An inducible long noncoding RNA amplifies DNA damage signaling (original) (raw)
. Author manuscript; available in PMC: 2017 Mar 26.
Published in final edited form as: Nat Genet. 2016 Sep 26;48(11):1370–1376. doi: 10.1038/ng.3673
Abstract
Long noncoding RNAs (lncRNAs) are prevalent genes with frequently exquisite regulation but mostly unknown functions. Here we demonstrate a role of lncRNAs in guiding organismal DNA damage response. DNA damage activates transcription of DINO (_D_amage _I_nduced _NO_ncoding) via p53. DINO is required for p53-dependent gene expression, cell cycle arrest, and apoptosis in response to DNA damage, and DINO expression suffice to activate damage signaling and cell cycle arrest in the absence of DNA damage. DINO binds to and promotes p53 protein stabilization, mediating a p53 auto-amplification loop. Dino knockout or promoter inactivation in mice dampens p53 signaling and ameliorates acute radiation syndrome in vivo. Thus, inducible lncRNA can create a feedback loop with its cognate transcription factor to amplify cellular signaling networks.
INTRODUCTION
Cells are constantly subject to an array of external stimuli. Signal transduction pathways must record diverse inputs, and integrate these with prior experiences of the cell to decide whether to proliferate, differentiate, or die. The DNA damage response is critical for normal cell proliferation and suppression of cancer, and relies on the transcription factor p53. In mammals, the p53-dependent response to DNA damage is complex and tissue specific1. While the p53-dependent, p21-mediated cell cycle arrest in response to DNA damage can protect against radiation-induced gastrointestinal, cardiac, and hematologic toxicity2–4, p53-dependent apoptosis drives radiation toxicity in hematopoietic cells in a p53 dose-dependent manner5. Indeed, _p53−/−_mice are strikingly resistant to myeloablative doses of ionizing radiation due to a failure to activate the intrinsic apoptosis pathway, while p53+/− mice have intermediate radiation sensitivity between that of p53+/+ and p53−/− mice. Thus, fine-tuning the choice and amplitude of different p53 target genes is a critical aspect of the DNA damage response.
Eurkaryotic genomes are pervasively transcribed to generate diverse lncRNAs, especially from highly regulated enhancers and promoters6,7. Recently, several lncRNAs have been identified that regulate specific subsets of the p53-dependent gene expression signature8,9. The DNA damage-induced lncRNA PANDA negatively regulates apoptosis by blocking the transcription factor NF-YA, while lincRNA-p21 recruits hnRNP-K to regulate p21 in cis10–12. Furthermore, the APELA RNA expressed in mouse embryonic stem cells binds to hnRNPL to block its interaction with p53 and permit p53 accumulation in the mitochondria to elicit apoptosis13. While p53 is known to bind RNA14, the role of the lncRNAs in regulating the p53 protein remains mostly unknown.
p53 signaling is modulated by the regulation of p53 protein abundance. During normal cell cycles, a low level of damage resulting from DNA replication transiently activates p53 but is insufficient to robustly induce p53-responsive genes. This is because p53 has a short half-life and exists in a conformation with limited DNA binding efficiency15,16. With sustained DNA damage, p53 is stabilized, and p53-responsive genes (such as CDKN1A, DDB2, PUMA) are activated, leading to either cell cycle arrest or apoptosis. While still poorly understood, sustained DNA damage is believed to invoke a signal amplification mechanism that requires an, as yet, unidentified co-activator of the p53 response16. Here we report that a p53-inducible lncRNA serves as a key gating mechanism in the DNA damage response.
RESULTS
DINO is a conserved, DNA damage-inducible lncRNA
We identified a DNA damage-induced transcript upstream of CDKN1A, hereafter named DINO (Damage Induced Noncoding), in a screen for transcribed regions in human cell cycle promoters10 (Fig. 1a and Supplementary Fig. 1a–d). RACE showed that DINO is a 951-base RNA transcribed divergently from CDKN1A. Codon substitution frequency (CSF score = −55.7) and in vitro translation suggests that DINO does not code for protein (Supplementary Fig. 1e). DINO is induced ~100-fold in primary human fibroblasts in response to sustained doxorubicin-induced DNA damage, peaking at 10–24 hrs post damage and reaching ~1,000 copies per cell (Supplementary Fig. 1f). In contrast, the known p53 target gene CDKN1A is induced 5–10 fold (Fig. 1b). DINO is also induced upon DNA damage in human cancer cell lines and by other stressors, albeit at lower levels (Supplementary Fig. 1g–h).
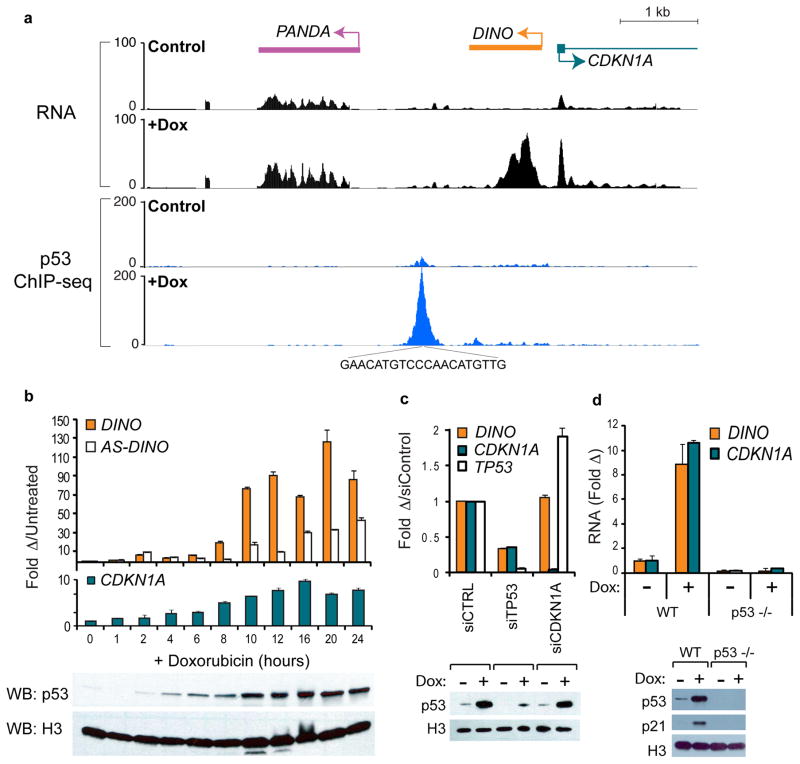
Figure 1. DINO encodes a p53-dependent DNA damage-induced transcript.
(a) Upper Panel: Transcription across the CDKN1A locus in human fibroblasts before or after 24 hours of doxorubicin (Dox) treatment, (Hung et al., 2011). Lower Panel: p53 ChIP-seq in untreated or doxorubicin treated U2OS cells.
(b) Time course of DINO and CDKN1A expression in human fibroblasts after DNA damage. Below: p53 and histone H3 immunoblot (WB). Mean +/− s.d are shown, n = 3.
(c) DINO expression and p53 immunoblot in fibroblasts treated with siCDKN1A or siTP53 24 hours prior to dox treatment. Mean +/− s.d. are shown, n = 3.
(d) DINO expression and p53 immunoblot in p53+/+ wildtype (WT) or p53−/− HCT116 cells +/− Dox (16hr). Mean +/− s.d are shown, n = 3.
See also Supplementary Figs. 1–2.
Because DINO is situated adjacent to a p53 binding site (Fig. 1a), we reasoned that its induction may require p53. Knockdown of p53 in human fibroblasts abrogated DINO induction (Fig. 1c). Wildtype HCT116 colon cancer cells induced DINO in response to doxorubicin, while isogenic p53−/− cells did not (Fig. 1d). Similarly, _p53_-null H1299 cells only induced DINO after complementation with wild type p53, but not with a p53 mutant derived from cancer-prone Li-Fraumeni syndrome (Supplementary Fig. 1i). DINO is not induced in p53 mutant tumor cell lines (Supplementary Fig. 1j).
One indication of a lncRNA’s functional significance is its evolutionary conservation. Functionally related lncRNAs between human, mouse and zebrafish can be identified by genomic synteny and conserved regions of microhomology, despite limited overall sequence identity17,18. Thus, we queried the mouse and zebrafish CDKN1A promoters for a DNA damage-inducible lncRNA immediately upstream of the first exon. A mouse transcript sense to Cdkn1a was observed at this position in mouse embryonic fibroblasts (MEFs) after doxorubicin treatment (Supplementary Fig. 2a). RACE, Northern blot, and RT-PCR established identity of the putative mouse Dino (Supplementary Fig. 2a–c). DNA damage induction of mouse Dino is also attenuated in p53−/− MEFs (Supplementary Fig. 2a). Similarly, RT-PCR and RACE analysis of zebrafish embryos revealed a UV-inducible sense-strand lncRNA at the precise syntenic location upstream of CDKN1A (Supplementary Fig. 2d–f). Several regions of microhomology were observed in human, mouse, and zebrafish DINO that are also conserved in the putative DINO encoding regions of five additional mammalian species (Supplementary Fig. 2g–i). Thus, DINO represents a potentially conserved transcriptional response to DNA damage.
DINO regulates the p53-dependent DNA damage response
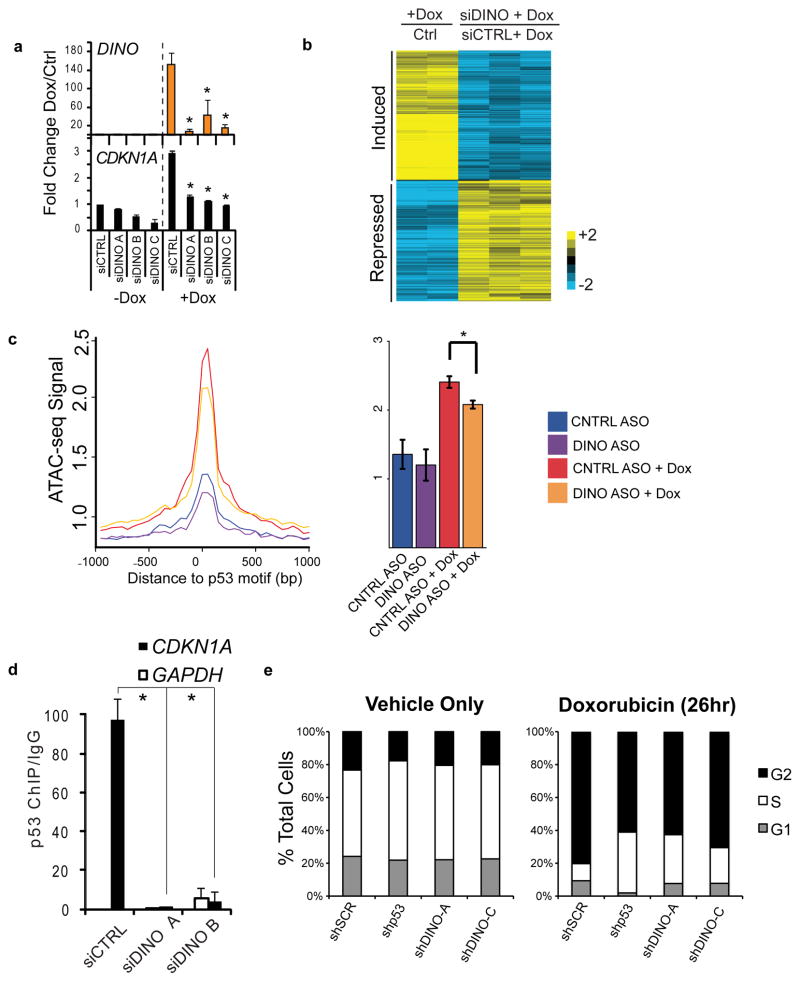
LncRNAs can regulate gene expression both in cis and in trans. Depletion of DINO by RNA interference revealed a key role in the DNA damage response. DINO depletion by multiple independent siRNAs in primary human fibroblasts blunted CDKN1A induction upon DNA damage (Fig. 2a). Microarray analysis showed that 215 of 417 genes normally regulated by DNA damage failed to respond after DINO depletion, including canonical p53-responsive genes CDKN1A, DDB2, GADD45α (Fig. 2b). KEGG pathway analysis revealed an enrichment of the p53 signaling pathway (P = 3.3 × 10−3, FDR < 0.05) (GO Terms: Supplementary Table 1, Gene Lists: Supplementary Table 2), and a majority of the genes affected by siDINO (60%) had a canonical p53-binding site. Assay of Transposase Accessible Chromatin by sequencing (ATAC-seq)19 revealed that chromatin accessibility at cognate p53 binding sites is induced by DNA damage genome-wide, but is reduced in DINO-depleted human fibroblasts (Fig. 2c, Supplementary Fig. 3b). Indeed, ChIP-qPCR showed DINO depletion caused a loss of p53 occupancy at its cognate target genes, such as at CDKN1A (Fig. 2d) and additional genes (Supplementary Fig. 3c–d). Finally, DINO is required for cell cycle arrest in response to DNA damage. Similar to p53 shRNA cells, DINO-depleted U2OS cells continued to divide following DNA damage when compared to control DINO-proficient cells (Fig. 2e and Supplementary Fig. 3e). Similar results were obtained with independent siRNAs that target DINO (Supplementary Fig. 3f).
Figure 2. Loss of DINO abrogates DNA damage-induced gene regulation and cell cycle arrest.
(a) DINO and CDKN1A expression in fibroblasts treated with the indicated siRNAs in the presence or absence of doxorubicin (24hr), as measured by qRT-PCR. Mean +/− s.d. are shown; *, P < 0.05 compared to siCTRL (students T-test), n = 3.
(b) Columns 1 and 2: heatmap of genes > 2 fold changed in fibroblasts upon DNA damage with doxorubicin (26hr) by microarray analysis. Same genes were represented as DINO knockdown + Dox relative to control + Dox in columns 3–5.
(c) Genome-wide chromatin accessibility at p53 binding elements in fibroblasts treated with doxorubicin and DINO knockdown as indicated (24hr), as measured by ATAC-seq. Mean +/− s.e.m are shown. * P = 1.2 × 10−4, Control ASO (n = 4), Control ASO + Dox (n = 4), DINO ASO (n = 6), DINO ASO + Dox (n = 7).
(d) p53 ChIP-qPCR at indicated promoters in control or DINO knockdown fibroblasts treated with doxorubicin (24hr). Mean +/− s.d are shown, n = 3.
(e) Cell cycle arrest following 26 hours of doxorubicin treatment in shControl and shDINO U2OS cells.
See also Supplementary Fig. 3 and Supplementary Tables 1–2.
DINO and p53 physically interact following DNA damage
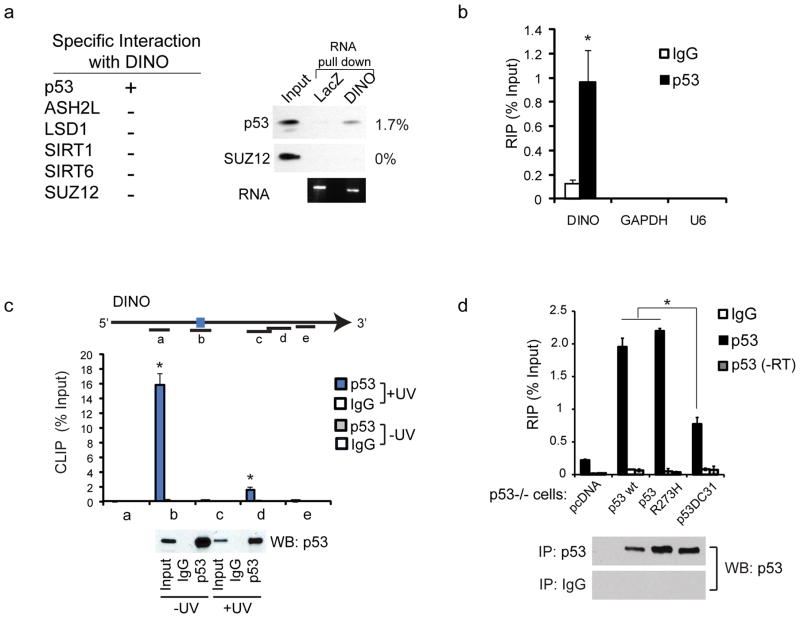
LncRNAs can act as modular scaffolds for chromatin modification complexes20, but their roles in mediating signal transduction are less understood. Therefore we asked whether DINO may physically interact with p53. In vitro transcribed DINO RNA preferentially retrieved p53 from the lysate of DNA damaged cells, but did not retrieve the Polycomb complex or several other control proteins (Supplementary Fig. 4a–b). DINO also had an increased affinity for recombinant purified p53 compared to PRC2 (Fig. 3a). In complementary experiments, immunoprecipitation of p53 from the chromatin of DNA damaged cells retrieved endogenous DINO RNA (Fig. 3b). Analysis of RNA fragments UV-crosslinked to endogenous p53 in DNA-damaged cells identified a major p53-binding site in DINO (Fig. 3c). Expression of wild type p53 or a DNA binding mutant R273H in _p53_-null cells enabled p53 retrieval of DINO, but a C-terminal deletion truncating a RNA-binding region of p53 (Ref. 14) decreased DINO interaction (Fig. 3d). Thus, the C-terminus of p53, which is also known to undergo extensive post-translational modifications, binds to a discrete domain of DINO. In order to investigate whether DINO localized to regulatory regions adjacent to DINO-dependent genes, we used ChIRP21 to map DINO’s chromatin occupancy. DINO RNA was specifically retrieved with orthogonal “even” and “odd” pools of biotinylated capture probes along with associated DNA; RNase-treated chromatin served as negative control (Supplementary Fig. 4c–d). We observed DINO occupancy at p53 binding elements in 7 of 10 p53-target genes examined, including CDKN1A, GADD45A, and _DDB_2; DINO occupancy was not detected at any of the 5 negative control loci (Supplementary Fig. 4e). These results suggest that DINO and p53 co-localize at multiple p53 target genes throughout the genome.
Figure 3. p53 is a DINO-binding protein.
(a) Left: Summary of RNA chromatography with DINO retrieval of proteins from whole cell lysate of doxorubicin (24hr) treated fibroblasts. Right: RNA chromatography using 6 pmol RNA and 1 ug of each recombinant protein as indicated.
(b) RNA IP of p53 in human fibroblasts followed by qRT-PCR of DINO, GAPDH and U6 snRNA. *, P < 0.05 compared to IgG control (students T-test). Mean +/− s.d. are shown, n = 3.
(c) UV crosslinking and immunoprecipitation (CLIP) of p53-bound DINO RNA in doxorubicin (24hr) treated human fibroblasts. qRT-PCR identifies the DINO region bound by p53 in vivo. Location of primer pairs along the DINO transcript indicated in the diagram above. Bottom: Immunoblot of p53 retrieved by CLIP. Mean +/− s.d. are shown, n = 3.
(d) p53 RIP of DINO with the indicated p53 constructs in p53−/− H1299 cells. Mean +/− s.d. are shown, n = 3.
See also Supplementary Fig. 4.
DINO stabilizes p53 and induces p53 target genes
Since DINO directly binds p53, we examined whether DINO expression regulates p53 protein following DNA damage. In response to DNA damage, p53 is stabilized by a series of post-translational modifications and inhibition of ubiquitination, leading to protein accumulation and transactivation of p53 targets. DINO depletion blocked the ability of DNA damage to induce p53 stabilization and p21 induction, despite preservation of p53 phosphorylation at Serine-9 (Fig. 4a), suggesting that DINO is required for p53 stability in the DNA damage response.
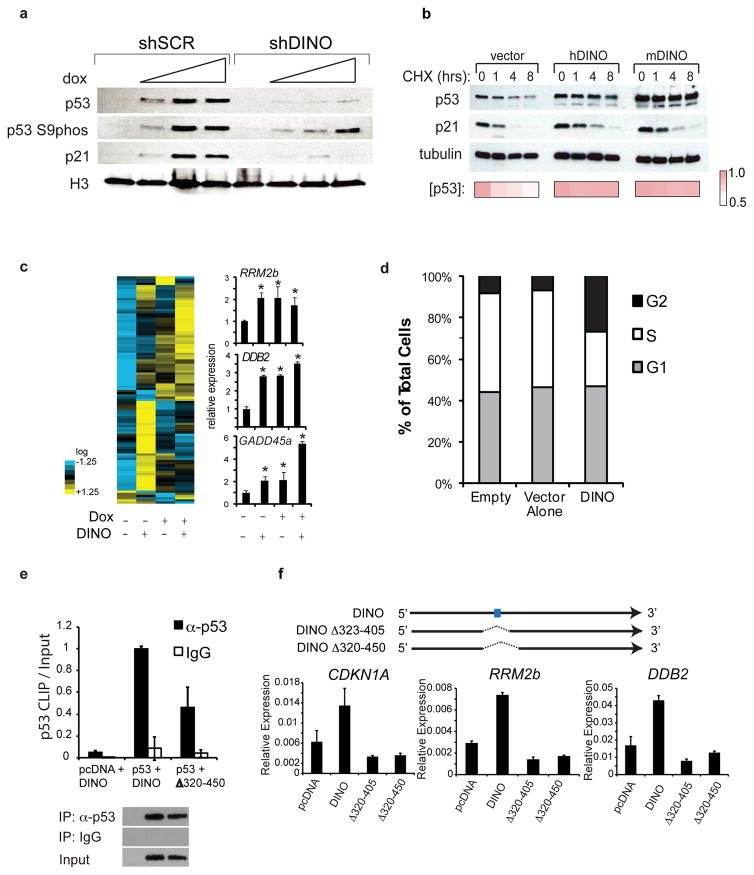
Figure 4. DINO stabilizes p53 and enhances regulation of its target genes.
(a) Immunoblot of p53, p53 phospho-S9, and p21 in shControl (shSCR) and shDINO U2OS cells. Doxorubicin (12hr) was used at the following concentrations: 0, 0.05, 0.1, 0.2 ug/ml.
(b) Human or mouse DINO expression stabilizes p53 in fibroblasts, as indicated by cycloheximide (CHX) chase analysis. p53 and p21 levels at indicated hours post cycloheximide addition. p53 densitometry values are indicated below.
(c) p53 target gene expression in 293 cells with empty vector, DINO overexpression, doxorubicin treatment (16hr), or DINO + doxorubicin. Left: nCounter quantification. Right: confirmation using qRT-PCR. Mean +/− s.d. are shown; *, P < 0.05 (students T-test), n = 3.
(d) Cell cycle analysis of BrdU/PI labeled 293 cells transfected with DINO.
(e) p53 retrieval of wildtype DINO or a DINO mutant lacking the putative p53 binding site, measured by p53 CLIP-qRT-PCR. Mean +/− s.d. are shown, n = 3.
(f) qRT-PCR for p53 target genes in 293 cells transfected with vector, full length DINO, or DINO deletion mutants DINOΔ323–405 or DINOΔ320–450 that lack the CLIP RNA fragment. Blue box indicates the primary p53 binding site identified by UV-CLIP-qPCR. Mean +/− s.d. are shown, n = 3.
See also Supplementary Fig. 5.
If DINO binding directly stabilizes p53 protein, overexpression of DINO in the absence of DNA damage may be sufficient to stabilize p53 and activate DNA damage signaling. Cycloheximide chase revealed that p53 protein was rapidly degraded in control cells, whereas enforced human or mouse DINO expression, in the absence of DNA damage, caused p53 stabilization and increased p21 protein level (Fig. 4b). DINO over-expression induced a large panel of p53 target genes including RRM2b, DDB2, and GADD45a, as measured by nCounter and qRT-PCR assays, and also caused substantial G2 cell cycle arrest, a well known p53-dependent checkpoint22 (Fig. 4c–d). These results indicate that DINO can act in trans when expressed separately from the CDKN1A locus, and highlight the functional conservation of human and mouse DINO.
Although DINO directly binds p53, it is possible that DINO regulates p53 in a manner independent of p53 binding. To examine this possibility, we generated DINO mutation constructs with focal deletion of the p53-binding motif. This motif forms a stem loop structure in vitro (Supplementary Fig. 5a–b), and deletion of this motif abrogated p53 binding in vivo (Figure 4e). Overexpression of DINO mutants failed to induce p53 target genes (Fig. 4f) or stabilize p53 protein (Supplementary Fig. 5c), confirming that DINO binding to p53 is important to activate the DNA damage response.
Dino knockout mice are deficient in p53 pathway functions
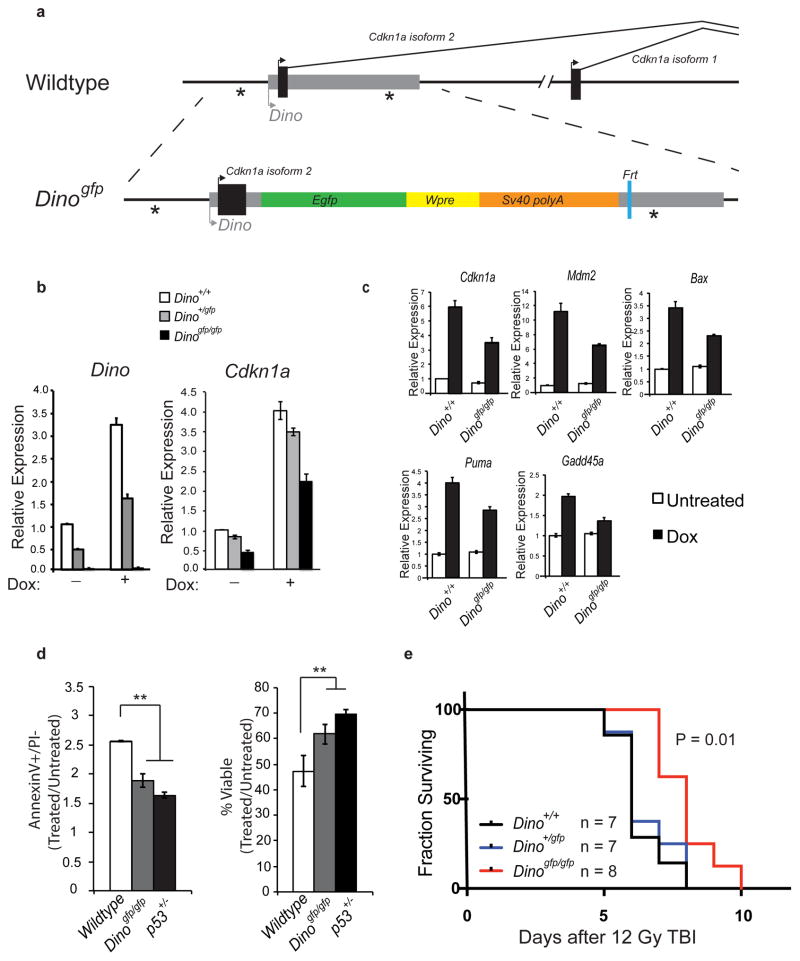
We generated C57BL/6 mice with genetic modification of Dino in order to characterize the role of the mouse lncRNA in the p53 response (Fig. 5a). The knockin allele Dinogfp was designed to achieve two purposes: (1) report the activity of the putative Dino promoter, and (2) disrupt the function of Dino by replacing the bulk of Dino sequence with GFP. We designed the targeting construct to avoid removal of endogenous p53 response elements within the Cdkn1a promoter (*). FRT flanked neomycin selection cassettes were removed by recombination in ES cells prior to blastocyst injection. Dinogfp/gfp mice are born with the expected M:F Mendelian ratio, and are viable, fertile, and have no apparent developmental defects.
Figure 5. Germline disruption of mouse Dino impairs the p53-dependent, DNA damage-induced apoptosis.
(a) Design of the Dinogfp allele.
(b) DNA damage induces Dino and Cdkn1a expression in MEFs of indicated genotype 16 hours after 0.2 ug/mL doxorubicin, measure by qRT-PCR. Mean +/− s.d. are shown, n = 3.
(c) qRT-PCR of DNA damage inducible, p53 responsive genes in MEFs of indicated genotype under conditions of physiologic oxygen (2%). Mean +/− s.d. are shown, n = 3.
(d) Relative apoptotic and viable fractions of thymocytes of indicated genotypes 6 hours after 1 Gy irradiation. Mean +/− s.d. are shown, ** P < 0.01 (students T-test), n = 3.
(e) Kaplan Meier survival curve of mice treated with a single fraction of lethal total body irradiation of 12 Gy. P < 0.01 (log-rank test for trend).
See also Supplementary Fig. 6.
MEFs isolated from Dino+/gfp E13.5 embryos induce GFP expression in response to DNA damage (Supplementary Fig. 6a), indicating that we did capture the Dino promoter. While Dino+/gfp MEFs retain 90% of the basal and DNA damage induced Cdkn1a expression relative to Dino+/+ MEFs, the expression of Cdkn1a in homozygous Dinogfp/gfp MEFs is decreased by nearly 50% (Fig. 5b). That C_dkn1a_ expression remains largely intact in Dino+/gfp MEFs relative to Dinogfp/gfp MEFs suggests that a single wildtype allele of Dino is sufficient to maintain near-wildtype levels of Cdkn1a. This supports a model of Dino regulation of gene expression in trans. We next examined whether mouse Dino, like human DINO, regulates p53-dependent gene expression in response to DNA damage. We cultured MEFs under low physiologic oxygen tension (2%), avoiding hyperoxic conditions that can activate p53 and induces senescence23. Dinogfp/gfp MEFs exhibit significantly dampened induction of canonical p53 target genes that control cell cycle, apoptosis, and DNA repair, including Cdkn1a, Mdm2, Bax, Puma, and Gadd45a (Fig. 5c).
Since gene expression analysis indicated a role for mouse Dino in the regulation of apoptosis signaling, we next examined Dinogfp mice for defects in DNA damage induced apoptosis. Ex vivo irradiation of thymocytes showed diminished apoptosis and enhanced survival in Dinogfp/gfp thymocytes relative to Dino+/+ thymocytes (Supplementary Fig. 5b). Since p53-dependent, radiation-induced apoptosis differs amongst thymocyte subsets24, we specifically examined the subset most sensitive to radiation, CD4+/CD8+ thymocytes. We observed reduced levels of apoptosis in the CD4+/CD8+ thymocytes of Dinogfp/gfp relative to Dino+/+ mice 6 hours after in vivo irradiation with 5Gy total body irradiation (TBI, Supplementary Fig. 6c). The significant but partial defect in apoptosis in Dinogfp/gfp thymocytes is similar to the defect observed in heterozygous p53+/− thymocytes (Fig. 5d)24. Such partial abrogation of DNA damage-induced apoptosis has also been observed in mice mutated in other important mediators of p53-induced apoptosis, such as Perp−/− and Puma+/− 25,26.
p53 modulates the organismal response to DNA damage and is a major determinant of tissue injury following lethal irradiation. Therefore, we examined whether loss of Dino altered the sensitivity of mice to lethal irradiation. Following exposure to a single, high dose fraction of 12Gy TBI, Dinogfp/gfp mice lived significantly longer than Dino+/+ and Dino+/gfp littermates (P = 0.01, Fig. 5e). Heterozygous Dino+/gfp mice had radiation sensitivity that was indistinguishable from wildtype mice. The organismal resistance to radiation toxicity in Dinogfp/gfp is reminiscent of Puma-deficient mice, encoding a key mediator of p53-induced apoptosis26–28. Furthermore, because _Cdkn1a_−/− animals have increased radiation sensitivity by several regimes2,4 that is the opposite of the Dino phenotype, our findings indicate that Dino’s role in DNA damage response must extend beyond cis regulation of Cdkn1a.
Dino regulates p53 signaling independent of p21
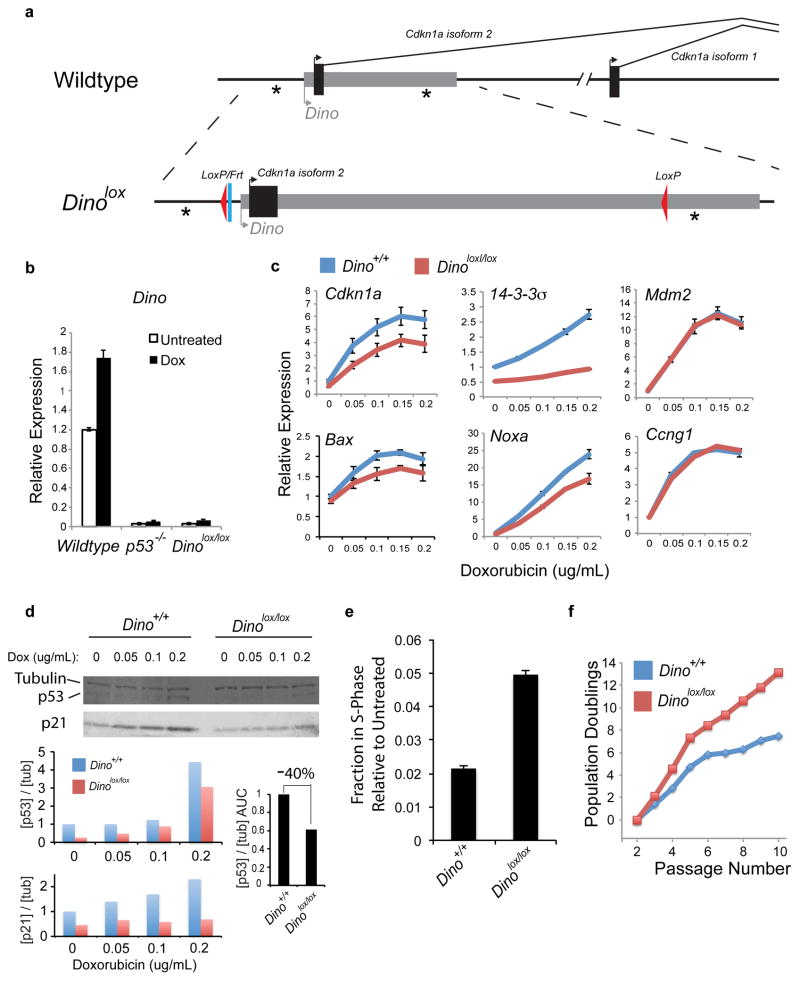
While the Dinogfp allele created a robust gene knockout and was ideally suited to investigate the role of Dino in regulating genes in trans, we created a second mouse with a focal modification of the Dino promoter in order to study the activity of DINO both in cis and in trans (Fig. 6a). The Dinolox allele contains ~150bp of exogenous DNA consisting of frt and loxP recombination sites and was intended to be used as a conditional knockout. This targeting strategy resulted in the smallest alteration of the Dino locus, and did not disturb any known transcription factor binding sites or Cdkn1a exons. Unexpectedly_,_ Dino expression is nearly completely attenuated in Dinolox/lox MEFs even in the absence of Cre-mediated recombination, and DNA-damage induced Dino is abrogated to a level to similar to p53−/− MEFs (Fig. 6b). The Dinolox/lox allele thus serves as a Dino promoter knockout.
Figure 6. Dino regulates the DNA damage response in both a p21-dependent and p21-independent manner.
(a) Design of Dinolox allele.
(b) Dinolox/lox inactivates basal and DNA-damage inducible expression of Dino lncRNA. Dino expression in MEFs with indicated genotypes +/− 0.2ug/mL dox for 8 hours is measured by qRT-PCR. p53−/− MEFs is shown for comparison. Mean +/− s.d. are shown, n = 3.
(c) qRT-PCR of p53-dependent transcripts in MEFs of indicated genotype 8 hours after treatment with the indicated doses of doxorubicin. Mean +/− s.d. are shown, n = 3.
(d) p53 protein abundance following DNA damage with dox. Top: Immunoblot. Bottom: Quantification of p53 and p21 protein abundance normalized to beta-tubulin. Summed area under the curve [AUC] shows 40% reduction of p53 protein accumulation in Dinolox/lox cells.
(e) Cell cycle analysis of doxorubicin-treated Dino+/+ and Dinolox/lox MEFs. Mean +/− s.d. are shown, n = 3.
(f) Population doublings of serially passaged primary Dino+/+ and Dinolox/lox MEFs.
See also Supplementary Fig. 7.
Dinolox/lox MEFs exhibit significant defect in the activation of a subset of DNA damage-inducible genes, including _Cdkn1a, Bax, 14-3-_3σ, and Noxa (Fig. 6c), despite p53 mRNA being at or above wild-type levels (Supplementary Fig. 7a–b). Furthermore, Cdkn1a−/− MEFs have no defect in the induction of these p53 target genes, demonstrating that Dino regulates DNA damage-inducible genes independently of p21 (Supplementary Fig. 7c). Importantly, both isoforms of Cdkn1a are impaired in Dinolox/lox MEFs (Supplementary Fig. 7d). Furthermore, DNA damage induced expression of Mdm2 exhibits a greater Dino-dependence when MEFs are cultured in 0.1% serum, when proliferative stress is diminished compared to standard growth conditions with 10% serum (Supplementary Fig. 7d). We observed diminished levels of p53 protein in Dinolox/lox MEFs compared to Dino+/+ MEFs in response to increasing doses of DNA damage (Fig. 6d), indicating that Dino is required for robust accumulation of mouse p53 protein following DNA damage, especially at a moderate dose of DNA damage. Similar results were observed in Dinogfp/gfp MEFs (Supplementary Fig. 7e). This defect in Dinolox/lox cells also translates to a substantial reduction in damage-induced accumulation of p21 protein, as an example of a p53 target. These results are analogous to findings of human DINO on human p53 stabilization. The defect in p53 protein abundance in Dinolox/lox MEFs occurs despite a slightly elevated level of p53 mRNA, and p53 protein abundance is rescued by the proteasome inhibitor MG-132 (Supplementary Fig. 7f), demonstrating that p53 protein is efficiently translated but is relatively unstable in Dinolox/lox MEFs. Finally, p53 CLIP in mouse MEFs specifically retrieves Dino, confirming that Dino and p53 directly interact in mouse cells (Supplementary Fig. 7g).
Consistent with the defects in DNA damage-inducible gene expression, Dino promoter knockout cells exhibit attenuated cell-cycle arrest following DNA damage. Doxorubicin-treated Dinolox/lox MEFs showed an increase in the proportion of S-phase populations relative to Dino+/+ MEFs (Fig. 6e). Moreover, Dinolox/lox MEFs failed to undergo replicative senescence and continued to proliferate well after Dino+/+ MEFs have entered crisis (Fig. 6f).
DISCUSSION
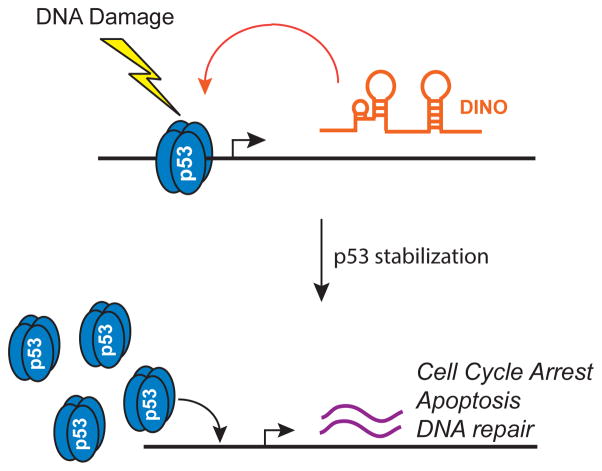
Our studies have revealed the lncRNA DINO as a new component of the DNA damage response, providing a feed-forward mechanism that amplifies p53 activity in response to DNA damage (Fig. 7). In the absence of DINO expression, p53 protein remains destabilized. However, induction of DINO by DNA damage or by enforced expression of DINO from a heterologous plasmid enhanced p53 protein stability and transactivation of p53 targets. Thus, a feed-forward loop of DINO and p53 stabilization may serve as a filter, ensuring that the DNA damage response is activated only after surpassing a threshold of damage. Moreover, these results highlight the concept of a direct physical feed back loop between transcription factor and its target inducible lncRNA for coordinating cell fate. Because proteins are synthesized in cytoplasm (thus away from chromatin, where transcription factors act), inducible lncRNAs are uniquely suitable to provide direct feedback in situ to the transcription factor regarding the status of the signaling pathway. The co-occupancy of DINO with p53 protein at the p53-response element of multiple p53-responsive loci is consistent with this model.
Figure 7.
Model of DINO function in p53 signaling.
These findings reinforce the importance of p53 protein dosage in the DNA damage response. Like p53+/− cells, Dino knockout cells contain diminished p53 protein levels both at baseline and following DNA damage. While the remaining p53 protein is able to elicit cell cycle arrest and apoptosis following DNA damage, these responses are dampened. The moderate reduction in p53 protein abundance can significantly impact animal survival following lethal irradiation. Furthermore, the observations that Dino knockout mice and cells are phenotypically similar to p53+/− mice in the acute DNA damage response raises the intriguing possibility that Dino knockout mice may be tumor prone, similar to p53+/− and epi-allelic p53 hypomorphs29, and is the subject of ongoing study.
That DINO’s interaction can functionally regulate p53 protein suggest a possible broader role for RNAs in the regulation of p53 signaling. The maternally imprinted lncRNA MEG3 can also enhance p53 activity, suggesting that multiple lncRNAs can regulate p5330. Additionally, the fact that lncRNAs are expressed in highly tissue- and context-specific manner31 may provide cells with unique opportunities to confer specificity on otherwise global processes.
ONLINE METHODS
Cell Culture and Treatments
Human cells were obtained from ATCC free of mycoplasma and maintained under standard growth conditions with growth medium formulated according to ATCC recommendations for each cell line. Unless otherwise specified, doxorubicin was used at a concentration of 0.2ug/mL. Unless specifically noted, the duration of doxorubicin treatment was 12 hours for ChIP experiments, 16–26 hours for gene expression experiments, and 26 hours for cell cycle analyses. For in vitro thymocyte apoptosis analysis, freshly isolated thymocytes were treated with indicated dose of radiation from a Cs137 source and cultured for 6 hours before AnnexinV/PI staining. In cycloheximide chase experiments, growth medium was supplemented with 25ug/mL cycloheximide (Sigma) for indicated times.
RNA interference
Human fetal lung fibroblasts were transfected with 3 independent 50nM ON-TARGETPlus siRNAs (Dharmacon) targeting DINO using Lipofectamine 2000 or 3 independent 100nM antisense oligonucleotides targeting DINO (Isis Pharmaceuticals). Large scale transfections were performed using the Amaxa Nucleofector NHDF kit. SiRNA and ASO sequences provided in Supplementary Table 3.
Gene Expression Analyses
Total RNA was extracted using TRIzol (Invitrogen) and the RNeasy Mini Kit (Qiagen), and genomic DNA was eliminated using TURBO DNA-free (Ambion). RT- PCR using 50 ng of total RNA was performed using the One-Step RT-PCR Master Mix (Applied Biosystems) or TaqMan Gene Expression Assays and normalized to either GAPDH (human) or β-actin (mouse). Strand-specific qRT-PCR for DINO was performed using One-Step RT-PCR MasterMix SYBR Green (Stratagene).
For microarray analysis, total RNA was profile using the Illumina Expression BeadChip Kit. Using SAM analysis, the human DNA damage gene set (n = 417) was defined as the set of genes that were at least 2 fold induced or repressed upon DNA damage in multiple biological replicates each of fibroblasts and p53+/+ HCT116.
3-seq libraries were generated from 50ng of total RNA using the TruSeq RNA Access Library Prep kit (Illumina) according to the manufacturers instructions. The p53-dependent, DNA damage responsive gene signature was previously described32.
The KEGG pathway analysis was performed using DAVID and significant enrichments (P < 0.05, FDR < 0.05, Benjamini-Hochberg test to correct for multiple hypothesis testing) are reported.
nCounter assays using the p53 Virtual Pathway Gene Set (Nanostring) were performed using isolated RNA according to the manufacturer’s instructions. Data was normalized to the geometric mean of the 6 internal references.
Crosslinking Immunoprecipitation (CLIP) and RNA Immunoprecipitation (RIP)
The CLIP method was performed as previously described33 with the following modifications: human fetal lung fibroblasts or MEFs were treated with doxorubicin for 16 hours hours, and either UV crosslinked (254nm) once at 4000mJ/cm2 or not crosslinked (no UV control). Lysates were prepared as previously indicated, but sonication was used to fragment the RNA to 200–400 nt and RNAses were not used. RIP was performed in H1299 cells transiently transfected with indicated plasmids using Fugene6. RIP was performed using RNA ChIP-IT (Active Motif). DO-1 (Sigma) was used for endogenous p53 immunoprecipitation.
ChIRP and ATACseq
ChIRP was performed using biotinylated probes according to previously described methods21 except that cells were crosslinked in 3% formaldehyde + 1mM EGS (ThermoFisher). See supplemental materials for probe sequences and qPCR primer pairs. Independent even and odd probe pools were used to ensure DINO-specific retrieval and RNase pre-treated chromatin served as negative controls. ATAC-seq libraries were generated using 50,000 primary human fibroblasts according to previously published protocol19 with 2 technical replicates and 2 biological replicates per condition. See supplementary information for details of data analyses.
Animal Models
All experiments were performed in accordance with the Stanford Institutional Animal Care and Use Committee. Dinogfp and Dinolox mice were generated by the Howard Hughes Medical Institute. C57BL/6 ES cells were selected with neomycin after introduction of the targeting construct. Following confirmation of anticipated recombination, the neomycin selection cassette was removed using Flip recombinase prior to blastocyst injection. E13.5 MEFs were isolated from timed pregnant mice according to standard protocols and genotypes confirmed by PCR.
Total body irradiation experiments were performed using a Kimtron Model #IC-224 small animal X-ray irradiator. Littermate mice were treated with a single fraction of 5Gy TBI at 6–8 weeks of age for in vivo thymocyte apoptosis analysis. For survival studies, 9 week old littermates were treated with a single fraction 12Gy TBI and observed twice daily until death. No animals treated with TBI were removed from analysis. Littermate offspring of two heterozygous parents were genotyped and assigned in roughly equal proportions to treatment groups in order to achieve cohorts that are nearly 50% male, 50% female for each group. Investigators were not blinded to the genotype.
Drugs and treatments
Doxorubicin, etoposide, methane methanylsulfonate (MMS) and hydrogen peroxide (H2O2) were purchased from Sigma. Treatment conditions, unless otherwise specified, were doxorubicin (0.2ug/mL), etoposide (10uM), MMS (1mM), H2O2 (500uM). Unless specifically noted, the duration of doxorubicin was 12hr for ChIP experiments, 16–26hrs for gene expression experiments, and 26hrs for cell cycle analyses. For MMS and H2O2, cells were washed after 1 hour of treatment and fresh medium replaced.
RNA chromatography
RNA chromatography was performed as previously described10. Recombinant RNA chromatography was performed with the following modifications: bead conjugated RNAs were incubated with 1 ug of recombinant p53 (Active Motif) in Binding Buffer (50 mM TrisCl 7.9, 10% Glycerol, 100 mM KCl, 5 mM MgCl2, 10 mM β-ME 0.1% NP-40) for 1 hour at 25C, then washed 5x with binding buffer.
qRT-PCR
Total RNA was extracted using TRIzol (Invitrogen) and the RNeasy Mini Kit (Qiagen), and genomic DNA was eliminated using TURBO DNA-free (Ambion). RT-PCR using 50 ng of total RNA was performed using the One-Step RT-PCR Master Mix (Applied Biosystems) or TaqMan Gene Expression Assays and normalized to GAPDH (human) or β-actin (mouse). The following assays were used: CDKN1A assay (Hs00355782_m1), DDB2 (Hs03044953_m1), TP53 (Hs99999147_m1), GADD45a (Hs00169255_m1), GADD45b (Hs00169587_m1), and GAPDH (Hs99999905_m1). Strand-specific qRT-PCR for DINO was performed using the One-Step RT-PCR Master Mix SYBR Green (Stratagene) using indicated primer pairs (Supplementary Table 3).
For zebrafish studies, 50 Zebrafish embryos were treated with 1500 mJ UV at 24 hpf and collected at 30 hpf in TRIzol. qRT-PCR was performed using custom (listed below) and previously described primers34.
ATAC-seq Data Analysis
For ATAC-Seq data preprocessing, paired end reads were trimmed for Illumina adapter sequences and transposase sequences using an in-house script and mapped to hg19 using Bowtie v0.12.935 with parameters –S –X2000 –m1. Duplicate reads were discarded with Samtools v0.1.1836. Peak calling using ZINBA was as described37. Chromosomal regions with a posterior probability of >0.99 were identified as peaks. For analysis of the ATAC-Seq signal intensity around p53 binding sites, we used p53 ChIP-Seq data downloaded from GEO accession number GSE46641 and CTCF ChIP-Seq from human lung fibroblasts was downloaded from GEO accession number GSE41048. Peak calling of the p53 and CTCF binding sites were performed by MACS238. MEME39 was used to identify p53 motif sites within the p53 binding sites. A 2kb window centered on the first base of the motif was divided into 40 equal sized bins of 50 bp. The number of unique-mapped and properly paired ATAC-seq tags overlapping each bin was counted. The ATAC-Seq signal around CTCF binding sites is affected by neither DINO knockout nor doxorubicin treatment, therefore the ATAC-Seq signal should have similar distribution around CTCF binding sites among all of the cell lines tested when accounting for sequencing depth. To normalize the ATAC-Seq signal intensity around P53 binding sites for all cell lines, the average fragment count plotted in each bin was first normalized to the ATAC-Seq signals surrounding CTCF binding sites, and then normalized to the total 10 millions of reads. The number of ATAC-Seq tags in the peak summit was compared in four different cell lines by T-test. The heat maps of ATAC-Seq at all the P53 motifs were generated using Java TreeView 3.0.
nCounter Assay
nCounter assays using the p53 Virtual Pathway Gene Set (Nanostring) were performed using isolated RNAs according to manufacturer’s instructions. Data was normalized using the geometric mean of the 6 internal references.
Statistical analyses
A two-tailed student T-test was used for analysis of statistical significance with a P < 0.05 considered significant. For mouse survival studies, the log-rank test was used. We determined that a sample size of at least 7 animals per genotype was required to observe a 2 day increase in time to event (death) between genotypes following lethal irradiation, with a power of 0.9.
DINO cloning and sequence analysis
DINO was initially identified using 5 bp-resolution high density tiling arrays covering the promoters of CDKN1A and multiple other genes10. 3′ and 5′ RACE was performed using the FirstChoice RLM-RACE Kit (Ambion) for human DINO and GeneRacer (Life Technologies) for mouse Dino. RNA was extracted from 0.2ug/ml doxorubicin (Sigma)-treated human fetal lung fibroblasts or MEFs, poly(A) selected using the Poly(A)Purist MAG kit (Ambion) and RACE was performed according to the standard manufacturer’s protocol.
DINO Evolutionary Conservation Analyses and Statistics
DINO syntetic sites and regions of microhomology were identified as previously described18. Briefly, the iterative process included: (1) beginning with empirically defined, strand-specific sequences for human, mouse, and zebrafish DINO, followed by (2) a search for putative DINO orthologs in marmoset, rat, rabbit, elephant, and horse using synteny and conserved motifs identified by MEME39, then (3) trim and re-orient putative DINO orthologs based on the strand of conserved motifs and re-run MEME.
MEME reports an estimate of the statistical significance of each motif it finds, the “motif E-value”, as well as an estimate of how well each occurrence, “site P-value”, matches the motif. The motif E-value is an estimate of how likely it is that the motif is not just a statistical artifact. The site P-values should only be used as relative indications of how well each site matches the motif. MEME uses an algorithm called Expectation Maximization to find short patterns of nucleic acids or amino acids that occur more frequently in the input sequences than would be expected by chance. Because these patterns need not be exact matches, they are described using a matrix: the Position Specific Scoring Matrix (PSSM). Once MEME has generated the PSSM, it uses a second algorithm, MAST, to find the best matches to that PSSM in the input data. The P-value tells you how well a particular site matches the PSSM found by MEME. The smaller the P-value indicates a more significant match.
DINO overexpression
For overexpression assays, the Lipofectamine 2000 (Invitrogen), Fugene 6 reagent (Roche) or Cell Line Nucleofector Kit V Kit (Amaxa) was used according to the manufacturer’s protocol. The following plasmids were used: pcDNA-human DINO, pcDNA-human DINOΔ320–450, pcDNA-human DINOΔ323–405, pcDNA-mouse DINO, pcDNA, pcDNA-Flag-p53 (Addgene #10838), pcDNA-Flag-p53 R273H, and C-terminus truncation pcDNA-Flag-p53 1–362 (ΔC31).
Antibodies
The following antibodies were used for p53 analysis: DO-1 (Sigma), 1C12 (Cell Signaling Technologies). Other antibodies for western analysis: anti-β-tubulin (Abcam ab6046), anti-H3 (ab1791), anti-LSD1 (ab17721), anti-SUZ12 (abcam ab12073), anti-p21 (Santa Cruz Biotech C-19), β-actin (Abcam).
RNA interference
Human fetal lung fibroblasts were transfected with 3 independent 50 nM ON-TARGETPlus siRNAs (Dharmacon) or 100nM ASOs40 targeting DINO using the Lipofectamine 2000 reagent. Large scale transfections were performed using the Amaxa Nucleofector NHDF kit. siRNAs for mRNAs (Ambion) were used as a pool of two. U2OS cells were infected with shRNAs targeting DINO using the pGIPZ shRNAmir lentiviral system.
Cycloheximide chase
Cells were reverse transfected (Lipofectamine 2000, Invitrogen) for 24 hours, then media was supplemented with 25 ug/ml cycloheximide (Sigma). At the indicated times post cycloheximide addition, cells were collected for western blot analysis: snap freeze over night, 30 minute lysis on ice in NET buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.1% Nonidet P40, freshly added 1 mM phenylmethylsulphonyl fluoride, protease inhibitors), sonicated with a Bioruptor (Diagenode) for 5 min (30-s ‘on’ and 30-s ‘off’), then centrifuged for 15 minutes at 13k RPM. Lysates were normalized by BCA and resolved on SDS-PAGE.
Crosslinking Immunoprecipitation (CLIP) and RNA Immunoprecipitation (RIP)
The CLIP method was modified from previous methods33 for CLIP-qRT-PCR: human fetal lung fibroblasts or U2OS cells were treated with doxorubicin for 16 hours, and either UV crosslinked (254nm) once at 4000mJ/cm2 or not crosslinked (no UV control). Lysates were prepared as previously indicated, but sonication was used to fragment the RNA to 200–400 nt and RNAses were not used. RIP was performed in H1299 cells transiently transfected with indicated plasmids using Fugene6. RIP was performed using RNA ChIP-IT (Active Motif). DO-1 (Sigma) was used for endogenous p53 immunoprecipitation.
SHAPE
SHAPE analysis was performed as described41. NMIA (13mM final concentration) or DMSO was used as modification or mock modification reactions, respectively. ddGTP and ddTTP were used in two separate sequencing reactions. cDNA extensions were visualized by phosphorimaging (STORM, Molecular Dynamics). cDNA bands were integrated with SAFA42. SHAPE reactivities were normalized to a scale spanning 0 to 1.5, where 1.0 is defined as the mean intensity of highly reactive nucleotides43. RNA secondary structures were predicted using RNA structure software44 (Reuter et al., 2010). SHAPE data and RNA structure data were reconciled using SeqFOLD45.
Northern Blot
Northern blot was performed using the NorthernMax kit (Ambion) with 2.5–5ug of poly(A) selected RNA for each sample. Anti-sense RNA probes to human DINO nucleotides 203–716 and mouse DINO nucleotides 291–842 were synthesized using T7 MegaScript (Ambion) with incorporation of UTP-α-P32.
Supplementary Material
1
2
3
4
5
Acknowledgments
Supported by NIH R01-ES023168, R01-CA118750, R01-HG004361 to H.Y.C., NIH DP1-HD075622 to J.K.C., and ASCO Conquer Cancer Foundation/Susan G. Komen Young Investigator Award and Stanford Radiation Oncology Kaplan Funds to A.M.S.
Footnotes
DATABASES
Microarray and sequencing datasets are available on GEO under accession numbers GSE42368 and GSE76420. Human DINO and mouse Dino sequences are deposited in GenBank under accession numbers JX993265 and JX993266 respectively
AUTHOR CONTRIBUTIONS
H.Y.C. conceived and supervised the study. A.M.S., T.H., and H.Y.C. designed the experiments. A.M.S., J.T.G., T.H., R.A.F., A.Y.P., A.P.S., R.B. performed experiments. A.M.S.,J.T.G., T.H., R.A.F., Y.S., and K.Q. performed statistical analyses and analyzed the data. D.K.B, S.G., J.K.G., and L.D.A. contributed materials. A.M.S., T.H., and H.Y.C. wrote the paper.
Competing Financial Interests
S.G. is an employee of Ionis Pharmaceuticals. The other authors declare no competing financial interests.
References
- 1.Komarova EA, et al. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene. 2004;23:3265–71. doi: 10.1038/sj.onc.1207494. [DOI] [PubMed] [Google Scholar]
- 2.Kirsch DG, et al. p53 controls radiation-induced gastrointestinal syndrome in mice independent of apoptosis. Science. 2010;327:593–6. doi: 10.1126/science.1166202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CL, et al. p53 functions in endothelial cells to prevent radiation-induced myocardial injury in mice. Sci Signal. 2012;5:ra52. doi: 10.1126/scisignal.2002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson SM, et al. Mitigation of hematologic radiation toxicity in mice through pharmacological quiescence induced by CDK4/6 inhibition. J Clin Invest. 2010;120:2528–36. doi: 10.1172/JCI41402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westphal CH, et al. Loss of atm radiosensitizes multiple p53 null tissues. Cancer Res. 1998;58:5637–9. [PubMed] [Google Scholar]
- 6.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium, F. et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez Y, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leveille N, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrova N, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–90. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, et al. An Apela RNA-Containing Negative Feedback Loop Regulates p53-Mediated Apoptosis in Embryonic Stem Cells. Cell Stem Cell. 2015;16:669–83. doi: 10.1016/j.stem.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley KJ, Maher LJ., 3rd p53 RNA interactions: new clues in an old mystery. RNA. 2007;13:1825–33. doi: 10.1261/rna.673407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–55. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 16.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinn JJ, et al. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016;30:191–207. doi: 10.1101/gad.272187.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giono LE, Manfredi JJ. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol. 2006;209:13–20. doi: 10.1002/jcp.20689. [DOI] [PubMed] [Google Scholar]
- 23.Parrinello S, et al. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–9. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 25.Ihrie RA, et al. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr Biol. 2003;13:1985–90. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers JR, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 27.Qiu W, et al. PUMA regulates intestinal progenitor cell radiosensitivity and gastrointestinal syndrome. Cell Stem Cell. 2008;2:576–83. doi: 10.1016/j.stem.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibowitz BJ, et al. Ionizing irradiation induces acute haematopoietic syndrome and gastrointestinal syndrome independently in mice. Nat Commun. 2014;5:3494. doi: 10.1038/ncomms4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–42. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 31.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenzelmann Broz D, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–31. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu TX, et al. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003;5:903–14. doi: 10.1016/s1534-5807(03)00339-3. [DOI] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–87. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–6. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 42.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semiautomated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–54. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gherghe C, et al. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci U S A. 2010;107:19248–53. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang Z, Snyder MP, Chang HY. SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 2013;23:377–87. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5