylmalonylcarnitine calibration curve (N.B. Because we generate calibration curves from standardized calibrants, it is not necessary for us to use a heavy-labeled internal standard for each compound measured. We empirically determine the response relationship between each acylcarnitine and its internal standard using a multiple-point calibration curve constructed with standardized calibrants and internal standards). Other peaks in the inset figure are succinylcarnitine (Peak 30), glutaroylcarnitine (Peak 32) and Peak 31, the C8:1 acylcarnitine _cis_-3,4-methylene-heptanoylcarnitine [40]. Peak 31a is another C8:1 acylcarnitine, presumed to be the trans- isomer. Fig. 5Ab is from the initial analysis of urine diluted 1:5. Here, the methylmalonylcarnitine peak (Peak 34) is nearly as large as the d3-glutaroylcarnitine internal standard peak (Peak 33). When comparing the peak shape of methylmalonylcarnitine with d3-glutaroylcarnitine or glutaroylcarnitine (Peak 32), it is apparent that methylmalonyl-L-carnitine is much wider, less symmetrical, and shows chromatographic peak splitting. This is because methylmalonylcarnitine is actually composed of two diastereomeric forms (_R_-methylmalonyl- and _S_-methylmalonyl-L-carnitine). In this case, these diastereomers are not completely chromatographically resolved. However, we observe that both diastereomers are present in this sample in approximately equal amounts. Fig. 5B shows, in a separate chromatogram, the analysis of free carnitine and butyrobetaine for this sample. Fig. 5C is the total carnitine chromatogram. The urinary excretion of total carnitine and free carnitine is 50 times the normal value. The excretion of butyrobetaine (the biosynthetic precursor of carnitine [55]), at 1944 μmol/g creatinine, is 400 times the normal value. Table 1 shows the “balance study” for this analysis, with 80% of the calculated acylcarnitine pool accounted for by the sum of the quantified individual acylcarnitines. The full analysis report is provided in the Supplemental Material.
Fig. 5.
UHPLC-MS/MS chromatograms of the short- and medium-chain acylcarnitines, free carnitine, butyrobetaine, and total carnitine from analysis of a urine specimen provided by CAP with the expected diagnosis of methylmalonic academia (MMA). Peak identities and colors are the same as in Fig. 1. A) Acylcarnitine chromatogram from a 1:50 dilution of the urine. Peak 6 is propionylcarnitine. The inset chromatograms a) accentuate methylmalonylcarnitine and b) 10x more concentrated urine accentuating methylmalonylcarnitine. Peak 34 is methylmalonylcarnitine. B) Free carnitine and butyrobetaine chromatogram. Peak Identities: 101) free carnitine, 102) d3-carnitine, 103) butyrobetaine, 104) d3-butyrobetaine. C) Total carnitine chromatogram. Peak Identities: 105) total carnitine, 106) d3-carnitine.
With these enormous excretion values, the patient is undoubtedly being treated with carnitine. These results illustrate the utility of urine acylcarnitine analysis for therapeutic monitoring and perhaps carnitine dose adjustment. Ideally, the carnitine treatment dose in MMA should be sufficient to prevent sequestration of free CoASH, which is required for normal metabolism, while limiting the negative consequences of high dose carnitine therapy [56,57]. Therapeutic carnitine treatment facilitates excretion of the accumulated excess acyl groups in the form of acylcarnitines, i.e. propionyl- and methylmalonylcarnitine, and the patient’s dose should be sufficient to promote this. However, the carnitine dose can be too high. The excretion of acetylcarnitine is very high (3243 μmol/g creatinine, normal value < 44.15) and 47% as much as the propionylcarnitine. Because the bioavailability of oral carnitine is only 10–20%, the high butyrobetaine value is consistent with some of the carnitine dose being degraded by colonic bacteria to butyrobetaine [58]. These values suggest that the patient’s carnitine dose may be too high and could be reduced. However, if the acetylcarnitine excretion were to become low despite carnitine treatment, then as a consequence of MMA, propionic acid would accumulate, leading to CoASH sequestration and to carnitine insufficiency. Therefore, the careful monitoring of butyrobetaine and acetylcarnitine excretion, in addition to propionylcarnitine, methylmalonylcarnitine, and free carnitine excretion, can be used to optimize the patient’s therapeutic carnitine dose levels, potentially leading to improved patient care.
In another example of urine analysis, we participated in a metabolomics study in which untargeted metabolomics identified changes in acylcarnitine concentrations [59]. We validated these untargeted findings with our targeted acylcarnitine analysis. Our collaborators then showed that acylcarnitines vary as a function of both kidney cancer status and cancer grade. The study concluded with the observation that urine contains possible acylcarnitine biomarkers for kidney cancer. These results were obtainable only because of our ability to accurately and reproducibly quantify acylcarnitines in urine.
3.5 Rat Heart Acylcarnitines for Research: Fatty Acid Chain Elongation in Palmitate-Perfused Working Rat Heart
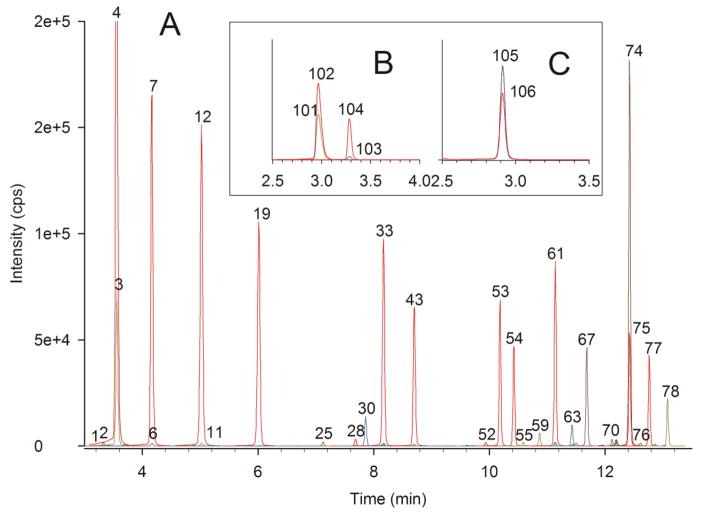
Using acylcarnitine analysis for research protocols requires accuracy, precision, and flexibility in sample type choices and analysis strategies. We conducted studies in fatty acid perfused working rat hearts, looking for the source of the two-carbon units for fatty acid chain elongation [46]. We measured acylcarnitines as a proxy for their respective acyl-CoA derivatives formed from both β-oxidation and chain-elongation of palmitic acid. Powdered rat heart (44.4 mg) was homogenized in 750 μL of acetonitrile / 2-propanol (3+1) + 250 μL of 0.1 M KH2PO4 (pH 6.7) [60]. The sample was centrifuged (5 min, 16,000x_g_), and the supernatant was transferred to a new clean tube. Total carnitine, free carnitine, and butyrobetaine were quantified using 10 μL of the supernatant, and acylcarnitines were quantified using a second 10 μL aliquot. Fig. 6 shows UHPLC-MS/MS chromatograms resulting from acylcarnitine, free carnitine, butyrobetaine, and total carnitine analysis of palmitate-perfused rat heart. In Fig. 6, a large amount of palmitoylcarnitine (Peak 74, reflecting palmitoyl-CoA) and myristoylcarnitine (Peak 67, from β-oxidation of palmitoyl-CoA and reflecting myristoyl-CoA) were observed as expected. The surprising result was the large amount of stearoylcarnitine (Peak 78), reflecting chain elongation of palmitoyl-CoA [61]. Table 1 shows the “balance study” for this analysis, with 93% of the calculated acylcarnitine pool accounted for by the sum of the quantified individual acylcarnitines. The full analysis report is provided in the Supplemental Material.
Fig. 6.
UHPLC-MS/MS chromatograms of acylcarnitines, free carnitine, butyrobetaine, and total carnitine from analysis of a rat heart perfused with palmitic acid. Peak identities and colors are the same as in Fig. 1 and Fig. 5. The chromatogram displays prominent increases in myristoyl- (Peak 67), palmitoyl- (Peak 74), and stearoylcarnitine (Peak 78). Analysis was performed using 10 μL of a 1 mL homogenate of 44.4 mg of powdered rat heart. A) acylcarnitine chromatogram B) free carnitine and butyrobetaine chromatogram. C) total carnitine chromatogram.
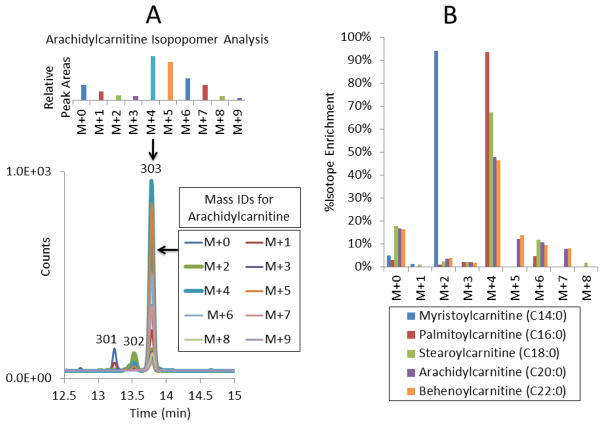
In a separate experiment to further characterize chain elongation, rat heart was perfused with [1,2,3,4-13C4]-palmitic acid, and a rat heart homogenate was prepared and analyzed [46]. The mass spectrometer acquisition method was modified to collect data for isotopomer analysis, and the chromatogram was extended to observe C20:0 and C22:0 acylcarnitines (arachidyl- and behenoylcarnitine). We found evidence of both chain elongation and β-oxidation in stearoyl-, arachidyl-, and behenoylcarnitine (reflecting their respective acyl-CoAs), and this can be seen in the complex isotope enrichment pattern of arachidylcarnitine (Peak 303) shown in Fig. 7A. Fig. 7A also shows the necessity for the chromatographic separation of the C20:2 (Peak 301) and C20:1 (Peak 302) from C20:0 acylcarnitine, since the mass of the M+0 C20:0 chromatographic peak is identical to and indistinguishable from the M+2 peak of C20:1 and M+4 peak of C20:2. This study would not be possible with tandem MS “profiles” (beyond the problem of analyzing tissue homogenates by that technique) because, for example, the M+0 C20:0 value would be incorrectly quantified due to contributions from M+2 of C20:1 and M+4 of C20:2. Without chromatographic separation of saturated from unsaturated acylcarnitines, the mass spectral results in this study would be uninterpretable due to the multiple overlapping isomeric contributions from the unsaturated isomers of myristoyl-, palmitoyl-, stearoyl-, arachidyl-, and behenoylcarnitine.
Fig. 7.
Perfused Rat Heart with Isotopomer Analysis. Rat heart was perfused for 60 min with [1,2,3,4-13C4]-palmitic acid in the working heart mode. A) The chromatographic separation and the determination of the mass isotopomer distribution in arachidylcarnitine is shown. Peak Identities: 301) C20:2 acylcarnitine, 302) C20:1 acylcarnitine, 303) C20:0 acylcarnitine (arachidylcarnitine) B) The % isotope enrichment compared to unlabeled myristoyl-, palmitoyl-, stearoyl-, and arachidylcarnitine are presented. The isotopomer patterns of stearoyl-, arachidyl-, and behenoylcarnitine show evidence of both chain elongation and β-oxidation of their analogous acyl-CoAs.
Comparison of the unlabeled and labeled analyses was done using the enrichment calculator IsoPat2 [62]. With the unlabeled isotope pattern of each acylcarnitine used as a standard, the relative enrichment of its analogous labeled acylcarnitine was then determined (see Fig. 7B). For palmitoylcarnitine, 94% of the enrichment was in the M+4 isotope, as expected following perfusion with [1,2,3,4-13C4]-palmitic acid, conversion to palmitoyl-CoA, and then reflected in palmitoylcarnitine. Myristoylcarnitine was determined to have 94% enrichment in the M+2 isotope, consistent with β-oxidation of M+4 palmitoyl-CoA to M+2 myristoyl-CoA, and then reflected in M+2 myristoylcarnitine. However, stearoyl-, arachidyl-, and behenoylcarnitine were enriched in M+6 as well as M+4 isotopes. The formation of M+4 and M+6 stearoylcarnitine from M+4 palmitate reflects chain elongation of M+4 palmitate using M+0 and M+2 two-carbon chain extender units derived from β-oxidation of M+4 palmitoyl-CoA. This effect is further demonstrated in the isotopomer patterns observed in arachidyl- and behenoylcarnitine (Fig. 7B). Arachidyl- and behenoylcarnitine also showed apparent enrichment in M+5 and M+7 isotopes. We believe this is an artifact resulting from M+4 and M+6 signal spilling into M+5 and M+7, respectively, and correctable with more precise mass designations for MRM isolation and sampling.
3.6 Human Cerebral Spinal Fluid: High Sensitivity Accurate Analysis
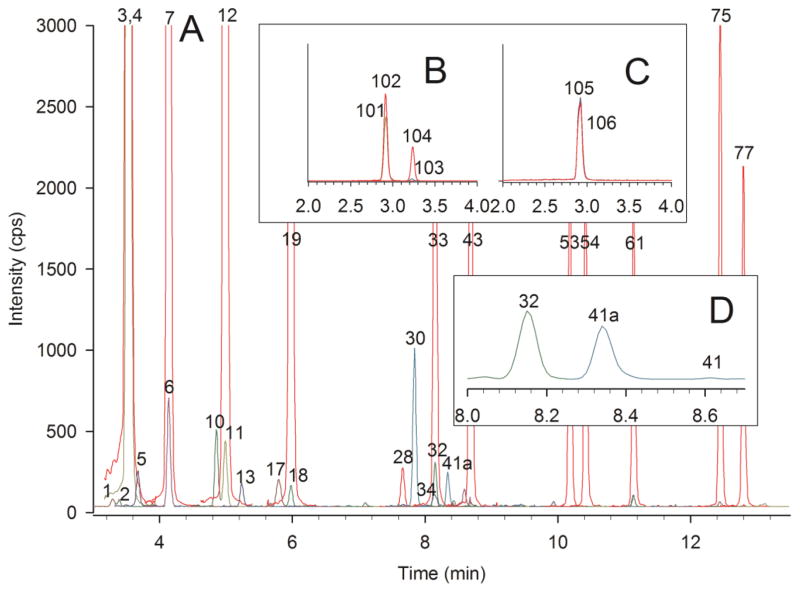
We purchased human CSF from a biological products company and analyzed this sample using our standard procedure. Since the concentration values for carnitine and acylcarnitines in CSF were significantly lower as compared to plasma, urine, and tissues, we diluted our calibrants and internal standards 1:10 to accommodate the lower concentrations. Fig. 8 shows UHPLC-MS/MS chromatograms of acylcarnitine, free carnitine, butyrobetaine, and total carnitine analysis of a normal CSF sample. Acetylcarnitine (Peak 3) accounts for 71% of the acylcarnitine pool (see Supplemental Material). Propionylcarnitine (Peak 6) is also quantified. There are no long-chain acylcarnitines in this specimen, but there are quantifiable amounts of isobutyryl-, butyryl-, tigloyl-, 2-methyl-butyryl-, 3-hydroxy-isovaleryl-, succinyl-, methylmalonyl-, and glutaroylcarnitine. The C5 dicarboxylic acylcarnitines are specifically highlighted in Fig. 8D. The peak labeled 41a is isomeric with glutaroyl- and ethylmalonylcarnitine but chromatographs between them; Peak 41a is presumed to be methylsuccinylcarnitine. These three isomeric forms of C5 dicarboxylic acylcarnitine were clearly chromatographically separated, and definitive compound assignments were made based on co-chromatography with standard compounds. Methylsuccinylcarnitine was observed in all the CSF control samples we analyzed. Again, tandem MS “profiles” do not resolve isomeric acylcarnitines and would report all these compounds together as C5 dicarboxylic acylcarnitine.
Fig. 8.
UHPLC-MS/MS chromatograms of acylcarnitines, free carnitine, butyrobetaine, and total carnitine from analysis of a human cerebral spinal fluid (CSF) specimen. Analysis was performed using 10 μL of CSF with calibrants and internal standards diluted 1:10 to accommodate the low concentration of carnitine and acylcarnitines in CSF. Peak identities and colors are the same as in Fig. 1 and Fig. 5. A) acylcarnitine chromatogram B) free carnitine and butyrobetaine chromatogram. C) total carnitine chromatogram. D) Expanded range showing only C5 dicarboxylic acylcarnitines. Peak 32 is glutaroylcarnitine, Peak 41 is ethylmalonylcarnitine, and Peak 41a is presumed to be methylsuccinylcarnitine.
This CSF sample was quantified using 10 μL of sample. Analysis was performed without any modification to the sample isolation protocol, but to accommodate the lower concentrations, the calibrant and internal standard concentrations were reduced. The analysis remains highly accurate; Table 1 also shows the “balance study” for this analysis. The entire calculated acylcarnitine pool (102%) is accounted for by the sum of the quantified individual acylcarnitines.
4. Conclusions
We have shown the application of our validated UHPLC-MS/MS second-tier method for quantification of total carnitine, free carnitine, butyrobetaine, and acylcarnitines to CAP plasma testing samples, a CAP urine sample, research rat heart homogenates, and a CSF sample. We have demonstrated quantitative accuracy in diagnosing malonic acidemia (MA), which is problematic with tandem MS “profiles”. Since tandem MS “profiles” cannot distinguish among isomeric acylcarnitine species, elevated C4 acylcarnitine could be diagnosed as either SCADD or IBD. In contrast, with our method, we unambiguously determined the correct diagnosis as SCADD. In a second example, elevated HO-C5 and C5:1 acylcarnitines could be diagnosed as either 3-MCC or BKT as tandem MS “profiles”. However, with our method, we unambiguously determined the correct diagnosis as BKT. In analyzing urinary acylcarnitines in a CAP sample from a patient with MMA, we demonstrated the flexibility of our second-tier system to accurately quantify a urine sample with no modification to our method or sample workup. This provided valuable information for follow-up to treatment with carnitine. Urine is not typically analyzed by tandem MS. For perfused rat hearts, we provided both accurate quantification and isotopomer analysis, demonstrating the robustness of both the sample preparation scheme and the chromatographic and mass spectral analysis capabilities. In a high sensitivity application, we analyzed a CSF sample without modification to our sample preparation protocol. In all of these examples, we show that going beyond tandem MS acylcarnitine “profiles” is both feasible and advantageous using our validated second-tier UHPLC-MS/MS method. The accuracy and precision of the procedure should make it the method of choice for clinical and basic research projects, including treatment protocol studies, acylcarnitine biomarker studies, and metabolite studies using tissue, urine, or other sample matrices. Due to these unique capabilities, numerous collaborations and publications [59,63,64,65,66] have resulted from the application of this validated acylcarnitine analysis method.
Supplementary Material
supplement
Highlights.
- Applied our UHPLC-MS/MS acylcarnitine method to clinical and research samples
- Absolute quantification of 66 acylcarnitine species in 14 min chromatograms
- Examined plasma malonic acidemia, SCADD vs. IBD, 3-MCC vs. BKT, and urine MMA
- Show labeled fatty acid perfused rat heart isotopomers and CSF research samples
- These results are not possible when using tandem MS acylcarnitine “profiles”
Acknowledgments
We wish to thank Bruce A. Barshop, MD, PhD and the ACMG/CAP Biochemical and Molecular Genetics Resource Committee, and Pamela Provax MT (ASCP) of the College of American Pathologists for permission to publish the CAP data. We also wish to thank Lori Hezel for the urinary organic acid analysis. Sarah Stewart and Edward J. Lesnefsky, MD performed the working heart perfusions; their contributions were supported by the National Institutes of Health Grant PO1 AG15885 Project 3 and Cores B and D.
This work was supported by the Center for Mitochondrial Diseases in the Department of Pharmacology at Case Western Reserve University School of Medicine, in conjunction with the Center for Inherited Disorders of Energy Metabolism (CIDEM), University Hospitals Case Medical Center, Cleveland, OH. Portions of these results were presented at the Annual Meeting of the Society for Inherited Metabolic Disorders, Charlotte, North Carolina, (April 2012), Mass Spectrometry: Applications to the Clinical Laboratory (MSACL) 4th Annual Conference & Exhibits, San Diego, CA (January 2012), the 60th ASMS Conference on Mass Spectrometry and Allied Topics in Vancouver, Canada (May 2012), and the 62nd ASMS Conference on Mass Spectrometry and Allied Topics in Baltimore MD (June 2014).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schulze A, Lindner M, Kohlmüller D, Olgemöller K. Expanded newborn screening for inborn errors of metabolism by electrospray ionization-tandem mass spectrometry: results, outcome, and implications. Pediatrics. 2003 Jun;111(6 Pt 1):1399–1406. doi: 10.1542/peds.111.6.1399. [DOI] [PubMed] [Google Scholar]
- 2.Rashed MS, Bucknall MP, Little D, Awad A, Jacob M, Alamoudi M, Alwattar M, Ozand PT. Screening blood spots for inborn errors of metabolism by electrospray tandem mass spectrometry with a microplate batch process and a computer algorithm for automated flagging of abnormal profiles. Clin Chem. 1997;43:1129–1141. [PubMed] [Google Scholar]
- 3.Lehotay DC, Hall P, Lepage J, Eichhorst JC, Etter ML, Greenberg CR. LC-MS/MS progress in newborn screening. Clin Biochem. 2011;44:21–31. doi: 10.1016/j.clinbiochem.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 4.De Jesús VR, Chace DH, Lim TH, Mei JV, Hannon WH. Comparison of amino acids and acylcarnitines assay methods used in newborn screening assays by tandem mass spectrometry. Clin Chim Acta. 2010;411:684–689. doi: 10.1016/j.cca.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 5.Millington DS, Kodo N, Norwood DL, Roe CR. Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J Inherit Metab Dis. 1990;13:321–324. doi: 10.1007/BF01799385. [DOI] [PubMed] [Google Scholar]
- 6.Moore S, Spackman DH, Stein WH. Chromatography of Amino Acids on Sulfonated Polystyrene Resins. An Improved System. Anal Chem. 1958;30:1185–1190. [Google Scholar]
- 7.Dietzen DJ, Weindel AL, Carayannopoulos MO, Landt M, Normansell ET, Reimschisel TE, Smith CH. Rapid comprehensive amino acid analysis by liquid chromatography/tandem mass spectrometry: comparison to cation exchange with post-column ninhydrin detection. Rapid Commun Mass Spectrom. 2008;22:3481–3488. doi: 10.1002/rcm.3754. [DOI] [PubMed] [Google Scholar]
- 8.AB SCIEX Technical Note. Rapid and Reproducible Amino Acid Analysis of Physiological Fluids for Clinical Research Using LC/MS/MS with the aTRAQ™ Kit. available from http://www.absciex.com/
- 9.Waters Technical Note 720001837en. UPLC Amino Acid Analysis Solution. available from http://www.waters.com/
- 10.Agilent Application Note 5990–4547EN. Improved Amino Acid Methods using Agilent ZORBAX Eclipse Plus C18 Columns for a Variety of Agilent LC Instrumentation and Separation Goals. available from http://www.chem.agilent.com/
- 11.Millington DS, Norwood DL, Kodo N, Roe CR, Inoue F. Application of fast atom bombardment with tandem mass spectrometry and liquid chromatography/mass spectrometry to the analysis of acylcarnitines in human urine, blood, and tissue. Anal Biochem. 1989;180:331–339. doi: 10.1016/0003-2697(89)90441-7. [DOI] [PubMed] [Google Scholar]
- 12.Ozben T. Expanded newborn screening and confirmatory follow-up testing for inborn errors of metabolism detected by tandem mass spectrometry. Clin Chem Lab Med. 2013;51:157–176. doi: 10.1515/cclm-2012-0472. [DOI] [PubMed] [Google Scholar]
- 13.Bennett MJ, Coates PM, Hale DE, Millington DS, Pollitt RJ, Rinaldo P, Roe CR, Tanaka K. Analysis of abnormal urinary metabolites in the newborn period in medium-chain acyl-CoA dehydrogenase deficiency. J Inherit Metab Dis. 1990;13:707–715. doi: 10.1007/BF01799572. [DOI] [PubMed] [Google Scholar]
- 14.Maeda Y, Ito T, Suzuki A, Kurono Y, Ueta A, Yokoi K, Sumi S, Togari H, Sugiyama N. Simultaneous quantification of acylcarnitine isomers containing dicarboxylic acylcarnitines in human serum and urine by high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:799–806. doi: 10.1002/rcm.2905. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer I, Ruiz-Sala P, Vicente Y, Merinero B, Pérez-Cerdá C, Ugarte M. Separation and identification of plasma short-chain acylcarnitine isomers by HPLC/MS/MS for the differential diagnosis of fatty acid oxidation defects and organic acidemias. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860:121–126. doi: 10.1016/j.jchromb.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Gucciardi A, Pirillo P, Di Gangi IM, Naturale M, Giordano G. A rapid UPLC-MS/MS method for simultaneous separation of 48 acylcarnitines in dried blood spots and plasma useful as a second-tier test for expanded newborn screening. Anal Bioanal Chem. 2012;404:741–751. doi: 10.1007/s00216-012-6194-1. [DOI] [PubMed] [Google Scholar]
- 17.Chace DH, Lim T, Hansen CR, Adam BW, Hannon WH. Quantification of malonylcarnitine in dried blood spots by use of MS/MS varies by stable isotope internal standard composition. Clin Chim Acta. 2009;402:14–18. doi: 10.1016/j.cca.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 18.Lim TH, De Jesús VR, Meredith NK, Sternberg MR, Chace DH, Mei JV, Hannon WH. Proficiency testing outcomes of 3-hydroxyisovalerylcarnitine measurements by tandem mass spectrometry in newborn screening. Clin Chim Acta. 2011;412:631–635. doi: 10.1016/j.cca.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Available at: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm386366.pdf
- 20.Karnes HT, Shiu G, Shah VP. Validation of bioanalytical methods. Pharm Res. 1991;8:421–426. doi: 10.1023/a:1015882607690. [DOI] [PubMed] [Google Scholar]
- 21.Hirche F, Fischer M, Keller J, Eder K. Determination of carnitine, its short chain acyl esters and metabolic precursors trimethyllysine and gamma-butyrobetaine by quasi-solid phase extraction and MS/MS detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2158–2162. doi: 10.1016/j.jchromb.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 22.Vernez L, Wenk M, Krähenbühl S. Determination of carnitine and acylcarnitines in plasma by high-performance liquid chromatography/electrospray ionization ion trap tandem mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:1233–1238. doi: 10.1002/rcm.1470. [DOI] [PubMed] [Google Scholar]
- 23.Kivilompolo M, Öhrnberg L, Orešič M, Hyötyläinen T. Rapid quantitative analysis of carnitine and acylcarnitines by ultra-high performance-hydrophilic interaction liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2013;1292:189–194. doi: 10.1016/j.chroma.2012.12.073. [DOI] [PubMed] [Google Scholar]
- 24.Peng M, Fang X, Huang Y, Cai Y, Liang C, Lin R, Liu L. Separation and identification of underivatized plasma acylcarnitine isomers using liquid chromatography-tandem mass spectrometry for the differential diagnosis of organic acidemias and fatty acid oxidation defects. J Chromatogr A. 2013;1319:97–106. doi: 10.1016/j.chroma.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 25.Cederblad G, Lindstedt S. A method for the determination of carnitine in the picomole range. Clin Chim Acta. 1972;37:235–243. doi: 10.1016/0009-8981(72)90438-x. [DOI] [PubMed] [Google Scholar]
- 26.Hoppel CL. In: Techniques in Diagnostic Human Genetics: A Laboratory Manual. Hommes FA, editor. Wiley-Liss; New York: 1991. pp. 309–326. [Google Scholar]
- 27.Ingalls ST, Minkler PE, Hoppel CL, Nordlander JE. Derivatization of carboxylic acids by reaction with 4′-bromophenacyl trifluoromethanesulfonate prior to determination by high-performance liquid chromatography. J Chromatogr. 1984;299:365–376. doi: 10.1016/s0021-9673(01)97852-5. [DOI] [PubMed] [Google Scholar]
- 28.Minkler PE, Ingalls ST, Kormos LS, Weir DE, Hoppel CL. Determination of carnitine, butyrobetaine, and betaine as 4′-bromophenacyl ester derivatives by high-performance liquid chromatography. J Chromatogr. 1984;336:271–283. doi: 10.1016/s0378-4347(00)85150-6. [DOI] [PubMed] [Google Scholar]
- 29.Minkler PE, Hoppel CL. Determination of free carnitine and ‘total’ carnitine in human urine: derivatization with 4′-bromophenacyl trifluoromethanesulfonate and high performance liquid chromatography. Clin Chim Acta. 1992;212:55–64. doi: 10.1016/0009-8981(92)90137-f. [DOI] [PubMed] [Google Scholar]
- 30.Minkler PE, Ingalls ST, Hoppel CL. High-performance liquid chromatographic separation of acylcarnitines following derivatization with 4′-bromophenacyl trifluoromethanesulfonate. Anal Biochem. 1990;185:29–35. doi: 10.1016/0003-2697(90)90250-d. [DOI] [PubMed] [Google Scholar]
- 31.Minkler PE, Hoppel CL. Quantification of carnitine and specific acylcarnitines by high-performance liquid chromatography: application to normal human urine and urine from patients with methylmalonic aciduria, isovaleric acidemia or medium-chain acyl-CoA dehydrogenase deficiency. J Chromatogr. 1993;613:203–221. doi: 10.1016/0378-4347(93)80135-q. [DOI] [PubMed] [Google Scholar]
- 32.Minkler PE, Brass EP, Hiatt WR, Ingalls ST, Hoppel CL. Quantification of carnitine, acetylcarnitine, and total carnitine in tissues by high-performance liquid chromatography: the effect of exercise on carnitine homeostasis in man. Anal Biochem. 1995;231:315–322. doi: 10.1006/abio.1995.0057. [DOI] [PubMed] [Google Scholar]
- 33.Minkler PE, Kerner J, North KN, Hoppel CL. Quantitation of long-chain acylcarnitines by HPLC/fluorescence detection: application to plasma and tissue specimens from patients with carnitine palmitoyltransferase-II deficiency. Clin Chim Acta. 2005;352:81–92. doi: 10.1016/j.cccn.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Minkler PE, Ingalls ST, Hoppel CL. Strategy for the isolation, derivatization, chromatographic separation, and detection of carnitine and acylcarnitines. Anal Chem. 2005;77:1448–1457. doi: 10.1021/ac0487810. [DOI] [PubMed] [Google Scholar]
- 35.Minkler PE, Stoll MS, Ingalls ST, Yang S, Kerner J, Hoppel CL. Quantification of carnitine and acylcarnitines in biological matrices by HPLC electrospray ionization-mass spectrometry. Clin Chem. 2008;54:1451–1462. doi: 10.1373/clinchem.2007.099226. [DOI] [PubMed] [Google Scholar]
- 36.Minkler PE, Stoll MS, Ingalls ST, Kerner J, Hoppel CL. Validated Method for the Quantification of Free and Total Carnitine, Butyrobetaine, and Acylcarnitines in Biological Samples. Anal Chem. 2015;87:8994–9001. doi: 10.1021/acs.analchem.5b02198. [DOI] [PubMed] [Google Scholar]
- 37.Ziegler HJ, Bruckner P, Binon FJ. O-acylation of dl-carnitine chloride. Org Chem. 1967;32:3989–3991. doi: 10.1021/jo01287a057. [DOI] [PubMed] [Google Scholar]
- 38.Brendel K, Bressler R. The resolution of (plus or minus)-carnitine and the synthesis of acylcarnitines. Biochim Biophys Acta. 1967;137:98–106. doi: 10.1016/0005-2760(67)90012-4. [DOI] [PubMed] [Google Scholar]
- 39.Johnson DW. Synthesis of dicarboxylic acylcarnitines. Chem Phys Lipids. 2004;129:161–171. doi: 10.1016/j.chemphyslip.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Minkler P, Hoppel C. cis-3,4-Methylene-heptanoylcarnitine: characterization and verification of the C8:1 acylcarnitine in human urine. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;857:251–258. doi: 10.1016/j.jchromb.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 41.Kuster T, Torresani T, Kleinert P, Durka S, Neuheiser F, Heizmann CW, Troxler H. Filter paper cards contaminated with EMLA cream produce artefacts on acylcarnitine analysis. J Inherit Metab Dis. 2004;27:707–709. doi: 10.1023/b:boli.0000043024.08450.0c. [DOI] [PubMed] [Google Scholar]
- 42.Abdenur JE, Chamoles NA, Guinle AE, Schenone AB, Fuertes AN. Diagnosis of isovaleric acidaemia by tandem mass spectrometry: false positive result due to pivaloylcarnitine in a newborn screening programme. J Inherit Metab Dis. 1998;21:624–630. doi: 10.1023/a:1005424331822. [DOI] [PubMed] [Google Scholar]
- 43.Reindl BA, Lynch DW, Ramirez M, Valbracht M, Davis-Keppen L, Tams KC, Groeneveld S. Sani-cloth wipe mimics rare enzyme deficiency malonic aciduria on newborn screen. Pediatrics. 2012;130:2012–0569. doi: 10.1542/peds.2012-0569. [DOI] [PubMed] [Google Scholar]
- 44.Boemer F, Schoos R, de Halleux V, Kalenga M, Debray FG. Surprising causes of C5-carnitine false positive results in newborn screening. Mol Genet Metab. 2014;111:52–54. doi: 10.1016/j.ymgme.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Johnson DW. Inaccurate measurement of free carnitine by the electrospray tandem mass spectrometry screening method for blood spots. J Inherit Metab Dis. 1999;22:201–202. doi: 10.1023/a:1005443212817. [DOI] [PubMed] [Google Scholar]
- 46.Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL. Fatty acid chain elongation in palmitate-perfused working rat heart: mitochondrial acetyl-CoA is the source of two-carbon units for chain elongation. J Biol Chem. 2014;289(14):10223–10234. doi: 10.1074/jbc.M113.524314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baertling F, Mayatepek E, Thimm E, Schlune A, Kovacevic A, Distelmaier F, Salomons GS, Meissner T. Malonic aciduria: long-term follow-up of new patients detected by newborn screening. Eur J Pediatr. 2014;173:1719–1722. doi: 10.1007/s00431-014-2421-4. [DOI] [PubMed] [Google Scholar]
- 48.CAP Surveys. BGL-A ACMG/CAP Biochemical Genetics Participant Summary. 2012:8–9. [Google Scholar]
- 49.Corydon MJ, Vockley J, Rinaldo P, Rhead WJ, Kjeldsen M, Winter V, Riggs C, Babovic-Vuksanovic D, Smeitink J, De Jong J, Levy H, Sewell AC, Roe C, Matern D, Dasouki M, Gregersen N. Role of common gene variations in the molecular pathogenesis of short-chain acyl-CoA dehydrogenase deficiency. Pediatr Res. 2001;49:18–23. doi: 10.1203/00006450-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 50.Roe CR, Cederbaum SD, Roe DS, Mardach R, Galindo A, Sweetman L. Isolated isobutyryl-CoA dehydrogenase deficiency: an unrecognized defect in human valine metabolism. Mol Genet Metab. 1998;65:264–271. doi: 10.1006/mgme.1998.2758. [DOI] [PubMed] [Google Scholar]
- 51.CAP Surveys. BGL-B ACMG/CAP Biochemical Genetics Participant Summary. 2012:8–9. [Google Scholar]
- 52.McCandless SE, Chandrasekar R, Linard S, Kikano S, Rice L. Sequencing from dried blood spots in infants with “false positive” newborn screen for MCAD deficiency. Mol Genet Metab. 2013;108:51–55. doi: 10.1016/j.ymgme.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgartner MR, Almashanu S, Suormala T, Obie C, Cole RN, Packman S, Baumgartner ER, Valle D. The molecular basis of human 3-methylcrotonyl-CoA carboxylase deficiency. J Clin Invest. 2001;107:495–504. doi: 10.1172/JCI11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukao T, Yamaguchi S, Orii T. Molecular basis of beta-ketothiolase deficiency: mutations and polymorphisms in the human mitochondrial acetoacetyl-coenzyme A thiolase gene Hashimoto. Hum Mutat. 1995;5:113–120. doi: 10.1002/humu.1380050203. [DOI] [PubMed] [Google Scholar]
- 55.Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem J. 2002;361:417–429. doi: 10.1042/0264-6021:3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rebouche CJ. L-Carnitine. In: Erdman JW Jr, Macdonald IA, Zeisel SH, editors. Present Knowledge in Nutrition. 10. International Life Sciences Institute, John Wiley & Sons, Inc; 2012. pp. 391–404. [Google Scholar]
- 57.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, Didonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rebouche CJ, Chenard CA. Metabolic fate of dietary carnitine in human adults: identification and quantification of urinary and fecal metabolites. J Nutr. 1991;121:539–546. doi: 10.1093/jn/121.4.539. [DOI] [PubMed] [Google Scholar]
- 59.Ganti S, Taylor SL, Kim K, Hoppel CL, Guo L, Yang J, Evans C, Weiss RH. Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer. 2012;130(12):2791–2800. doi: 10.1002/ijc.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minkler PE, Kerner J, Ingalls ST, Hoppel CL. Novel isolation procedure for short-, medium-, and long-chain acyl-coenzyme A esters from tissue. Anal Biochem. 2008;376(2):275–276. doi: 10.1016/j.ab.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL. Fatty acid chain-elongation in perfused rat heart: synthesis of stearoylcarnitine from perfused palmitate. FEBS Lett. 2007;581(23):4491–4494. doi: 10.1016/j.febslet.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gruber CC, Oberdorfer G, Voss CV, Kremsner JM, Kappe CO, Kroutil W. An algorithm for the deconvolution of mass spectroscopic patterns in isotope labeling studies. Evaluation for the hydrogen-deuterium exchange reaction in ketones. J Org Chem. 2007;72(15):5778–5783. doi: 10.1021/jo070831o. [DOI] [PubMed] [Google Scholar]
- 63.Fouque D, Holt S, Guebre-Egziabher F, Nakamura K, Vianey-Saban C, Hadj-Aïssa A, Hoppel CL, Kopple JD. Relationship between serum carnitine, acylcarnitines, and renal function in patients with chronic renal disease. J Ren Nutr. 2006;16(2):125–131. doi: 10.1053/j.jrn.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, Hwang DH, Newman JW, Garvey WT. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Weeghel M, te Brinke H, van Lenthe H, Kulik W, Minkler PE, Stoll MS, Sass JO, Janssen U, Stoffel W, Schwab KO, Wanders RJ, Hoppel CL, Houten SM. Functional redundancy of mitochondrial enoyl-CoA isomerases in the oxidation of unsaturated fatty acids. FASEB J. 2012;26(10):4316–4326. doi: 10.1096/fj.12-206326. [DOI] [PubMed] [Google Scholar]
- 66.Murphy WJ, Steiber A, Connery GC, Carder J, Spry L, Hoppel C. Altered carnitine metabolism in dialysis patients with reduced physical function may be due to dysfunctional fatty acid oxidation. Nephrol Dial Transplant. 2012;27(1):304–310. doi: 10.1093/ndt/gfr334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplement