ALS mutations disrupt phase separation mediated by -helical structure in the TDP-43 low complexity C-terminal domain (original) (raw)
. Author manuscript; available in PMC: 2017 Sep 6.
Published in final edited form as: Structure. 2016 Aug 18;24(9):1537–1549. doi: 10.1016/j.str.2016.07.007
Summary
RNA-binding protein TDP-43 mediates essential RNA processing but forms cytoplasmic neuronal inclusions via its C-terminal domain (CTD) in amyotrophic lateral sclerosis (ALS). It remains unclear if aggregated TDP-43 is neurotoxic and if ~50 ALS-associated missense mutations in TDP-43 CTD promote aggregation, or if loss-of-normal-function plays a role in disease. Recent work points to the ability of related proteins to assemble into functional phase-separated ribonucleoprotein granules via their structurally-disordered “prion-like” domains. Here, we provide atomic details on the structure and assembly of the low-complexity CTD of TDP-43 into liquid-liquid phase-separated in vitro granules and demonstrate that ALS-associated variants disrupt interactions within granules. Using NMR spectroscopy, simulation, and microscopy, we find that a subregion cooperatively but transiently folds into a helix that mediates TDP-43 phase separation. ALS-associated mutations disrupt phase separation by inhibiting interaction and helical stabilization. Therefore, ALS-associated mutations can disrupt TDP-43 interactions, affecting function beyond encouraging aggregation.
Graphical Abstract
eTOC blurb
The C-terminal domain of TDP-43 forms inclusions in ALS but contains a helical region of unknown function harboring several ALS-associated mutations. Conicella et al. show that TDP-43 undergoes liquid-liquid phase separation (LLPS) in part via helical contacts that are disrupted by disease mutation.
Introduction
The biogenesis of mRNA in eukaryotes is mediated by a broad array of RNA-binding proteins including the heterogeneous ribonucleoprotein (hnRNP) family(Kwon et al., 2013). Notably, aggregates of TAR DNA-binding protein of 43 kDa (TDP-43) are found in the ~97% of all cases of amyotrophic lateral sclerosis (ALS) and ~45% of frontotemporal dementia (Ling et al., 2013), which may share common etiology but affect distinct nervous system regions.
TDP-43 is a multidomain protein containing a folded(Mompean et al., 2016) multimer-forming(Chang et al., 2012; Wang et al., 2013) N-terminal domain, tandem RNA-binding domains that bind (UG)-rich sequences(Lukavsky et al., 2013), and a predominantly disordered C-terminal domain (CTD) that is essential for hnRNP interactions and splicing activity(Buratti et al., 2005; D'Ambrogio et al., 2009). ALS-associated TDP-43 inclusions are primarily fragments including the entire CTD(Neumann et al., 2006). The domain is classified as “prion-like” due to a low complexity composition rich in polar and poor in aliphatic and charged residues, resembling the amino acid frequency in yeast prion domains(King et al., 2012). The domain is aggregation-prone both in vitro and in cell and organismal models of ALS (Budini et al., 2015; Johnson et al., 2009; Wegorzewska et al., 2009). However, the connection between TDP-43 aggregation and toxicity remains unclear(Arnold et al., 2013; D'Alton et al., 2014). Although the majority of cases of ALS have no known genetic cause, familial and sporadic ALS cases are associated with mutations in many genes, including dozens of missense mutations in TDP-43(Kwon et al., 2012). Of the 52 ALS-associated missense mutations in TDP-43, all but three are located in the CTD(Buratti, 2015). The mechanism connecting TDP-43 CTD and its mutations with ALS remains mysterious, and clear evidence for either loss of normal function or gain of toxic function in ALS is lacking(Ling et al., 2013).
The structure/function relationship for disordered domains of RNA-binding proteins is beginning to come into focus. These domains can encode dynamic self-assembly into intracellular membraneless organelles or puncta including nucleoli and ribonucleoprotein (RNP) granules (Kato et al., 2012; Mitrea and Kriwacki, 2016; Nott et al., 2015). Many RNP granules are liquid-liquid phase separated assemblies that flow, fuse, and return to spherical shape like viscous “droplets”(Brangwynne et al., 2009). Related RNA-binding proteins FUS and hnRNP A1, which also contain low-complexity domains, form liquid-like assemblies in vitro and in cells(Molliex et al., 2015; Patel et al., 2015). We recently showed that the low complexity domain of FUS retains significant structural disorder upon assembly into in vitro models of granules(Burke et al., 2015). Similarly, TDP-43 CTD is primarily disordered(Lim et al., 2016) and purified full length TDP-43 was assumed undergo in vitro LLPS though rapid conversion to aggregates prevented direct observation of liquid character(Molliex et al., 2015). Importantly, concentration of TDP-43 within dynamic, physiological phase-separated stress granules requires the C-terminal regions(Bentmann et al., 2012). Therefore, the molecular details of TDP-43 interactions within highly concentrated granules are of high importance for understanding the TDP-43 function/dysfunction. Although TDP-43 aggregation via its CTD and the effect of disease-associated mutations on aggregation has begun to be examined, TDP-43 structure within RNP granules remains unknown. Unlike FUS and hnRNP A1, TDP-43 CTD contains a conserved region of with both helical propensity and ALS mutations. Although some TDP-43 variants increase aggregation (Johnson et al., 2009), two mutations (Q331K and M337V) can induce motor neuron defects in mice in the absence of inclusion formation(Arnold et al., 2013; D'Alton et al., 2014) and do not effect TDP-43 aggregation(Jiang et al., 2016). Therefore, it is possible that ALS mutations disrupt normal TDP-43 function by altering the ability of the protein to participate in functional complexes mediated by phase separation. Here we elucidate the structural basis of dynamic granule formation of TDP-43 via its C-terminal domain.
Results
TDP-43 C-terminal domain contains a cooperatively formed, partially-populated α-helix
As expected for low complexity domains, sequence alignments of the TDP-43 C-terminal domain show incomplete sequence conservation even among vertebrates (Figure S1A). An exception is a perfectly conserved region spanning residues 319 to 341, rich in aliphatic residues (7 alanine, 5 methionine, 2 leucine) nearly absent in the remaining ~120 residues of the domain. Correspondingly, secondary structure prediction for TDP-43 C-terminal domain is disordered except for an α-helical segment (321–330) (Figure S1B) recently confirmed(Lim et al., 2016). We tested the hypothesis that the 321 to 330 region cooperatively forms α-helical structure.
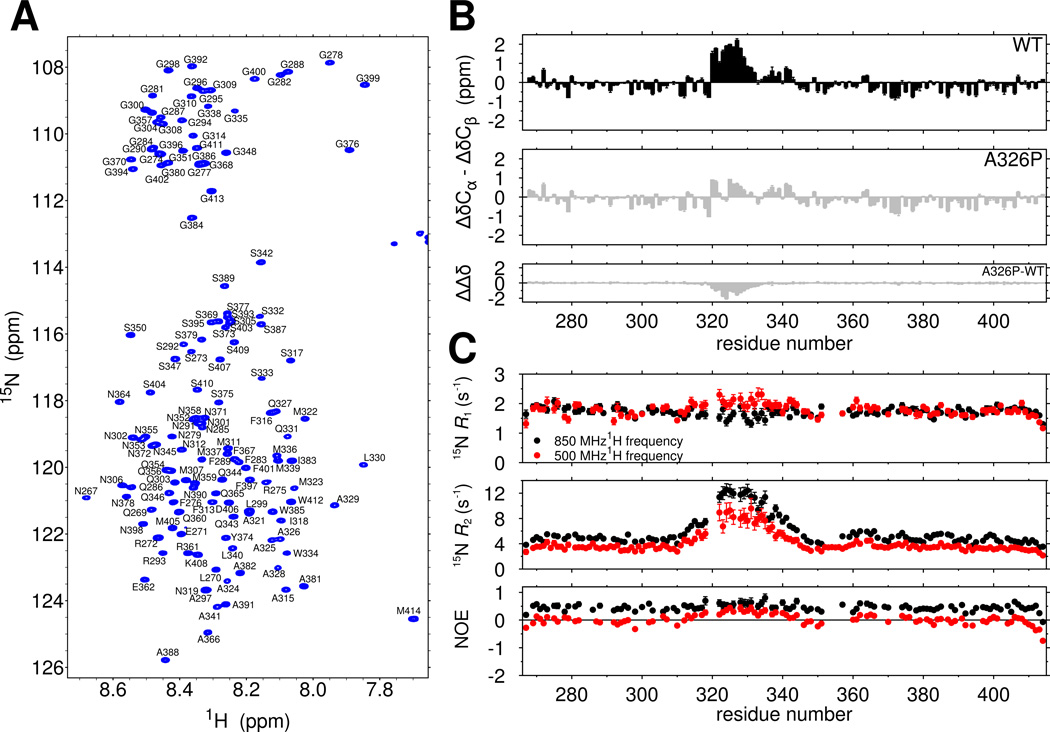
The two dimensional NMR spectrum of the TDP-43 C-terminal domain serves as a fingerprint of protein structure, with a narrow range of 1H chemical shifts consistent with predominant disorder (Figure 1A). We obtained complete chemical shift assignments of the domain to compute secondary chemical shift values that report on local secondary structure (Figure 1B, top). For the majority of the domain, the difference between the observed Cα and Cβ chemical shift values and those predicted for an entirely disordered protein, δΔCα-δΔCβ, do not exceed ±0.75 ppm, consistent with intrinsic disorder. However, consecutive positive secondary chemical shift values >0.8 ppm are observed for residues 321 to 330 consistent partial but significant α-helical secondary structure(Lim et al., 2016). Slightly positive values in the adjacent region (331 to 342) are consistent with minor helical population. Considering the maximum secondary chemical shifts for α-helices are on the order of ~5 ppm, our observations suggest residues 321 to 330 populate helical structure in ~50% of the conformational ensemble. The chemical shift index(Wishart et al., 1992), secondary structure propensity (SSP) scores (Marsh et al., 2006), and δ2D population predictions (Sormanni et al., 2015) are also consistent with partial helical structure in the region 321–330 (Figure S1C–E).
Figure 1.
The TDP-43 C-terminal domain contains a cooperatively formed α-helix. See also Figure S1. (A) The narrow assigned 1H-15N HSQC of TDP-43267–414 suggests the domain is primarily intrinsically disordered. (B) Secondary shifts (ΔδCα-ΔδCβ) confirm the location of a helical region from 321–330 that is disrupted by a single helix-breaking mutation, A326P, suggesting cooperative secondary structure formation. The difference in secondary shifts for A326P relative to WT is represented as ΔΔδ. (C) 15N spin relaxation parameters suggest slowed reorientation motions for the 321–330 region. Data are plotted as mean ± SD.
Partial α-helical structure could arise either from a transient, cooperative helix or from individual residues with propensity to form helical dihedral angles. Therefore, we introduced a structure-breaking proline residue into the center of the region (A326P), which we hypothesized would broadly disrupt cooperative α-helical structure. A326P nearly abolishes all helical chemical shifts (Fig 1B, middle and bottom). A similar proline variant outside the helical region (M337P) had little local or regional effect (Figure S1F), demonstrating that the reduction in secondary chemical shift values is sequence (and hence structure) dependent. Taken together, these experiments suggest that 321 to 330 cooperatively forms a transient α-helix.
Structure in an otherwise unstructured domain would also be expected to slow local reorientational motions. Therefore, we probed backbone ps-ns motions using 15N spin relaxation. The values of transverse relaxation rate constants, 15N _R_2, are sharply elevated, consistent with slowed motions in the 321–340 region (Figure 1C, middle). Deviation from the average longitudinal relaxation rate constants, _R_1 (Figure 1C, top), and heteronuclear nuclear Overhauser effect (NOE) values (Figure 1C, bottom) are also present. Local differences from the average in all three 15N spin relaxation parameters suggest that the region is locally rigid due to formation of structure. To confirm that increases in _R_2 result primarily from slower motion and not primarily from conformational exchange, we used backbone 15N Carr-Purcell-Meiboom-Gill (CPMG) experiments to rule out exchange on the 100 µs to ms timescales and observed no residues with relaxation dispersion > 2 s−1 at any position, although, broadening due to chemical exchange at more rapid timescales cannot be ruled out with this experiment. However, effectively identical J(0) values independently derived from spectral density mapping at two fields (Figure S1G) provide further evidence for slowed ps-ns motions in this region.
Structural characterization of TDP-43 helical domain by simulations
Attempts to directly visualize contacts by 1H-1H NOE experiments were hampered by the low concentration needed for this aggregation-prone protein. Therefore, we used a physics-based molecular simulation approach validated specifically for intrinsically disordered proteins(Zerze et al., 2015) to generate models of the structural diversity. We used parallel tempering metadynamics in well-tempered ensembles(Deighan et al., 2012) combined with protein and water molecular force fields that provide experimentally faithful cooperative secondary structure formation and disordered protein radii of gyration(Best et al., 2014). We used a TDP-43 subpeptide encompassing residues 310 to 350 including the helical region and >10 flanking disordered residues. Ensemble convergence was confirmed by structural metrics (Figure S2A). SPARTA+ derived simulated ensemble secondary chemical shift predictions(Shen and Bax, 2010) correlate well with those observed by experiment (Fig 2A, top) and the RMSD of 0.7 ppm is less than the individual SPARTA+ Cα and Cβ prediction uncertainties, suggesting good correspondence.
Figure 2.
Details of the TDP-43 helical subdomain monomeric structure by molecular simulation. See also Figure S2. (A) Correspondence between chemical shift deviations from random coil reference values (top) and 3JHNHα scalar coupling constants predicted from the simulation ensemble of TDP-43310–350 (red) and measured by experiment (black, open circles) demonstrate that the simulation faithfully captures structural character of the helical subdomain. (B) Based on regions of continuous backbone dihedral angles (ϕ,ψ), α-helical structure of TDP-43 peptide shows extended helical structure primarily in the 321 to 334 region. (i–v) Ensemble members and their total population (labeled %) representing populated regions of the α-helix map based on structural clustering. (C) Secondary structure sampled by each position in the TDP-43 simulated ensemble based on DSSP in three classes. Data are plotted as mean ± SEM for simulation data.
In order to further validate the simulation ensemble, we compared experimental 3JHNHα scalar coupling constants, sensitive to the backbone torsion angle ϕ, to those predicted from simulation. Experimental values (Figure 2A, bottom) for the 321–330 region were 4.8 ± 0.8 Hz, consistent with α-helical structure (Smith et al., 1996). 3JHNHα values for the remainder of the TDP-43 C-terminal domain averaged 6.3 ± 0.6 Hz, consistent with random coil. Given that the uncertainty in SPARTA+ predicted values is ~0.4 Hz (Vogeli et al., 2007), the rms deviation between experiment and simulation of 0.7 Hz over the entire peptide suggests good correspondence. Furthermore, sequence-specific differences in local dynamics observed by backbone relaxation experiments are recapitulated by the values calculated from a set of trajectories launched from the equilibrated ensemble (Figure S2C, left). Taken together, the values of the NMR observables calculated from the simulations are in excellent accord with the values measured by NMR, suggesting that the simulated structural ensemble can provide insight into the heterogeneous structural ensemble of the TDP-43 C-terminal domain. The agreement between simulation and experiment in a partially-ordered peptide system demonstrates recent improvements in protein forcefields and sampling algorithms.
Analysis of structural ensemble of TDP-43 α-helical region as probed by simulation
To characterize the simulated ensemble structural diversity, we measured secondary structure content at each residue position. Using DSSP (Kabsch and Sander, 1983)which measures hydrogen bond formation, we find that α-helical structure is present in greater than 50% of the ensemble for residues 321–328 (Figure 2C and Figure S2B) and to a lesser degree in residues 313–317, 329–334, and 340–343. No significant β-sheet structure is found (<5% at all sites). To provide insight into the position and lengths of the secondary structures populated, we computed secondary structure maps based on binning consecutive stretches of α-helical, β-sheet, and random coil torsion angles(Iglesias et al., 2013). Significant consecutive helical residues are apparent, including a ~25% populated 10-residue helix spanning 321 to 330. A longer 15-residue helix 320–334 is present at 12%. Minor populations of short helical conformations 3 to 6 residues in length are present at 313–318, 332–334, and 340–343 (Figure 2B). No significant population of β-strand or polyproline II conformation is found (Figure S2D).
TDP-43 C-terminal domain undergoes liquid-liquid phase separation
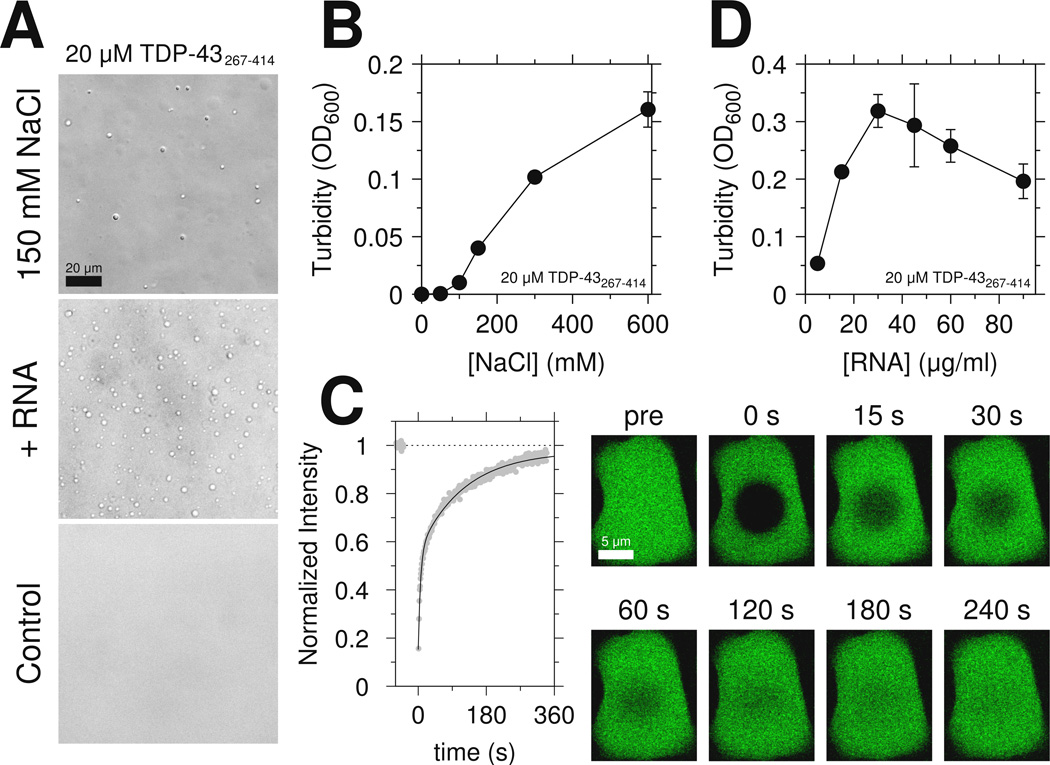
Because the low complexity domain of FUS is sufficient for LLPS(Burke et al., 2015), we tested if the C-terminal domain of TDP-43 is similarly sufficient. At low salt (0 M NaCl), 20 µM C-terminal domain remains monomeric. However, upon addition of 150 mM NaCl, the domain rapidly undergoes LLPS (Figure 3A top). These assemblies appear as dynamic, high protein concentration liquid droplets that flow and fuse and show effectively complete fluorescence recovery after photobleaching (FRAP), highlighting their liquid-like character (Figure 3C and Movie S1). Increasing salt concentration increased phase separation extent (Figure 3B) which is stable over ~20 minutes (see below). Unlike the low-complexity domain of FUS which contains no positively charged residues, TDP-43 CTD contains an RGG motif known to interact with RNA(Phan et al., 2011). Therefore we also tested if addition of RNA enhances LLPS even in the absence of a folded nucleic acid binding domain. Addition of yeast RNA as low as 3 µg/ml resulted in phase separation (Figure 3A middle). As previously observed for full-length FUS aggregation(Schwartz et al., 2013) and phase separation(Burke et al., 2015), TDP-43 C-terminal domain LLPS peaks in the presence of increasing amounts of RNA, possibly due to saturation of the protein-RNA interactions. Interestingly, monomeric protein remaining visible to solution NMR after addition of salt (Figure S3A) or RNA (Figure S3B) shows little change in chemical shifts (Figures S3C and S3D), suggesting that the disordered structure of the protein is not globally altered. Moreover, simple increase of protein concentration at 0mM NaCl is not sufficient for LLPS. Instead, >100 µM protein concentrations led to clear solutions that converted to solid/hydrogel-like samples with concomitant loss of NMR signal intensity over ~24 hrs. In summary, the C-terminal domain of TDP-43 is sufficient for phase separation at physiological salt concentrations.
Figure 3.
The region spanning residues 321 to 340 mediates TDP-43 C-terminal domain liquid phase separation. See also Figure S3 and Movie S1. (A) DIC micrographs of 20 µM TDP-43 CTD liquid-liquid phase separation in the presence of salt (150 mM NaCl, top) or yeast RNA extract (middle). No phase-separation is observed in the control condition. Scale bar is 20 µm. (B) The extent of phase separation increases with increasing salt concentration as monitored by turbidity. (C) Fluorescence recovery curve (and corresponding images) measured for a large TDP-43 C-terminal domain liquid droplet at a total sample concentration of 50 µM. Scale bar is 5 µm. Data are plotted as mean±SD.
TDP-43 C-terminal domain phase separation is mediated by the 321–340 region and altered by ALS mutation
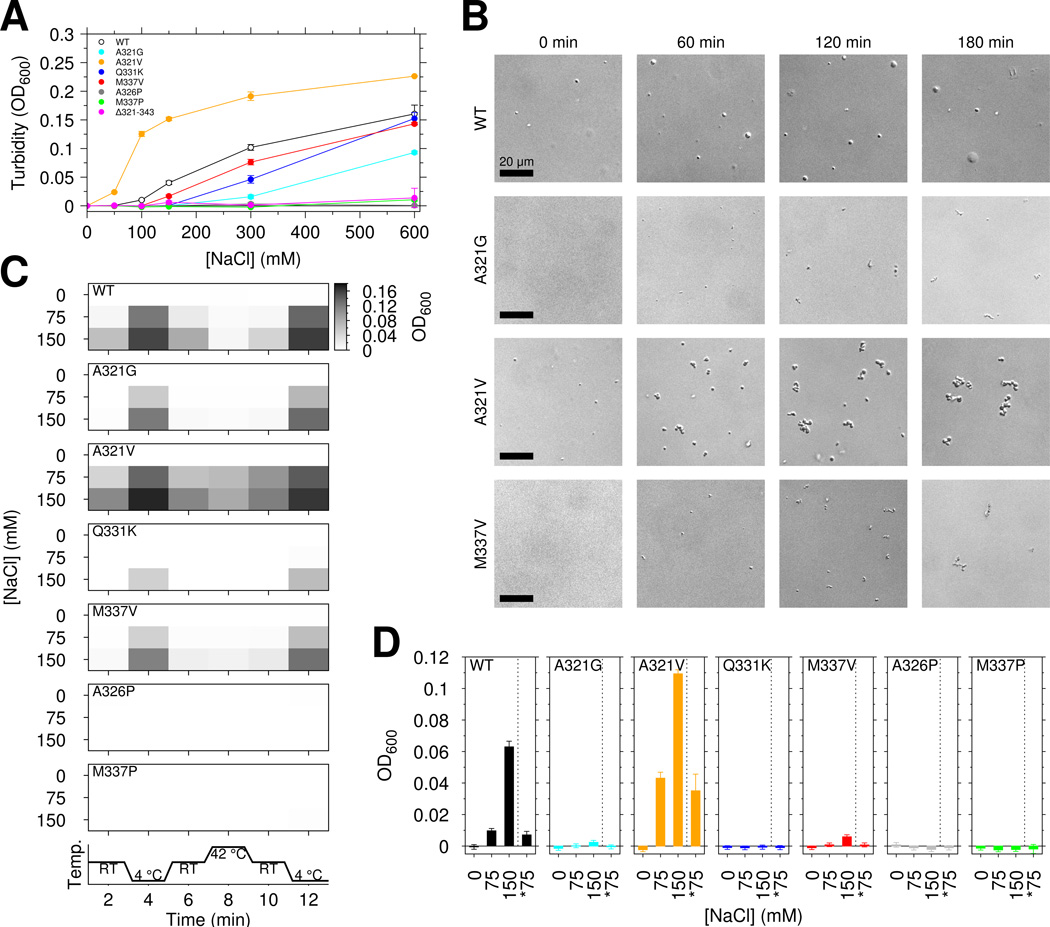
We next tested the effect of TDP-43 C-terminal domain ALS mutations on phase separation. Unexpectedly, for samples of 20 µM CTD in 150 mM NaCl, A321G and Q331K completely abrogated phase separation of the TDP-43 C-terminal domain while M337V decreased turbidity by ~50% (Figure 4A). Interestingly, both removal of the region harboring the α-helical structure and these mutations (Δ321–343) as well as introduction of the helix-breaking A326P resulted in loss of phase separation, suggesting that the helical region is important for LLPS. Furthermore, the proline substitution M337P that does not affect helical structure of monomeric TDP-43 (Figure S1F) also completely abrogates turbidity. Therefore, contacts outside the helical 321–330 region contribute to phase separation. Of the ALS mutations, only A321V showed increased turbidity. To determine if any of the observed turbidity arises from protein aggregation, we visualized the assemblies. Immediately after initial phase separation by addition of salt, the A321V assemblies are round and show FRAP consistent with liquid (Figure 4B left and Figure S4D), suggesting that A321V enhances phase separation, possibly due to increased hydrophobicity. Next we tested the reversibility of the turbidity. Because liquid-liquid phase separation is salt dependent (Figure 3A), we examined the effect of diluting salt on the turbidity. Turbid solutions of 20 µM CTD in 150 mM NaCl were created and, after 2 minute incubation, diluted 1:1 with 20 µM CTD in 0 mM NaCl (Figure 4D). These solutions showed no significant difference in turbidity with samples directly created at 75 mM NaCl, suggesting no irreversible aggregate formation. We note the turbidity of 20 µM CTD at 150 mM NaCl is stable over >20 minutes (Figure S4A, left). Next we tested the reversibility of temperature-dependent increases in liquid-liquid phase separation extent. For wild type and ALS variants, decrease in temperature resulted in increased turbidity that was reversible (Figure 4C and S4B). However after exposure to 42 °C for 2 minutes followed by cooling to room temperature, A321V showed increased turbidity (at 75 mM NaCl, OD of 0.0614±0.005 vs. 0.0997±0.005, p-value= 0.0008, see Figure 4C), concomitant with the formation of irregularly shaped aggregates visible by microscopy (Figure S4C).
Figure 4.
ALS mutations affect TDP-43 liquid-liquid phase separation, reversibility, and aggregation. See also Figure S4. (A) TDP-43 CTD ALS variants decrease the salt-dependent turbidity with respect to wild type, except increased turbidity for A321V. Data for wild type are repeated from Figure 3B with open circles for clarity. (B) DIC micrographs from aliquots taken at time points after addition of 150 mM salt to 20 µM CTD suggest that over time CTD ALS variants form assemblies that are morphologically distinct from liquid droplets. Scale bar is 20 µm. (C) Temperature cycling of wild type and mutant CTD immediately after addition of salt modulates sample turbidity, highlighting the dynamic, reversible character of CTD liquid phase separation. (D) CTD liquid-liquid phase separation is also reversible upon dilution of salt. Turbidity of 20 µM CTD was measured for samples prepared in 0, 75, and 150 mM NaCl as well samples prepared at 150 mM NaCl and then diluted to 75 mM NaCl at constant protein concentration (*75, e.g. 1:1 dilution of 20 µM CTD in 150 mM NaCl with addition of 20 µM CTD in 0 mM NaCl). Data are plotted as mean ± SD.
To further probe conversion to aggregated species, we examined phase separation as a function of time. Some but not all ALS associated mutations examined here are reported to alter aggregation of TDP-43(Jiang et al., 2016). In our hands, between 30 minutes and ~4 hours after LLPS induction, turbidity changes (Figure S4A, right). To determine if changes in turbidity are associated with conversion of liquid assemblies to solid aggregates, we attempted to use the amyloid fibril specific dye thioflavin T. However, no CTD variants showed strong thioflavin T signatures (Johnson et al., 2009). Instead, we used microscopy to characterize morphology. After >60 min incubation, A321V, A321G, and M337V assemblies are irregularly shaped (Figure 4B) where each µm-sized cluster travels as a solid unit without fusion. These structures also appear less dynamic by FRAP, with A321V and also wild type showing impaired recovery (Figure S3E). Taken together, these data suggest that wild type and ALS mutants form reversible assemblies at short incubation times that convert to aggregates, as observed previously for similar domains(Lin et al., 2015). In short, TDP-43 C-terminal domain liquid-liquid phase separation is mediated by intermolecular interactions that include contacts in the 321 to 340 region, and ALS mutations significantly disrupt phase separation (A321G, Q331K, and M337V) encouraging the conversion to aggregates.
ALS mutations alter assembly of TDP-43 C-terminal domain
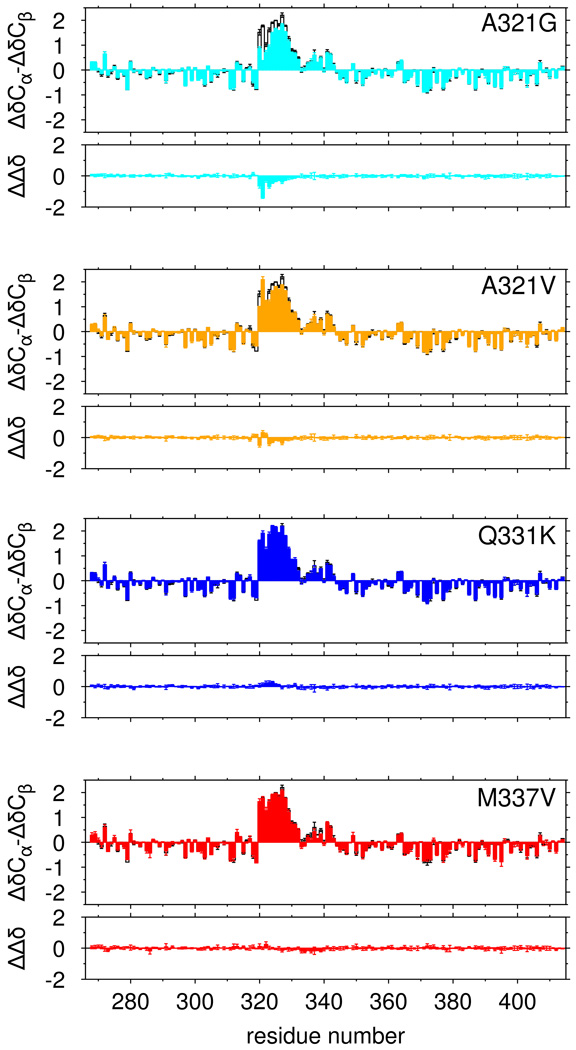
We tested the hypothesis that ALS mutations disrupt phase separation by disrupting the structure of the partially α-helical 321 to 330 region. In low salt (0 mM NaCl) conditions where no phase separation is observed for any variant, we measured the effect of ALS associated variants on the structure of TDP-43 C-terminal domain by comparing secondary chemical shifts to those of the wild type. Of these mutations, A321G resulted in the most significant disruption (Figure 5). A321V showed modest effects while Q331K and M337V showed nearly no change from wild type. All of these mutations had lower perturbation than A326P, suggesting that some ALS mutations only partially disrupt helical structure while others have no effect. Although structural changes are minor, we tested the ability of our simulations to capture these perturbations. Changes in 13C chemical shifts due to A321G are reproduced by the simulations (Figure S5A) and can be attributed to decreased helical population and shortening of alpha helical segments (Figure S5B,C) Taken together, although selected ALS variants cause small changes in population of helical population, disruption in phase separation cannot be accounted for solely by change in the monomeric structure.
Figure 5.
Only select ALS-associated mutations disrupt α-helical structure of TDP-43. Secondary shifts (ΔδCα-ΔδCβ), and the difference in secondary shift relative to WT (ΔΔδ) for ALS-associated mutations A321G (cyan) and A321V (orange) show minor local disruption of helical signatures compared to wild type (black, open bars) while Q331K (blue), and M337V (red) show no significant change in helical structure. Data are plotted as mean ± SD.
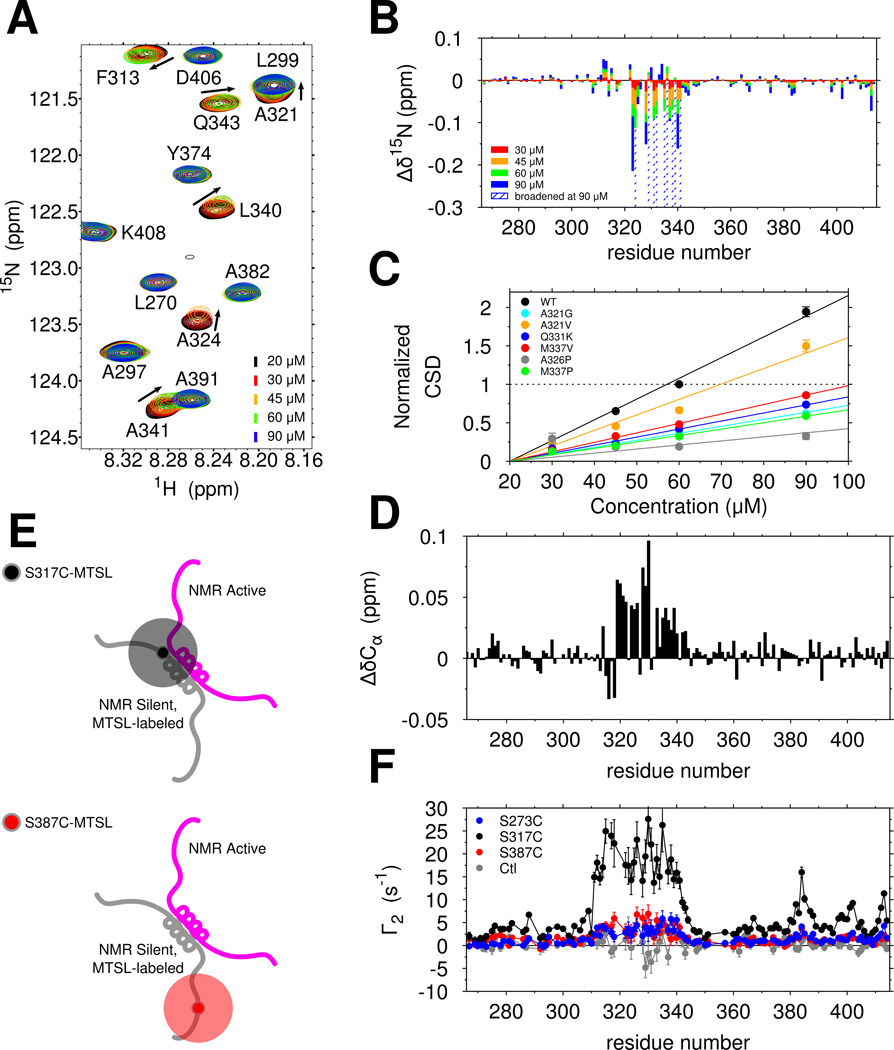
We next set out to test the hypothesis that ALS mutations disrupt phase separation by disrupting assembly mediated by 321–340 region contacts. When making samples for NMR experiments (in no salt buffer, hence in the absence of phase separation, see Figure 4A), we noticed that resonances of the wild-type TDP-43 C-terminal domain progressively shift and broaden (weaken) as a function of increasing concentration even in the absence of phase separation (Figure 6A). Furthermore, the broadened resonances are recovered by increasing temperature from 10°C to 25°C, and consequently increasing _k_ex, thus driving the kinetics into the fast exchange regime (Figure S6B). Therefore, a monomer-multimer (dimer or higher-order) equilibrium is established with a micromolar-range dissociation constant and shifted peaks represent a weighted average of positions in the monomeric and multimeric states. Resonances of the 321 to 330 helical region and the adjacent 331 to 340 region experience the greatest peak shifts and broadening (Figure 6B). The observed chemical shift perturbations are effectively linear with increasing concentration (Figure 6C, black line), consistent with unsaturated interactions. ALS mutations all had smaller concentration-dependent 1H and 15N chemical shift changes (Figure 6C and S6A), suggesting that these mutations suppress intermolecular interaction of TDP-43 CTD. The engineered helix-disrupting variant A326P also disrupts these interactions (Figure 6C). Interestingly, the engineered M337P variant, which disrupts phase separation (Figure 4A,C) also shows near complete disruption of intermolecular interaction despite having an intact 321 to 330 helical region (Figure S1F). Therefore, helical structure from 321 to 330 as well as additional contributions from 331 to 340 are essential for LLPS interactions.
Figure 6.
The TDP-43 C-terminal domain self-assembles via helix-helix contacts that are disrupted by ALS-associated mutations. See also Figure S4. (A) Overlay of 1H-15N HSQC spectra of TDP-43 C-terminal domain at increasing concentrations show chemical shift differences (arrows) and intensity reduction associated with intermolecular interactions. (B) Interactions between copies of TDP-43 C-terminal domain as probed by large 15N chemical shift differences (Δδ15N) are localized to region 321 to 340. (C) Normalized 15N chemical shift differences (CSDs) are fit to a line and demonstrate that ALS-associated mutations and structure-breaking variants disrupt domain interactions. (D) Positive Cα chemical shift differences (ΔδCα) between 60 µM and 20 µM wild type, consistent with enhanced helical structure, localize to the 321 to 340 region. (E) Labeling scheme for probing transient, intermolecular interactions by paramagnetic relaxation enhancement (PRE). (F) Intermolecular PRE values (Γ2) measured for mixtures of WT TDP-43 with MTSL-labeled S273C (blue), S317C (black) and S387C (red) confirm that 321 to 340 primarily interacts with itself and weakly interacts with other regions. A control experiment (gray, unconjugated MTSL mixed with 15N CTD) confirms that MTSL does directly interact with the 310–340 region.
Secondary structural characterization of TDP-43 interactions
In order to elucidate the contributions of the 321 to 340 region to TDP-43 CTD assembly, we sought to characterize the assembled structure. Efforts to push the equilibrium towards assembly by increasing the concentration were unsuccessful: >90 µM at 10°C and >60 µM at 25 °C led to rapid aggregation. Isolating the region of interest (residues 300–360) resulted in formation of an insoluble species.
Therefore, we next sought to characterize the secondary structure formed in the minor interacting species. We noticed that all large 15N chemical shifts perturbations induced by interaction (Figure 6B, nine residues positions) are upfield (i.e. negative), consistent with greater helical structure for every residue type (Wang and Jardetzky, 2002; Zhang et al., 2003), strongly suggesting increased helical structure upon assembly across the 321 to 340 region. For TDP-43 C-terminal domain, the 1H 13C HSQC is too overlapped to quantitate shifts and efforts to obtain residue-specific simultaneous 13Cα/13Cβ shifts using triple resonance experiments at elevated concentrations resulted in poor signal to noise. We therefore used the more sensitive HNCA experiment to compare peak positions in the monomeric reference state (20 µM) to those in the presence of a minor population of the complex (60 µM). Positive (downfield) 13Cα shift perturbations were observed from residue 321 to 341 (15 residues with shifts >0.025 ppm, Figure 6D), consistent with increased helical structure across the entire region. These data point to stabilization of helical structure upon interaction, as observed in previous heterodimeric systems(Mittal et al., 2013).
Contacts stabilizing TDP-43 intermolecular interactions
To characterize the transiently assembled state, we used intermolecular paramagnetic relaxation enhancement (PRE) NMR to probe the dynamic structural contacts (Figure 6E). We attached a small stabilized nitroxide compound (MTSL, 184 Da), which generates measurable relaxation of 1H positions even in minor population, to the NMR-silent TDP-43 C-terminal domain at single engineered cysteine positions. By mixing with NMR-active 15N-labeled (no spin label) CTD, we mapped the intermolecular contacts formed. When the label is placed immediately N-terminal to the helical region (S317C-MTSL), we observed relaxation enhancement distributed evenly across 310–340 (Figure 6F, black). Because relaxation enhancements are not sharply peaked, TDP-43 interactions are likely best described as a dynamic encounter complex ensemble(Tang et al., 2006) rather than a single rigid structure. An additional peak near residues 382–385, corresponding to a short alanine-rich region with ALS-associated mutations (A382T, A382P, I383V, G384R, W385G) that also shows significant chemical shift perturbations (Figure 6B), suggests that the region near residue 317 can transiently interact with 382–385 in vitro. A label placed in this vicinity (S387C-MTSL) showed modest enhancements. Lower enhancements were observed for S273C-MTSL. These data suggest that self-interactions mediated by the 321 to 340 region contribute significantly to chemical shift differences observed upon increasing concentration. Additional contacts mediated by the 382 to 385 region are observed while contributions from additional regions may be difficult to observe due to disorder.
TDP-43 C-terminal domain complex formation
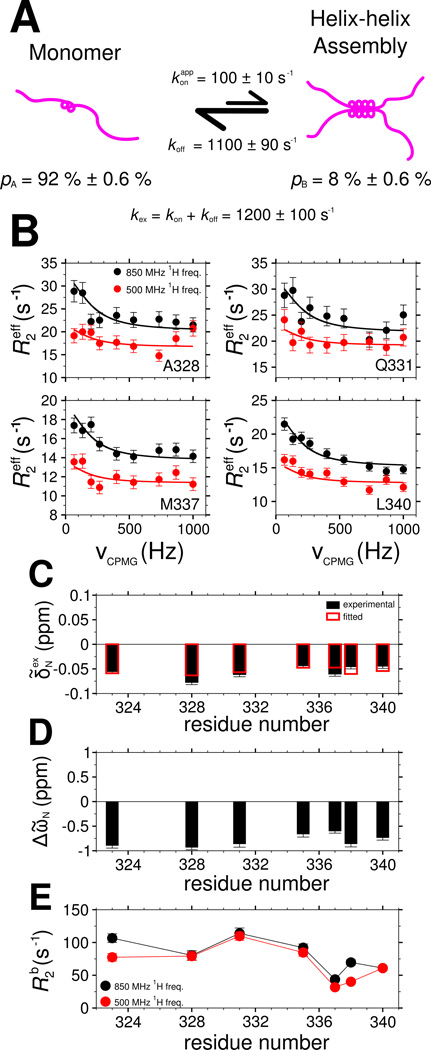
As noted above, resonance broadening for the 321 to 340 region suggest intermolecular TDP-43 interactions are formed/unformed in an exchange process on the intermediate chemical shift timescale (i.e. _k_ex is ~1 µs−1 to ~1 ms−1). We used relaxation dispersion NMR experiments (Wang et al., 2001) to characterize the structure and equilibrium kinetics of the C-terminal domain interactions. From the observed relaxation rate of each resonance, the global exchange rate, _k_ex=_k_on + _k_off, and assembled state population, _p_B, as well as the residue-specific chemical shift differences between the monomeric and assembled states, ΔῶN, and the values of the transverse relaxation rate constants, _R_2b, in the assembled state can be extracted (Figure 7A).
Figure 7.
Characterization of TDP-43 C-terminal domain interaction dynamics by 15N CPMG relaxation dispersion. See also Figure S5. (A) A two-state model for TDP-43 C-terminal domain interaction at 45 µM in which a population of monomers (_p_A) is in equilibrium with a multimeric assembled state (_p_B) governed by a global exchange rate _k_ex. (B) Relaxation dispersion profiles and (C) exchange-induced 15N chemical shifts measured (circles, B; filled boxes, C) and calculated (lines, B; open boxes, C) by simultaneous optimization. Best-fit values of model parameters: (D) 15N chemical shift differences (ΔῶN) between the assembled and monomeric state and (E) 15N transverse relaxation in the assembled state (_R_2b). Data are plotted as mean ± SD.
At concentrations low enough to have sufficiently intense resonances in the helical region (e.g 45 µM), significant relaxation dispersion is observed for seven non-overlapped residues in 321–340 (Figure 7B and Figure S7A). To confirm that the observed dispersion is a result of assembly, dispersion profiles were also measured at 20 µM. No significant dispersion was observed for any peak position, suggesting that the observed _R_2eff at 20 µM can serve as values for the intrinsic _R_2 of the monomeric state (_R_2a) in our analysis (Figure S7E). To quantitate exchange kinetics, the relaxation dispersion profiles at 45 µM were simultaneously fit with exchange induced chemical shifts (Figure 7C), as previously described(Libich et al., 2013), to the two-state model of chemical exchange. At 45 µM, the best-fit rate of exchange, _k_ex, is 1200 s−1 ± 100 s−1, the population in the assembled state, _p_B, is 8.0% ± 0.6% and the rate constant of disassembly, _k_off, is 1100 ± 90 s−1. The chemical shifts differences (ΔῶN) between the monomer and assembled state range from −0.6 and −1 ppm (Figure 7D) where the sign of the difference is extracted by direct observation of the sign of the peak shifts as a function of concentration. These values are of a magnitude consistent with increased helical structure(Wang and Jardetzky, 2002). In addition, the assembled state spin-relaxation rate values, _R_2b, are on the order of 100 s−1, consistent with a slowtumbling multimer (Figure 7E).
Given that we were unable to saturate the assembled state in our NMR titration before resonance broadening and rapid aggregation and that the _R_2b values are higher than expected for a dimer, we sought further confirmation of the best-fit parameters. One simple strategy is to alter the sample concentration, which should only change _p_B and _k_ex (and _k_on). If our two-state model of assembly holds, then we would recapitulate residue-specific exchange parameters (_R_2b and ΔῶN) as well as _k_off that we observed at 45 µM. Therefore, 15N CPMG dispersion profiles and exchange induced shifts at 30 µM were analyzed independent of the 45 µM data (Figure S7B). At 30 µM, best-fit value of _k_ex is 1000 s−1 ± 200 s−1, _p_B is 3% ± 0.9%, and _k_off is 970 ± 190 s−1. ΔῶN and _R_2b values are similar to those for 45 µM (Figure S7C,D), suggesting that the exchange between monomeric and multimeric TDP-43 CTD can be faithfully represented by a two-state model.
Discussion
TDP-43 C-terminal domain phase separation and cellular function
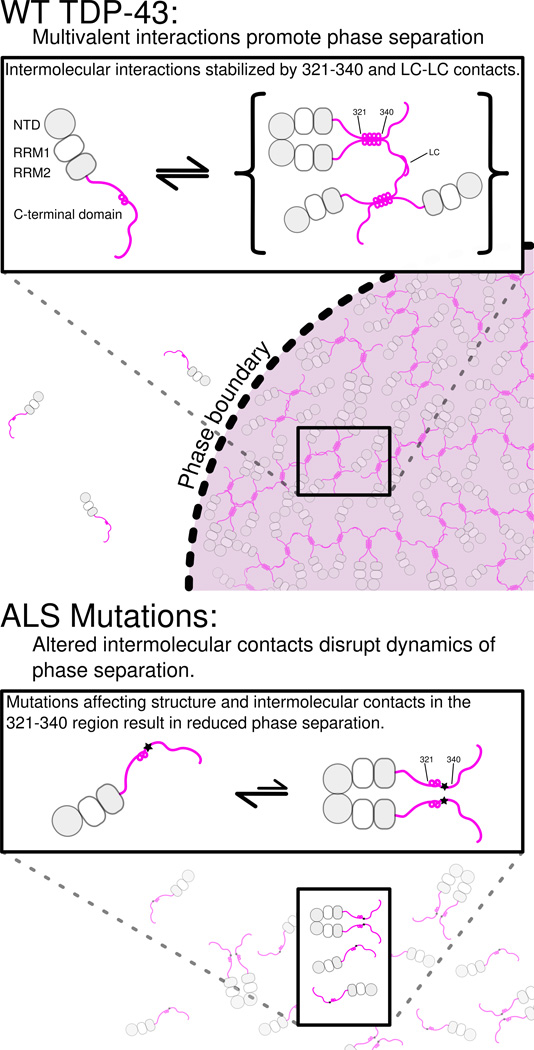
TDP-43 has been implicated in a wide array of cellular processes including regulation of mRNA splicing(Ayala et al., 2005; Buratti et al., 2001), RNA transport and localization(Alami et al., 2014), and partitioning into cytoplasmic translation-arresting stress granules(Colombrita et al., 2009; Jain et al., 2016). TDP-43 function has been shown to require protein-protein interactions mediated by the N-terminal(Mompean et al., 2016) and C-terminal(Buratti et al., 2005) domains and RNA interactions through the tandem RRMs(Lukavsky et al., 2013). Many of these functions are thought to take place in large protein/RNA assemblies known as ribonucleoprotein granules, though the structural and mechanistic details of TDP-43 incorporation and interaction in these complexes remains poorly understood. One mechanism proposed to give rise to granule structures is liquid-liquid phase separation through weak intermolecular interactions via disordered protein regions, which has been previously demonstrated for a variety of DNA- and RNA binding proteins. Consistent with these studies, our results demonstrate that the C-terminal domain of TDP-43 is sufficient for protein liquid-liquid phase separation (Figure 8). Additionally, LLPS is enhanced upon addition of RNA in the absence of folded RNA-binding domains, suggesting that interactions of RGG motifs in TDP-43 and other proteins may play a direct role in RNA-mediated phase separation. Intermolecular interactions at a conserved α-helix element and an adjacent region in the TDP-43 C-terminal domain are critical for LLPS. ALS-associated mutations disrupt interactions and inhibit LLPS, suggesting a molecular mechanism for the observed loss of function (Arnold et al., 2013) and alteration in granule assembly (Alami et al., 2014) linked to these same variants. Therefore, the effects of ALS mutations on TDP-43 LLPS observed here may provide insight into the mechanism by which functions of TDP-43 are compromised in cells. TDP-43 is highly abundant, present on average at ~1 µM in cells (Beck et al., 2011) and is further concentrated in the nucleus and in granules (Alami et al., 2014), suggesting that µM-affinity interactions between helical elements of TDP-43 C-terminal domain probed here will be populated in cells. Furthermore, TDP-43 forms interactions via its N-terminal domain (Wang et al., 2013) that serve to bring TDP-43 C-terminal domains into proximity, increasing the effective concentration and hence the likelihood of C-terminal contacts.
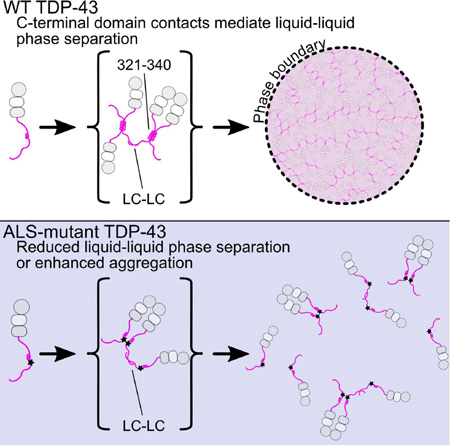
Figure 8. Model for TDP-43 phase separation driven by the C-terminal domain.
In the context of WT TDP-43 (top), the C-terminal domain mediates multimerization via intermolecular contacts in the 321–340 region that taken on additional α-helical structure upon assembly. These intermolecular contacts cooperate with additional interactions between the remainder of the aromatic-rich “prion-like” low-complexity (LC) domain to promote liquid phase separation of TDP-43 via multivalent (helix-helix and LC-LC) contacts. Additional interaction valency stimulating phase separation may be added by LC domain RNA-binding and, in full-length TDP-43, by N-terminal domain multimerization (gray circles) and RNA-binding by tandem RNA recognition motifs (RRM1 and RRM2, gray/white squares). ALS mutations (bottom) that affect structure or intermolecular contacts in the 321–340 region disrupt multivalent interactions and result in altered phase separation. Only the A321V mutation increases phase separation.
TDP-43 C-terminal domain phase separation is unusual because it depends on the integrity of a short but distinct, evolutionarily-conserved secondary structural element. In fact, ALS-associated mutations, engineered variants, and helical spectral signatures upon interaction all point to “folding-upon-assembly” where helical structure in the disordered region 331 to 340 is stabilized by intermolecular self-interaction. However, the interactions alone are insufficient to promote liquid-liquid phase separation in vitro, as no phase separation was observed at low salt concentrations. These interactions therefore cooperate with multivalent aromatic/polar-mediated contacts, possibly including transient β-sheet contacts (Xiang et al., 2015), to mediate a dynamic network leading to phase separation. ALS and engineered mutations in the 321–340 region that disrupt intermolecular contacts do so without a clear pattern of altering hydrophobicity or charge, underscoring the importance of structure in TDP-43 interactions. Importantly, the stabilization and assembly of structured helical elements in TDP-43 C-terminal domain phase separation is categorically distinct from proposed α-helix to β-sheet conversion in aggregation(Lim et al., 2016). Similar to other ALS-associated low complexity domains (Lin et al., 2015), our observations suggest that phase separated TDP-43 C-terminal domain can convert from liquid forms to solid aggregates and that ALS mutations enhance that conversion. Given that TDP-43 contains dozens more ALS-associated mutations including a cluster in the region 382 to 385 can interact with the 321 to 340 region, it will be important to determine if these variants also have effect phase separation.
Our mutational analysis suggests that small perturbations to the amino acid sequence of the TDP-43 C-terminal domain can result in dramatic differences in LLPS. Given that the TDP-43 C-terminal domain harbors numerous confirmed sites of post-translational modifications (Dammer et al., 2012; Kametani et al., 2009; Nonaka et al., 2016), such modifications may alter TDP-43 self-interaction by self and/or RNA interactions.
The role of TDP-43 phase separation in neurodegenerative disease
TDP-43 remains a primary target in neurodegenerative disease research because of its aggregate pathology and role in ALS, frontotemporal dementia(Neumann et al., 2006), and Alzheimer’s disease (Amador-Ortiz et al., 2007). Consequently, a number of studies have focused on the aggregation propensity of TDP-43 in the context of disease mutations (Budini et al., 2012; Jiang et al., 2013; Mompean et al., 2014), although the effects of TDP-43 aggregates on the severity of the disease phenotype in vivo are unclear(Ash et al., 2010; D'Alton et al., 2014; Estes et al., 2011). Here, we demonstrate here that the tested ALS-causing mutations result in perturbations to liquid phase separation via alteration of interactions of a uniquely well-conserved segment of the C-terminal domain. The in vitro granules we analyze here are composed only of the C-terminal domain in isolation which is required for robust, but only sufficient for partial, granule localization in cells(Bentmann et al., 2012). Therefore, our results suggest ALS associated mutations may not simply favor aggregated conformers but may also destabilize normal TDP-43 self-interactions, leaving the remainder of the chain available for aggregation.
One common theme that emerges from in vivo TDP-43 neurodegenerative disease models is misregulation of TDP-43 cellular RNA target splicing(Arnold et al., 2013) or transport/localization(Alami et al., 2014), both of which have been shown to involve dynamic protein granules. Moreover, TDP-43 is known to interact with a number of heterogeneous ribonucleoproteins involved in RNA splicing or translation including FUS, hnRNP A2, and hnRNP A1(Freibaum et al., 2010) which also partition into phase separated granules, form inclusions in ALS and related disease, and carry disease mutations in aggregation-prone, low complexity domains. Our data therefore support a model for TDP-43 cellular toxicity in which disruption of contacts within functional granules by disease mutation can contribute to misregulation of cellular RNA processes dependent on RNP granules.
Experimental Procedures
Cloning, expression, and purification of TDP-43267–414 and ALS variants
Human TDP-43 DNA sequence was codon-optimized for bacterial expression. ALS and control mutations were introduced with site-directed mutagenesis. The C-terminal domain of TDP-43 (residues 267–414) was subcloned into optimized vectors(Peti and Page, 2007), expressed in E. coli, and purified using standard immobilized metal ion affinity chromatography (IMAC) procedures, and was digested and purified to remove histidine tag. To measure paramagnetic relaxation enhancement (PRE) rates, the paramagnetic compound MTSL was conjugated to a series of single engineered cysteine at positions. See Supplemental Information for detailed procedures.
Turbidity Measurements
WT TDP-43267–414 and mutant variants were diluted to a final concentration of 20 µM in a 100 µl volume of 20 mM MES, pH 6.1 Turbidity was assessed by measuring the optical density at 600 nm. In order to assess changes to turbidity over extended periods of time, WT TDP267–414 and mutant variants were diluted to a final concentration of 20 µM in 20 mM MES, pH 6.1, 1 mM DTT supplemented with 0 to 500 mM NaCl. Turbidity measurements were recorded in triplicate in sealed 96-well clear-bottom plates. In order to assess salt reversibility of CTD liquid droplets, an additional sample was prepared comprising a 1:1 mixture of 20 µM protein prepared in 150 mM salt with 20 µM protein prepared in 0 mM salt. See Supplemental Information for detailed procedures.
Microscopy
Differential interference contrast (DIC) images and FRAP of WT TDP-43267–414 and mutant variants were conducted in 20 mM MES pH 6.1 with 150 mM NaCl or 15 µg/ml torula yeast RNA extract (WT DIC only). Control samples were prepared by omitting NaCl. Protein samples were then spotted onto glass coverslips. For fluorescence images and FRAP of TDP-43 C-terminal domain phase separation and fusion events, the N-terminal primary amine of WT TDP-43267–414 was conjugated to DyLight 488 NHS ester (Thermo Scientific). Excess unconjugated dye was removed by desalting through two 0.5 ml Zeba Spin Desalting columns. Fluorescently labeled TDP-43267–414 was added at a final concentration of 5 nM to 50 µM unlabeled TDP-43267–414 in the presence of 150 mM NaCl as described above.
NMR Spectroscopy
NMR experiments were recorded on Bruker Avance 850 MHz or 500 MHz 1H Larmor frequency spectrometers with HCN TCI z-gradient cryoprobes at 10 °C or 25 °C. Experimental sweep widths, acquisition times, and the number of transients were optimized for the necessary resolution, experiment time, and signal-to-noise ratio. Backbone amide resonance assignments for WT and mutant TDP-43267–414, backbone dynamics, and scalar couplings were obtained using standard experiments. Intermolecular PREs were quantified by measuring backbone amide 1HN _R_2 rates for paramagnetic and diamagnetic samples. For a complete list of NMR experiments and parameters, see Supplemental Experimental Procedures.
TDP-43 Ensemble Simulations
We performed parallel tempering metadynamics in well-tempered ensemble simulations of TDP-43310–350 in TIP4P/2005 water using the Amber ff03ws protein force field (Best et al., 2014). Simulation ensembles were validated against experimental secondary shifts and 3JHNHα scalar coupling constants. Secondary structure content was assigned using DSSP (Kabsch and Sander, 1983). Structure-based clustering was performed using GROMOS (Daura et al., 1999) based on backbone RMSD. See Supplemental Experimental Procedures for detailed protocols.
Supplementary Material
1
2
Highlights.
Cooperative helix formed by TDP-43 residues 321–330.
Helix and conserved residues 331–340 essential for liquid-liquid phase separation.
ALS mutations within 321–340 alter phase separation and disrupt self-interaction.
Helical structure enhanced by self-interaction of 321–340 region.
Acknowledgments
We thank Frank Shewmaker, Michael Clarkson, and Veronica Ryan for discussions. We thank Geoff Williams at the Leduc Bioimaging Facility at Brown University for microscopy assistance. Research reported in this publication was supported in part by the National Institute Of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under Award Number R01GM118530 (to N.L.F) and a subproject as part of an Institutional Development Award (IDeA) from NIGMS (P20 GM104937). A.E.C. was supported in part by an NIGMS training grant to the graduate program in Molecular Biology, Cell Biology and Biochemistry (MCB) at Brown University (T32 GM07601). Work at Lehigh University was supported by the U.S. Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES), Division of Material Sciences and Engineering, under Award DE-SC0013979. Use of the high-performance computing capabilities of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (NSF) Grant TG-MCB-120014, is gratefully acknowledged. This research is based in part on data obtained at the Brown University Structural Biology Core Facility supported by the Division of Biology and Medicine, Brown University. We thank Christoph Schorl and the Brown Genomics Core Facility supported by NIGMS P30GM103410, NCRR P30RR031153, P20RR018728 and S10RR02763, National Science Foundation EPSCoR 0554548. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Accession numbers for NMR chemical shift assignments in this paper for TDP-43267–414 WT, A321G, A321V, Q331K, M337V, A326P, M337P are BMRB: 26823, 26826, 26827, 26829, 26830, 26828, and 26831, respectively.
Contributions
A.E.C., G.H.Z., J.M., and N.L.F. conceived of, performed, and analyzed experiments, and wrote manuscript. A.E.C. and N.L.F. performed in vitro experiments. G.H.Z. and J.M. performed computational experiments.
References
- Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SS, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, Graff-Radford NR, Hutton ML, Dickson DW. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Molecular Systems Biology. 2011;7 doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) The Journal of biological chemistry. 2012;287:23079–23094. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Zheng W, Mittal J. Balanced Protein-Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. J Chem Theory Comput. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Budini M, Romano V, Avendano-Vazquez SE, Bembich S, Buratti E, Baralle FE. Role of selected mutations in the Q/N rich region of TDP-43 in EGFP-12xQ/N-induced aggregate formation. Brain Res. 2012;1462:139–150. doi: 10.1016/j.brainres.2012.02.031. [DOI] [PubMed] [Google Scholar]
- Budini M, Romano V, Quadri Z, Buratti E, Baralle FE. TDP-43 loss of cellular function through aggregation requires additional structural determinants beyond its C-terminal Q/N prion-like domain. Hum Mol Genet. 2015;24:9–20. doi: 10.1093/hmg/ddu415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E. Functional Significance of TDP-43 Mutations in Disease. Adv Genet. 2015;91:1–53. doi: 10.1016/bs.adgen.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Wu TH, Wu CY, Chiang MH, Toh EK, Hsu YC, Lin KF, Liao YH, Huang TH, Huang JJ. The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem Biophys Res Commun. 2012;425:219–224. doi: 10.1016/j.bbrc.2012.07.071. [DOI] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- D'Alton S, Altshuler M, Cannon A, Dickson DW, Petrucelli L, Lewis J. Divergent phenotypes in mutant TDP-43 transgenic mice highlight potential confounds in TDP-43 transgenic modeling. PLoS One. 2014;9:e86513. doi: 10.1371/journal.pone.0086513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Fallini C, Gozal YM, Duong DM, Rossoll W, Xu P, Lah JJ, Levey AI, Peng J, Bassell GJ, et al. Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One. 2012;7:e38658. doi: 10.1371/journal.pone.0038658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide folding: When simulation meets experiment. Angew Chem Int Edit. 1999;38:236–240. [Google Scholar]
- Deighan M, Bonomi M, Pfaendtner J. Efficient Simulation of Explicitly Solvated Proteins in the Well-Tempered Ensemble. J Chem Theory Comput. 2012;8:2189–2192. doi: 10.1021/ct300297t. [DOI] [PubMed] [Google Scholar]
- Estes PS, Boehringer A, Zwick R, Tang JE, Grigsby B, Zarnescu DC. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum Mol Genet. 2011;20:2308–2321. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias J, Sanchez-Martínez M, Crehuet R. SS-map. Intrinsically Disordered Proteins. 2013;1:e25323. doi: 10.4161/idp.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LL, Che MX, Zhao J, Zhou CJ, Xie MY, Li HY, He JH, Hu HY. Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J Biol Chem. 2013;288:19614–19624. doi: 10.1074/jbc.M113.463828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LL, Zhao J, Yin XF, He WT, Yang H, Che MX, Hu HY. Two mutations G335D and Q343R within the amyloidogenic core region of TDP-43 influence its aggregation and inclusion formation. Sci Rep. 2016;6:23928. doi: 10.1038/srep23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kametani F, Nonaka T, Suzuki T, Arai T, Dohmae N, Akiyama H, Hasegawa M. Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochem Biophys Res Commun. 2009;382:405–409. doi: 10.1016/j.bbrc.2009.03.038. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MJ, Baek W, Ki CS, Kim HY, Koh SH, Kim JW, Kim SH. Screening of the SOD1, FUS, TARDBP, ANG, and OPTN mutations in Korean patients with familial and sporadic ALS. Neurobiol Aging. 2012;33:1017 e1017–1017 e1023. doi: 10.1016/j.neurobiolaging.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- Libich DS, Fawzi NL, Ying J, Clore GM. Probing the transient dark state of substrate binding to GroEL by relaxation-based solution NMR. Proc Natl Acad Sci U S A. 2013;110:11361–11366. doi: 10.1073/pnas.1305715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Wei Y, Lu Y, Song J. ALS-Causing Mutations Significantly Perturb the Self-Assembly and Interaction with Nucleic Acid of the Intrinsically Disordered Prion-Like Domain of TDP-43. PLoS Biol. 2016;14:e1002338. doi: 10.1371/journal.pbio.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S-C, Polymenidou M, Cleveland DW. Converging Mechanisms in ALS and FTD: Disrupted RNA and Protein Homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain FH. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha-and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal J, Yoo TH, Georgiou G, Truskett TM. Structural ensemble of an intrinsically disordered polypeptide. J Phys Chem B. 2013;117:118–124. doi: 10.1021/jp308984e. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompean M, Buratti E, Guarnaccia C, Brito RM, Chakrabartty A, Baralle FE, Laurents DV. Structural characterization of the minimal segment of TDP-43 competent for aggregation. Arch Biochem Biophys. 2014;545:53–62. doi: 10.1016/j.abb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Mompean M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, Laurents DV. The TDP-43 N-Terminal Domain Structure at High Resolution. FEBS J. 2016 doi: 10.1111/febs.13651. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Suzuki G, Tanaka Y, Kametani F, Hirai S, Okado H, Miyashita T, Saitoe M, Akiyama H, Masai H, et al. Phosphorylation of TAR DNA-binding Protein of 43 kDa (TDP-43) by Truncated Casein Kinase 1δ Triggers Mislocalization and Accumulation of TDP-43. Journal of Biological Chemistry. 2016 doi: 10.1074/jbc.M115.695379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee, Hyun O, Jawerth L, Maharana S, Jahnel M, Hein, Marco Y, Stoynov S, Mahamid J, Saha S, Franzmann, Titus M, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein expression and purification. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LJ, Bolin KA, Schwalbe H, MacArthur MW, Thornton JM, Dobson CM. Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J Mol Biol. 1996;255:494–506. doi: 10.1006/jmbi.1996.0041. [DOI] [PubMed] [Google Scholar]
- Sormanni P, Camilloni C, Fariselli P, Vendruscolo M. The s2D method: simultaneous sequence-based prediction of the statistical populations of ordered and disordered regions in proteins. J Mol Biol. 2015;427:982–996. doi: 10.1016/j.jmb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- Vogeli B, Ying J, Grishaev A, Bax A. Limits on variations in protein backbone dynamics from precise measurements of scalar couplings. J Am Chem Soc. 2007;129:9377–9385. doi: 10.1021/ja070324o. [DOI] [PubMed] [Google Scholar]
- Wang C, Grey MJ, Palmer AG., 3rd CPMG sequences with enhanced sensitivity to chemical exchange. J Biomol NMR. 2001;21:361–366. doi: 10.1023/a:1013328206498. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT, Kuo PH, Chiang CH, Liang JR, Chen YR, Wang S, Shen JC, Yuan HS. The truncated C-terminal RNA recognition motif of TDP-43 protein plays a key role in forming proteinaceous aggregates. J Biol Chem. 2013;288:9049–9057. doi: 10.1074/jbc.M112.438564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerze GH, Best RB, Mittal J. Sequence- and Temperature-Dependent Properties of Unfolded and Disordered Proteins from Atomistic Simulations. J Phys Chem B. 2015;119:14622–14630. doi: 10.1021/acs.jpcb.5b08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2