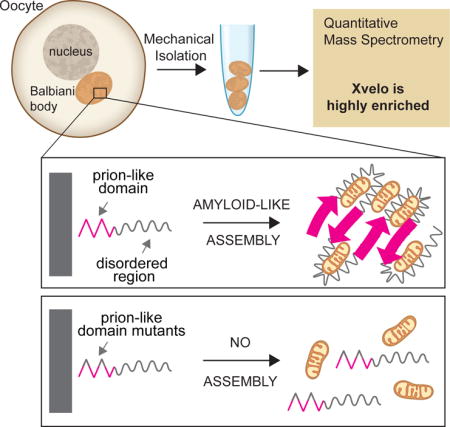
Amyloid-like Self-assembly of a Cellular Compartment (original) (raw)
. Author manuscript; available in PMC: 2017 Jul 28.
SUMMARY
Most vertebrate oocytes contain a Balbiani body; a large, non-membrane bound compartment packed with RNA, mitochondria and other organelles. Little is known about this compartment, though it specifies germ-line identity in many non-mammalian vertebrates. We show Xvelo, a disordered protein with an N-terminal prion-like domain, is an abundant constituent of Xenopus Balbiani bodies. Disruption of the prion-like domain of Xvelo, or substitution with a prion-like domain from an unrelated protein interferes with its incorporation into Balbiani bodies in vivo. Recombinant Xvelo forms amyloid-like networks in vitro. Amyloid-like assemblies of Xvelo recruit both RNA and mitochondria in binding assays. We propose that Xenopus Balbiani bodies form by amyloid-like assembly of Xvelo, accompanied by co-recruitment of mitochondria and RNA. Prion-like domains are found in germ plasm organizing proteins in other species, suggesting that Balbiani body formation by amyloid-like assembly could be a conserved mechanism that helps oocytes function as long-lived germ cells.
Graphical abstract
INTRODUCTION
Female germ cells, oocytes, are highly specialized cells. In many species, oocytes are long-lived, and lie dormant for months or years before they are activated prior to fertilization (Li and Albertini, 2013). They ensure the continuity of the species by providing the female genome and mitochondria, along with most of the housekeeping machinery and nutrients the early embryo will need after fertilization. Oocytes have a unique subcellular organization, with a large nucleus, called the germinal vesicle, and a large cytoplasm. In many species, the cytoplasm of the early oocytes contains a highly specialized compartment called the Balbiani body. It is non-membrane bound, and is densely packed with mitochondria, RNA, ER and Golgi. The Balbiani body assembles early in oocyte formation (Lei and Spradling, 2016), disappears in late-stage oocytes in mammals (Pepling et al., 2007) and forms dispersed isles at the vegetal pole in late stage oocytes of Xenopus and zebrafish (Bontems et al., 2009; Kloc et al., 2004).
How, or why, the Balbiani body forms is largely mysterious (Gupta et al., 2010; Heim et al., 2014; Kloc and Etkin, 1995; Kloc et al., 1993; Marlow and Mullins, 2008). In some species, including frogs and fish, but not newts and mammals, one function of the Balbiani body is to serve as an embryonic determinant that specifies germ line identity by forming germ plasm (Lesch and Page, 2012; Richardson and Lehmann, 2010). Germ plasm, a special part of oocyte cytoplasm, protects specific maternal RNAs from degradation and is believed to host healthy mitochondria during development to pass on to future generations (Houston and King, 2000; Kogo et al., 2011). Germ line specification is the only known function of the Balbiani body, but it is notable that oocytes from species that use inductive processes to specify their germ line, including mammals, still contain a Balbiani body, whose ultrastructure is similar to those of organisms that use germ plasm (Hertig, 1968). We do not know what functions the Balbiani body serves in all vertebrates. We speculate their most conserved function is to protect the quality of mitochondria and other organelles during long periods of oocyte dormancy, which can extend for decades in humans.
An elegant maternal effect screen in zebrafish identified only one gene, bucky ball, required for Balbiani body formation (Dosch et al., 2004). In buc mutants, the Balbiani body did not form, and oocyte polarity was disrupted. Overexpression of Bucky ball protein stimulated numerous Balbiani bodies and ectopic germ cell formation (Bontems et al., 2009; Heim et al., 2014; Marlow and Mullins, 2008). Bucky ball is proposed to be a structural organizer of zebrafish Balbiani bodies, but the biochemical basis of this proposed function has been unclear.
Xenopus laevis provides complementary advantages for analysis of Balbiani bodies because oocytes are abundant and accessible, easy to manipulate due to their large size, and amenable to live imaging. Xenopus oocytes are classified in 6 different stages according to Dumont, 1972. Stage I oocytes contain a clearly visible Balbiani body (Figure 1A). The Balbiani body gradually disappears during oocyte development to give rise to dispersed germ plasm islands at the vegetal pole of the stage VI oocytes during oocyte maturation (Kloc et al., 2004).
Figure 1. A Balbiani body is a non-membrane bound compartment packed with membranous organelles.
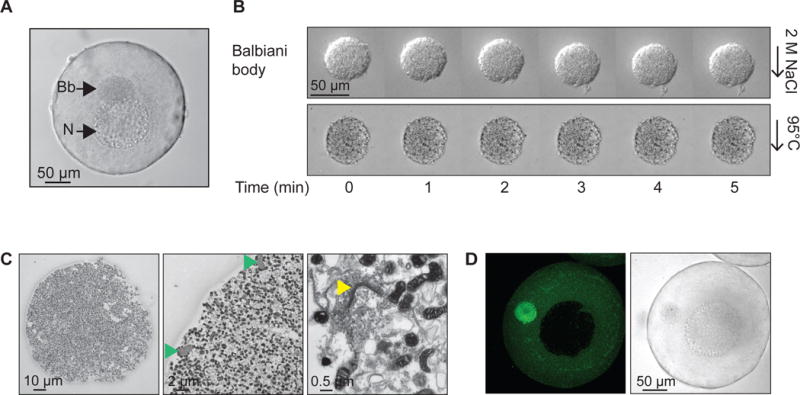
(A) Phase contrast image of a stage I Xenopus oocyte. Bb, Balbiani body; N, nucleus, or germinal vesicle.
(B) Balbiani body immobilized in perfusion chambers. 2M NaCl (first panel) or 95 °C 50mM Hepes, 100mM KCl, pH 7.6 buffer (second panel) was perfused into the chambers.
(C) Thin-section electron microscope (EM) images of isolated Balbiani bodies from stage I Xenopus oocytes. Mitochondria (dark spots), RNP particles (green arrow head) and Golgi stacks (yellow arrow head) are clearly visible.
(D) Stage I oocytes were incubated in 10 μM Thioflavin T in 1× MMR for 10 minutes and washed twice with 1X MMR.
See also Figure S1, Table S1 and Movie S1
Here, we report that the Balbiani body forms via amyloid-like self-assembly of Xvelo, the Xenopus homolog of Bucky ball. Amyloids and amyloid-like proteins have largely been studied in the context of neurodegenerative diseases, such as Alzheimer’s disease, amyotrophic lateral sclerosis, or prion diseases (Haass and Selkoe, 2007; Koo et al., 1999; Polymenidou and Cleveland, 2011). Several examples of amyloid-like aggregation mechanisms with normal biological functions have recently emerged (Berchowitz et al., 2015; Fowler et al., 2006; Hou et al., 2011; Li et al., 2012). Both pathological and physiological amyloid-forming proteins contain low complexity regions, which are intrinsically disordered, but can undergo conformational conversions into an amyloid-like fibrillar state (Kato et al., 2012). These regions are also found in yeast prion proteins, a class of infectious proteins that generate heritable phenotypic diversity in clonal population of yeast cells (Alberti et al., 2009; Halfmann et al., 2012; Wickner et al., 2013). Protein regions with sequence compositions similar to yeast prions are called prion-like domains (PLDs) (Alberti et al., 2009; Si et al., 2003).
RESULTS
A Balbiani body has a rigid structure that stains with Thioflavin T
We began our studies on Balbiani bodies by manually dissecting them from Xenopus laevis stage I oocytes, which are transparent and 50–300 μm in diameter (Figure 1A). Stage I oocytes were placed in a glass bottom dish together with a cytoskeleton stabilizing buffer, and mechanically disrupted using tweezers, leaving the Balbiani body intact. The Balbiani body preserved its circular shape and did not disintegrate under the considerable shear forces applied during isolation (Movie S1). Balbiani bodies also retained their structure under harsh conditions including high salt, 2 M NaCl (Figure 1B, first panel) and high temperature (up to 95 °C, Figure 1B, second panel). Confirming previous work on intact oocytes, thin section electron microscopy of the isolated Balbiani bodies revealed densely packed mitochondria, ER and Golgi stacks as well as compact fibrillar elements that others have shown to be made of ribonucleoprotein (RNP) (Figure 1C) (Al-Mukhtar and Webb, 1971; Balinsky and Devis, 1963; Billett and Adam, 1976).
To probe the molecular properties of the Balbiani body, we introduced a number of small molecules into the oocytes. Thioflavin T (ThT), a dye that stains the beta sheet-rich structures of amyloid (Alberti et al., 2010; Nilsson, 2004), strongly accumulated in the Balbiani body (Figure 1D), suggesting it is rich in beta-sheet structures, which is a hallmark of amyloids.
Xvelo is highly concentrated in Balbiani bodies
To analyze the composition of Balbiani bodies, we used quantitative mass spectrometry (McAlister et al., 2014; Wühr et al., 2014). This revealed that the most enriched Balbiani body proteins that are not part of organelles are Velo1 (commonly known as Xvelo) and fetal hemoglobin subunits, Hba1 and Hbg1 (Figure S1A–S1C). We focused on Xvelo because it is homologous to zebrafish Bucky ball, which plays a crucial role in Balbiani body organization (Bontems et al., 2009). Computational analysis of Xvelo sequence predicted an intrinsically disordered protein with no known domains, apart from a C-terminal positively charged region that could bind RNA.
Quantitative Western blotting using a peptide antibody against the C-terminus of Xvelo (Figure S2A) provided an estimate of Xvelo concentration in oocytes of ~ 8 μM (Figure S2B, S2C). If all the Xvelo was concentrated in the Balbiani body, and assuming an average late stage I oocyte diameter is 250 μm and Balbiani body diameter is 60 μm, this corresponds to ~560 μM in the Balbiani body. Compared with the published concentrations of proteins in a frog egg, where the most concentrated proteins were alpha-1-antitrypsin (Serpina1) and actin (Actg1) at 17.6 and 14.3 μM, respectively (Wühr et al., 2014), one can appreciate the exceptionally high concentration of Xvelo in Balbiani bodies. This high local concentration of Xvelo, together with its homology to the zebrafish protein Bucky ball, makes it a likely candidate for a structural organizer of the Balbiani body.
Xvelo forms a stable matrix in Balbiani bodies
To investigate Xvelo function in Xenopus, we began by injecting mRNAs encoding for Xvelo tagged with GFP (Xvelo-GFP) into the oocyte cytoplasm. Xvelo-GFP mainly localized to the Balbiani body with little soluble protein present in the cytoplasm (Figure 2A). More detailed analysis at higher magnification showed that Xvelo-GFP filled the gaps between the mitochondria in the Balbiani body (Figure 2B). These results suggest that Xvelo may form a matrix in which mitochondria and other organelles are embedded.
Figure 2. Xvelo forms a stable matrix.
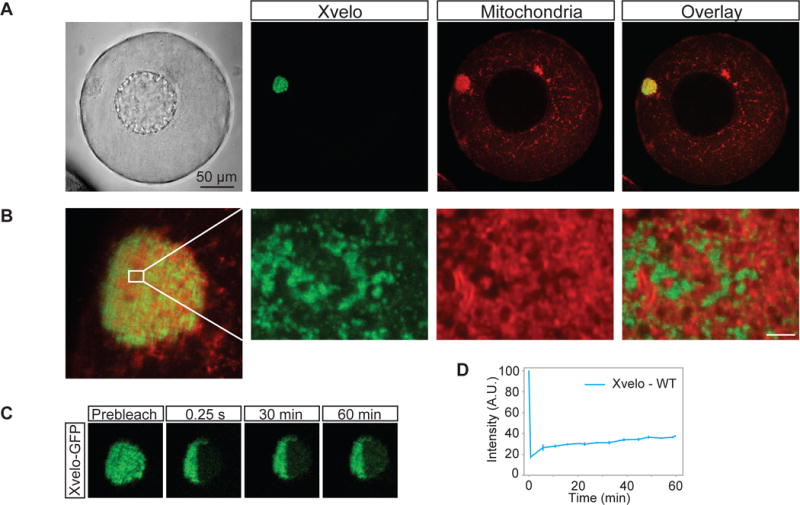
(A) mRNA encoding for Xvelo-GFP was microinjected into stage I oocytes. Mitotracker Deep Red was used to label mitochondria. Oocytes were imaged live with a scanning confocal microscope with a 40× Water immersion objective.
(B) Magnification of the Balbiani body in (A).
(C) Internal rearrangement of fluorescent Xvelo-GFP particles after half bleach over time.
(D) The fluorescent recovery of the half-bleached Xvelo-GFP in the Balbiani body in (C) and two other biological replicates are shown by quantification of fluorescence in bleached region over time. Fluorescent intensity changes in the bleached region per pixel over time were plotted after it was normalized for photobleaching by using an unbleached neighbouring area and background subtraction.
See also Figure S2
To test whether Xvelo was part of a stable matrix, we used FRAP (Fluorescence Recovery After Photobleaching) to probe its dynamics. We again injected mRNA encoding for Xvelo-GFP, and after overnight incubation, imaged the oocytes. After photobleaching Xvelo-GFP, maximum recovery of approximately 20 % was seen after one hour (Figure 2C, 2D). We conclude that Xvelo matrix is highly stable, with slow turnover, consistent with a role as a structural matrix.
Xvelo association with the Balbiani Body requires its prion-like domain
We next investigated how the Xvelo matrix is formed and held together. Despite the lack of any conventional domains, Xvelo has a prion-like domain (PLD) at its N-terminus, which is detected by several prion detection algorithms (Figure 3A, Figure S3A) (Alberti et al., 2009; Lancaster et al., 2014; Toombs et al., 2012). It also has a lysine/arginine rich region at its C-terminus that might act as an RNA binding domain (Figure 3A). To assess the behavior of these different parts of Xvelo in vivo, we performed a structure-function analysis by dividing the protein into four fragments (See Supplemental Experimental Procedures for details, Data S1A).
Figure 3. Xvelo self-assembly is dependent on its prion-like domain.
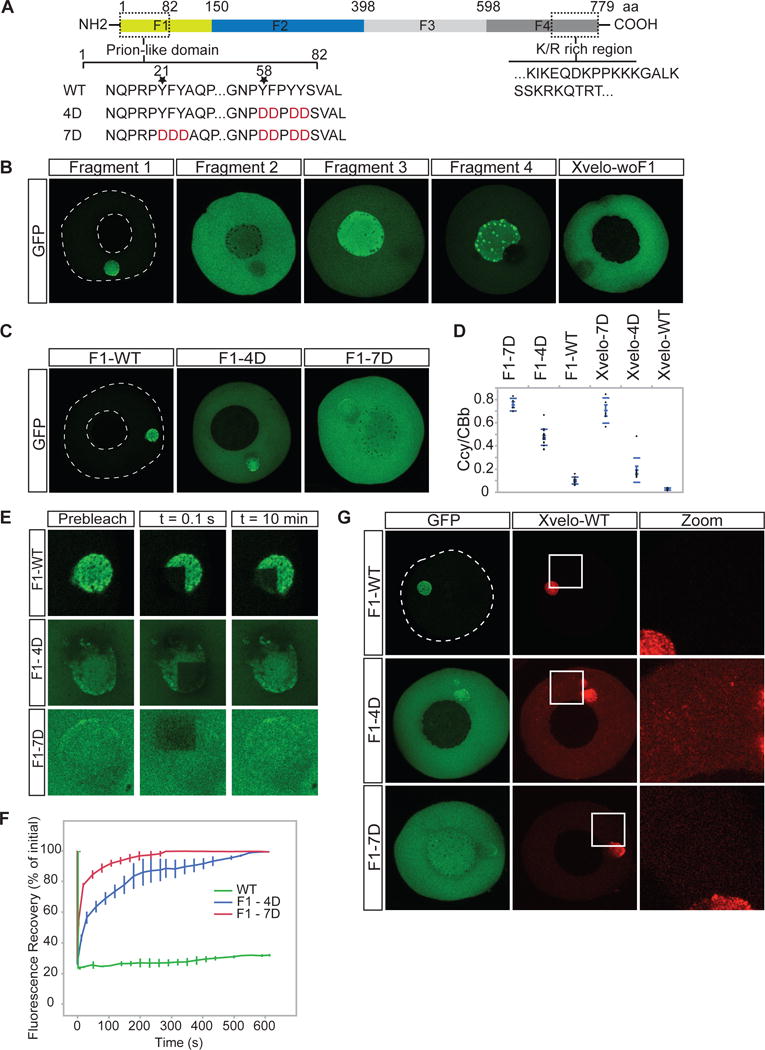
(A) Diagram of the known structural elements of Xvelo. Prion-like domain, mutants (4D and 7D) and the fragments of Xvelo (F1 to F4) are marked in the figure.
(B) mRNAs encoding for Xvelo fragments shown in (A) and Xvelo without fragment 1 (Xvelo-woF1) are in vitro synthesized and micro-injected into stage I oocytes. Oocytes were imaged after overnight incubation in Oocyte Culture Medium (OCM).
(C) mRNAs encoding for wild type and PLD mutants of Fragment1-GFP were microinjected into oocytes. Oocytes were incubated overnight and imaged.
(D) Ratio of GFP concentration in the oocyte cytoplasm (Ccy) to the Balbiani body (CBb) in oocytes injected with mRNAs encoding for indicated proteins. Relative concentrations were calculated by using oocyte or the Balbiani body volume from z-stacks and the fluorescent intensity of GFP. Mean values and standard errors of 10 oocytes are plotted.
(E) Internal rearrangement of fluorescent wild-type or mutant F1-GFP particles after photobleaching over time.
(F) The fluorescent recovery of photobleached wild-type or mutant F1-GFP in Balbiani bodies in (E) and two other biological replicates for each are shown by quantification of fluorescence in bleached region over time normalized by an unbleached neighbouring region.
(G) mRNAs encoding for full length Xvelo-mCherry wild-type, and GFP tagged fragments, F1-WT-GFP, F1-4D-GFP and F1-7D-GFP, were in vitro synthesized. F1-WT-GFP or mutants were mixed with equal amounts of full length Xvelo-mCherry mRNA and microinjected into the oocytes. After overnight incubation, the oocytes were imaged by scanning confocal microscopy.
See also Figure S3
We in vitro synthesized mRNAs encoding for the four Xvelo fragments tagged with GFP, and injected them into oocytes at equal concentrations. Each fragment of Xvelo localized differently (Figure 3B). The C-terminal F4 fragment, which carries the positively charged region, localized to nucleoli. Nucleoli are RNA-rich, thus this localization may reflect predicted RNA binding activity of the F4 fragment (Figure 3B). The N-terminal F1 fragment, which carries the PLD, localized to the Balbiani body. Its localization pattern was indistinguishable from the full-length protein (Figure 2A, 3B). An additional fragment lacking the F1 fragment but retaining the rest of the Xvelo protein (Xvelo-woF1), did not localize to the Balbiani body (Figure 3B). Thus, we conclude that the N-terminus, which contains the PLD, is the key region that targets Xvelo to the Balbiani body.
We next designed mutants that would disrupt the propensity of Xvelo for amyloid-like self-assembly. Charged amino acids are strongly disfavored in PLDs, as they interfere with the formation of a hydrophobic nucleus (Alberti et al., 2009; de la Paz and Serrano, 2004; Serio et al., 2000). Thus, to create defective PLD mutants of Xvelo, we mutated either 4 or 7 aromatic residues in its PLD to a negatively charged amino acid: aspartate (Figure 3A). We call these 4D and 7D mutants, respectively (Figure 3A). The mutants no longer scored positive in prion detection algorithms (Figure S3B, S3C). We injected mRNAs encoding for wild-type or mutant versions of Xvelo tagged with GFP into the oocytes and imaged after overnight incubation. We used mRNAs encoding for both Fragment 1 (the PLD) and full-length Xvelo to observe the effects of the mutants in the oocytes in case the full-length protein interferes with the assembly pattern of the mutants. Fragment 1 proteins carrying the 4D and 7D mutations still exhibited a partial localization to the Balbiani body but they also showed a diffuse signal in the cytoplasm, which was not observed for the wild-type protein (Figure 3C, 3D). The effect was much stronger for the 7D mutant, which was barely enriched in the Balbiani body (Figure 3C, 3D). Full-length proteins behaved similar to their F1 counterparts (Figure 3D and S3D). Thus, we conclude that the PLD of Xvelo is essential, and sufficient for Balbiani body localization and the two conserved motifs enriched for aromatic residues provide an essential structural role in Xvelo targeting.
To investigate whether the mutations in the Xvelo PLD change the association dynamics of the GFP-tagged constructs with the endogenous Xvelo matrix, we performed FRAP on wildtype and mutants. Fragment 1 had a slow turnover rate, similar to the full-length protein (Figure 2C–D, 3E–3F and Figure S3E–S3F). However, the mutants recovered from photobleaching significantly faster than wild-type; the 100 % recovery time was 10 minutes for 4D and 3 minutes for 7D mutants after photobleaching (Figure 3E, 3F) (This should be compared to the wild-type, which recovers to 20 % after one hour in Figure 2C–D). The recovery times of the full-length mutants were similar to the F1 fragments (Figure S3F). These results suggest that the PLD drives the association of Xvelo with the pre-assembled Xvelo in the matrix.
If the mutants change the association dynamics of Xvelo with the matrix, they might impose a dominant negative effect – i.e inhibit the assembly of wild-type Xvelo into the matrix. To test this, we co-injected mRNAs encoding for full-length Xvelo-WT-mCherry and GFP-tagged F1 wild type and mutants at equal concentrations and imaged by live confocal microscopy the next day. Wild-type Xvelo co-localized with the F1 fragment in the Balbiani body and there was little signal in the cytoplasm (Figure 3G). However, in the presence of the 4D mutant, the wild-type Xvelo also formed discrete small aggregates in the cytoplasm (Figure 3G). The 7D mutant also increased the amount of diffusely localized wild-type Xvelo but it did not cause the punctate localization pattern seen with the 4D mutant (Figure 3G). The difference between the mutants may be due to the fact that the 4D mutant binds more strongly to wild-type Xvelo because of its remaining hydrophobic motif. The same patterns were observed when the experiments were repeated with full-length mutants (Figure S3G). Thus, we conclude that mutant PLDs can partially inhibit the self-assembly of wild-type Xvelo, most likely by binding to it and blocking growth into larger assemblies.
Recombinant Xvelo forms micron scale networks in vitro
To test whether Xvelo can form amyloid-like fibers on its own, we analyzed recombinant Xvelo-GFP in vitro. Xvelo-GFP did not express at all in bacteria (data not shown) but expressed well in baculovirus-infected insect Sf9 cells. Purification of Xvelo-GFP from insect cells was challenging due to its strong tendency to aggregate, but we found that addition of 300 mM arginine to Xvelo-GFP solubilized it in an otherwise physiological buffer (Figure S4A). The guanidino fragment of arginine may function similar to guanidinium ion as a solubilizing agent (England and Haran, 2011; Tsumoto et al., 2004).
We purified soluble Xvelo-GFP in 300 mM arginine, then diluted into 30 mM arginine which induced self-assembly (Figure 4A, first panel). Xvelo-GFP self-assembled first into small, then large, micron-scale networks over time (Figure 4A, first panel). To test if network assembly required the PLD of Xvelo, we expressed and purified the F1 fragment containing the PLD. F1-WT-GFP also formed large networks over time, with striking similarity to networks formed by full-length Xvelo, showing that network assembly is driven by the PLD. We confirmed the critical role of the PLD by showing that two full-length PLD mutants, Xvelo-4D-GFP and Xvelo-7D-GFP did not form networks (Figure 4A, lower panels) (Figure 4B).
Figure 4. Xvelo forms micron scale networks in vitro.
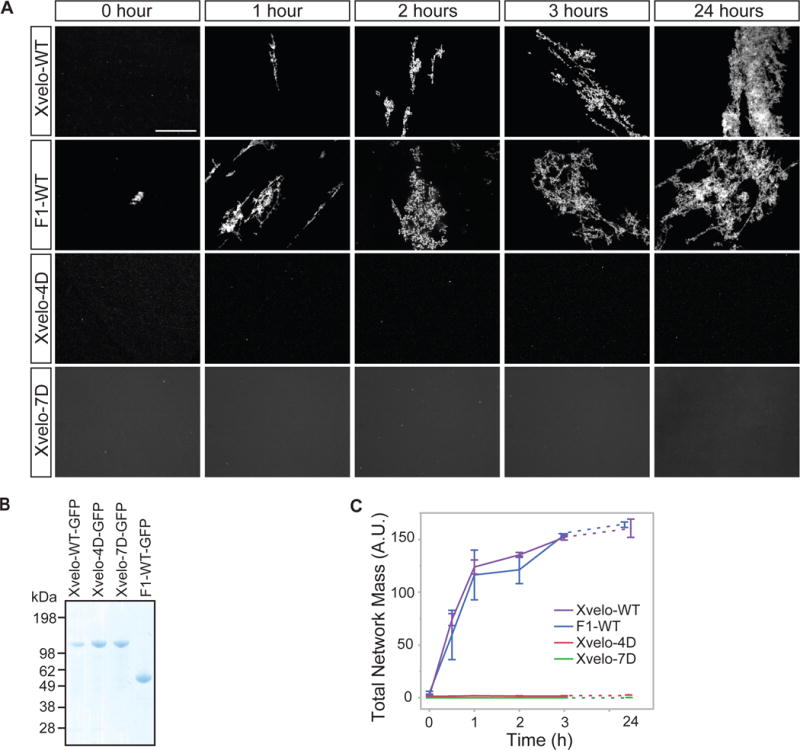
(A) 15 μM of recombinant Xvelo-GFP, F1-WT-GFP, and full length mutants, Xvelo-4D-GFP and Xvelo-7D-GFP, were diluted into a low arginine buffer (30 mM) to promote their self-assembly. The reaction mixtures were incubated at 25 °C for the indicated time intervals, and squashed under a coverslip to be imaged by spinning confocal microscopy.
(B) Coomassie stained gels depicting recombinant Xvelo-GFP, Xvelo-4D-GFP, Xvelo-7D-GFP and F1-WT-GFP.
(C) Quantification of networks in (A). 20 images were taken and stitched together, a threshold was applied and the network intensities were measured. The integrated intensity of networks per sample at each time point (total network mass) is plotted. Means and standard errors of three biological replicates are shown.
See also Figure S4
To quantify Xvelo self-assembly in vitro, we tested Xvelo concentrations similar to its physiological concentration (Figure S2C). To balance between the aggregation propensity of Xvelo at high concentrations and the measured physiological concentration of 8 μM in oocytes if it was uniformly distributed, we chose a stock concentration of 15 μM for each recombinant protein so that after 10-fold dilution, the Xvelo concentration in the mixture was 1.5 μM. We quantified total network mass by summing the fluorescense intensity of networks in an area corresponding to a total of 20 images (Figure 4B, S4B). This analysis further confirmed that the F1 and full-length Xvelo-GFP have indistinguishable kinetics of network formation while the PLD mutants could not form any networks (Figure 4C). Xvelo-4D-GFP formed small precipitates after dilution out of high arginine, but these precipitates remained in solution without forming networks after overnight incubation (Figure 4A, third panel). Xvelo-7D-GFP was completely soluble with no precipitates upon dilution, indicating its complete inability to self-assemble. We also found that the assembly kinetics of Xvelo networks were dependent on Xvelo concentration (Figure S4C). We conclude that Xvelo can form networks on its own in vitro and that the Xvelo PLD is essential and sufficient for the formation of these networks.
Xvelo networks exhibit amyloid-like properties in vitro and in vivo
We next tested whether the Xvelo networks we see in vitro have amyloid-like properties. For this purpose we tested a number of different criteria. First we showed that both Xvelo-WT and F1-GFP networks were resistant to SDS treatment (Figure 5A). We noticed a partial solubilization of the Xvelo network by SDS (Figure 5A, Figure S5A), suggesting the presence of a more stable fiber backbone, which is decorated with more loosely associated Xvelo that can be released by treatment with SDS. Next we monitored acquisition of Thioflavin T (ThT) fluorescence, using RFP-tagged Xvelo, Fragment 1 and mutants Xvelo-4D and Xvelo-7D, and the yeast Mot3 prion as a rapidly-aggregating positive control (Alberti et al., 2009). F1 and full-length Xvelo networks quickly acquired ThT fluorescence in a PLD-dependent manner (Figure 5B). We supported these findings by showing that full-length and F1 networks were recognized by an anti-amyloid fibril antibody, whereas the mutants or an unrelated protein (EB1-RFP) were not (Figure 5C). Finally, negative stain TEM revealed that Xvelo-WT forms fibers reminiscent of amyloids in vitro in a PLD-dependent manner (Figure 5D).
Figure 5. Xvelo shows amyloid-like features in vivo and in vitro.
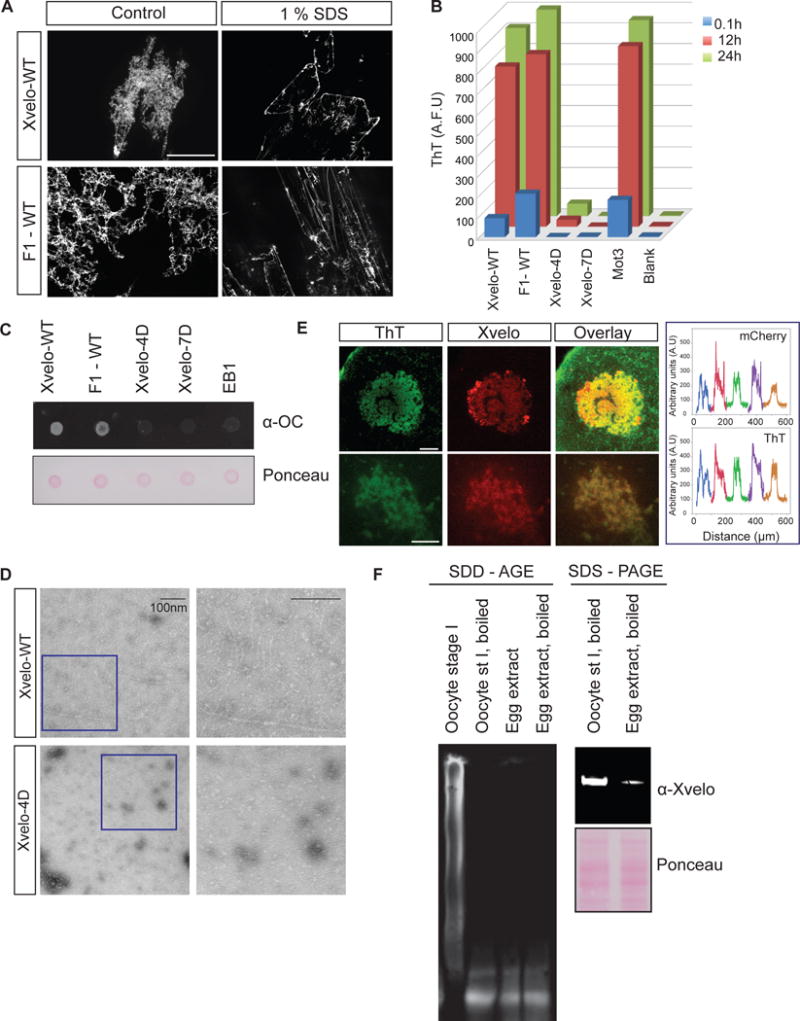
(A) SDS was added to Xvelo and F1-WT-GFP networks to a final 1 % concentration, and the reactions were incubated at room temperature for 15 minutes. The resulting mixtures were squashed under a coverslip and imaged by a spinning-disc confocal microscope.
(B) A final concentration of 5 μM of Thioflavin T was added to the wildtype, F1 and mutant network reactions at the indicated time points. Yeast prion Mot3 was used as a positive control, whereas blank was only buffer and ThT. ThT fluorescence was measured (arbitrary units) by a fluorescence plate reader.
(C) 1 μg of RFP tagged wild-type, F1 and mutant recombinant proteins were dot-blotted on a nitrocellulose membrane and assayed for reactivity with α-amyloid fibril OC. EB1-RFP was used as a negative control.
(D) Negative stain electron microscopy images of the untagged Xvelo-WT and Xvelo-4D self-assembly reactions (Scale bars 100 nm).
(E) Stage I oocytes were injected with mRNA coding for Xvelo-mCherry, and incubated overnight. The oocytes were incubated in 10 μM ThT, washed twice and imaged by confocal microscopy. Lower panel: Zoomed in images. Line scans showing the co-localization of Xvelo-mCherry and ThT stain from 5 Balbiani bodies were plotted. Each colour represents the line scan of a different Balbiani body. We speculate that the outer rim Xvelo-mCherry signal belongs to the newly translated Xvelo-mCherry protein that has just started to form a new, immature matrix, and does not yet stain with ThT.
(F) SDD-AGE detects SDS-resistant Xvelo aggregates in vivo. Equal amounts of cytoplasmic extracts of stage I oocytes and mature eggs were loaded onto SDS-PAGE. Five times more amount of egg extracts were loaded for SDD-AGE gels to make Xvelo concentrations comparable between the oocyte and egg extract lanes. Xvelo was detected by an anti-Xvelo antibody.
See also Figure S5
To confirm that the Xvelo matrix behaves like an amyloid in vivo, we examined whether Xvelo staining coincides with the strong staining of the Balbiani body with ThT. For this purpose, we injected oocytes with mRNA encoding for Xvelo-mCherry, incubated the oocytes with ThT and imaged them by live confocal microscopy. Xvelo and ThT signals overlapped strongly (Figure 5E). Considering Xvelo is the only enriched protein in the Balbiani body with a PLD, and that the endogenous Xvelo concentration exceeds 500 μM in the Balbiani body, the majority of the ThT signal likely comes from endogenous Xvelo.
Next, we used semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) to check the solubility of Xvelo. SDD-AGE allows the resolution of a wide size range of SDS-resistant aggregates (Alberti et al., 2010; Bagriantsev et al., 2006). We collected stage I oocytes and used egg extracts as a comparison. Indeed, Xvelo formed SDS-resistant aggregates in vivo detected by the SDD-AGE gel. Moreover, it did not form any detectable SDS-resistant aggregates in the mature egg (Figure 5F).
Therefore, because Xvelo forms SDS-resistant, filamentous assemblies that bind ThT in a manner that depends on the presence of its prion-like domain, we conclude that Xvelo forms amyloid-like networks in stage I oocytes and in vitro. Although Xvelo is present in the eggs at a detectable, albeit a much lower, concentration of ~ 80 nM (Figure 5F, Figure S2D), it does not form SDS resistant assemblies in the eggs (Figure 5F). Thus, the amyloid-like characteristics of Xvelo are transient, and regulated during development.
Prion-Like Domain Specificity for targeting to the Balbiani Body
To examine whether other proteins with prion-like domains might also be involved in organizing the Balbiani body, we first looked in our mass spectrometry list for other proteins with a PLD. However, apart from the common contaminant yolk protein, vitellogenin, none of the other enriched proteins in the Balbiani body were predicted to contain a prion-like domain.
To investigate whether Xvelo is unique in its ability to form a stable matrix in the Balbiani body, we selected 5 RNA binding proteins with prion-like domains, namely CPEB3, Dazap1, FUS, hnRNPA1 and Tia1, as well as the aggregation-prone mutant of FUS, FUS-156E, and injected mRNAs encoding for GFP-tagged versions of these proteins into the oocytes. Apart from CPEB3, all these proteins are present naturally in Xenopus eggs and oocytes (Wühr et al., 2014). Amongst these proteins, hnRNPA1, CPEB3, FUS and FUS156E did not localize to the Balbiani body (Figure 6A). Tia1 and Dazap1 localised to the Balbiani body (as well as the cytoplasm, and nucleus, respectively). However, fast turnover rates of both Tia1 and Dazap1 after photobleaching strongly suggest that they do not incorporate into a stable matrix (Figure S6A–B). Both Tia1 and Dazap1 are implicated in translational repression by binding to 3′UTRs of mRNAs (Dixon et al., 2003; Steger, 2001) and thus, their localization pattern can be explained by their binding to the repressed mRNAs in the Balbiani bodies upon overexpression.
Figure 6. Xvelo has unique properties for forming a stable matrix.
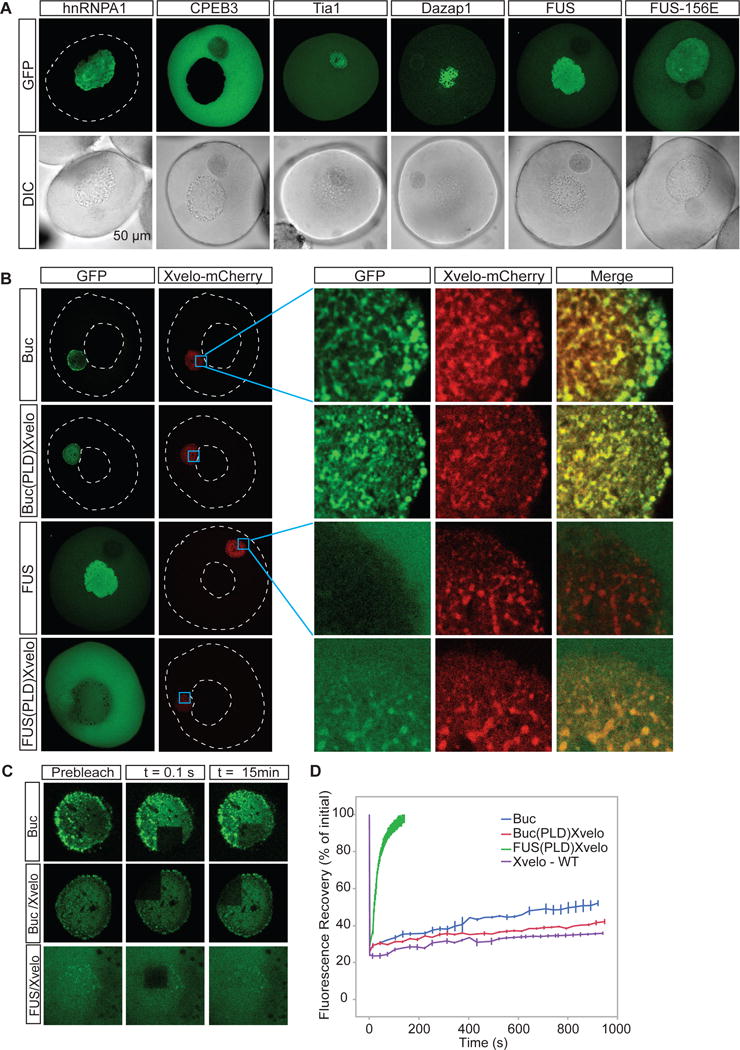
(A) mRNAs encoding for GFP tagged hnRNPA1, CPEB3, Tia1, Dazap1, FUS and FUS156E were in vitro synthesized and microinjected into the oocytes. After overnight incubation, the oocytes were imaged by scanning confocal microscopy.
(B) mRNAs encoding for Xvelo-mCherry and GFP tagged Bucky ball (Buc), FUS, and the PLD-swap versions of Xvelo, in which the PLD of Xvelo was replaced either by the PLD of Buckyball (BucPLDXvelo) or the PLD of FUS (FUSPLDXvelo) were injected into the oocytes at equal concentrations and imaged after overnight incubation.
(C) Internal rearrangement of fluorescent Buckyball-GFP (Buc), and PLD-swap versions of Xvelo, Buc(PLD)Xvelo-GFP and FUS(PLD)Xvelo-GFP after photobleaching over time.
(D) The fluorescent recovery of photobleached constructs in Balbiani bodies in (C), as well as Xvelo-WT, and two other biological replicates for each are shown by quantification of fluorescence in bleached region over time normalized by an unbleached neighbouring region.
See also Figure S6
Our experiments suggest that the PLD of Xvelo has specific features to target to and form a stable matrix in the Balbiani body. Part of the evidence that Xvelo structurally organizes Balbiani bodies is its sequence homology to zebrafish Bucky ball, whose genetics pointed to such a function (Bontems et al., 2009). To test if the two proteins exhibit similar sub-cellular dynamics, we expressed Bucky ball in Xenopus oocytes as a GFP fusion and looked at the characteristics of incorporation of Bucky ball into Xenopus Balbiani bodies. Bucky ball targeted to the Balbiani body, co-localizing with Xvelo (Figure 6B). Bucky ball turnover time after photobleaching was still a little faster than Xvelo, but of a similar order of magnitude (Figure 6C, 6D). Thus, we conclude that sequence features conserved in the PLDs of Buckyball and Xvelo are required for targeting to the Balbiani bodies in oocytes. This experiment also provides strong evidence that Bucky ball and Xvelo are functional homologues.
To further investigate the specificity of the Xvelo PLD for targeting to the Balbiani body, we swapped the PLD of Xvelo with the PLD of FUS, an unrelated prion-like RNA binding protein, and with the PLD of Bucky ball. The resulting chimeric proteins were named FUS(PLD)Xvelo and Buc(PLD)Xvelo, respectively (Figure S6C). Buc(PLD)XVelo localized to the Balbiani body, with a FRAP time in between Xvelo-WT and Buckyball-WT (Figure 6C, 6D). FUS(PLD)Xvelo localized to the cytoplasm, and weakly to the Balbiani body (Figure 6B). The Balbiani body-localized protein recovered quickly after photobleaching, with a half-life of approximately one minute, much faster than that observed for Buc(PLD)Xvelo (~2 hours) (Figure 6C, 6D). Taken together, these data provide strong evidence that the prion-like domains of functional homologs Xvelo and Bucky ball have unique features that target and form a stable matrix in the Balbiani body.
We next examined the specificity of PLD interactions in vitro with an aggregation assay that compared the assembly properties of Xvelo and FUS. We repeated previous reports showing that FUS-WT forms liquid droplets in vitro, whereas the aggregation prone mutant FUS-156E forms aggregates (Patel et al., 2015). We mixed Xvelo-RFP with either FUS-GFP or FUS-156E-GFP in vitro in a high arginine, high salt buffer and then diluted out the arginine and salt to initiate aggregation. Xvelo-RFP networks and FUS-WT-GFP droplets or FUS-156E-GFP aggregates formed in the vicinity of each other but clearly were separate, and did not interact with one another (Figure S6D). Thus, we conclude that PLDs do not aggregate with one another randomly, even when they are in close proximity at high concentration.
Xvelo binds RNA and clusters mitochondria
A key biological function of Xvelo during Balbiani body formation is likely to be the binding and concentration of organelles and RNA through its amyloid-like self-assembly. We looked for direct evidence that Xvelo can form a network that is sufficient to bind and concentrate mitochondria and RNA. To test whether Xvelo-GFP networks can bind RNA, we used a non-specific control mRNA coding for mCherry and an RNA that is enriched in Balbiani bodies, the Xenopus homolog of nanos, xcat-2 (Zhou and King, 1996). Both of the RNAs bound to the networks (only mCherry-RNA results are shown). In contrast, the F1 fragment that lacks the C-terminal motif did not bind to either of the RNAs. Thus, Xvelo networks can sequester RNA in a manner that requires a putative RNA-binding domain at the C-terminus of Xvelo (Figure 7A), but apparently without any strong RNA sequence preference in vitro.
Figure 7. Xvelo-GFP aggregates bind to RNA and cluster mitochondria.
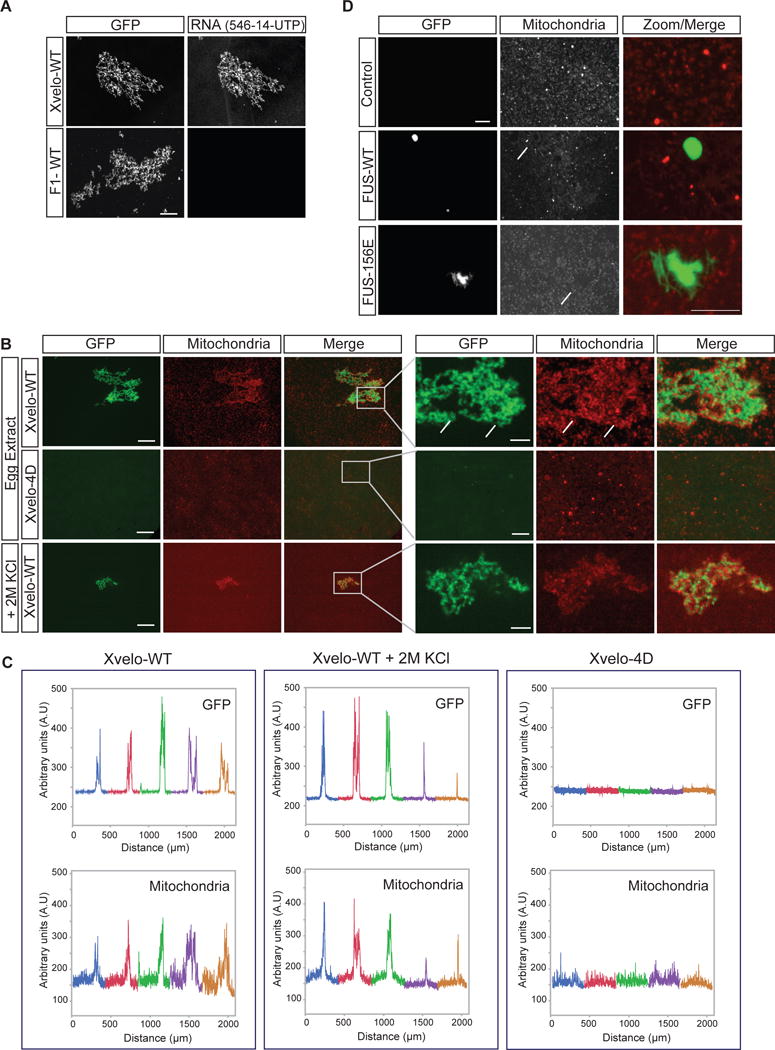
(A) Labeled RNAs, Xenopus nanos homolog xcat-2, and an mRNA encoding for mCherry protein were prepared using the MEGAscript SP6 kit with ChromaTide Alexa Fluor 546-14-UTP. RNAs were added to pre-assembled networks and imaged with a spinning-disc confocal microscope.
(B) Recombinant Xvelo-WT-GFP or the prion-like domain mutant, Xvelo-4D-GFP were added to Xenopus egg extracts with intact actin. Xvelo-GFP fills the gaps between mitochondria (arrows, compare to Figure 2B). Mitochondria were labeled with MitoTracker Deep Red. Images were taken with a spinning-disc confocal microscope. Lower panel: The extracts were diluted with KCl so that the final KCl concentration was 2M.
(C) Line scans of Xvelo-WT-GFP and Xvelo-4D-GFP and mitochondria in Xenopus egg extracts. Five images were stitched together to have an area spanning larger than 2 mm2. Each colour represents a different field.
(D) Recombinant FUS-WT-GFP or the aggregation prone mutant, FUS-G156E-GFP were added to Xenopus egg extracts with intact actin. Arrows point to the exclusion zones of mitochondria in presence of FUS structures.
See also Figure S7
Germ cells receive a pool of organelles from cyst cells to form Balbiani bodies and become oocytes in mice (Lei and Spradling, 2016). In early stage oocyte development, these organelles are clustered into Balbiani bodies by an unknown mechanism. We asked whether Xvelo can cluster organelles on its own in a cell free system, and thus, stimulate aspects of Balbiani body reconstruction in vitro. For this, we used an established cell-free system, cytoplasmic extracts from Xenopus eggs with intact actin, which contain abundant organelles and RNA, like the germ cell environment prior to Balbiani body formation (Field et al., 2014; Lei and Spradling, 2016). We labeled the mitochondria with Mitotracker and added Xvelo-GFP to the Xenopus egg extracts to 1.5 μM final concentration. Xvelo-GFP self-assembled as expected, and induced co-clustering of mitochondria (Figure 7B). Mitochondrial clustering activity depended on the PLD of Xvelo (Figure 7B, second panel). When we treated these clustered mitochondria with 2M KCl, mitochondria were still stably bound to, and entrapped by, the Xvelo network (Figure 7B, third panel). We used line-scans to quantify co-clustering of Xvelo assemblies and mitochondria over 5 frames, in an area extending 2 mm2 (Figure 7C). Note the similar line-scans for Xvelo-WT and mitochondria, demonstrating co-clustering, and the lack of mitochondrial clustering with the 4D mutant, which does not self-assemble. Although Fragment 1 can still form networks similar to full length networks in egg extracts, these networks cannot recruit mitochondria (Figure S7A). This allows us to speculate that entrapment of mitochondria is not based on the geometrical properties of Xvelo networks.
As a control, we introduced FUS-WT and FUS-156E to egg extracts. Both FUS-WT and FUS-156E behaved as expected in the egg extracts; WT formed droplets and 156E formed aggregates. Strikingly, FUS structures were avoided by mitochondria in the egg extracts, as opposed to the clustering we see by Xvelo (Figure 7D). Thus, we conclude that mitochondrial clustering in egg extracts is a specific property of Xvelo that requires its amyloid-like self-assembly.
DISCUSSION
Here, we show that Xvelo, a highly enriched protein in Xenopus Balbiani bodies, forms a mitochondria-embedding, SDS resistant matrix in vivo pervading the entire volume of the Balbiani body in early stage oocytes. Xvelo has a prion-like domain in its N terminus, which is sufficient and necessary to target and incorporate into the Balbiani body. Pure protein experiments further support our in vivo data that Xvelo has amyloid-like properties. Our key functional experiment, namely clustering of mitochondria by Xvelo in egg extracts is a reconstruction of aspects of Balbiani body formation. The fact that Xvelo can cluster mitochondria but the prion-like domain mutant does not, strongly suggests that this clustering is dependent on its prion-like domain. Thus, these data suggest that the organelle-rich Balbiani body is organized by a functional amyloid, into a dense matrix that sequesters mitochondria and other organelles.
The Balbiani body changes its properties during development: it is a stable structure in the early oocyte, and either disappears in mammals (Hertig and Adams, 1967; Pepling et al., 2007) or dissociates into small dispersed isles, called germ plasm, in the mature oocytes of germ plasm containing species. This suggests that its formation and dispersal are regulated. We did not detect any SDS-resistant Xvelo aggregates in egg extracts (Figure 5F), suggesting Xvelo does not form amyloid-like structures in the egg. How this transformation occurs remains unclear. Our preliminary data suggest that Xvelo is extensively phosphorylated in the egg, and most of these sites are not phosphorylated in the oocyte. Regulation by phosphorylation could be a mechanism determining the dispersal of the Balbiani body, perhaps by kinases or phosphatases that are activated at fertilization. Understanding the regulation of the physical state of Xvelo as the oocyte matures will be important to elucidate the fate of the organelles residing in the Balbiani body.
Balbiani bodies are found in most vertebrates; mouse Balbiani bodies were identified only recently, but Balbiani bodies in humans were observed decades ago, although they are almost untouched in the literature (Hertig and Adams, 1967; Pepling et al., 2007). Could proteins similar to Xvelo be required for the formation of Balbiani-like structures in other organisms? In zebrafish, bucky ball was identified in a maternal effect mutant screen as the only gene that is essential for Balbiani body formation (Dosch et al., 2004). Although the sequence similarity between Xvelo and Bucky ball proteins are poor (Data S1B), these proteins have long patches of intrinsically disordered regions, and score positive in prion detection programs (Data S1C). Oskar is a key protein required to organize Drosophila pole plasm (Ephrussi et al., 1991), but no homologs of Oskar have been identified in vertebrates. We found that Oskar also is a disordered protein with a predicted PLD (Data S1D). This suggests that amyloid-like self-assembly of a disordered protein could be an evolutionary conserved mechanism for Balbiani body formation. Strikingly, a recent paper has also linked the formation of large amyloid-like aggregates to gametogenesis in yeast, suggesting amyloid-like mechanisms may be involved in germ line specification across kingdoms (Berchowitz et al., 2015).
Despite the low sequence conservation of Xvelo and Bucky ball (Data S1B), key residues in their PLDs are conserved, suggesting that these residues are structurally important and underlie the observed specificity of assembly. Xvelo does not interact with other proteins with PLDs, such as FUS, upon self-assembly in vitro. This is in contrast to the promiscuous behavior of many disease-causing amyloids, which often show cross-seeding interactions and promote mutual nucleation events. We attribute this to the fact that Xvelo self-assembly does not involve an intermediate liquid-like state. We speculate that fast assembly into an inert amyloid-like structure prevents aberrant interactions with other prion-like proteins, thus reducing the likelihood of a disease condition. We propose that rapid assembly kinetics and high specificity are important driving forces underlying the evolution of functional amyloids.
What are the potential advantages of using an amyloid-like mechanism to form the Balbiani body? One could imagine packing away germ line components in amyloid-like structures is protective. The tightly packed structures could prevent other proteins from diffusing into them, such as regulators, thus keeping the organelles in a dormant state. It could act to slow down the diffusion of small toxic molecules generated by mitochondria, which could be damaging. It also provides a novel way to organize the cytoplasm, forming a rigid, giant ‘body’, in which the organelles are clustered together into one place and kept there for many years. Future studies are likely to provide mechanistic insight into the central question of how the germ line of an organism provides young cytoplasm with its complement of organelles in every generation, while the somatic cells age and die.
EXPERIMENTAL PROCEDURES
Detailed methods are available in the Supplemental Experimental Procedures.
Oocyte Handling and Injections
All experiments using Xenopus and zebrafish were done with approval of the HMS animal care review board. Ovaries were surgically removed from adult female Xenopus laevis frogs and treated with 2 mg/mL collagenase 1A (Sigma) in 1X MMR by gentle rocking, until most of the oocytes were clearly dissociated. Oocytes were later injected with mRNAs encoding for indicated proteins by using a FemtoJet express microinjector (Eppendorf).
Xvelo Protein Purification from Insect Cells
Recombinant versions of MBP-Xvelo-GFP, -RFP and no-fluorescent tag (for negative stain EM studies) were expressed in Sf9 insect cells using the baculovirus expression system. Insect cells were harvested in lysis buffer (50mM HEPES pH 7.6, 100 mM KCl, 1 M Arginine). The MBP (Maltose Binding Protein) tag was captured using Dextrin Sepharose Resin and cleaved off using HRV 3C Protease (MPI-CBG, in house) by incubation overnight on ice.
Microscopy
Differential interference contrast (DIC) and phase contrast microscopy for microinjections, perfusion chambers and Balbiani body isolations were performed using a standard wide-field epifluorescence Nikon inverted microscope equipped with a Hamamatsu Orca CCD camera and 4×, 10× and 20× dry objectives. Live confocal microscopy with oocytes was performed using Nikon A1R Laser Scanning confocal equipped with 10× dry and 40× water immersion objectives. Spinning disc confocal images were taken with a Nikon Ti inverted microscope with Yokagawa CSU-×1 spinning disk confocal with Spectral Applied Research Aurora Borealis modification, equipped with 20× dry, 40× and 60× oil immersion objectives.
Semi-denaturing Detergent-agarose Gel Electrophoresis (SDD-AGE)
Stage I oocyte and egg extracts were prepared according to (Field et al., 2014) with intact actin. SDD-AGE was adapted from Alberti et al., 2009. The protein concentrations of the lysates were adjusted and protein samples were mixed with 4× sample buffer (80mM Tris, 40mM acetic acid, 2mM EDTA, 20 % (v/v) glycerol, 3 % (w/v) SDS, bromophenol blue) and incubated at room temperature for 15 minutes before loading onto a 1.8 % agarose gel containing 1× TAE and 0.1 % SDS. The gel was run in running buffer (1× TAE, 0.1 % SDS) at 90 V, followed by wet-transfer to nitrocellulose membranes (Amersham Biosciences). Xvelo protein was detected by an anti-Xvelo antibody.
Supplementary Material
1
10
11
12
2
3
4
5
6
7
8
9
Highlights.
- The organelle content of the Balbiani body is held together by an Xvelo matrix.
- Xvelo forms amyloid-like networks in vitro, which can recruit RNA and mitochondria.
- Prion-like domain of Xvelo dictates specificity in amyloid assembly.
- Amyloid-like polymerization is conserved among vertebrate Balbiani body organizers.
Acknowledgments
We thank members of T.J.M and A.A.H labs, especially Avinash Patel for helpful discussions, Andrei Pozniakovski for cloning, and Christine Field for help in making extracts. We’d like to thank Doris Richter and Sonja Kroschwald for technical assistance. We are grateful to the Protein Expression, Electron Microscopy and Image Processing facilities of the MPI-CBG for their support. We would like to thank Nikon Imaging Center at Harvard Medical School for microscopy support. Proteomic analysis was supported by NIH Grant R01 GM103785 (PI Marc Kirschner). M.W. was supported by the Charles A. King Trust Postdoctoral Fellowship. This work was supported by the MaxSynBio consortium, which is jointly funded by the Federal Ministry of Education and Research of Germany and the Max Planck Society to A.A.H. and NIH grant GM39565 to T.J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
E.B. and T.J.M. conceived the project together. E.B. designed the experiments with A.A.H., S.A. and T.J.M. All experiments were performed by E.B., except for mass spectrometry analysis by M.W. and S.P.G and electron microscopy by M.C. and M.R. Protein expression and purification were performed by M.R., D.D., and R.L. The manuscript was written by E.B. with input from A.A.H., S.A. and T.J.M.
References
- Al-Mukhtar KA, Webb AC. An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. Journal of embryology and experimental morphology. 1971;26:195–217. [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, Lindquist S. Biochemical, cell biological, and genetic assays to analyze amyloid and prion aggregation in yeast. Methods in enzymology. 2010;470:709–734. doi: 10.1016/S0076-6879(10)70030-6. [DOI] [PubMed] [Google Scholar]
- Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods in enzymology. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Balinsky B, Devis RJ. Origin and differentiation of cytoplasmic structures in the oocytes of Xenopus laevis. Acta Embryol Morphol Exp. 1963;6:55–108. [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billett F, Adam E. The structure of the mitochondrial cloud of Xenopus laevis oocytes. Journal of embryology and experimental morphology. 1976;36:697–710. [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Current Biology. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- de la Paz ML, Serrano L. Sequence determinants of amyloid fibril formation. Proceedings of the National Academy of Sciences. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, Prescott SM. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. The Journal of experimental medicine. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Developmental cell. 2004;6:771–780. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- England JL, Haran G. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annual review of physical chemistry. 2011;62:257. doi: 10.1146/annurev-physchem-032210-103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Field CM, Nguyen PA, Ishihara K, Groen AC, Mitchison TJ. Xenopus egg cytoplasm with intact actin. Methods Enzymol. 2014;540:399–415. doi: 10.1016/B978-0-12-397924-7.00022-4. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS biology. 2006;4:100. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta T, Marlow FL, Ferriola D, Mackiewicz K, Dapprich J, Monos D, Mullins MC. Microtubule actin crosslinking factor 1 regulates the Balbiani body and animal-vegetal polarity of the zebrafish oocyte. PLoS Genet. 2010;6:e1001073–e1001073. doi: 10.1371/journal.pgen.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nature reviews Molecular cell biology. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim AE, Hartung O, Rothhamel S, Ferreira E, Jenny A, Marlow FL. Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development. 2014;141:842–854. doi: 10.1242/dev.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig AT. The primary human oocyte: some observations on the fine structure of Balbiani’s vitelline body and the origin of the annulate lamellae. American Journal of Anatomy. 1968;122:107–137. doi: 10.1002/aja.1001220107. [DOI] [PubMed] [Google Scholar]
- Hertig AT, Adams EC. Studies on the human oocyte and its follicle I. Ultrastructural and histochemical observations on the primordial follicle stage. The Journal of cell biology. 1967;34:647–675. doi: 10.1083/jcb.34.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Current topics in developmental biology. 2000;50:155–IN152. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Etkin LD. The Balbiani body and germ cell determinants: 150 years later. Current topics in developmental biology. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- Kloc M, Spohr G, Etkin LD. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- Kogo N, Tazaki A, Kashino Y, Morichika K, Orii H, Mochii M, Watanabe K. Germ-line mitochondria exhibit suppressed respiratory activity to support their accurate transmission to the next generation. Developmental biology. 2011;349:462–469. doi: 10.1016/j.ydbio.2010.11.021. [DOI] [PubMed] [Google Scholar]
- Koo EH, Lansbury PT, Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proceedings of the National Academy of Sciences. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, King OD. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu310. btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Spradling AC. Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science. 2016;352:95–99. doi: 10.1126/science.aad2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Page DC. Genetics of germ cell development. Nature Reviews Genetics. 2012;13:781–794. doi: 10.1038/nrg3294. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nature reviews Molecular cell biology. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Developmental biology. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Analytical chemistry. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Pepling ME, Wilhelm JE, O’Hara AL, Gephardt GW, Spradling AC. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proceedings of the National Academy of Sciences. 2007;104:187–192. doi: 10.1073/pnas.0609923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nature reviews Molecular cell biology. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anatomy and embryology. 2001;203:323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. Proceedings of the National Academy of Sciences. 2012;109:6519–6524. doi: 10.1073/pnas.1119366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumoto K, Umetsu M, Kumagai I, Ejima D, Philo JS, Arakawa T. Role of arginine in protein refolding, solubilization, and purification. Biotechnology progress. 2004;20:1301–1308. doi: 10.1021/bp0498793. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman DA, Kelly AC, Gorkovskiy A, Dayani Y, Zhou A. Amyloids and yeast prion biology. Biochemistry. 2013;52:1514–1527. doi: 10.1021/bi301686a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Freeman RM, Presler M, Horb ME, Peshkin L, Gygi SP, Kirschner MW. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Current Biology. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, King ML. Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development. 1996;122:2947–2953. doi: 10.1242/dev.122.9.2947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
10
11
12
2
3
4
5
6
7
8
9