Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen-activated protein kinase signaling to late endosomes (original) (raw)
Abstract
Signaling pathways in eukaryotic cells are often controlled by the formation of specific signaling complexes, which are coordinated by scaffold and adaptor proteins. Elucidating their molecular architecture is essential to understand the spatial and temporal regulation of cellular signaling. p14 and MP1 form a tight (_K_d = 12.8 nM) endosomal adaptor/scaffold complex, which regulates mitogen-activated protein kinase (MAPK) signaling. Here, we present the 1.9-Å crystal structure of a biologically functional p14/MP1 complex. The overall topology of the individual MP1 and p14 proteins is almost identical, having a central five-stranded β-sheet sandwiched between a two-helix and a one-helix layer. Formation of the p14/MP1 heterodimer proceeds by β-sheet augmentation and yields a unique, almost symmetrical, complex with several potential protein-binding sites on its surface. Mutational analysis allowed identification of the p14 endosomal adaptor motif, which seems to orient the complex relative to the endosomal membrane. Two highly conserved and hydrophobic protein-binding sites are located on the opposite “cytoplasmic” face of the p14/MP1 heterodimer and might therefore function as docking sites for the target proteins extracellular regulated kinase (ERK) 1 and MAPK/ERK kinase 1. Furthermore, detailed sequence analyses revealed that MP1/p14, together with profilins, define a protein superfamily of small subcellular adaptor proteins, named ProflAP. Taken together, the presented work provides insight into the spatial regulation of MAPK signaling, illustrating how p14 and MP1 collaborate as an endosomal adaptor/scaffold complex.
The process by which extracellular signals are transmitted from the plasma membrane to specific intracellular sites is an essential feature of cellular regulation. Regulatory mechanisms exist to influence where and when signaling enzymes exert their function in the cell. The identification of scaffold and adaptor proteins, which help to recruit enzymes into localized signaling complexes, provides a conceptual framework for the spatial and temporal coordination of signal transduction. Despite their overall importance, little is known about the molecular mechanisms that are used by scaffold and adaptor proteins to orchestrate signaling entities and to guarantee the translation of extracellular signals into specific biological responses (1–3).
A well studied signaling pathway is the mitogen-activated protein kinase (MAPK) cascade, which underlies the regulation of many cellular processes (4). The basic signaling unit of this enzymatic cascade is a three-kinase module allowing the transmission of an extracellular cue to specific target proteins by sequential phosphorylation events: A MAPK kinase kinase activates a MAPK kinase, which in turn activates a MAPK that finally is capable of activating multiple target proteins. Interestingly, activation of a particular MAPK yields different biological outputs. Therefore the individual MAPK pathways have acquired sophisticated regulatory mechanisms that mainly depend on certain scaffold and adaptor proteins (2, 5, 6).
The prototype of a scaffold protein is the protein Ste5, which is critical for the mating pheromone response in Saccharomyces cerevisiae. Ste5 binds all three members of the Ste11–Ste7–Fus3 MAPK cascade, thereby facilitating signal transmission within the three-kinase module. Additionally, it positions the Ste11–Ste7–Fus3 complex in close proximity to the plasma membrane, allowing activation by membrane-associated upstream regulators (7, 8). Although the general setup of the MAPK cascade is well conserved throughout eukaryotic evolution, no Ste5p is present in higher eukaryotes.
Higher eukaryotes use different scaffolding complexes. Kinase suppressor of Ras (KSR), connector enhancer of KSR (CNK), and MAPK/ERK kinase 1 (MEK1) partner (MP1) are known to regulate and specify signal transduction within the extracellular regulated kinase (ERK) cascade (9, 10). The ERK cascade is one of the four different MAPK pathways in higher eukaryotes (4) and is critically involved in the regulation of cell proliferation, differentiation, and survival, depending on the extracellular stimuli (6). Its three-kinase module is formed by Raf, MEK, and ERK. The existence of three Raf (Raf-1, B-Raf, and A-Raf), two MEK (MEK1 and MEK2), and two ERK (ERK1 and ERK2) isoforms introduces additional complexity in mammalian cells. KSR proteins combine the properties of scaffolding and adaptor proteins. They regulate the assembly and activation of the Ras–MEK–ERK module upon Ras activation at the plasma membrane. In addition, KSR interacts with several regulatory proteins that control membrane attachment of the KSR–MAPK signaling complex (11–13). The MP1 scaffold was initially identified by its capability to bind to the proline-rich region of MEK1 and to activate ERK1 but not ERK2 signaling. Thus it was proposed that MP1 facilitates ERK signaling by specifically mediating MEK1–ERK1 interactions (14).
The existence of adaptor proteins that determine the localization of scaffold proteins provides additional flexibility in regulating the efficiency and the specificity of the MAPK cascade (5). As shown by Teis et al. (16), p14 functions as an endosomal adaptor for MP1. p14 is required to attach MP1 to late endosomes, since depletion of p14 resulted in mislocalization of MP1 to the cytoplasm and in defective ERK signaling (15, 16). Interestingly, only endosomal ERK activation was affected by short interfering RNA of either p14 or MP1 (16). In contrast, the p14/MP1 complex appears dispensable for the initial phase of MAPK signaling at the plasma membrane, which requires KSR (12). These findings raise the intriguing possibility that KSR assembles a signaling complex at the plasma membrane and generates an ERK signal that is different from a p14/MP1-coordinated ERK signal emanating from endosomes (17). However, the molecular mechanisms of compartmentalized ERK signaling remain to be elucidated.
Here, we show in atomic detail how p14 and MP1 form an adaptor/scaffold complex and how this complex is tethered to late endosomes. Using specific mutations, we identified the endosomal adaptor motif of p14, which determines the relative orientation of the p14/MP1 heterodimer relative to the endosomal membrane.
Materials and Methods
Cloning, Expression, and Protein Purification. For bicistronic protein expression, the DNA sequences encoding murine p14 and MP1 were amplified by PCR and subcloned separately into pET28 (Novagen) with MP1 being expressed as a C-terminal His-tag protein. The MP1 construct was subcloned together with its ribosomal-binding site into the pET28 vector expressing p14. The resulting bicistronic plasmid pET-p14MP1 allowed coexpression of p14 and MP1 in rather equal amounts. The plasmid pET-p14MP1 was transformed into the Escherichia coli strain BL21(DE3). Cells were grown at 37°C in LB medium and induced at OD600 = 0.6 by adding 1 mM isopropyl β-d-thiogalactoside. Protein induction was prolonged overnight at 37°C. The cultures were harvested by centrifugation, resuspended in 50 mM sodium/potassium phosphate (pH 7.5)/300 mM NaCl, and lysed by freeze–thawing and subsequent sonification on ice. The protein complex was purified by affinity chromatography using the nickel nitrilotriacetate resin from Qiagen. A Superdex 75 column (Amersham Pharmacia Biosciences) was instrumental for further polishing and to adjust the protein buffer to 10 mM Hepes (pH 7.5)/150 mM NaCl/2 mM 2-mercaptoethanol. Before crystallization, p14/MP1 was concentrated by ultrafiltration to 7 mg/ml by using Centripreps (YM-3, Amicon).
The p14Δb3 and the p14b3* mutants were generated by using a PCR mutagenesis protocol. The blunt-end PCR products where subcloned into the pCR-blunt vector from Invitrogen. Generation of the Xpress-tagged mutants of p14 and MP1 proceeded as for the respective WT cDNA (15, 16). Cells were transfected with the different constructs by using the Lipofectamine Plus system (Invitrogen), following the manufacturer's suggestions.
Crystallization and Structure Solution. Crystallization was carried out at 19°C by using the sitting-drop vapor diffusion method. The keys for obtaining well diffracting crystals were the use of selenomethionine-substituted protein and the nanoliter setup of crystallization trials. Crystals of p14/MP1 were grown in sitting drops at 19°C by mixing 200 nl of protein with 100 nl of a crystallization solution containing 25% (wt/vol) polyethylene glycol 4000, 0.1 M Tris (pH 8.5), and 0.2 M MgCl2. Crystal trials were setup in Greiner plates with a reservoir volume of 100 μl. Orthorhombic crystals appeared overnight with dimensions of 50 × 50 × 200 μm3. For cryo measurements, crystals were transferred from the crystallization drop to the mother liquor supplied with 15% 2-methyl-2,4-pentanediol as cryoprotectant and rapidly frozen in a 100-K stream of nitrogen gas. All crystals belonged to space group _P_212121 with cell constants a = 50.1 Å, b = 62.5 Å, and c = 73.0 Å, corresponding to a solvent content of 33% for one p14/MP1 heterodimer in the asymmetric unit. Single anomalous diffraction data of the selenomethionine derivative were collected at the Deutsches Elektronen-Synchrotron, Hamburg (beamline BW6). The diffraction data were processed and scaled with the programs denzo and scalepack (18). Subsequently, difference Fourier analyses performed with shelx enabled us to identify four of the six theoretical selenium sites. Refinement of heavy atom parameters and phase calculation were done with sharp (19) and solvent flattening was with solomon (20). Energy-restrained crystallographic refinement was carried out with maximum likelihood algorithms implemented in cns (21), using the protein parameters of Engh and Huber (22). Refinement, model rebuilding with the program o (23), and water incorporation proceeded smoothly by means of rigid body, positional, and later _B_-factor optimization. The entire structure was checked by using simulated annealing composite omit maps. Some terminal protein segments were hardly visible in these maps and were therefore omitted from the model. After the addition of solvent molecules, the refinement converged at an R factor of 21.3% (_R_free = 25.9%). The data collection, phasing, and refinement statistics are summarized in Table 1, which is published as supporting information on the PNAS web site. All graphical presentations were prepared by using the programs molscript (24), raster3d (25), and dino (www.dino3d.org). Molecular surfaces and electrostatic potential were calculated by using msms (26) and mead (27), respectively.
Coimmunoprecipitation, Pulldown Experiments, and Immunofluorescence. Experiments where performed as described in previous studies by using the corresponding antibodies (15, 16).
Phosphorylation Reaction. HeLa cells were serum starved overnight and stimulated with epidermal growth factor (EGF) for 10 min. A total cell lysate was generated as described (16). The phosphorylation reaction contained 40 μl of cell lysate from EGF-stimulated HeLa cells and 0, 20, 100, 500, and 2,000 pg of purified p14/MP1 complex, kinase reaction buffer (50 mM Hepes, pH 7.4/10 mM MgCl2/1 mM DTT), and 5 μM ATP in a final volume of 60 μl, which was incubated for 30 min at 30°C. The total reaction was subjected to SDS/PAGE and probed with the indicated antibodies.
Endosome Purification from BHK Cells. BHK (baby hamster kidney) cells were washed three times with PBS. Cells were pelleted at 200 × g for 5 min. The cell pellet was rebuffered in HB buffer (250 mM sucrose/3 mM imidazole, pH 7.4/1 mM EDTA/protease inhibitors) and again pelleted at 2,500 rpm for 10 min. The cell pellet was resuspended in HB buffer and homogenized by four passes through a 22-gauge needle. Postnuclear supernatant was obtained by centrifugation at 2,500 rpm for 15 min at 4°C. The postnuclear supernatant was diluted 1:1.2 in 62% sucrose, loaded on the bottom of a Beckman SW41 tube, and overlaid with 4 ml of 35% sucrose and 3 ml of 25% sucrose. Early and late endosomal fractions were obtained in different fractions by centrifugation at 100,000 × g for 3 h at 4°C.
Biosensor Measurements. To determine quantitatively the interactions between the purified p14 protein (purified from E. coli as GST-fused protein) and MP1 (purified from Sf9 insect cells as His-tagged protein), the surface plasmon resonance technique was applied. All biosensor measurements were carried out on a BIAcore-X system (Biacore, Uppsala) at 25°C. For that purpose, the biosensor chip CM5 was first loaded with anti-GST antibody by using covalent derivatization. Purified GST-tagged p14 was injected in biosensor buffer (10 mM Hepes, pH 7.4/150 mM NaCl/0.01% Nonidet P-40) at a flow rate of 10 μl/min, resulting in a deposition of ≈600–650 response units. Next the purified MP1 was injected at increasing concentrations (10–250 nM) at a flow rate of 10 μl/min. The values for unspecific binding measured in the reference cell were subtracted. The evaluation of the kinetic parameters for MP1 binding to p14 was performed by nonlinear fitting of binding data by using the biaevaluation 2.1 analysis software. The binding constants were derived from the differential binding curves, assuming an A + B ⇄ AB association type.
Results and Discussion
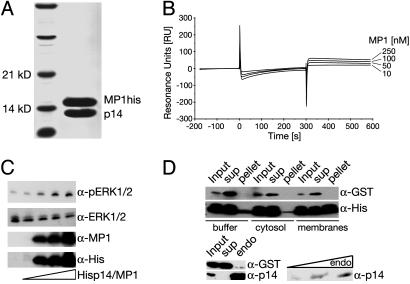
Biochemical Characterization of the Recombinant p14/MP1 Complex. Separate expression and purification of full-length MP1 or p14 in E. coli yielded low protein amounts, which were rather insoluble (solubility <1 mg/ml). Coexpressing the two proteins with a bicistronic vector enabled us to purify the physiological complex in large quantities and in high concentration (>15 mg/ml). His-tagging one of the two components allowed for isolating the p14/MP1 complex by Ni-affinity chromatography. Further polishing was achieved by gel filtration chromatography. Homogeneity and 1:1 stoichiometry were validated by SDS/PAGE (Fig. 1_A_) and MS (data not shown). The real-time stability of the p14/MP1 complex was investigated by using the Biacore technique. GST-p14 was immobilized on the CM5 chip coated with anti-GST antibody. Purified MP1 was injected in the concentration range of 10–250 nM (Fig. 1_B_). The apparent association rate constant for MP1 binding to GST-p14 was 4.68 × 104 M–1·s–1, and the dissociation rate constant was 5.97 × 10–4 s–1, thus leading to a _K_d of 12.8 nM that reflects the high-affinity association of p14 and MP1.
Fig. 1.
In vitro properties of the recombinant p14/MP1 complex. (A) SDS/PAGE of the purified p14/MP1 complex. (B) Quantitative Biosensor analysis of MP1 association with p14. The CM5 sensor chip was loaded with GST-tagged p14 and purified MP1 was injected at the indicated concentration (10–250 nM). (C) The recombinant p14/MP1 complex enhances ERK1/2 activation (phosphorylation). Increasing amounts of purified p14/MP1 were added to a cell lysate from EGF-stimulated HeLa cells and incubated at 30°C for 30 min. Cell lysates were separated by SDS/PAGE and probed with the indicated antibodies (α-). (D) Recombinant p14/MP1 binds endosomal membranes in a concentration-dependent manner. (Upper) p14/MP1 complex and GST were incubated with buffer, cytosol, or total cell membranes. Only p14/MP1 copelleted with total cell membranes. (Lower) p14/MP1 complex and GST were incubated with increasing amounts of purified endosomes. Only p14/MP1 copelleted with purified endosomes. Input (before 100,000 × g centrifugation) membrane pellets and supernatant were separated by SDS/PAGE and probed with the indicated antibodies.
We have previously shown that p14 and MP1 are required for MAPK signaling and synergize to enhance ERK1/2 activation (16). Therefore we next tested whether the purified p14/MP1 would enhance ERK1/2 activation in a cell lysate. The addition of increasing concentrations of the p14/MP1 complex resulted in enhanced activation of ERK1/2 in a cell lysate from EGF-stimulated HeLa cells (Fig. 1_C_). Another essential feature of p14/MP1 for biological function is its endosomal localization (15, 16). Therefore we next investigated whether the recombinant p14/MP1 complex would bind to cell membranes in vitro (Fig. 1_D_). The p14/MP1 complex and GST, which we used as a control having a similar size and molecular weight, were incubated with buffer alone, cytoplasm, and total cell membranes. A high-speed centrifugation at 100,000 × g was used to pellet membranes and membrane-bound proteins. GST was always found in the supernatant regardless of whether it was incubated with buffer, cytosol, or membranes. Incubation of p14/MP1 with buffer alone or cytoplasm did not result in copelleting, whereas incubation of p14/MP1 with total cell membranes resulted in efficient copelleting with membranes. To confirm the specificity of membrane binding, we used purified endosomes (15) instead of total cell membranes. The p14/MP1 complex did copellet efficiently with endosomes, whereas GST was recovered only in the supernatant after 100,000 × g centrifugation. Moreover copelleting of the p14/MP1 complex with endosomes was dependent on the amount of endosomes present in the binding assay. Taken together, p14 and MP1 form a tight complex that is capable of enhancing ERK1/2 activation in a cell lysate and binding to endosomal membranes. These results show that the isolated recombinant p14/MP1 complex mirrors the properties of the complex in vivo and should allow reconstruction of the biologically relevant structure.
Structure of p14 and MP1. Crystals suitable for x-ray diffraction were obtained by the vapor diffusion method but only with nanoliter setups. The brick-shaped crystals belonged to space group _P_212121, having unit cell constants of a = 50.1 Å, b = 62.5 Å, and c = 73.0 Å. A selenomethionine-substituted p14/MP1 complex was purified and crystallized, and the structure was determined by single-wavelength anomalous dispersion. The experimental electron density map was of excellent quality (Fig. 2_A_) and enabled us to build the entire model with the exception of a few terminal residues. The model was refined to a crystallographic R factor of 21.3% and a free R factor of 25.9% at 1.9-Å resolution and exhibited good stereochemistry (see Table 1).
Fig. 2.
Structure of p14/MP1. (A) Experimental electron density map of a representative region of the p14/MP1 complex overlaid with the final model. The map was calculated at 1.9-Å resolution and is contoured at 1.2σ. (B) Ribbon diagram of the individual MP1 (green) and p14 (white) monomers. A color ramp was used to illustrate the course of the polypeptide. All secondary structure elements and the termini are given. (C) Stereoview of the superposition of MP1 (green) and p14 (white). The most divergent regions (Left) and some of the prominent surface pockets (Right) are labeled. (D) Ribbon diagram of the heterodimer with MP1 in green and p14 in white. On the basis of the constituting helix layers, two different faces of the p14/MP1 complex can be distinguished, i.e., the two- and four-helix sides, which are presented in orthogonal mode. The nomenclature and location of possible protein–protein interaction sites are given.
The p14 protein folds as a compact domain and consists of a central five-stranded antiparallel β-sheet sandwiched by α-helix b on one side and the terminal helices a and c on the other side (Fig. 2_B_). The electron density for the loop connecting αb and β3 was not well defined, and this b3 loop (loop nomenclature according to connected secondary structure elements) should be considered as very flexible. Furthermore, the seven C-terminal residues of p14 could not be traced in the electron density map. MP1 obtains a homologous fold built around a central five-stranded β-sheet with the same strand order 2 1 5 4 3. The terminal helices a and c flank one side of the sheet, whereas the α-helices b and b2, which together equal the p14 helix b, and the 310-helix b1 shield the other side of the β-sheet. The entire MP1 structure was well defined by electron density with the exception of seven C-terminal residues. Several potential protein-binding pockets can be distinguished in both proteins. Two pockets are located on either side of helix b (Fig. 2_C_). The 2b pocket is lined by the N terminus of helix b, loops 2b and 34, whereas the b3 pocket is organized around the central part of helix b, loops 12 and 45. A third pocket, the ac pocket, is formed by the terminal helices a and c.
Both proteins adopt a strikingly homologous domain structure, with very similar shape, dimensions, and relative orientations of secondary structure elements (Fig. 2_C_). The least-square superposition aligns 95 Cα atoms with rms deviations of 1.82 Å. The first region of divergence comprises the N and C termini. In MP1, the C-terminal extension of helix c is folded as a helical segment that further runs across and reaches over the edge of the curved β-sheet. In p14 the corresponding segment points away from the protein body, protruding into the surroundings. Furthermore, the N terminus of p14 approaches its C terminus, thereby extending with Met-1 and Leu-2 the hydrophobic surface of the ac pocket, a feature that is absent from MP1. Loop b3 represents the second divergent region, which is composed of residues 57–70 in p14 and 59–68 in MP1. In both cases this loop is not attached by any interaction to the body of the protein and thus appears to be rather flexible. It protrudes into the protein periphery but with different orientations. The third region of divergence is also situated on the helix b layer side and is composed of the N terminus of helix b and its preceding loop 2b. In p14, a wide hydrophobic 2b pocket can be observed that is partially blocked in MP1 by helix b1 and a differently folded loop 2b. Furthermore, in MP1 the remainder of the corresponding 2b pocket is filled by several bulky residues such as His-43, Phe-49, Tyr-75, and Tyr-78, which have smaller counterparts in p14.
The p14/MP1 Complex Is an Almost Symmetrical Twin Couple. In the p14/MP1 complex, the domains reveal an extensive interactions surface, allowing for high-affinity binding. As shown in Fig. 2_D_, the complex exhibits a striking symmetric appearance that has no structural homolog in the Protein Data Bank. p14 and MP1 associate to form a continuous, antiparallel 10-stranded β-sheet with strand order 2 1 5 4 3 3* 4* 5* 1* 2* (the asterisk denotes MP1). The inner pair of β-strands, i.e., 3 and 3*, interact with each other by nine main-chain hydrogen bonds. One side of the central β-sheet is covered by two two-helix bundles, ac and a*c* (the four-helix side), whereas the other side is covered by the bb* two-helix bundle (the two-helix side). The b helices are coupled to each other by a zipper-like arrangement of several intertwining hydrophobic residues, including Ala-59, Tyr-56, Ile-52, Ile-48, Phe-49*, Thr-52*, Phe-53*, Ala-56*, Gln-59* (aliphatic part), and Leu-63*. This hydrophobic cluster is extended by several residues of β-strands 3 and 3*, including Leu-69, Ile-72, Met-74, Cys-76, Ile-71*, Cys-73*, and Tyr-75*, thereby forming a prominent hydrophobic core in the center of the heterodimer with almost equal contributions from p14 and MP1. Furthermore, loop b3* of MP1 fits precisely in the complementary-shaped 2b pocket of p14. Here, the two MP1 leucines, Leu-63* and Leu-65*, interact with the p14 residues Ile-48, Ile-52, Met-74, Val-81, Ile-83, and Tyr-94 in a lock-and-key fashion. The other face of the β-sheet, the four-helix side, is rather polar. On this side, heterodimer stabilization is achieved by several hydrogen bonds and salt bridges formed between Asp-75–Lys-69*, Lys-70–Asn-76*, Asp-67–Thr-77*, and Glu-78–Ser-66*. The latter two interactions seem to seal both sides of the mixed β-sheet 33*. In the center of the four-helix side, a deep polar canyon penetrates the entire heterodimer, running along β-strand 3* of MP1. The walls of this midline canyon are lined with several basic residues (Arg-80, Lys-105, Lys-70, Lys-69*, Arg-85*, and Lys-108*) and several residues with H-donor groups (Gln-109, Asn-76*, Ser-70*, Tyr-74*, Gln-79*, Thr-100*, and Ser-105*).
Using 100 mM phosphate as crystallization additive, we obtained a second crystal form that provided some interesting features. Here, the flexible b3 loop of p14 was clearly defined by electron density and was rather rigid as indicated by its average crystallographic thermal motion factor (see Fig. 5_A_, which is published as supporting information on the PNAS web site). Most interestingly, loop b3 obtains a different conformation than in the previously determined structure. It is bent toward MP1 and is in van der Waals contact with a few residues of the MP1 2b* pocket. Furthermore, one of its side chains, Phe-64, protrudes outwards into the hydrophobic ac pocket of a neighboring p14 molecule, where it undergoes ring-stacking interactions with Tyr-114 (see Fig. 5_B_). The ability to attain different conformations and to establish crystal contacts by means of a solvent-exposed hydrophobic residue highlights the potential of the p14 loop b3 to mediate protein–protein interactions.
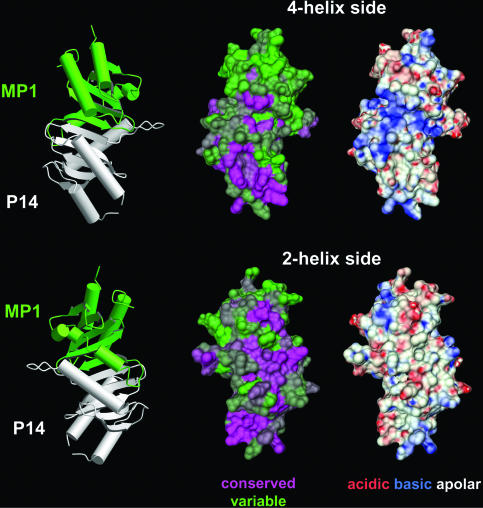
The Surface Properties of MP1/p14 Point to Several Potential Protein-Binding Sites. To identify potential sites for interactions with other molecules, we analyzed the crystal contacts, the electrostatic properties, and the conservation pattern of the p14/MP1 heterodimer (Fig. 3). Both p14 and MP1 have many solvent-exposed apolar residues that constitute distinctive hydrophobic pockets on the two- and the four-helix sides of the heterodimer.
Fig. 3.
Surface properties of the p14/MP1 complex. The four-helix side (Upper) and the two-helix side (Lower) are shown. Left shows Relative orientation of the heterodimer, whereas Center and Right show the p14/MP1 molecular surfaces of the corresponding orientations. (Center) Conservation pattern calculated with al2co (29) is mapped on the surface, with magenta indicating the most-conserved areas and green indicating the least-conserved areas. (Right) Electrostatic potential calculated with mead is mapped on the p14/MP1 surface. Red indicates negatively charged and blue indicates positively charged regions.
On the four-helix side, only one hydrophobic pocket can be observed, which corresponds to the p14 ac pocket, i.e., at the interface of helices a and c. Residues that line this hydrophobic pocket are highly conserved in the p14 family, and thus the p14 ac pocket should be considered as potential protein-binding pocket. Consistently, this pocket is also involved in establishing crystal contacts, binding a phenylalanine residue of a molecular neighbor (see Fig. 5_B_). A second remarkable feature of the four-helix side is a strikingly electropositive channel that penetrates the entire heterodimer at the p14/MP1 midline. Some of the residues that are mainly responsible for the basic rendering of this channel are strictly conserved, whereas for other residues conservative exchanges are allowed, which will not change the overall electrostatic surface potential. Thus, the electropositive channel seems to be a common feature of p14/MP1 complexes and could be of functional importance. It is also notable that MP1 exhibits a rather weak conservation pattern on the four-helix side, suggesting that this face of MP1 plays only a minor role in binding to target proteins.
In contrast, MP1 has a potential protein interaction patch on the two-helix side. Several highly conserved apolar residues are clustered around the invariant DG motif setting up the b3* pocket. The hydrophobic part of this pocket is constructed by Val-30*, Pro-31*, Val-32*, Val-120*, and Val-121*. In both crystal forms, this part is occupied by the side chain of Tyr-37 that originates from a symmetry-related p14 molecule (Fig. 5_B_). Furthermore the OH function of this tyrosine interacts with the carboxylate group of Asp-26, which is held in place by the highly invariant DG motif of MP1. On the two-helix side, an additional prominent hydrophobic pocket is situated beneath loop l2 of p14. This loop approaches and runs along helix b because of a special backbone conformation of its DG motif (Glu-28–Gly-29). The close helix-loop packing is stabilized by several van der Waals interactions and leads to the opening up of the hydrophobic core of p14. As a consequence, residues Met-1, Leu-2, Leu-7, Leu-11, Leu-24, Leu-32, Val-86, Leu-89, Pro-112, and Leu-115 form a huge hydrophobic b3 pocket, with several apolar residues being completely exposed to the solvent. Most of these residues constitute the most highly conserved stretch of amino acids in p14. Thus the b3 pocket of p14 might represent a protein-binding surface. Remarkably, sequence and structural comparisons further demonstrate the importance of the 2b and b3 pockets for mediating protein–protein interactions (for details, see Supporting Results and Discussion and Fig. 6, which are published as supporting information on the PNAS web site). Whereas the 2b pockets of p14 and MP1 are exploited for heterodimer formation, the respective b3 pockets are poised for binding partner proteins (Fig. 2_D_).
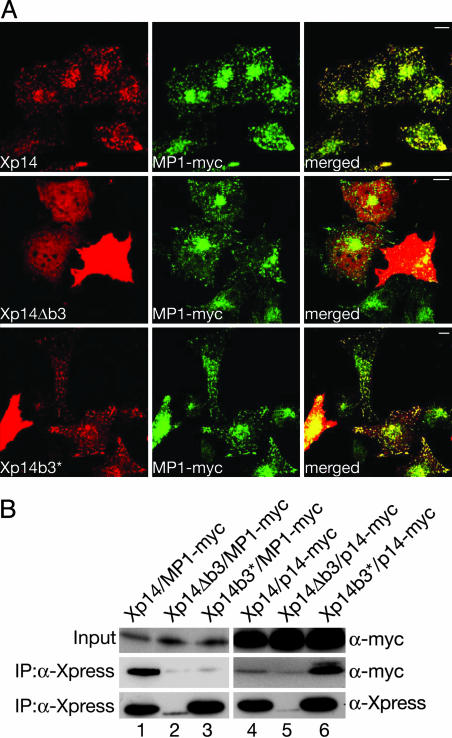
Mutational Analysis of p14. The structural analysis of the p14/MP1 complex implicates an important role for the b3 loops of p14 and MP1 in defining the respective interaction partners. We therefore investigated the function of this loop by generating specific mutations in p14. The loop b3 was either deleted (p14Δb3: p14 amino acid residues 57–70 were replaced with Gly-Ala) or exchanged with the corresponding b3* loop from MP1 (p14b3*: p14 amino acid residues 57–70 replaced with MP1 amino acid residues 59–68). To study the effects of these mutations, Xpress-tagged p14 (Xp14) mutants were coexpressed with myc-tagged MP1 (MP1-myc) in HeLa cells (Fig. 4).
Fig. 4.
Mutations affecting the b3 loop of p14 alter the subcellular distribution of p14/MP1. (A) Xpress-tagged WT p14, p14Δb3, and p14b3* proteins were coexpressed with MP1-myc. Subsequently, cells were subjected to indirect immunofluorescence analysis using anti-Xpress and anti-myc antibodies. Xp14 (red) and MP1-myc (green) colocalize on late endosomes (yellow). Xp14Δb3 was localized in the cytoplasm and did not colocalize with MP1 on late endosomes. Xp14b3* was found to colocalize with MP1-myc on late endosomes but also partially in the cytoplasm. (B) Xp14, Xp14Δb3, and Xp14b3* were coexpressed with MP1-myc or myc-tagged p14. The Xpress-tagged proteins were immunoprecipitated (IP) with an anti-Xpress antibody. To demonstrate equal MP1-myc or myc-p14 expression, cell lysates were probed with anti-myc antibodies (Input). Probing p14 immunoprecipitates with anti-myc (MP1 and p14) antibodies detected interaction of Xp14 with MP1-myc (lane 1) and of Xp14b3* with myc-p14 (lane 6).
Immunofluorescence and confocal image analysis were used to determine the subcellular location of the p14 mutants. Xp14 did colocalize with MP1-myc in 81% of all cells on late endosomes (n = 100). Only in cells that strongly overexpress both Xp14 and MP1-myc were the two proteins found in the cytoplasm. Remarkably, the Xp14Δb3 was never found to colocalize with MP1-myc on late endosomes and was always observed in the cytoplasm (n = 100). Obviously, loop b3 is required to define the endosomal localization of p14. However, in 28% of all p14Δb3-expressing cells (n = 100), the mutated protein was found in subcellular protein aggregates that did not colocalize with MP1-myc. Moreover, it was not possible to express Xp14Δb3 in amounts equal to Xp14 or Xp14b3* protein levels (Fig. 4_B_). These results indicate that cells do not tolerate high levels of the cytoplasmic, mislocalized Xp14Δb3 protein.
Surprisingly, the Xp14b3* mutant showed endosomal localization in 64% (n = 100) of all Xp14b3*-positive cells. According to the p14/MP1 crystal structure, loop b3 of MP1 is essential to interact with p14. Thus the Xp14b3* mutant could have acquired the capability to interact with endogenous p14, forming an Xp14b3*/p14 pseudohomodimer. To test this prediction, the interactions of Xp14, Xp14Δb3, and Xp14b3* with MP1-myc or p14-myc were investigated by using co-immunoprecipitation experiments. MP1-myc was co-immunoprecipitated only in the presence of Xp14 (Fig. 4, lane 1), whereas Xp14Δb3 and the Xp14b3* mutants showed no significant interactions with MP1 (lanes 2 and 3). Interestingly, p14-myc was co-immunoprecipitated only by the Xp14b3* (lane 6) mutant and not by Xp14 (lane 4) or Xp14Δb3 (lane 5). These findings confirm that the p14b3* mutant is indeed able to form a p14b3*/p14 dimer, demonstrating the important role of the MP1 loop b3 to establish and stabilize the interaction with p14. Taking the results together, the mutational analysis demonstrates that the b3 loop of p14 is required to target p14 to late endosomes and that the b3* loop of MP1 defines the interaction of MP1 with p14.
p14/MP1 Acting as a MAPK-Scaffolding Complex Anchored to Late Endosomes. The endosomal localization of the p14/MP1-MAPK scaffold complex is essential for efficient signaling in the ERK cascade upon EGF simulation. Whereas MP1 acts as the corresponding scaffold protein, p14 functions as an adaptor localizing the MP1–MAPK module to endosomes (16). The present structural and biochemical data allow the proposal of a model: How p14 and MP1 collaborate in endosomal MAPK signal transduction.
Regardless of its small size, several potential protein-binding sites are located on the p14 adaptor protein. Mutational analysis demonstrated the importance of its b3 loop acting as endosomal adaptor motif. It appears likely that this loop targets the complex to late endosomes by specific interaction with a so far unknown target protein. Further work is needed to identify such a partner. Interestingly, the ac pocket of p14 and the positively charged midline canyon are located on the same side of the heterodimer as loop b3. Thus it is tempting to speculate that the latter two features contribute to defining the localization of the p14/MP1 complex relative to the endosomal membrane. We believe that the four-helix side of heterodimer will face the endosomal membrane, thereby providing access for cytoplasmic partners to the two-helix side of the heterodimer. Especially the conservation pattern of surface-exposed MP1 residues supports this idea. Whereas the four-helix side exhibits a rather weak degree of conservation, the two-helix side is much more conserved. Here, a potential protein-binding site is organized around the highly conserved DG motif. The corresponding surface patch is characterized by a highly electronegative region enclosed by hydrophobic residues. Remarkably, all MAPKs use a negatively charged region, the so-called conserved docking domain (sequence of the ERK2 motif: LEQYYDPTDEPVA) to interact with substrates, regulators, and activators. It has been shown that activators of MAPKs such as MEK1/2 bind to the conserved docking domain by a positively charged region, the so-called docking site (sequence of MEK1: MPKKKPTPIQLNPNP) (28). We propose that the acidic patch clustered around the highly conserved DG motif of MP1 might be used to establish a similar interaction to MEK1. Interestingly, the prominent b3 pocket of p14 is located on the same side of the heterodimer and might thus function as binding site for a second protein (Fig. 2_D_). The two p14- and MP1-binding sites could act in concert to facilitate the interaction between MEK1 and ERK1. Consistently, biochemical data indicate a synergistic mode of action for mediating p14/MP1-dependent MAPK signaling (16). More studies are necessary to investigate this model. The present structure will be instrumental to construct site-specific mutants that might interfere with p14/MP1 dimerization and/or binding to the MAPK partner proteins MEK1 and ERK1.
Supplementary Material
Supporting Information
Acknowledgments
The Institute for Molecular Pathology is funded by Boehringer Ingelheim. Parts of this work were supported by the Fonds zur Förderung der Wissenschaftlichen Forschung (SFB021 to L.A.H. and P15037 to S.M.-S.), the Austrian Genome Program (GENAU; to F.E. and L.A.H.), the Deutsche Forschungsgemeinschaft (SFB487 to M.H.), and bioinformatics contract study 2002–2004 for the Bundesministerium für Wirtschaft und Arbeit.
Abbreviations: MAPK, mitogen-activated protein kinase; EGF, epidermal growth factor; KSR, kinase suppressor of Ras; ERK, extracellular regulated kinase; MEK, MAPK/ERK kinase; MP1, MEK1 partner; MP1-myc, myc-tagged MP1.
Data deposition: The atomic coordinates and structure factors for the two p14/MP1 complex structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1VET and 1VEU, respectively).
References
- 1.Pawson, T. & Scott, J. D. (1997) Science 278**,** 2075–2080. [DOI] [PubMed] [Google Scholar]
- 2.Morrison, D. K. & Davis, R. J. (2003) Annu. Rev. Cell Dev. Biol. 19**,** 91–118. [DOI] [PubMed] [Google Scholar]
- 3.Pawson, T. (2004) Cell 116**,** 191–203. [DOI] [PubMed] [Google Scholar]
- 4.Chang, L. & Karin, M. (2001) Nature 410**,** 37–40. [DOI] [PubMed] [Google Scholar]
- 5.Garrington, T. P. & Johnson, G. L. (1999) Curr. Opin. Cell Biol. 11**,** 211–218. [DOI] [PubMed] [Google Scholar]
- 6.Kolch, W. (2000) Biochem. J. 351**,** 289–305. [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, K. Y., Satterberg, B., Lyons, D. M. & Elion, E. A. (1994) Cell 78**,** 499–512. [DOI] [PubMed] [Google Scholar]
- 8.Elion, E. A. (2001) J. Cell Sci. 114**,** 3967–3978. [DOI] [PubMed] [Google Scholar]
- 9.Therrien, M., Wong, A. M. & Rubin, G. M. (1998) Cell 95**,** 343–353. [DOI] [PubMed] [Google Scholar]
- 10.Schaeffer, H. J. & Weber, M. J. (1999) Mol. Cell. Biol. 19**,** 2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison, D. K. (2001) J. Cell Sci. 114**,** 1609–1612. [DOI] [PubMed] [Google Scholar]
- 12.Muller, J., Ory, S., Copeland, T., Piwnica-Worms, H. & Morrison, D. K. (2001) Mol. Cell 8**,** 983–993. [DOI] [PubMed] [Google Scholar]
- 13.Roy, F., Laberge, G., Douziech, M., Ferland-McCollough, D. & Therrien, M. (2002) Genes Dev. 16**,** 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer, H. J., Catling, A. D., Eblen, S. T., Collier, L. S., Krauss, A. & Weber, M. J. (1998) Science 281**,** 1668–1671. [DOI] [PubMed] [Google Scholar]
- 15.Wunderlich, W., Fialka, I., Teis, D., Alpi, A., Pfeifer, A., Parton, R. G., Lottspeich, F. & Huber, L. A. (2001) J. Cell Biol. 152**,** 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teis, D., Wunderlich, W. & Huber, L. A. (2002) Dev. Cell 3**,** 803–814. [DOI] [PubMed] [Google Scholar]
- 17.Teis, D. & Huber, L. A. (2003) Cell. Mol. Life Sci. 60**,** 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276**,** 307–326. [DOI] [PubMed] [Google Scholar]
- 19.La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276**,** 472–494. [DOI] [PubMed] [Google Scholar]
- 20.Abrahams, J. P. & Leslie, A. G. W. (1996) Acta Crystallogr. D 52**,** 30–42. [DOI] [PubMed] [Google Scholar]
- 21.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54**,** 905–921. [DOI] [PubMed] [Google Scholar]
- 22.Engh, R. A. & Huber, R. (1991) Acta Crystallogr. A 47**,** 392–400. [Google Scholar]
- 23.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47**,** 110–119. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24**,** 946–950. [Google Scholar]
- 25.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277**,** 505–524. [DOI] [PubMed] [Google Scholar]
- 26.Sanner, M. F., Olson, A. J. & Spehner, J. C. (1996) Biopolymers 38**,** 305–320. [DOI] [PubMed] [Google Scholar]
- 27.Bashford, D. & Gerwert, K. (1992) J. Mol. Biol. 224**,** 473–486. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue, T., Adachi, M., Moriguchi, T. & Nishida, E. (2000) Nat. Cell Biol. 2**,** 110–116. [DOI] [PubMed] [Google Scholar]
- 29.Pei, J. & Grishin, N. V. (2001) Bioinformatics 17**,** 700–712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information