Stress granules at the intersection of autophagy and ALS (original) (raw)
. Author manuscript; available in PMC: 2017 Oct 15.
Abstract
Amyotrophic lateral sclerosis (ALS) is a progressive, fatal disease caused by loss of upper and lower motor neurons. The majority of ALS cases are classified as sporadic (80-90%), with the remaining considered familial based on patient history. The last decade has seen a surge in the identification of ALS-causing genes – including TARDBP (TDP-43), FUS, MATR3 (Matrin-3), C9ORF72 and several others – providing important insights into the molecular pathways involved in pathogenesis. Most of the protein products of ALS-linked genes fall into two functional categories: RNA-binding/homeostasis and protein-quality control (i.e. autophagy and proteasome). The RNA-binding proteins tend to be aggregation-prone with low-complexity domains similar to the prion-forming domains of yeast. Many also incorporate into stress granules (SGs), which are cytoplasmic ribonucleoprotein complexes that form in response to cellular stress. Mutant forms of TDP-43 and FUS perturb SG dynamics, lengthening their cytoplasmic persistence. Recent evidence suggests that SGs are regulated by the autophagy pathway, suggesting a unifying connection between many of the ALS-linked genes. Persistent SGs may give rise to intractable aggregates that disrupt neuronal homeostasis, thus failure to clear SGs by autophagic processes may promote ALS pathogenesis.
Keywords: Amyotrophic lateral sclerosis, autophagy, motor neuron diseases, neurodegeneration, neuromuscular diseases, P-bodies, protein degradation pathways, rapamycin, stress granules
1. Introduction
ALS is the most common adult-onset motor neuron disorder, typically striking in the fifth to seventh decades of life, though juvenile disease also exists. It is characterized by rapid degeneration of motor neurons, and subsequent atrophy of innervated muscle groups. Death is generally secondary to failure of respiratory muscles (Ravits and La Spada, 2009; Turner et al., 2013). ALS occurs globally in all races, ethnic and socioeconomic groups. There are no pharmacological interventions for the underlying molecular pathogenesis (Miller et al., 2012).
Neurodegenerative diseases often share two clinicopathological properties. First, by definition, they affect highly translationally-active neurons preferentially to other cell types; second, they are often associated with mutations in components of protein-quality control (PQC) (Hetz et al., 2009; Kabashi and Durham, 2006). It is perhaps not surprising that perturbations to PQC pathways would have significant impact on cells that are both especially translationally active and long-lived. In the case of ALS, a number of different proteins and metabolic pathways have been linked to pathogenesis, but issues of proteostasis (e.g. protein folding, aggregation and quality-control) appear to be the most common pathogenic theme (Andersen and Al-Chalabi, 2011; Renton et al., 2014). Many ALS-associated proteins have intriguing properties with regard to self-association, aggregation-propensity, and interaction with cytoplasmic stress granules (SGs) (Bosco et al., 2010; Colombrita et al., 2009; Daigle et al., 2013; Dewey et al., 2012; Guo et al., 2011; Liu-Yesucevitz et al., 2010; McDonald et al., 2011; Sun et al., 2011; Vance et al., 2013). Other ALS-associated proteins have explicit functions in PQC pathways, including autophagy. Below we discuss the intersections between protein aggregation, SGs and autophagy in ALS pathogenesis.
2. Stress Granules – Discrete Stress-Induced Cytoplasmic Sites of Ribonucleoprotein Accumulation
Ribonucleoprotein (RNP) granules are cellular sites dedicated to RNA processing. Well-characterized types of RNP granules include transport RNPs, processing bodies (P-bodies) and stress granules (SGs); all of which have distinct roles in mRNA regulation (Anderson and Kedersha, 2008; Kedersha et al., 2005). Transport RNPs ensure localized neuronal translation of RNAs by facilitating their transport along cytoskeletal elements while maintaining temporary translational repression (Kiebler and Bassell, 2006). SGs and P-bodies are phenotypically similar, non-membrane-bound, discrete cytoplasmic structures visible by light microscopy (Buchan and Parker, 2009; Guil et al., 2006). They contain many of the same proteins, but each has exclusive constituents; P-bodies are enriched for proteins involved in RNA degradation, while SGs are preferentially composed of translation initiation factors (Reineke and Lloyd, 2013). Thus, P-bodies are classified as foci of RNA breakdown and turnover, and SGs are thought to be sites of paused translation initiation and global translation repression (Anderson and Kedersha, 2008; Li et al., 2013; Parker and Sheth, 2007; Thomas et al., 2011). Both SGs and P-bodies have the ability to exchange mRNAs with bulk cytoplasm depending on cellular conditions (Decker and Parker, 2012).
The formation of SGs is believed to be a conserved, protective response to various cell stresses. Some example stresses include: oxidative (Anderson and Kedersha, 2002; Anderson and Kedersha, 2008; Bosco et al., 2010; Daigle et al., 2013), mitochondrial (Buchan et al., 2011; Chalupnikova et al., 2008; Stoecklin et al., 2004), proteasomal (Fournier et al., 2010; Mazroui et al., 2007) and viral (Emara and Brinton, 2007; Raaben et al., 2007). Interestingly, many external stimuli/stressors do not induce SG formation in mammalian cells, suggesting SGs are a specific response not common to all stress (Kedersha et al., 1999). There are several proposed means by which they exert their protection. SGs may offer direct protection for certain mRNAs from damaging stressors (Kedersha and Anderson, 2002). Alternatively, SGs may sequester unwanted mRNAs, preventing their translation, such as viral RNAs during infection (Beckham and Parker, 2008), or less critical mRNAs during stress conditions (Li et al., 2013; Unsworth et al., 2010; Wolozin, 2012). Thus, SGs may offer prioritization of specific protein products (Scheu et al., 2006). More generally, SGs may decrease protein stress through the global repression of translation by binding mRNAs that would otherwise be translated. The apparently causal role of phosphorylated eIF2α in facilitating SG formation supports this hypothesis, as eIF2α has an established role in translation repression (Kedersha et al., 1999).
SGs contain polyadenylated mRNAs, translation initiation factors, small ribosome subunits and several RNA-binding proteins (Anderson and Kedersha, 2008; Daigle et al., 2016). Putative SG functions all demand the intimate association of these components within a discrete cytosolic space, removed from the majority of cellular machinery. Importantly, RNPs containing specific mRNAs are critical for transport and localized translation in neuronal dendrites. Different types of RNPs (SG, P-body, transport) share similar components (Decker and Parker, 2012), thus neurons may be particularly sensitive to disruption of RNP homeostasis.
SGs are assembled and disassembled through the formation and dissolution of a “liquid-liquid phase-separated state”, in which the components that form SGs “demix” from the bulk solution to create a unique micro-environment. This transient phase-separated state presumably allows for a rapid, reversible response to stress (Elbaum-Garfinkle et al., 2015; Lin et al., 2015; Molliex et al., 2015). Several proteins, as well as mRNA, are implicated in driving the physical phase separation (Zhang et al., 2015). The protein TIA1, for example, is critical to the early stages of SG assembly. TIA1 has three amino-terminal RNA recognition motifs and a carboxy-terminal domain, which has low-complexity composition similar to the intrinsically-disordered domains that drive yeast prion proteins to form self-propagating amyloid fibrils. In fact, a peculiarity about many of the proteins that are both linked to ALS and SG formation is they possess yeast prion-like domains (discussed below). Substitution of this domain of TIA1 with the actual prion-forming domain of yeast prion protein Sup35, results in a restoration of SG formation, which is lost following native TIA1 prion-like domain deletion (Gilks et al., 2004).
The evolutionary conservation of SGs in eukaryotic cells indicates that they serve critical cellular functions. However, the promiscuous, en masse sequestration of mRNA transcripts in cytosolic granules would clearly have dramatic implications for cell survival. As with any metabolic pathway, SG formation must be balanced with mechanisms to ensure disassembly. Intracellular component turnover relies on multiple pathways, including autophagy and ubiquitin-mediated proteolysis (Ciechanover, 1994; Cuervo et al., 2005; Glickman and Ciechanover, 2002; Levine et al., 2008; Reed, 2003).
3. Autophagy – a Mechanism for Clearing Protein Aggregates
Autophagy is a well-studied system for disposal of a variety of intracellular species. First identified in the context of hormone studies in rats, it has since been appreciated as a mechanism for nearly all eukaryotic cells to dispose of a wide variety of intracellular components deemed unnecessary or maladaptive (Deter et al., 1967; Gomes and Scorrano, 2013). Autophagy involves an autophagosome, a double-membrane bound structure that forms from extant membrane-bound organelles (Chan and Tang, 2013). The autophagosome engulfs regions of the cytosol and fuses with the lysosome to become the autophagolysosome where its contents are catabolized (Deter et al., 1967; Gomes and Scorrano, 2013). This membrane-enclosed mechanism is sometimes more specifically called macroautophagy to distinguish it from other types of autophagic processes. Not all autophagy processes employ the formation of a new autophagosome. Chaperone-mediated autophagy involves the direct targeting of substrates to the lysosome via chaperone intermediates and then active translocation of the substrates across the lysosomal membrane. Microautophagy involves the direct engulfment of cytoplasmic content by invagination of the lysosomal membrane (Mizushima et al., 2008).
Autophagy has historically been considered a non-selective process of cellular digestion, in contrast to the ubiquitin-proteasome system (Gomes and Scorrano, 2013; Meijer et al., 2007). In general, less selectivity may be a consequence of the extremely diverse homeostatic functions of autophagy (Cuervo et al., 2005). Gross protein turnover via autophagic pathways, for instance, is likely a critical means to balance anabolic pathways with a free source of available amino acids (Harris et al., 2004; Meijer, 2009). This last point is strongly supported by the observation that amino acid deprivation is a very potent inducer of autophagocytic pathways in mammalian cells (Ghislat et al., 2015; Mortimore and Schworer, 1977). However recent findings suggest a greater degree of specificity and overlap with the proteasome system (discussed below).
a. Selective Macroautophagy
Despite the historical perspective of autophagy being a generic pathway, recent work suggests importance of a selective function with implications for human disease (De Duve and Wattiaux, 1966; Gomes and Scorrano, 2013). Specifically, ubiquitin tagging may have a role in targeting selective autophagy (Heo et al., 2015; Kirkin et al., 2009). Moreover, selective autophagy is particularly well-suited to clearance of large (> 2 um) protein aggregates and RNP granules that need to be disassembled, in part because large protein aggregates and persistent RNP granules are known to be resistant to ubiquitin-proteasome degradation (Venkatraman et al., 2004).
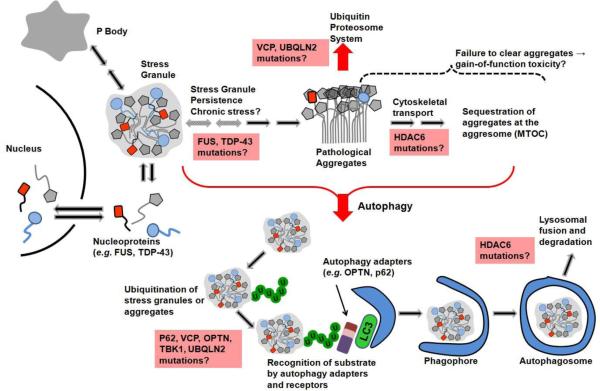
The general model of selective autophagic engulfment of substrate is shown in Figure 1. Initially, the phospholipid-conjugated LC3 protein (Atg8 in yeast) facilitates the formation of membranous phagophore structures that recognize the targeted material through various acceptor and receptor proteins. The phagophore envelops the substrate to form an autophagosome that fuses with the lysosome, forming the autophagolysosome (Klionsky et al., 2014). This structure hosts proteolysis, facilitating amino acid export to the cytosol for re-use (Harris et al., 2004; Klionsky et al., 2014; Meijer, 2009).
Figure 1.
Schematic showing the connection between stress granules, protein aggregation and autophagy. Together, these structures and processes are commonly dysfunctional in ALS. Boxes in pink indicate mutant genes that could be affecting the indicated process.
Many ubiquitinating factors and adapter proteins enable specificity in selective macroautophagy. For example, ubiquillin-2 is implicated in facilitating interaction between an autophagosome and its target (N'Diaye et al., 2009; Rothenberg et al., 2010). Proteins p62 and NBR1 function as selective cargo receptors, linking ubiquitin tags with autophagosome receptors (Lamark et al., 2009; Pankiv et al., 2007). NBR1 in particular has been shown to mediate selective autophagy in plant cells dealing with otherwise intractable protein aggregates (Zhou et al., 2013a). Many of these adaptor proteins recognize the phagophore through “LC3-interacting region” motifs in their peptide sequence (Birgisdottir et al., 2013). Moreover, mutations in several adapters are linked to sub-types of ALS (discussed below).
b. Autophagic Clearance of Protein Aggregates and Stress Granules
Autophagy has been linked to the clearing of the large protein aggregates that feature prominently in several neurodegenerative diseases. For example, the protein alpha-synuclein forms intracellular fibrils in neurons of patients with Parkinson’s disease (Baba et al., 1998). Formation of these fibrous aggregates correlates with a massive increase in the presence of autophagosomes in cultured cells (Shintani and Klionsky, 2004; Stefanis et al., 2001). This could be a consequence of increased formation or impaired clearance of autophagosomes (Shintani and Klionsky, 2004). In a similar fashion, aggregates of mutant huntingtin protein, which are linked to Huntington’s disease, are also associated with an abundance of autophagosomes (Kegel et al., 2000). In the case of both huntingtin and alpha-synuclein, promotion of autophagy via pharmacological stimulation by rapamycin results in an increase in autophagic degradation of the two proteins, as well as a corresponding reduction in toxicity (Ravikumar et al., 2002; Shintani and Klionsky, 2004; Webb et al., 2003).
Similarly, autophagy is involved in the clearance of RNP granules. Buchan and colleagues demonstrated that SG breakdown in particular was dependent on selective autophagy. Strikingly, and very similarly to previous experiments with huntingtin and alpha-synuclein, rapamycin administration promoted SG clearance, while selective autophagy inhibitors had the opposite effect (Buchan et al., 2013). Likewise, when autophagy-initiating proteins (Atg proteins) were partially deleted in yeast, cytosolic SG persistence was dramatically increased. Beyond the canonical Atg family, an additional protein of interest is valosin-containing protein (VCP/p97), which appears to specifically target SGs to autophagic pathways for degradation (Buchan et al., 2013; Ju et al., 2009). When VCP is silenced with siRNA or with specific chemical inhibitors in stressed HeLa cells, SGs accumulate (Buchan et al., 2013).
Other mechanisms are linked to SG clearance, such as decapping SG mRNAs, returning sequestered mRNAs to active translation, and direct 5’ to 3’ digestion of constituent mRNA molecules. Such fates likely involve dynamic handling of mRNAs among P-bodies, SGs, and polysomes (Bhattacharyya et al., 2006; Brengues et al., 2005; Buchan et al., 2013; Sheth and Parker, 2003). Intriguingly, Buchan and colleagues showed that upon inactivation of these pathways via genetic knockout, targeting of SGs to autophagy appeared to increase (Buchan et al., 2013). The metabolic and homeostatic mechanisms of mRNA translation and degradation likely reflect a very dynamic, complex cycle, of which autophagy is a critical player.
c. Autophagy in Animal Models of ALS
Connections between autophagy and ALS have been observed in animal models. The first gene linked to familial ALS was superoxide dismutase 1 (SOD1). Induction of the autophagy pathway has been observed in a transgenic mouse model of SOD1-ALS, as well as spinal cord tissues from SOD1-ALS patients, supporting the idea that pathogenic mutations in SOD1 impair, or require, autophagy pathways (Morimoto et al., 2007; Sasaki, 2011). Since autophagy is critical to managing the burden of misfolded and toxic proteins, it is not unexpected that perturbed autophagy has also been observed in the postmortem tissues of sporadic ALS patients (Morimoto et al., 2007). Retinoic acid has been demonstrated to act on PQC pathways including autophagy, and interestingly, control rats maintained on a retinoid-free diet show ALS-like symptoms (Anguiano et al., 2013; Castillo et al., 2013; Corcoran et al., 2002; Kolarcik and Bowser, 2012; Rajawat et al., 2010; Rajawat et al., 2011; Riancho et al., 2015; Riancho et al., 2016). Furthermore, treatment with Bexarotene, a retinoid-X receptor agonist, or induction of autophagy in a SOD1 mutant mouse model drastically delayed motor symptoms and ALS pathology suggesting that targeting PQC machinery could be a good therapeutic target for SOD1-ALS (Castillo et al., 2013; Crippa et al., 2010; Riancho et al., 2015; Riancho et al., 2016).
Autophagic dysfunctions have also been implicated in subtypes of ALS caused by mutations in TDP-43 and FUS (discussed more below). It has been suggested that TDP-43 is involved in regulating autophagy in general and particularly autophagosomal and lysosomal biogenesis (Bose et al., 2011; Filimonenko et al., 2007; Xia et al., 2016; Ying et al., 2016). Interestingly, TDP-43 is managed by both proteasome and autophagy pathways where the soluble form of TDP-43 is degraded by the proteasome; and oligomeric and aggregated forms of TDP-43 are cleared by autophagy (Xia et al., 2016). Given that TDP-43 is a highly aggregation-prone protein, it is not surprising to observe that impaired turn-over of TDP-43 directly correlates with motor dysfunctions and reduced autophagy-related proteins in a mouse model of TDP-43 proteinopathy (Caccamo et al., 2015). Similarly, pathogenic mutations in FUS have been shown to impair the autophagy pathway in cellular models and ALS patient cells (Soo et al., 2015). Importantly, pharmacological induction of autophagy ameliorates neurodegenerative symptoms as well as TDP-43 protein mislocalization in Drosophila and rodent models of TDP-43 proteinopathy, further supporting a notion that autophagy could be a potential therapeutic target for ALS (Barmada et al., 2014; Caccamo et al., 2009; Cheng et al., 2015; Wang et al., 2012).
4. Intersections Between ALS, Stress Granules and Autophagy
A curious feature of many SG proteins is low-complexity domains resembling the prion-forming domains of certain yeast proteins. Moreover, many SG proteins are also found in the pathological inclusions in ALS patients’ motor neurons. In fact, many ALS-associated proteins have significant structural or functional overlap with SG proteins. This predicted overlap is due to the presence of RNA-binding domains and prion-like domains (Arai et al., 2006; Ju et al., 2011; Kwiatkowski et al., 2009; Maekawa et al., 2009; Neumann et al., 2006; Udan and Baloh, 2011; Vance et al., 2009), as well as observed functional interactions in experimental models (Acosta et al., 2014; Baron et al., 2013; Daigle et al., 2016; Dewey et al., 2012; Di Salvio et al., 2015; Lenzi et al., 2015; McDonald et al., 2011; Parker et al., 2012; Sama et al., 2013). These observations encourage the idea that RNA homeostasis and prion-like mechanisms of protein self-association are critical to ALS pathogenesis.
Prion-like mechanisms have been implicated in a variety of neurodegenerative diseases, most notoriously the transmissible spongiform encephalopathies (TSEs) (Gielbert et al., 2015). In TSEs, auto-catalytic processes drive the propagation of misfolded protein isoforms within the nervous tissue of infected animals. This propagation results in the accumulation of insoluble protein aggregates that spatially correlates with cell death (Jeffrey et al., 1995). Soluble misfolded oligomeric species, resistant to proteolytic degradation, presumably precede the formation of macroscopic aggregates (Bessen et al., 1997; Jeffrey et al., 1995). Together, these oligomeric and larger aggregates are thought to exert cellular toxicity. Thus, the fact that ALS-linked SG proteins have prion-resembling domains that are aggregation-prone suggests that ALS could have a prion-like mechanism in which toxic misfolded proteins self-propagate.
Given the potential that prion-like mechanisms underlie disease pathologies, as well as SG dynamics, interest in the intersection between disease and SG homeostasis is unsurprisingly robust (Acosta et al., 2014; Baron et al., 2013; Bentmann et al., 2013; Daigle et al., 2016; Dewey et al., 2010; Dewey et al., 2012; Di Salvio et al., 2015; Lenzi et al., 2015; Li et al., 2013; McDonald et al., 2011; Parker et al., 2012; Sama et al., 2013; Walker et al., 2013) . The high local concentration of proteins with prion-like domains in SGs is thought to facilitate the transient phase-separated granule structure. However, self-association between these domains is not static (Burke et al., 2015). Instead, under certain conditions, the normal internal interactions are hypothesized to evolve into terminally-aggregated species that are not easily cleared by the cell. Specifically, an inappropriate persistence, or excessive mRNA binding, of SGs may be a critical element in neurodegeneration. If SG pathology overwhelms autophagy pathways, cellular pathology may follow. Potential consequences of ALS-associated genetic mutations are listed in Table 1 and discussed below.
Table 1.
Summery of ALS linked proteins, their broad functions and potential link to the disease
| Gene | Protein Function | Proposed Link to Disease | Stress Granule Component | Present in Pathological Inclusions |
|---|---|---|---|---|
| ANG | Neuroprotection and angiogenesis | Enhanced susceptibility to cellular stress | ||
| ATXN2 | Unknown | Persistent cytosolic aggregates Cytotoxic dipeptides, | Yes | Yes |
| C9ORF72 | Facilitates intracellular endosomal trafficking | impaired autophagy, RNA-mediated aberrant protein sequestration | Ambiguous2 | |
| CHMP2B | Facilitation of endosomal trafficking | Impaired autophagy | ||
| FUS | DNA/RNA homeostasis | Persistent cytotoxic RNP aggregates, loss of genome integrity | Yes | Yes |
| HDAC6 | Facilitates autophagolysosome maturation | Impaired autophagy | Yes | |
| MATR3 | RNA/DNA homeostasis | Persistent cytotoxic RNP aggregates, loss of genome integrity | Yes | |
| NEFH | Structural neurofilament | Loss of neuronal function | ||
| OPTN | Facilitates autophagy | Impaired autophagy | Yes | |
| P62/SQSTM1 | Autophagy adapter | Impaired autophagy | Yes | |
| PFN-1 | Actin binding protein | Persistent cytotoxic RNP aggregates, impaired cytoskeletal transport | Yes | Yes |
| PRPH | Structural neurofilament | Loss of neuronal function | ||
| SOD1 | Reducing enzyme for superoxide radicals | Persistent cytotoxic aggregates | Yes | |
| TBK-1 | Facilitates autophagy | Impaired autophagy | Ambiguous1 | |
| TDP-43 | DNA/RNA homeostasis | Persistent cytotoxic RNP aggregates, loss of genome integrity | Yes | Yes |
| UBQLN-2 | Facilitates autophagy/proteasome degradation | Impaired autophagy/proteasome function | Yes | |
| VAPB | Facilitates unfolded protein response in protein quality control | Persistent cytotoxic aggregates | Yes | |
| VCP | Facilitates autophagy | Impaired autophagy | Yes |
a. FUS, TDP-43, Matrin-3 and hnRNPA2/A1 as Stress Granule-Associated Proteins
The proteins fused-in-sarcoma (FUS) and TAR DNA binding protein-43 kDa (TDP-43) are well-established DNA/RNA-binding proteins that are implicated in similar functions, including transcription regulation and RNA splicing (Lagier-Tourenne et al., 2010; Lagier-Tourenne et al., 2012). They have similar structures, including nucleic acid binding domains and prion-like domains of low intrinsic complexity. Mutations of both proteins, characteristically in domains regulating subcellular localization, are correlated with development of ALS. In diseased cells, a characteristic pattern of cytoplasmic foci and structural association with SGs has been widely reported in diverse organisms (Acosta et al., 2014; Arai et al., 2006; Barmada et al., 2010; Baron et al., 2013; Bentmann et al., 2013; Dewey et al., 2012; Di Salvio et al., 2015, Daigle, 2016 #305; Johnson et al., 2008; Ju et al., 2011; Kwiatkowski et al., 2009; Lenzi et al., 2015; Li et al., 2010; Maekawa et al., 2009; McDonald et al., 2011; Neumann et al., 2006; Parker et al., 2012; Sama et al., 2013; Sun et al., 2011; Udan and Baloh, 2011; Vance et al., 2009; Walker et al., 2013).
Both FUS and TDP-43 have default nuclear localization consistent with their RNA-related functions. They also demonstrate an ability to regulate their own expression (Avendano-Vazquez et al., 2012; Budini and Buratti, 2011; D'Alton et al., 2015; Dormann and Haass, 2011; Zhou et al., 2013b). An abundance of intranuclear FUS causes exon 7 skipping in FUS transcription, driving FUS transcripts to nonsense mediated decay and preventing pathological protein buildup (Zhou et al., 2013b); an accumulation of FUS outside of the nucleus could thus result in self-perpetuating high expression, exacerbating the potential for misfolding in the cytoplasm. FUS is also implicated in DNA repair. It accumulates at sites of DNA damage and is implicated in regulating DNA repair pathways (Hicks et al., 2000; Lagier-Tourenne et al., 2010; Mastrocola et al., 2013; Patel et al., 2015a; Rulten et al., 2014; Sama et al., 2013; Wang et al., 2013). This self-association is likely mediated by its prion-like domain, leading to intranuclear phase-separated droplets that facilitate DNA repair by recruiting specific proteins (Patel et al., 2015a; Rulten et al., 2014).
Despite the intranuclear roles described above, there is very clearly a cytosolic role for both proteins, as evidenced by the presence of a nuclear export sequence (Lorenzo-Betancor et al., 2014). This cytoplasmic shuttling follows various types of cell stress (Sama et al., 2013), perhaps facilitating physiological SG metabolism. FUS, for example, localizes to the cytoplasm following post-translational modifications, including methylation and phosphorylation (Deng et al., 2014; Scaramuzzino et al., 2013; Tradewell et al., 2011). Normally, an intact nuclear localization signal (NLS) likely ensures a prompt return to the nucleus, especially in the context of SG disassembly (Sama et al., 2013). In the case that the proteins contain an NLS mutation, persistent cytosolic localization of FUS or TDP-43 results (Acosta et al., 2014; Barmada et al., 2010; Dormann and Haass, 2011). The subsequent cytosolic co-localization with SGs is perhaps a consequence of FUS/TDP-43 having prion-like domains similar to the domains found in many SG constituent proteins (Gilks et al., 2004; Udan and Baloh, 2011). Co-localization may modify the properties of SGs. Both FUS and TDP-43 have domains that would presumably bind resident mRNAs (Schwartz et al., 2013; Ugras and Shorter, 2012). Thus, cytoplasmic localization of both proteins may drive inappropriate interactions within SGs. Subsequent modification of SG homeostasis, such as excessively promiscuous SG-mRNA binding or inappropriate persistence of SGs, may overwhelm regulatory processes and lead to intractable aggregation. Such a model is especially attractive in light of the biophysical properties of liquid-liquid phase-separated FUS droplets (Burke et al., 2015; Patel et al., 2015a; Zhang et al., 2015), which have been shown in vitro to evolve into irreversible aggregates in a time-dependent manner. If FUS and SGs behave so similarly in isolation, such mechanisms may drive pathology when driven into cytosolic juxtaposition by ALS-associated mutations.
The interaction of FUS and TDP-43 with cytosolic SGs represents a putative cytosolic gain-of-function toxicity pathway. This hypothesis is supported by a variety of studies (Ju et al., 2011; Kryndushkin et al., 2011; Lanson et al., 2011; Scaramuzzino et al., 2013; Sharma et al., 2016; Sun et al., 2011). In model systems, ectopic expression of human FUS or TDP-43 results in cytoplasmic aggregation into discrete FUS or TDP-43-positive foci, as well as cellular toxicities. Toxicity can be partially ameliorated by disruption of either the prion-like domain or the RNA-binding domains (Daigle et al., 2013; Sun et al., 2011), suggesting that both of these domains are critical to exerting toxicity. Interruption of the prion-like domain may interfere with protein association, while the RNA-binding interruption may prevent RNA from being irreversibly sequestered in the aggregates. Moreover, disruption of RNA-binding capacity may hamper further protein association by preventing RNA-nucleation of protein interactions (Schwartz et al., 2013). Importantly, in a transgenic FUS mouse line, postnatal knockdown of FUS did not promote motor neuron degeneration, whereas expression of FUS mutants was dominant negative for motor neuron survival ((Sharma et al., 2016).
While gain-of-function toxicity appears to be a major part of the FUS/TDP-43 story, deletion of these proteins has also been shown to have deleterious consequences for mammalian cells (Ayala et al., 2008; Chiang et al., 2010; Fiesel and Kahle, 2011; Hicks et al., 2000; Kuroda et al., 2000; Orozco et al., 2012; Sama et al., 2014; Shan et al., 2010). Thus, a loss of intranuclear function must be considered as a contributor to pathology. First, it may lead to a loss of nucleic-acid regulation as well as other intranuclear functions ascribed to both FUS and TDP-43 (Avendano-Vazquez et al., 2012; Budini and Buratti, 2011; D'Alton et al., 2015; Dormann and Haass, 2011; Hicks et al., 2000; Lagier-Tourenne et al., 2010; Mastrocola et al., 2013; Patel et al., 2015a; Rulten et al., 2014; Sama et al., 2014; Wang et al., 2013; Zhou et al., 2013b). Second, the loss of autoregulation of expression could lead to unchecked transcription, cytosolic translation, and continued cytoplasmic aggregation with SGs as described above. Specifically, TDP-43-linked ALS cases were recently shown to exhibit an abundance of so-called cryptic exons in nervous system tissue (Pochet et al., 2015). TDP-43 has a role, similar to FUS autoregulation, to repress expression of the cryptic exons. Loss of intranuclear TDP-43 to cytoplasmic SGs/aggregates may lead to excessive cryptic exon expression, driving nervous system pathology in ALS patients (Ling et al., 2015).
Additional ALS-linked proteins Matrin-3 and hnRNPA2/A1 have DNA/RNA-binding properties. Matrin-3 is, like FUS and TDP-43, a resident of the nucleus and is implicated in various RNA-related functions. Matrin-3 was shown to interact with TDP-43 in cytosolic aggregates in spinal neurons. Also, ALS-linked mutations appear to modulate the degree of this interaction (Gallego-Iradi et al., 2015; Johnson et al., 2014; Millecamps et al., 2014). While less well characterized than FUS and TDP-43, the potential for Matrin-3 to interact with SGs, perhaps even via a direct association with TDP-43, would have obvious functional implications.
In the case of hnRNPA2/A1, we again see nucleic acid binding capacity joined by the presence of a prion-like domain. Proteins harboring mutations linked to ALS phenotypes display an increased in vitro aggregation propensity relative to wild-type, capacity to propagate as a yeast prion when swapped with the native Sup35 prion-domain in yeast, as well as an enhanced recruitment to stress granules (Kim et al., 2013). All of these observations strongly support the importance of SG association and protein self-association in facilitating pathways of toxicity.
While the roles of FUS, TDP-43, Matrin-3, and hnRNPA2/A1 in ALS will likely continue to be clarified, there are intriguing hints of a link between these proteins and SGs and autophagy. Indeed, the clearance of FUS and TDP-43 aggregates by autophagy has been reported (Caccamo et al., 2009; Ryu et al., 2014; Wang et al., 2010). Recent reports suggest that autophagy of FUS aggregates can be enhanced by overexpression of the critical autophagy protein Rab1 (Soo et al., 2015). This suggests aggregates are overwhelming autophagic machinery in degenerating motor neurons.
b. C9ORF72 and VCP as Regulators of Autophagy
Beyond the prion-like nucleic acid binding proteins discussed thus far, a number of other proteins have been associated with ALS pathogenesis. These proteins also offer pathogenetic connections with SGs and autophagy. In fact, the most common genetic abnormality linked with familial ALS is a disordered intron in the nucleic acid sequence located on chromosome 9, open reading frame 72 (C9ORF72). Specifically, ALS has been associated with a hexanucleotide repeat expansion (HRE) GGGGCC inherited in an autosomal dominant fashion (DeJesus-Hernandez et al., 2011; Renton et al., 2011). The potential avenues by which the HRE may drive neurotoxicity are numerous, and include both nucleic acid-driven and protein-driven explanations.
It is yet not entirely clear how HRE causes motor neuron degeneration associated with ALS. It has been demonstrated that the C9 expanded hexanucleotides abnormally bind and sequester RNA binding proteins in nuclear foci in ALS patient neurons (Donnelly et al., 2013). Expression of the HRE is enough to cause the formation of SGs under stress conditions. These SGs sequester other disease-associated proteins such as hnRNPA1 and hnRNPA2/B1 (Farg et al., 2014). Moreover, repeat expression leads to elevated levels of a protein called p62 in motor neurons differentiated from C9 induced pluripotent stem cells (Almeida et al., 2013). Importantly, p62 has been implicated as an adaptor protein for selective autophagy of protein aggregates (Pankiv et al., 2007). In addition to this, C9 hexanucleotide repeat causes increased sensitivity to cellular stress induced by autophagy inhibitors, further supporting the role of autophagy in C9ORF72 mediated neurodegeneration (Almeida et al., 2013).
In addition to RNA foci, C9ORF72 repeat transcripts can produce unconventional, repeat-associated non-ATG mediated (RAN) translation products. These RAN peptides, translation of which occurs without a start codon, have been observed in ALS patient tissues as well as in different experimental models (Gendron et al., 2013; Mori et al., 2013; Wen et al., 2014). RAN translation can occur in all possible reading frames, producing dipeptide repeat proteins in the sense and antisense direction (antisense: poly-PR and poly-PA; sense: poly-GA and poly-GR; both sense: poly-GP). These C9 RAN products have been examined in cellular and animal models where it has been shown that GR and PR dipeptides are extremely toxic as well as being especially aggregation-prone (Mizielinska et al., 2014; Wen et al., 2014). Additionally, GA proteins have been shown to form neuronal inclusions that stain positive for the autophagy-mediator p62 (Schludi et al., 2015). Proteins known to sequester in SGs such as PABP-1 localize to TDP-43 positive inclusions more frequently in patients carrying expansions in C9ORF72 (McGurk et al., 2014).
Interestingly, it has been demonstrated that the C9ORF72 translation product colocalizes with proteins implicated in regulating autophagy and endocytic transport such as Rab1, Rab5, Rab7 and Rab11 in mammalian neuronal cells and human spinal cord motor neurons. Perhaps most intriguing, depletion of C9ORF72 led to an increase in autophagosome dysregulation, specifically by disrupting function of receptors Trk-beta and LC3. Moreover, pharmacological inhibition of the 26S proteasome in addition to C9ORF72 overexpression led to an accumulation of SGs (Farg et al., 2014). These findings suggest that C9ORF72 is a regulator of endosomal trafficking, functions with critical importance to autophagy. This role in regulating endosomal and vesicular trafficking is further evident from the fact that C9ORF72 colocalizes with other ALS-linked proteins in ubiquilin-2 and LC3-positive vesicles. Protein interaction by C9ORF72 also includes heterogeneous nuclear ribonucleoproteins, Matrin 3, hnRNPA2/B1 and hnRNPA1 (Donnelly et al., 2013; Farg et al., 2014; Haeusler et al., 2014).
The link between dysregulated autophagy, SGs, and development of an ALS phenotype is further enhanced by studies of Valosin-containing protein, VCP. Ju and colleagues showed the importance of intact VCP in allowing for normal autophagic pathways (Ju et al., 2009). Specifically, this study demonstrated the accumulation of both autophagosomes arrested before association with lysosomes, and TDP-43 positive protein inclusions in cells depleted of wild type VCP (Ju et al., 2009). Moreover, Buchan et al. showed that VCP helps target SGs to the autophagosome for disassembly, while Cherkasov and workers showed that this disassembly was chaperone mediated (Buchan et al., 2013; Cherkasov et al., 2013). Surprisingly, another report showed that impaired VCP led to a reduction in the size and formation of SGs, suggesting that VCP may play an important role in both SG assembly and targeting to the autophagosome (Seguin et al., 2014). This same study showed a concomitant accumulation of ubiquitinated proteins with a reduction in SG formation.
c. Optineurin, TBK1 and Ubiquilin-2 as Modulators of Stress Granule Clearance by Autophagy
Optineurin is an additional protein implicated in both ALS (Maruyama et al., 2010) and autophagic pathways, specifically by binding to the autophagosome receptor LC3 (N'Diaye et al., 2009; Rothenberg et al., 2010; Wong and Holzbaur, 2014). Optineurin has been shown to act as an adaptor for selective autophagy, similarly to p62 previously described (Pankiv et al., 2007; Wild et al., 2011). It is well documented that optineurin binds LC3, and that this association facilitates mitophagy (autophagy of mitochondria). A protein called Parkin, which has been associated with Parkinson’s disease, enhanced the stability of the association between optineurin and mitochondria (Wong and Holzbaur, 2014). Moreover, Shen and colleagues demonstrated that overexpression of wild-type optineurin facilitated the autophagy-mediated clearance of cytosolic aggregates containing the aggregation-prone protein mutant huntingtin (Shen et al., 2015).
Optineurin has a robust aggregation propensity in addition to its ability to regulate autophagy (Kryndushkin et al., 2012). The case of optineurin actually offers a very compelling example of how complicated is the interaction between aggregation propensity and autophagy. Mutations in the ubiquitin-binding domain of optineurin have been shown to reduce its ability to perform its autophagic function (Shen et al., 2015). Mutations in this domain include the E478G mutation linked to ALS (Maruyama et al., 2010). Optineurin E478G, despite its reduced ability to bind ubiquitin, still has the potential to associate with and trap wild-type optineurin in the cytosol. This prevents the association of wild-type optineurin to autophagic machinery, and therefore the appropriate maturation of autophagosomes. The authors used LC3 turnover as a measure of autophagic flux to show that this wild-type optineurin trapping effect of E478G led to measurable decreases in autophagy for the cell, as well as a subsequent increase the abundance of cytosolic aggregates (Shen et al., 2015).
As described above, optineurin may offer an example of how aggregation propensity and dysregulation of autophagy are directly related in the cell. If a key function of autophagy is to clear intracellular protein aggregates, it makes sense that major regulators of autophagy would have the capacity to interact with protein aggregates. It is perhaps via this property of self-association that proteins are able to physically interact with aggregates, and then modulate autophagy via other protein domains.
Optineurin is also implicated in ALS pathogenesis as a result of interaction with kinase pathways. Phosphorylation is perhaps the most widely used cellular signal for countless metabolic processes. Unsurprisingly, kinases like TBK1 have been implicated in diverse human pathologies from hepatic dysfunction to neurodegeneration (Bonnard et al., 2000; Freischmidt et al., 2015). Indeed, TBK1 was first linked to ALS in 2015 (Cirulli et al., 2015). Subsequent studies showed that TBK1-linked ALS was likely due to haploinsufficiency of this kinase, an observation that builds on many of the ideas so far discussed in this review (Freischmidt et al., 2015). Perhaps the simplest explanation for TBK1-mediated ALS pathogenesis arises from the observation that TBK1 phosphorylates optineurin in cell models (Heo et al., 2015; Morton et al., 2008; Wild et al., 2011). Indeed, this phosphorylation has been shown to enhance autophagy in the context of autophagic anti-microbial innate immunity (Pilli et al., 2012; Radtke et al., 2007; Wild et al., 2011). Despite the differing context, there is clearly the potential for insufficient phosphorylation of optineurin to impair autophagosome formation and maturation.
Optineurin and TBK1 have also been discovered co-localizing in protein aggregates, and the phosphorylation of optineurin at serine 177 has been shown to be critical to its function in mediating clearance of aggregated proteins via autophagy (Korac et al., 2013). Again, the aggregation propensity of optineurin may facilitate its role as a modulator of autophagy-mediated aggregate clearance, and TBK1 association with optineurin in aggregates may be an additional factor necessary for this process. Specifically, the phosphorylation event may drive the conversion of optineurin from simply a ubiquitin/aggregate-associated protein to an LC3-mediated autophagy receptor (N'Diaye et al., 2009; Rothenberg et al., 2010; Wong and Holzbaur, 2014). This would specifically facilitate the autophagy of ubiquitinated and aggregated proteins, precisely the role of selective autophagy (Lee et al., 2010).
The above model makes sense in the context of disease-associated optineurin mutations. For instance, open-angle glaucoma has been associated with an E50K mutation in optineurin. This mutation has been shown to enhance the TBK1-optineurin interaction, as well as reducing appropriate intracellular solubility of optineurin (Kryndushkin et al., 2012; Minegishi et al., 2013). A reduced solubility of the TBK1-optineurin complex could be expected to deplete the pool of available phosphorylated optineurin to act as an autophagy adaptor. Moreover, insufficient availability of TBK1 would clearly impair autophagy, and may simply lead to the uncontrolled association of optineurin with aggregates. This itself may actually promote more and more promiscuous cytosolic binding of cytosolic SG-associated proteins.
Like optineurin, mutations in the X-linked protein ubiquilin-2 are associated with familial ALS (Deng et al., 2011). Ubiquilin-2 has been well characterized for its function in delivering ubiquitin-tagged proteins to the proteasome for degradation (Deng et al., 2011; Walters et al., 2004) (Williams et al., 2012; Zhang et al., 2014). It has also been implicated in delivering polyubiquitinated substrates to the autophagy receptor LC3 (Rothenberg et al., 2010). Reduction in ubiquilin-2 is associated with concomitant reduction in autophagosome formation (Rothenberg et al., 2010). Given its role in protein turnover, the simplest explanation for the association of ubiquilin-2 and ALS would be a loss of function and the resulting accumulation of intracellular aggregates. SGs are enriched for ubiquitinated species (Kwon et al., 2007), thus failure to identify these species by ubiqulin-2 could reduce SG turnover by autophagy, leading to SG persistence.
d. Other Proteins Linked to ALS and Autophagy
The final step in autophagy is the fusion of the autophagosome with the lysosome. This organelle maturation appears to be mediated by the cytosolic protein HDAC6, which contains ubiquitin-binding and dynein-interacting domains (Kawaguchi et al., 2003; Lee et al., 2010). It has been implicated as a regulator of diverse cellular processes through deacetylating and destabilizing microtubules, mediating the transport of ubiquitinated proteins into aggresomes and facilitating autophagosome-lysosome fusion (Kawaguchi et al., 2003; Lee et al., 2010; Pandey et al., 2007a; Pandey et al., 2007b). Interestingly, HDAC6 physically interacts with G3BP, a known component of SGs, and is sequestered into SGs under different stress conditions. Moreover, genetic or pharmacological inhibition of HDAC6 impairs SG formation, suggesting that HDAC6 is essential for the assembly of SGs (Kwon et al., 2007).
The impairment of HDAC6 has been further implicated in pathogenesis of certain types of ALS (Gal et al., 2013), though its precise effect remain unclear (Gal et al., 2013; Taes et al., 2013). In a mouse model of SOD1 disease, HDAC6 promoted increased life span and autophagolysosome formation (Chen et al., 2015). Recent studies have shown that the autophagy-adapter p62 localizes with HDAC6 and controls its deacetylase activity (Yan et al., 2013). Mutations in p62 itself have also been linked to cases of ALS in multiple studies (Fecto et al., 2011; Teyssou et al., 2013). While their relationship to autophagy and protein aggregation in driving ALS pathogenesis is likely complicated, their implication serves to further bolster the importance of autophagy-related proteins in motor neuron disease.
5. Pharmacological Modulators of Autophagy in ALS Models
Numerous studies have found autophagy to be involved in the degradation of misfolded proteins associated with many neurodegenerative diseases. Several autophagy-modulating molecules have been evaluated for their specific effects on ALS models (Cipolat Mis et al., 2016). In a cell model of TDP-43-linked ALS, the autophagy-inhibitor 3-methyladenine was found to significantly inhibit the degradation of TDP-43 species (Scotter et al., 2014). Likewise, activation of autophagy by rapamycin, decreased the severity of motor dysfunction in a mouse model of TDP-43/ALS (Wang et al., 2012) and locomotive defects in a TDP-43/ALS Drosophila model (Cheng et al., 2015). Similar observations were made in cultured neuronal cells expressing mutant ALS-causing FUS; rapamycin reduced FUS-positive SGs, as well as neurite fragmentation and cell death in neurons expressing mutant FUS under oxidative stress (Ryu et al., 2014). Also, inducing autophagy with rapamycin was shown to improve in vivo pathology of transgenic mice harboring a disease-causing VCP mutation (Nalbandian et al., 2015). Other autophagy modifiers have also been shown to have varying success in ALS models, including trehalose, lithium and Withaferin A (Castillo et al., 2013; Fornai et al., 2008; Patel et al., 2015b). Small molecule library screens have also yielded authopagy-modulating compounds that show promise in reducing ALS-linked cytotoxicity in primary neuronal models (Barmada et al., 2014). Thus, in conclusion, autophagy enhancement remains a promising strategy for future ALS therapeutics.
6. Discussion and Future Directions
The proposed pathways described here relating to RNP formation, autophagy, and protein aggregation in the context of ALS are admittedly hypothetical and complex. It appears likely that autophagy is critical to modulating ALS, at least in part, because it facilitates RNP granule turnover. With every newly-identified ALS-related protein, it becomes clear that many different quality-control pathways play critical disease-related roles. Understanding why certain quality-control pathways are central to any given neurodegeneration model suggests the opportunity to predict novel proteins that may be associated with various neurodegenerative diseases beyond ALS, as well as identifying new therapeutic targets. Exploring a metabolically basic function like autophagy can also have a multiplier effect because so many related neurodegenerative diseases could benefit from properly regulated autophagic processes. Thus, understanding important players in autophagic pathways will help identify therapeutic targets that could have wide applications.
Highlights.
- ALS is a fatal adult-onset motor neuron disease for which no cure is available
- Pathogenic mutations in ALS-associated genes have been shown to perturb autophagy pathways
- Many ALS-associated proteins accumulate in cytoplasmic stress granules
- Mutant ALS-causing proteins such as FUS, TDP-43, and SOD1 have been shown to be cleared by autophagy in cellular and human stem-cell derived neurons
Acknowledgments
This work was supported by the National Institute of Health R01 Grant (NS081303), R21 grant (NS094921) and the Robert Packard Center for ALS at Johns Hopkins to UBP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta JR, Goldsbury C, Winnick C, Badrock AP, Fraser ST, Laird AS, Hall TE, Don EK, Fifita JA, Blair IP, Nicholson GA, Cole NJ. Mutant human FUS Is ubiquitously mislocalized and generates persistent stress granules in primary cultured transgenic zebrafish cells. PLoS One. 2014;9:e90572. doi: 10.1371/journal.pone.0090572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, Seeley WW, Boxer AL, Petrucelli L, Miller BL, Gao FB. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–99. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–15. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–34. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9:374–82. doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–11. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Avendano-Vazquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, Baralle FE. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 2012;26:1679–84. doi: 10.1101/gad.194829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D'Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–85. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–84. [PMC free article] [PubMed] [Google Scholar]
- Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–49. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–85. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Kaushansky LJ, Ward CL, Sama RR, Chian RJ, Boggio KJ, Quaresma AJ, Nickerson JA, Bosco DA. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener. 2013;8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–12. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E, Haass C, Dormann D. Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. Febs J. 2013;280:4348–70. doi: 10.1111/febs.12287. [DOI] [PubMed] [Google Scholar]
- Bessen RA, Raymond GJ, Caughey B. In situ formation of protease-resistant prion protein in transmissible spongiform encephalopathy-infected brain slices. J Biol Chem. 1997;272:15227–31. doi: 10.1074/jbc.272.24.15227. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–21. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126:3237–47. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, Itie A, Wakeham A, Shahinian A, Henzel WJ, Elia AJ, Shillinglaw W, Mak TW, Cao Z, Yeh WC. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. Embo J. 2000;19:4976–85. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr., Sapp P, McKenna-Yasek D, Brown RH, Jr., Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–75. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JK, Huang CC, Shen CK. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem. 2011;286:44441–8. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–9. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon JH, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124:228–39. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–74. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budini M, Buratti E. TDP-43 autoregulation: implications for disease. J Mol Neurosci. 2011;45:473–9. doi: 10.1007/s12031-011-9573-8. [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell. 2015;60:231–41. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Majumder S, Deng JJ, Bai Y, Thornton FB, Oddo S. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J Biol Chem. 2009;284:27416–24. doi: 10.1074/jbc.M109.031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Shaw DM, Guarino F, Messina A, Walker AW, Oddo S. Reduced protein turnover mediates functional deficits in transgenic mice expressing the 25 kDa C-terminal fragment of TDP-43. Hum Mol Genet. 2015;24:4625–35. doi: 10.1093/hmg/ddv193. [DOI] [PubMed] [Google Scholar]
- Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B, Hetz C. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–20. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem. 2008;283:35186–98. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SN, Tang BL. Location and membrane sources for autophagosome formation - from ER-mitochondria contact sites to Golgi-endosome-derived carriers. Mol Membr Biol. 2013;30:394–402. doi: 10.3109/09687688.2013.850178. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang XJ, Li LX, Wang Y, Zhong RJ, Le W. Histone deacetylase 6 delays motor neuron degeneration by ameliorating the autophagic flux defect in a transgenic mouse model of amyotrophic lateral sclerosis. Neurosci Bull. 2015;31:459–68. doi: 10.1007/s12264-015-1539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CW, Lin MJ, Shen CK. Rapamycin alleviates pathogenesis of a new Drosophila model of ALS-TDP. J Neurogenet. 2015;29:59–68. doi: 10.3109/01677063.2015.1077832. [DOI] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452–62. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A. 2010;107:16320–4. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Cipolat Mis MS, Brajkovic S, Frattini E, Di Fonzo A, Corti S. Autophagy in motor neuron disease: Key pathogenetic mechanisms and therapeutic targets. Mol Cell Neurosci. 2016;72:84–90. doi: 10.1016/j.mcn.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, Ren Z, Keebler J, Han Y, Levy SE, Boone BE, Wimbish JR, Waite LL, Jones AL, Carulli JP, Day-Williams AG, Staropoli JF, Xin WW, Chesi A, Raphael AR, McKenna-Yasek D, Cady J, Vianney de Jong JM, Kenna KP, Smith BN, Topp S, Miller J, Gkazi A, Consortium FS, Al-Chalabi A, van den Berg LH, Veldink J, Silani V, Ticozzi N, Shaw CE, Baloh RH, Appel S, Simpson E, Lagier-Tourenne C, Pulst SM, Gibson S, Trojanowski JQ, Elman L, McCluskey L, Grossman M, Shneider NA, Chung WK, Ravits JM, Glass JD, Sims KB, Van Deerlin VM, Maniatis T, Hayes SD, Ordureau A, Swarup S, Landers J, Baas F, Allen AS, Bedlack RS, Harper JW, Gitler AD, Rouleau GA, Brown R, Harms MB, Cooper GM, Harris T, Myers RM, Goldstein DB. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–61. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- Corcoran J, So PL, Maden M. Absence of retinoids can induce motoneuron disease in the adult rat and a retinoid defect is present in motoneuron disease patients. J Cell Sci. 2002;115:4735–41. doi: 10.1242/jcs.00169. [DOI] [PubMed] [Google Scholar]
- Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, Galbiati M, Fontana E, Marino M, Carra S, Bendotti C, De Biasi S, Poletti A. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS) Hum Mol Genet. 2010;19:3440–56. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining "clean" cells. Autophagy. 2005;1:131–40. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- D'Alton S, Altshuler M, Lewis J. Studies of alternative isoforms provide insight into TDP-43 autoregulation and pathogenesis. Rna. 2015;21:1419–32. doi: 10.1261/rna.047647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Lanson NA, Jr., Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Krishnamurthy K, Ramesh N, Casci I, Monaghan J, McAvoy K, Godfrey EW, Daniel DC, Johnson EM, Monahan Z, Shewmaker F, Pasinelli P, Pandey UB. Pur Alpha Ameliorates FUS Toxicity and Regulates Cytoplasmic Stress Granule Dynamics. 2016 doi: 10.1007/s00401-015-1530-0. Submitted, under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–92. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, Ito D, Murray ME, Dickson DW, Seyfried NT, Kukar T. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci. 2014;34:7802–13. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–6. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P, 3rd, Good SK, Johnson BA, Herz J, Yu G. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2010;31:1098–108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvio M, Piccinni V, Gerbino V, Mantoni F, Camerini S, Lenzi J, Rosa A, Chellini L, Loreni F, Carri MT, Bozzoni I, Cozzolino M, Cestra G. Pur-alpha functionally interacts with FUS carrying ALS-associated mutations. Cell Death Dis. 2015;6:e1943. doi: 10.1038/cddis.2015.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Haass C. TDP-43 and FUS: a nuclear affair. Trends Neurosci. 2011;34:339–48. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–94. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104:9041–6. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–95. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, Donkervoort S, Ajroud-Driss S, Sufit RL, Heller SL, Deng HX, Siddique T. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, Kahle PJ. TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. Febs J. 2011;278:3550–68. doi: 10.1111/j.1742-4658.2011.08258.x. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, Lazzeri G, Spalloni A, Bellio N, Lenzi P, Modugno N, Siciliano G, Isidoro C, Murri L, Ruggieri S, Paparelli A. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2008;105:2052–7. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MJ, Gareau C, Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 2010;10:12. doi: 10.1186/1475-2867-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, Marroquin N, Nordin F, Hubers A, Weydt P, Pinto S, Press R, Millecamps S, Molko N, Bernard E, Desnuelle C, Soriani MH, Dorst J, Graf E, Nordstrom U, Feiler MS, Putz S, Boeckers TM, Meyer T, Winkler AS, Winkelman J, de Carvalho M, Thal DR, Otto M, Brannstrom T, Volk AE, Kursula P, Danzer KM, Lichtner P, Dikic I, Meitinger T, Ludolph AC, Strom TM, Andersen PM, Weishaupt JH. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–6. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- Gal J, Chen J, Barnett KR, Yang L, Brumley E, Zhu H. HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J Biol Chem. 2013;288:15035–45. doi: 10.1074/jbc.M112.431957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Iradi MC, Clare AM, Brown HH, Janus C, Lewis J, Borchelt DR. Subcellular Localization of Matrin 3 Containing Mutations Associated with ALS and Distal Myopathy. PLoS One. 2015;10:e0142144. doi: 10.1371/journal.pone.0142144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislat G, Patron M, Rizzuto R, Knecht E. Withdrawal of essential amino acids increases autophagy by a pathway involving Ca2+/calmodulin-dependent kinase kinase-beta (CaMKK-beta) J Biol Chem. 2015;287:38625–36. doi: 10.1074/jbc.M112.365767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielbert A, Thorne JK, Hope J. Pyroglutamyl-N-terminal prion protein fragments in sheep brain following the development of transmissible spongiform encephalopathies. Front Mol Biosci. 2015;2:7. doi: 10.3389/fmolb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–98. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim Biophys Acta. 2013;1833:205–12. doi: 10.1016/j.bbamcr.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–58. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, Liu J, Xu M, Yang Y, Wang C, Zhang D, Bigio EH, Mesulam M, Shen Y, Xu Q, Fushimi K, Wu JY. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–30. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Joshi M, Jeoung NH. Mechanisms responsible for regulation of branched-chain amino acid catabolism. Biochem Biophys Res Commun. 2004;313:391–6. doi: 10.1016/j.bbrc.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White EK, Koury MJ, Oltz EM, Van Kaer L, Ruley HE. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–9. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Goodbrand IA, Goodsir CM. Pathology of the transmissible spongiform encephalopathies with special emphasis on ultrastructure. Micron. 1995;26:277–98. doi: 10.1016/0968-4328(95)00004-n. [DOI] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105:6439–44. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, Gibbs JR, Nalls MA, Morgan S, Shoai M, Hardy J, Pittman A, Orrell RW, Malaspina A, Sidle KC, Fratta P, Harms MB, Baloh RH, Pestronk A, Weihl CC, Rogaeva E, Zinman L, Drory VE, Borghero G, Mora G, Calvo A, Rothstein JD, Consortium I, Drepper C, Sendtner M, Singleton AB, Taylor JP, Cookson MR, Restagno G, Sabatelli M, Bowser R, Chio A, Traynor BJ. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–6. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–88. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, Bosco DA, Hayward LJ, Brown RH, Jr., Lindquist S, Ringe D, Petsko GA. A Yeast Model of FUS/TLS-Dependent Cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Durham HD. Failure of protein quality control in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1038–50. doi: 10.1016/j.bbadis.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–38. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–9. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–78. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–90. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I'm confused. Autophagy. 2014;10:549–51. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarcik CL, Bowser R. Retinoid signaling alterations in amyotrophic lateral sclerosis. Am J Neurodegener Dis. 2012;1:130–45. [PMC free article] [PubMed] [Google Scholar]
- Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126:580–92. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB, Shewmaker F. FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis. Protein Cell. 2011;2:223–36. doi: 10.1007/s13238-011-1525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Ihrke G, Piermartiri TC, Shewmaker F. A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol Microbiol. 2012;86:1531–47. doi: 10.1111/mmi.12075. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, de Rooij DG, Akhmedov A, Ashley T, Ron D. Male sterility and enhanced radiation sensitivity in TLS(−/−) mice. Embo J. 2000;19:453–62. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–94. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, Wancewicz E, Kim AS, Watt A, Freier S, Hicks GG, Donohue JP, Shiue L, Bennett CF, Ravits J, Cleveland DW, Yeo GW. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–90. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- Lanson NA, Jr., Maltare A, King H, Smith R, Kim JH, Taylor JP, Lloyd TE, Pandey UB. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 2010;29:969–80. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi J, De Santis R, de Turris V, Morlando M, Laneve P, Calvo A, Caliendo V, Chio A, Rosa A, Bozzoni I. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis Model Mech. 2015;8:755–66. doi: 10.1242/dmm.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, 3rd, Fushimi K, Wu JY. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 2010;107:3169–74. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–72. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–19. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–5. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Betancor O, Ogaki K, Soto-Ortolaza A, Labbe C, Vilarino-Guell C, Rajput A, Rajput AH, Pastor P, Ortega S, Lorenzo E, Strongosky AJ, van Gerpen JA, Uitti RJ, Wszolek ZK, Ross OA. Analysis of nuclear export sequence regions of FUS-Related RNA-binding proteins in essential tremor. PLoS One. 2014;9:e111989. doi: 10.1371/journal.pone.0111989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Leigh PN, King A, Jones E, Steele JC, Bodi I, Shaw CE, Hortobagyi T, Al-Sarraj S. TDP-43 is consistently co-localized with ubiquitinated inclusions in sporadic and Guam amyotrophic lateral sclerosis but not in familial amyotrophic lateral sclerosis with and without SOD1 mutations. Neuropathology. 2009;29:672–83. doi: 10.1111/j.1440-1789.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–41. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18:2603–18. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–10. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- McGurk L, Lee VM, Trojanowksi JQ, Van Deerlin VM, Lee EB, Bonini NM. Poly-A binding protein-1 localization to a subset of TDP-43 inclusions in amyotrophic lateral sclerosis occurs more frequently in patients harboring an expansion in C9orf72. J Neuropathol Exp Neurol. 2014;73:837–45. doi: 10.1097/NEN.0000000000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ. Autophagy research: lessons from metabolism. Autophagy. 2009;5:3–5. doi: 10.4161/auto.5.1.7207. [DOI] [PubMed] [Google Scholar]
- Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Millecamps S, De Septenville A, Teyssou E, Daniau M, Camuzat A, Albert M, LeGuern E, Galimberti D, Brice A, Marie Y, Le Ber I. Genetic analysis of matrin 3 gene in French amyotrophic lateral sclerosis patients and frontotemporal lobar degeneration with amyotrophic lateral sclerosis patients. Neurobiol Aging. 2014;35:2882. doi: 10.1016/j.neurobiolaging.2014.07.016. e13-5. [DOI] [PubMed] [Google Scholar]
- Miller RG, Mitchell JD, Moore DH. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND) Cochrane Database Syst Rev. 2012;3:CD001447. doi: 10.1002/14651858.CD001447.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Iejima D, Kobayashi H, Chi ZL, Kawase K, Yamamoto T, Seki T, Yuasa S, Fukuda K, Iwata T. Enhanced optineurin E50K-TBK1 interaction evokes protein insolubility and initiates familial primary open-angle glaucoma. Hum Mol Genet. 2013;22:3559–67. doi: 10.1093/hmg/ddt210. [DOI] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, Cleverley K, Nicoll AJ, Pickering-Brown S, Dols J, Cabecinha M, Hendrich O, Fratta P, Fisher EM, Partridge L, Isaacs AM. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–4. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, Abe K. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–7. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–6. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- N'Diaye EN, Debnath J, Brown EJ. Ubiquilins accelerate autophagosome maturation and promote cell survival during nutrient starvation. Autophagy. 2009;5:573–5. doi: 10.4161/auto.5.4.8312. [DOI] [PubMed] [Google Scholar]
- Nalbandian A, Llewellyn KJ, Nguyen C, Yazdi PG, Kimonis VE. Rapamycin and chloroquine: the in vitro and in vivo effects of autophagy-modifying drugs show promising results in valosin containing protein multisystem proteinopathy. PLoS One. 2015;10:e0122888. doi: 10.1371/journal.pone.0122888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Orozco D, Tahirovic S, Rentzsch K, Schwenk BM, Haass C, Edbauer D. Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO Rep. 2012;13:759–64. doi: 10.1038/embor.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Batlevi Y, Baehrecke EH, Taylor JP. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 2007a;3:643–5. doi: 10.4161/auto.5050. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007b;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker SJ, Meyerowitz J, James JL, Liddell JR, Crouch PJ, Kanninen KM, White AR. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int. 2012;60:415–24. doi: 10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015a;162:1066–77. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Patel P, Julien JP, Kriz J. Early-stage treatment with Withaferin A reduces levels of misfolded superoxide dismutase 1 and extends lifespan in a mouse model of amyotrophic lateral sclerosis. Neurotherapeutics. 2015b;12:217–33. doi: 10.1007/s13311-014-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T, Deretic V. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–34. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochet R, Nicaise C, Mitrecic D. Translation of the focus toward excellence in translational science: comment on "TDP-43 Repression of Nonconserved Cryptic Exons is Compromised in ALS-FTD". Croat Med J. 2015;56:493–5. doi: 10.3325/cmj.2015.56.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 2007;9:2218–29. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MX. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajawat Y, Hilioti Z, Bossis I. Autophagy: a target for retinoic acids. Autophagy. 2010;6:1224–6. doi: 10.4161/auto.6.8.13793. [DOI] [PubMed] [Google Scholar]
- Rajawat Y, Hilioti Z, Bossis I. Retinoic acid induces autophagosome maturation through redistribution of the cation-independent mannose-6-phosphate receptor. Antioxid Redox Signal. 2011;14:2165–77. doi: 10.1089/ars.2010.3491. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravits JM, La Spada AR. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology. 2009;73:805–11. doi: 10.1212/WNL.0b013e3181b6bbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4:855–64. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- Reineke LC, Lloyd RE. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–67. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riancho J, Ruiz-Soto M, Berciano MT, Berciano J, Lafarga M. Neuroprotective Effect of Bexarotene in the SOD1(G93A) Mouse Model of Amyotrophic Lateral Sclerosis. Front Cell Neurosci. 2015;9:250. doi: 10.3389/fncel.2015.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riancho J, Berciano MT, Ruiz-Soto M, Berciano J, Landreth G, Lafarga M. Retinoids and motor neuron disease: Potential role in amyotrophic lateral sclerosis. J Neurol Sci. 2016;360:115–20. doi: 10.1016/j.jns.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–32. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F, Hafezparast M, Caldecott KW. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–14. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, Lee JA. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging. 2014;35:2822–31. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Sama RR, Ward CL, Kaushansky LJ, Lemay N, Ishigaki S, Urano F, Bosco DA. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol. 2013;228:2222–31. doi: 10.1002/jcp.24395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama RR, Ward CL, Bosco DA. Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro. 2014:6. doi: 10.1177/1759091414544472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2011;70:349–59. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- Scaramuzzino C, Monaghan J, Milioto C, Lanson NA, Jr., Maltare A, Aggarwal T, Casci I, Fackelmayer FO, Pennuto M, Pandey UB. Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS One. 2013;8:e61576. doi: 10.1371/journal.pone.0061576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheu S, Stetson DB, Reinhardt RL, Leber JH, Mohrs M, Locksley RM. Activation of the integrated stress response during T helper cell differentiation. Nat Immunol. 2006;7:644–51. doi: 10.1038/ni1338. [DOI] [PubMed] [Google Scholar]
- Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130:537–55. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–25. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotter EL, Vance C, Nishimura AL, Lee YB, Chen HJ, Urwin H, Sardone V, Mitchell JC, Rogelj B, Rubinsztein DC, Shaw CE. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci. 2014;127:1263–78. doi: 10.1242/jcs.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, Carra S. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 2014;21:1838–51. doi: 10.1038/cdd.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci U S A. 2010;107:16325–30. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Lyashchenko AK, Lu L, Nasrabady SE, Elmaleh M, Mendelsohn M, Nemes A, Tapia JC, Mentis GZ, Shneider NA. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun. 2016;7:10465. doi: 10.1038/ncomms10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Li HY, Chen GC, Chern Y, Tu PH. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy. 2015;11:685–700. doi: 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–8. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo KY, Halloran M, Sundaramoorthy V, Parakh S, Toth RP, Southam KA, McLean CA, Lock P, King A, Farg MA, Atkin JD. Rab1-dependent ER-Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS. Acta Neuropathol. 2015;130:679–97. doi: 10.1007/s00401-015-1468-2. [DOI] [PubMed] [Google Scholar]
- Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–60. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. Embo J. 2004;23:1313–24. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular Determinants and Genetic Modifiers of Aggregation and Toxicity for the ALS Disease Protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taes I, Timmers M, Hersmus N, Bento-Abreu A, Van Den Bosch L, Van Damme P, Auwerx J, Robberecht W. Hdac6 deletion delays disease progression in the SOD1G93A mouse model of ALS. Hum Mol Genet. 2013;22:1783–90. doi: 10.1093/hmg/ddt028. [DOI] [PubMed] [Google Scholar]
- Teyssou E, Takeda T, Lebon V, Boillee S, Doukoure B, Bataillon G, Sazdovitch V, Cazeneuve C, Meininger V, LeGuern E, Salachas F, Seilhean D, Millecamps S. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol. 2013;125:511–22. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–34. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2011;21:136–49. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- Turner MR, Hardiman O, Benatar M, Brooks BR, Chio A, de Carvalho M, Ince PG, Lin C, Miller RG, Mitsumoto H, Nicholson G, Ravits J, Shaw PJ, Swash M, Talbot K, Traynor BJ, Van den Berg LH, Veldink JH, Vucic S, Kiernan MC. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12:310–22. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan M, Baloh RH. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugras SE, Shorter J. RNA-Binding Proteins in Amyotrophic Lateral Sclerosis and Neurodegeneration. Neurol Res Int. 2012;2012:432780. doi: 10.1155/2012/432780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yague E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. Faseb J. 2010;24:3370–80. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Scotter EL, Nishimura AL, Troakes C, Mitchell JC, Kathe C, Urwin H, Manser C, Miller CC, Hortobagyi T, Dragunow M, Rogelj B, Shaw CE. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet. 2013;22:2676–88. doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Walker AK, Soo KY, Sundaramoorthy V, Parakh S, Ma Y, Farg MA, Wallace RH, Crouch PJ, Turner BJ, Horne MK, Atkin JD. ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One. 2013;8:e81170. doi: 10.1371/journal.pone.0081170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim Biophys Acta. 2004;1695:73–87. doi: 10.1016/j.bbamcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Wang IF, Guo BS, Liu YC, Wu CC, Yang CH, Tsai KJ, Shen CK. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109:15024–9. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–91. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan H, Ying Z, Li B, Wang H, Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469:112–6. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–13. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, Pasinelli P, Ichida JK, Trotti D. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–25. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Warraich ST, Yang S, Solski JA, Fernando R, Rouleau GA, Nicholson GA, Blair IP. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527. doi: 10.1016/j.neurobiolaging.2012.05.008. e3-10. [DOI] [PubMed] [Google Scholar]
- Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–48. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, Ren H, Chen D, Han J, Ying Z, Wang G. TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. EMBO J. 2016;35:121–42. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Seibenhener ML, Calderilla-Barbosa L, Diaz-Meco MT, Moscat J, Jiang J, Wooten MW, Wooten MC. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS One. 2013;8:e76016. doi: 10.1371/journal.pone.0076016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z, Xia Q, Hao Z, Xu D, Wang M, Wang H, Wang G. TARDBP/TDP-43 regulates autophagy in both MTORC1-dependent and MTORC1-independent manners. Autophagy. 2016;12:707–8. doi: 10.1080/15548627.2016.1151596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–30. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KY, Yang S, Warraich ST, Blair IP. Ubiquilin 2: a component of the ubiquitin-proteasome system with an emerging role in neurodegeneration. Int J Biochem Cell Biol. 2014;50:123–6. doi: 10.1016/j.biocel.2014.02.018. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013a;9:e1003196. doi: 10.1371/journal.pgen.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu S, Liu G, Ozturk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013b;9:e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]