cis-Regulatory and Protein Evolution in Orthologous and Duplicate Genes (original) (raw)
Abstract
The relationship between protein and regulatory sequence evolution is a central question in molecular evolution. It is currently not known to what extent changes in gene expression are coupled with the evolution of protein coding sequences, or whether these changes differ among orthologs (species homologs) and paralogs (duplicate genes). Here, we develop a method to measure the extent of functionally relevant _cis_-regulatory sequence change in homologous genes, and validate it using microarray data and experimentally verified regulatory elements in different eukaryotic species. By comparing the genomes of Caenorhabditis elegans and C. briggsae, we found that protein and regulatory evolution is weakly coupled in orthologs but not paralogs, suggesting that selective pressure on gene expression and protein evolution is quite similar and persists for a significant amount of time following speciation but not gene duplication. Additionally, duplicates of both species exhibit a dramatic acceleration of both regulatory and protein evolution compared to orthologs, suggesting increased directional selection and/or relaxed selection on both gene expression patterns and protein function in duplicate genes.
The relative importance of coding sequence change versus regulatory sequence change in evolution has vexed evolutionary geneticists for over 50 years. Given recent genomic analyses showing the conservation of many proteins among distantly related taxa, it has been proposed that regulatory changes play a key role in generating the great morphological diversity present in multicellular species. However, little is known about the evolution of gene regulation or its relationship to protein evolution. Do highly conserved genes also show conserved expression patterns? Or can gene expression evolve independently from protein function? The former pattern is expected if strong stabilizing selection acts on genes as integrated units in which protein sequence and expression pattern are not dissociable. At the same time it has been argued that “developmental systems drift” may result in reorganization of regulatory systems as long as general developmental patterns are preserved (True and Haag 2001). If so, a high turnover of gene regulatory elements may uncouple _cis_-element-mediated gene expression and protein evolution.
Differences in gene expression between species (or between duplicate genes) may entail changes in gene expression levels under the same conditions at the same developmental times, as well as gene expression changes in spatial, temporal, and environmental dimensions. Hereafter, we refer to the former changes as changes in expression magnitude and the latter as changes in relative expression. First attempts to find a correlation between the evolution of relative expression and protein evolution in yeast (using only duplicates) yielded contradictory results; one study argued that expression differences are not correlated with protein evolution (Wagner 2000), whereas more recent work suggests a weak correlation (Gu et al. 2002). However, a recent review (Wolfe and Li 2003) concluded that a wider analysis of regulatory and protein evolution is necessary. In particular, the dynamics of protein versus _cis_-regulatory evolution in duplicate genes, which are thought to play a central role in the evolution of novel molecular functions and the generation of genetic diversity (Haldane 1932; Ohno 1970), are still poorly understood.
Although there is some evidence that duplicate genes undergo an increased rate of protein evolution (Lynch and Conery 2000; Kondrashov et al. 2002; Nembaware et al. 2002), a systematic analysis of _cis_-regulatory versus coding sequence change in orthologous and duplicate genes has not been carried out. This deficiency is due in large part to the lack of a biologically relevant measure of _cis_-regulatory evolution that relates directly to gene expression. The identity of _cis_-acting regulatory motifs is generally unknown, and such motifs are sparsely scattered within noncoding DNA that is under little or no selective constraint. It has thus been almost impossible to discriminate between functionally relevant and stochastic changes in putative regulatory DNA without time-consuming gene-by-gene experiments.
Accordingly, we set out to develop a method to quantify functional regulatory changes in the regulatory regions of homologous genes that does not depend on knowledge of experimentally characterized or computationally predicted DNA binding sites. We began by identifying orthologous genes between the genomes of Caenorhabditis elegans and Caenorhabditis briggsae. Next we identified duplicate genes within each genome and calculated rates of protein evolution in both duplicates and orthologs by maximum likelihood (Yang 1997). Finally, we used duplicate gene pairs in conjunction with microarray expression data to develop a method to measure regulatory evolution called the shared motif method (SMM), and used it to measure _cis_-regulatory evolution in duplicate and orthologous genes. These data are summarized in Table 1, and the list of genes is provided as Supplemental material.
Table 1.
Comparison of Mean Rates of Protein Evolution (_d_N, _d_S) and Regulatory Evolution (_d_SM) in Orthologous and Duplicate Genes
| Number of pairs | _d_S | _d_N | _d_SM | |
|---|---|---|---|---|
| Orthologs between species | 2,150 | 1.11 (0.31) | 0.07 (0.06) | 0.59 (0.22) |
| Duplicates within C. elegans | 869 | 0.57 (0.43) | 0.17 (0.15) | 0.61 (0.30) |
| Duplicates within C. briggsae | 542 | 0.60 (0.41) | 0.22 (0.20) | 0.64 (0.31) |
RESULTS AND DISCUSSION
Regulatory Sequence Evolution
Because small intrachromosomal rearrangements resulting in changes in _cis_-element order, orientation, and spacing can occur over moderate stretches of evolutionary time, while leaving gene expression patterns intact (Ludwig et al. 2000), we detected “shared motifs” (regions of high local similarity) between the upstream regions of homologous genes. We define these motifs as conserved segments between sequences without respect to their order, orientation, or spacing (Fig. 1). By examining the cumulative fraction of shared motifs between sequences we defined a measure of functional regulatory sequence evolution called shared motif divergence (_d_SM). By definition, _d_SM is the fraction of both sequences that does not contain a region of significant local similarity by these criteria. For example, a _d_SM of 0 indicates a complete sharing of motifs between the sequences, whereas a _d_SM of 1 indicates an absence of shared motifs. Note that this measure is similar to a distance metric but has a maximum value of 1. Values of _d_SM = 1 are not necessarily equally divergent and should not be compared because they are “saturated” with sequence differences. In this study, the mean _d_SM was 0.59 between species and 0.61 and 0.64 among duplicate genes in the C. elegans and C. briggsae genomes, respectively (Table 1).
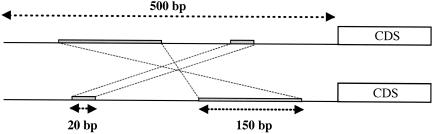
Figure 1.
Illustration of the shared motif method (SMM). The SMM discovers regions of local similarity between DNA sequences without respect to their order, orientation, or spacing. In this example, two 500-bp noncoding sequences, upstream from homologous coding sequences (CDS), are compared. After iterative local alignment in both their native and inverted sequence orientations (Methods), two regions of significant local similarity between the sequences were discovered. One region is 150 bp long but has been inverted in one of the sequences. The other is 20 bp long but has been translocated. The fraction of “shared motifs” between these sequences is simply (20 + 150) / 500, or 0.34. We define shared motif divergence (_d_SM) as one minus this fraction, or 1 – 0.34 = 0.66. Shared motif divergence is thus the fraction of the two sequences that does not contain a region of significant local alignment without respect to order, orientation, or spacing. Note, this example is a simplified caricature. Real sequence comparisons often exhibit more complex patterns of shared motif conservation (Supplemental Fig. 1).
Because genome-wide expression data for C. briggsae are not available, we validated our measure of regulatory sequence evolution using pairs of duplicate genes within the C. elegans genome. Because little is known about the average size of regulatory regions in C. elegans, we looked for shared motifs 100, 500, and 1000 bp upstream from annotated translation start sites. Among genes with annotated transcription start sites in C. elegans, we found no significant difference in _d_SM when calculated from transcription start versus translation start (Supplemental material).
Divergence between upstream sequences of each duplicate pair was measured by the shared motif method and was compared with (1) differences in the magnitude of expression across the life cycle of C. elegans in absolute numbers of transcripts as assessed by Affymetrix microarrays (Hill et al. 2000), and with (2) differences in relative expression, first across eight developmental stages (Hill et al. 2000) and then across 553 cDNA microarray experiments that included different nutrient conditions, developmental stages, and mutants (Kim et al. 2001). Data from cDNA microarrays describe only relative changes in gene expression, and therefore differences in gene expression magnitude cannot be determined from these results. Note also that all estimates of expression level are for genes that were reliably detected (Methods), and these are likely to be moderately to highly expressed.
We found only a marginally significant correlation between _d_SM and difference in relative expression using Affymetrix expression data (rs = 0.23, P < 0.07, Spearman rank correlation), and no significant correlation using the cDNA microarray data (Supplemental material; Kim et al. 2001). In contrast, we observed a highly significant correlation between _d_SM and difference in gene expression magnitude (rs = 0.47, P < 10–3) for upstream sequences of 500 bp (Fig. 2). Shorter and longer upstream sequences were less correlated with expression difference (data not shown). Importantly, no significant correlation was detected between gene similarity (estimated by _d_N) and expression difference (rs = –0.10, P = 0.18) which is expected if cross-hybridization of transcripts on Affymetrix arrays is a significant phenomenon. Although a weak correlation of synonymous substitution rate (_d_S) and expression difference was detected (rs = 0.23, P = 0.02), it disappeared after correcting for the relationship between _d_SM and _d_S (rs = 0.40, P < 0.001) in a multiple regression analysis (expression difference vs. _d_S, B = 52.78, P = 0.34, multiple regression formula: expression difference ∼_d_SM + _d_N + _d_S). These results taken together imply that _d_SM correlates with a functional difference in the gene expression magnitude between genes which is not a simple consequence of gene similarity (time since divergence or duplication event). Considering that (1) the measure _d_SM does not take into account differences in _trans_-acting factors or other mechanisms that mediate gene expression, and (2) that its performance is based on whole-animal assays, where changes in spatial expression will act to decrease the correlation between _d_SM and expression difference, the correlation between dSM and expression difference is remarkably high. Moreover, it is the first predictor available for estimating the expression difference between genes based on comparative sequence data alone.
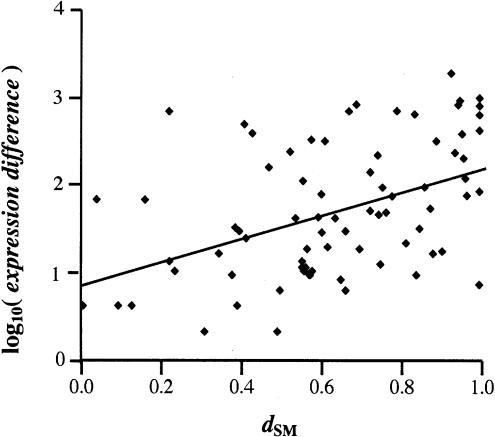
Figure 2.
Correlation between _d_SM and difference in magnitude of gene expression. We found a significant positive correlation between expression difference and shared motif divergence (_d_SM) in sequences 0–500 bp upstream of translation start between duplicate genes in duplicate families with two to five members (n = 76, rs = 0.47, P < 10–3; Spearman rank correlation). The log(expression difference) is linearly correlated with d_SM (R = 0.46; P ≪ 10–3; Pearson linear correlation); a linear fit of the data is also plotted (y = 0.85 + 1.37_x). Similar results were obtained with strict duplicate pairs or duplicate gene families of up to 10 members (data not shown).
As a further negative control, we examined the _d_SM of orthologous sequences in regions located further upstream of the translation start (1–1.5 kb) where the density of functional motifs is presumably lower. Orthology of these noncoding regions was inferred if both species exhibited syntenic conservation of the adjacent gene. The 1–1.5-kb upstream region was significantly more diverged on average than the 0–500-bp upstream test region used in the analysis (mean _d_SM = 0.78 versus 0.60, n = 362, P ≪ 10–4, Wilcoxon _U_-test). Further, the control region showed a distribution of _d_SM similar to that obtained between random sequences (mean _d_SM = 0.89) compared with the 0–500-bp test region (Supplemental Fig. 1). This result suggests that the conservation detected by the SMM is not a simple consequence of historical identity.
Although the SMM is not a motif discovery algorithm, the conserved blocks of sequences discovered by the method should include a high fraction of _cis_-acting elements that are experimentally known to be involved in gene regulation. We analyzed in detail the upstream sequence of a gene known to be up-regulated in response to heat shock in C. elegans (F44E5.5), which contains experimentally characterized _cis_-regulatory elements conserved in C. briggsae (GuhaThakurta et al. 2002). The conserved sequences identified by the SMM contained eight of nine experimentally verified _cis_-elements shared between the species. We found similar results (Supplemental Fig. 2) for experimentally verified motifs at the even-skipped locus in Drosophila melanogaster / D. pseudoobscura (Ludwig et al. 2000), for Apetala-3 in Arabidopsis thaliana / Brassica oleracea (Koch et al. 2001), and for CKM in Homo sapiens / Mus musculus (Wasserman et al. 2000). As a further positive control, we analyzed the upstream regions of a large set of human genes for which there was one or more experimentally characterized binding sites and a known orthologous gene in mouse (Supplemental material). Of 79 experimentally verified motifs among 20 different orthologous genes, 62 of 79 motifs (78%) were contained within conserved blocks discovered by the SMM (Supplemental material; Supplemental Table 2). The mean number of verified regulatory motifs in these 1-kb upstream regions was 3.95 and, on average, 3.10 motifs were found using the SMM.
_cis_-Regulatory and Protein Evolution are Weakly Coupled in Orthologs
We observed a positive correlation between functional regulatory evolution (_d_SM) and protein evolution (_d_N) in orthologs (Table 2, Fig. 3). As similarities in local mutation rate, or similar divergence times, may lead to the observed correlation between protein coding and noncoding change, we carried out multiple regressions involving _d_N, _d_SM, and _d_S, using _d_S as a simple measure of age/mutation rate.
Table 2.
Correlation Between Protein and Regulatory Evolution: Acceleration of Protein and Regulatory Evolution in Duplicate Genes
| Correlation between _d_N and _d_SM | _d_N/_d_Sa | _d_SM/_d_Sa | _d_N/_d_SMa | |
|---|---|---|---|---|
| Orthologs between species | rs = 0.16b,c | 0.05 | 0.53 | 0.09 |
| Duplicates within C. elegans | rs = 0.24b | 0.31 | 1.00 | 0.27 |
| Duplicates within C. briggsae | rs = 0.21b | 0.37 | 1.06 | 0.33 |
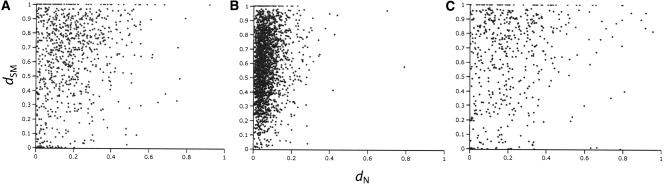
Figure 3.
Correlation between protein evolution (_d_N) and regulatory evolution (_d_SM) in (A) paralogous genes in C. elegans (r s = 0.24), (B) orthologous genes (rs = 0.16), and (C) paralogous genes in C. briggsae (rs = 0.21); P ≪ 10–4 for all tests. Multiple regressions that included _d_S revealed that the correlation between _d_N and _d_SM in paralogs is primarily a function of _d_S (i.e., duplicate age). No such effect was found in orthologs. Note that for some orthologs and duplicates, regulatory and protein evolution appear to be completely uncoupled.
Interestingly, there is a weak but significant correlation between protein and _cis_-regulatory evolution after controlling for the possibility that this correlation is a consequence of a similarity in local mutation rates (_d_SM vs. _d_N, B = 0.42, P < 10–6, multiple regression formula: _d_SM ∼ _d_N + _d_S), a consequence of poor gene prediction (Supplemental material), or due to inclusion of genes in operons (data not shown).
This implies that, for a given gene, there exists a significant coupling between rates of coding sequence and _cis_-regulatory sequence change—and by inference, a potential coupling of protein function and gene expression change. Such a correlation, which has been shown for very young duplicate genes in yeast (Gu et al. 2002), seems to hold for orthologous genes shared between the more distantly related C. elegans and C. briggsae. Thus many genes that are conserved at the protein level also show conserved _cis_-regulation as predicted by the hypothesis that strong stabilizing selection acts on genes as integrated units of evolution. If divergence reflects the action of purifying selection, a correlation in _cis_-regulatory and protein divergence implies that the selective consequences of a deleterious mutation in either the _cis_-regulatory or protein coding sequence of a given gene are similar. If so, many genes are “selectively important” for an organism in a manner that is not dissociable into protein product and expression pattern components, even over long stretches of evolutionary time.
On the other hand, the observation that coupling between regulatory and protein evolution is generally weak argues that some amount of “network drift” in _cis_-element-mediated gene expression may indeed occur. The maintenance of stable gene expression patterns in the face of coevolution of transcription factors and their _cis_-acting DNA binding sites (Shaw et al. 2002) as well as wholesale rearrangement of promoter architecture (Ludwig et al. 2000) has been experimentally demonstrated for a least two loci in Drosophila, bicoid and even-skipped, respectively. Such co-evolutionary drift may act to weaken the correlation between _cis_-regulatory and protein evolution across the genome, as expected under a general model of stabilizing selection. A definitive test of this hypothesis will require an independent assessment of gene expression patterns in C. briggsae in conjunction with the analysis of _cis_-regulatory evolution carried out here.
Note that the distinction between protein and _cis_-regulatory evolution is not entirely clear-cut. For example, coding sequences can contain motifs that act to enhance and silence mRNA splicing in constitutively and alternatively spliced exons (Blencowe 2000), which is an arguably regulatory function. Thus, the dynamic and interrelated nature of coding and noncoding sequence change must be kept in mind when interpreting these results.
_cis_-Regulatory and Protein Evolution Are Not Coupled in Paralogs
Although we also observed a correlation between functional _cis_-regulatory evolution (_d_SM) and protein evolution (_d_N) in paralogs (Table 2, Fig. 3), in contrast to orthologs, we found that the correlation between protein (_d_N) and regulatory evolution (_d_SM) is a result of their correlation with _d_S alone (_d_SM vs. _d_N, B = 0.046, P = 0.565, multiple regression formula = _d_SM ∼ _d_N + _d_S). In other words, _d_N and _d_SM increase together over time but are not themselves related. The observation that protein and regulatory evolution in paralogs is not coupled implies that these aspects of gene structure may evolve independently. It is interesting to note that this uncoupling can result from duplication events that do not encompass the entire regulatory region. Thus, shortly after duplication, _d_SM may immediately be very high whereas _d_N and _d_S are close to zero. This pattern can be observed for many genes (Figs. 3, 4) and explains the seemingly paradoxical result that regulatory and protein evolution are not coupled in duplicate genes, despite the fact that both are higher in duplicates versus orthologs.
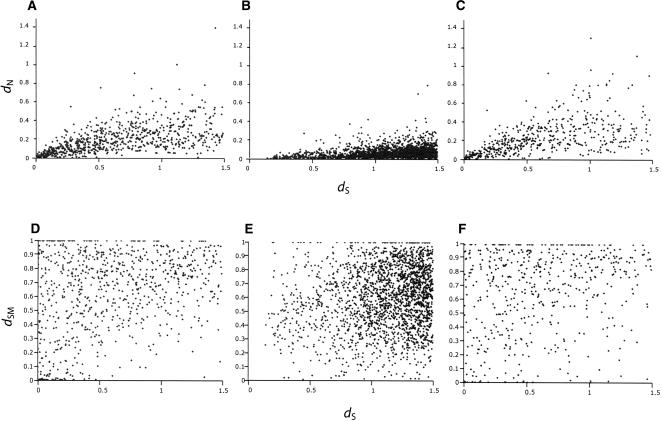
Figure 4.
Rates of protein evolution (_d_N/_d_S) in (A) paralogous genes in C. elegans, (B) orthologous genes between C. elegans and C. briggsae, and (C) paralogous genes in C. briggsae. Rates of regulatory evolution (_d_SM) in (D) paralogous genes in C. elegans, (E) orthologous genes between C. elegans and C. briggsae, and (F) paralogous genes in C. briggsae. In comparison with orthologs, duplicate genes in both the C. briggsae and C. elegans genomes exhibit a higher rate of amino-acid substitution and proximal _cis_-regulatory sequence evolution for the same amount of synonymous divergence.
Apparent independence between _cis_-regulatory and protein sequence change is not entirely unexpected, as it has long been suggested that duplicate genes may evolve new functions (Ohno 1970; Ohta 1987; Walsh 1995) or lose them in complementary ways (Hughes 1994; Force et al. 1999) through changes in their _cis_-regulatory sequence, protein sequence, or both. Accordingly, it has been predicted that accelerated protein and/or regulatory sequence evolution will occur in duplicated genes.
Accelerated Protein and Regulatory Evolution in Duplicated Genes
Our results also indicate that, in comparison with orthologs, duplicate genes in both the C. briggsae and C. elegans genomes exhibit a significantly accelerated rate of amino-acid replacement and _cis_-regulatory evolution for the same amount of synonymous mutation (Figs. 4, 5). Overall, rates of protein evolution (_d_N/_d_S) are substantially accelerated in duplicate genes in both the C. elegans and C. briggsae genomes, compared to orthologs between the species (P ≪ 10–4 for each test, Wilcoxon _U_-test, Table 2). Likewise, the mean amount of regulatory evolution (_d_SM/_d_S) in duplicate genes in the genomes of both species is dramatically accelerated compared to orthologs, even though many are much younger (P ≪ 10–4 for each test, Wilcoxon _U_-test, Table 2). This pattern of accelerated evolution is similar for tandem and nontandem duplicates (Supplemental material).
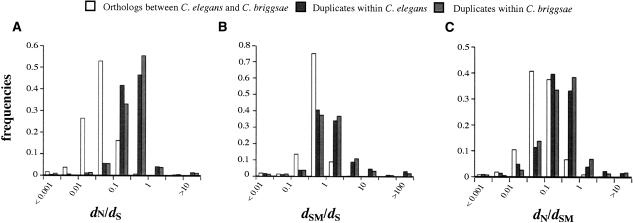
Figure 5.
Histogram of the rate of (A) protein evolution and (B) regulatory evolution in orthologs between C. elegans and C. briggsae vs. paralogs within C. elegans and C. briggsae. Both protein (_d_N) and regulatory evolution (_d_SM) are accelerated in paralogs compared to orthologs for the same amount of synonymous divergence. (C) Histogram of protein vs. regulatory evolution in orthologs and paralogs. For the same amount of regulatory divergence, paralogs have an accelerated rate of protein evolution compared to orthologs.
At least three nonmutually exclusive scenarios could be envisaged to explain the accelerated evolution of duplicate genes. First, duplicate genes could experience weaker purifying selection than orthologs following a speciation event, that is, relaxed selection. Second, duplicate genes could experience greater positive selection than orthologs. Third, duplicates may simply be older than orthologs, that is, they predate speciation and therefore have had more time to evolve amino-acid substitutions (_d_N). Under the last scenario, synonymous substitutions (_d_S) would be expected to increase over time at a similar rate as _d_N; however, if paralogs are more highly expressed than orthologs, they may be subject to high codon bias (Duret and Mouchiroud 1999; Castillo-Davis and Hartl 2002), which may result in underestimates of the synonymous substitution rate (_d_S)—even when likelihood methods are used (Dunn et al. 2001). We find, however, that orthologs are more highly expressed (Hill et al. 2000) than paralogs (P ≪ 10–4, Wilcoxon _U_-test; data not shown), and we calculate that at least 93% (502/542) of the duplicates examined in C. briggsae either post-date speciation or have undergone post-speciation gene conversion (gene conversion is expected to reduce synonymous and nonsynonymous divergence equally; Methods). Thus the accelerated rates of protein and regulatory evolution in duplicate genes must be due to either relaxed selection or the action of positive selection, or both.
Protein Evolution Appears to Outpace _cis_-Regulatory Evolution in Duplicate Genes
Finally, the distribution of _d_N/_d_SM (Fig. 5) among duplicates and orthologs indicates that, for a similar amount of regulatory divergence (_d_SM), the mean rate of amino-acid substitution (_d_N) is substantially higher in duplicate genes in both C. elegans and C. briggsae compared to orthologs between the two species (P ≪ 10–4, Wilcoxon _U_-test, Table 2). One possible explanation is that this pattern is due to an overall faster rate of saturation of _d_SM compared to _d_N; however, it should be noted that values of _d_SM are similar among both orthologs and duplicates, whereas values of _d_N differ greatly (Table 1). If not due to the saturation of _d_SM, the accelerated divergence observed in the protein-coding regions of duplicate genes may simply reflect the less deleterious consequences of changes in amino-acid sequence versus changes in gene expression in “redundant” genes (i.e., stronger purifying selection on gene regulation). Lastly, it is possible that the accelerated rate of protein evolution in duplicate genes is due to positive selection on coding regions either in both copies or only in one copy, as it has been recently posited (Conant and Wagner 2003; Kellis et al. 2004).
One indicator of positive selection is a _d_N/_d_S ratio significantly greater than one. We found no evidence for increased rates of positive selection in paralogs versus orthologs by this criterion (data not shown). Because positive selection across limited regions of a protein may occur without a global excess of amino-acid replacements (Nielsen and Yang 1998), we performed more sensitive likelihood-ratio tests for positive selection (Yang 2000) for all duplicate gene families of three to five members in both species (Methods). Although such tests have low power with small gene family sizes, we again found no evidence of positive selection.
The alternative explanation of the observed pattern of faster protein change in duplicates—namely that expression of a gene in the wrong tissue, cell, or at the wrong developmental timepoint may incur a high fitness cost compared to loss of protein function in one duplicate copy—therefore cannot be ruled out. Such a scenario is plausible, especially if gain-of-function mutations are more common in _cis_-regulatory sequences compared to protein coding sequences.
Conclusions
Taken together, these observations suggest that, until genes duplicate, selection on proximal _cis_-regulation is weakly coupled to selection on protein sequences; when genes duplicate, however, it appears that selection can act independently on gene regulation and protein sequences. Selective pressure on gene expression and protein function is therefore inferred to be quite similar and persists over long stretches of evolutionary time following divergence due to speciation but not necessarily gene duplication. Additionally, compared to orthologs, duplicate genes are unique in that they exhibit dramatically accelerated rates of both _cis_-regulatory and protein evolution, suggesting increased positive and/or relaxed selection on both gene expression patterns and protein sequence in duplicate genes. Although we found no evidence for the action of positive selection in duplicate genes, the observation of accelerated rates of protein evolution over _cis_-regulatory evolution in duplicate genes is noteworthy. Further analyses should help reveal whether this pattern is due to positive selection or merely reflects the greater selective consequences of gene mis-regulation, versus the abrogation of protein function, in redundant genes.
METHODS
Protein Sequence Analysis
Coding sequences (CDSs) of the genomes of both C. elegans (The C. elegans Sequencing Consortium 1998) and C. briggsae (The Sanger Institute and The Genome Sequencing Center, Washington University, St. Louis, unpubl.) were obtained from WormBase (http://wormbase.org). All CDS were mapped onto genomic locations using BLASTN (Altschul et al. 1997) and when available, annotations. Only one occurrence of overlapping CDS was retained. The method of reciprocal best hits (Tatusov et al. 1997) using BLASTN was used to establish a set of orthologs between the two species (E < 10–10 were considered significant matches). Orthologs obtained using only those genes not duplicated in either genome (1,765 / 2,150 = 82%) gave very similar results (data not shown). Duplicated genes within the _C. elegans_ and _C. briggsae_ genomes were obtained as follows. First a set of putative duplicate genes was obtained by significant BLASTN matches within each genome alone. Next each translated sequence of putative paralogs was globally aligned (Needleman and Wunsch 1970) against every other using the PAM250 matrix (Dayhoff et al. 1972), Gap(open) = –16 and Gap(ext) = –6. All scores were normalized using the length of the smallest of both sequences. Alignment scores >200 were considered significant. As a control, all translated C. elegans sequences were shuffled (Markov chains of order 0) and aligned in the same way. At this stringency, less than 0.001% of the random sequence alignments exhibit a significant score (data not shown). We considered families of five or fewer duplicate genes to avoid biases due to overrepresentation of very large gene families.
Next, all coding sequences were globally aligned by CLUSTALW (Thompson et al. 1994; default parameters) using the amino-acid translation of each sequence followed by back-translation into nucleotides. Maximum likelihood estimates of nonsynonymous substitution (_d_N) and synonymous substitution (_d_S) between pairwise alignments were obtained with PAML (Yang 1997) using a codon-based model of sequence evolution with _d_N, _d_S, and transition/transversion bias (κ) as free parameters and codon frequencies estimated from the data at each codon position (F3 × 4 model; Goldman and Yang 1994; Yang 1997). Based on simulations using random sequence pairs, pairs of sequences with κ > 8 or _d_S > 3 were excluded from analysis. Finally, because values of _d_S > 1.5 are prone to estimation error, we further restricted our dataset to orthologs and paralogs that exhibited a _d_S < 1.5.
To determine the minimum proportion of duplicate genes that post-date speciation, we determined the ancestry of all C. briggsae duplicate genes on the basis of significant BLASTN matches to genomes of both species (E < 10–10). Gene pairs that showed both of their closest matches within the C. briggsae genome were assumed to post-date speciation or have undergone gene-conversion post-speciation.
Likelihood ratio tests for positive selection within duplicate gene families were performed by comparing twice the log-likelihood difference of models M7 and M8 in PAML v3.13 (Yang 2000). This test compares the likelihood of the data under model M7 in which _d_N/_d_S among sites is constrained to be between 0 and 1, against model M8 where an additional category of sites with _d_N/_d_S > 1 is allowed. If the log-likelihood of the model allowing _d_N/_d_S > 1 (positive selection) is significantly greater, adaptive evolution may be inferred. Positive selection was inferred if 2(lnL1 – lnL2) ≥ 9.21, corresponding to P < 0.01 (∼χ2, df = 2) and if _d_N/_d_S was greater than one among at least one of the site classes. For these tests, duplicate gene family trees were constructed using PHYLIP (Felsenstein 1993) using translated sequences with default parameters under a maximum parsimony criterion.
Regulatory Sequence Analysis
Regulatory sequences are often located 5′ to proteins, comprising 5′ UTRs, promoter regions, and other regulatory elements such as enhancers. We define a shared motif as a region of high local similarity between two DNA sequences regardless of their order, orientation, or spacing (Fig. 1).
Our method is a derived implementation of the recursive local alignment algorithm described by Waterman and Eggert (1987). This method is based on local alignment by dynamic programming (Smith and Waterman 1981) and is guaranteed to find all optimal and suboptimal alignments between two sequences. Briefly, we find the best local alignment between two sequences, then mask off this particular alignment. Next, we search for next best subalignment between the sequences and continue this process iteratively until the next-best alignment score falls below a specified threshold (Supplemental material).
Alignments were performed between sequences in their native orientation and also by inverting one of the sequences. The scoring matrix used for alignment was an identity matrix: match = +4, mismatch = –4, match(N) = +1, Gap(open) = –4, Gap(extension) = –4. The symbol X is assigned a very negative score (–105) so that it can be used to mask sequences (Xs will not be aligned). Any non-A,C,G,T,X symbols were treated as N.
We define _d_SM as the fraction of both sequences that does not contain a region of significant local alignment, without respect to order or orientation (Fig. 1). In general, _d_SM can be thought of as the fraction of the aligned sequences that cannot be posited as homologous according to the above method. We found that upstream sequences of 500 bp and a minimum score of 48 (a combination of matches, mismatches, and gaps that sum to 48) was most predictive of expression difference between paralogs (Results), and we used these parameters for all subsequent analyses. Further details of the algorithm and its implementation can be found in the Supplemental material.
Computation of the first sequence alignment is O(n1 × n2) in memory and CPU time, where n = sequence length. Subsequent iterations remain memory-intensive but are much less CPU-intensive, as only part of the matrix needs to be recomputed. The main limitation of the SMM is its memory usage, which limits analysis to pairs of sequences typically <5kb. In comparison, other recent approaches such as the DBA method (Jareborg et al. 1999) are able to handle alignment of much larger sequences. However, the latter method and others do not detect contiguous shared regions if their order or orientation is not conserved.
C source code of the SMM software (sharmot) is freely available for download at: http://www.oeb.harvard.edu/faculty/wakeley/.
Expression Data
Expression data were obtained from Hill et al. (2000) in which Affymetrix oligonucleotide microarrays were used to examine mRNA expression at eight different stages of C. elegans development. We calculated the absolute value of the maximum difference in absolute number of transcripts for duplicate pairs through development where valid data for at least two timepoints for both genes was available [genes called “present” at least once by Hill et al. (2000)]. Similar results were found using mean expression difference through development (data not shown). Additionally, we measured changes in relative expression by computing Pearson's r correlation coefficient through development for duplicate pairs with valid data for ≥4 timepoints.
Data from 553 separate cDNA microarray experiments that included different nutrient conditions, developmental stages, and mutants (Kim et al. 2001) were also used to estimate differences in relative expression between pairs of duplicate genes. For each duplicate pair, we computed Pearson's r correlation coefficient across experiments on normalized log2[Cy3/Cy5] ratios for genes with valid data (>2-fold change) across >30 experiments.
Acknowledgments
We thank the anonymous reviewers and all members of the Wakeley and Hartl labs, as well as Eric Coissac, Eduardo P.C. Rocha, and Isabelle Gonçalves for their suggestions; Laura Garwin for her comments on an earlier version of the manuscript, and The Sanger Institute and the Genome Sequencing Center at Washington University for providing unfinished C. briggsae sequence. Special thanks to the Bauer Center for Genomics Research and Gordon L. Kindlmann at the University of Utah Scientific Computing and Imaging Institute for computational resources. G.A. was funded by La Fondation pour la Recherche Médicale.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2662504\. Article published online before print in July 2004.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**:** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J. 2000. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25**:** 106–110. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C.I. and Hartl, D.L. 2002. Genome evolution and developmental constraint in Caenorhabditis elegans. Mol. Biol. Evol. 19**:** 728–735. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282**:** 2012–2018. [DOI] [PubMed] [Google Scholar]
- Conant, G.C. and Wagner, A. 2003. Asymmetric sequence divergence of duplicate genes. Genome Res. 13**:** 2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M.O., Eck, R.V., and Park, C.M. 1972. A model of evolutionary change in protein sequences. In Atlas of protein sequence and structure, pp. 89–99, National Biomedical Research Foundation, Washington, D.C.
- Dunn, K.A., Bielawski, J.P., and Yang, Z. 2001. Substitution rates in Drosophila nuclear genes: Implications for translational selection. Genetics 157**:** 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L. and Mouchiroud, D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. 96**:** 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle, WA.
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151**:** 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N. and Yang, Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11**:** 725–736. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Nicolae, D., Lu, H.H., and Li, W.H. 2002. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18**:** 609–613. [DOI] [PubMed] [Google Scholar]
- GuhaThakurta, D., Palomar, L., Stormo, G.D., Tedesco, P., Johnson, T.E., Walker, D.W., Lithgow, G., Kim, S., and Link, C.D. 2002. Identification of a novel _cis_-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 12**:** 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J.B.S. 1932. The causes of evolution. Longmans and Green, London.
- Hill, A.A., Hunter, C.P., Tsung, B.T., Tucker-Kellogg, G., and Brown, E.L. 2000. Genomic analysis of gene expression in C. elegans. Science 290**:** 809–812. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. 1994. The evolution of functionally novel proteins after gene duplication. Proc. R Soc. Lond. B Biol. Sci. 256**:** 119–124. [DOI] [PubMed] [Google Scholar]
- Jareborg, N., Birney, E., and Durbin, R. 1999. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9**:** 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., Birren, B.W., and Lander, E.S. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428**:** 617–624. [DOI] [PubMed] [Google Scholar]
- Kim, S.K., Lund, J., Kiraly, M., Duke, K., Jiang, M., Stuart, J.M., Eizinger, A., Wylie, B.N., and Davidson, G.S. 2001. A gene expression map for Caenorhabditis elegans. Science 293**:** 2087–2092. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Weisshaar, B., Kroymann, J., Haubold, B., and Mitchell-Olds, T. 2001. Comparative genomics and regulatory evolution: Conservation and function of the Chs and Apetala3 promoters. Mol. Biol. Evol. 18**:** 1882–1891. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3**:** research0008.0001–0008.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, M.Z., Bergman, C., Patel, N.H., and Kreitman, M. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403**:** 564–567. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290**:** 1151–1155. [DOI] [PubMed] [Google Scholar]
- Needleman, S.B. and Wunsch, C.D. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48**:** 443–453. [DOI] [PubMed] [Google Scholar]
- Nembaware, V., Crum, K., Kelso, J., and Seoighe, C. 2002. Impact of the presence of paralogs on sequence divergence in a set of mouse–human orthologs. Genome Res. 12**:** 1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R. and Yang, Z. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148**:** 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Heidelberg.
- Ohta, T. 1987. Simulating evolution by gene duplication. Genetics 115**:** 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P.J., Wratten, N.S., McGregor, A.P., and Dover, G.A. 2002. Coevolution in bicoid-dependent promoters and the inception of regulatory incompatibilities among species of higher Diptera. Evol. Dev. 4**:** 265–277. [DOI] [PubMed] [Google Scholar]
- Smith, T.F. and Waterman, M.S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147**:** 195–197. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278**:** 631–637. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:** 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, J.R. and Haag, E.S. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3**:** 109–119. [DOI] [PubMed] [Google Scholar]
- Wagner, A. 2000. Decoupled evolution of coding region and mRNA expression patterns after gene duplication: Implications for the neutralist-selectionist debate. Proc. Natl. Acad. Sci. 97**:** 6579–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J.B. 1995. How often do duplicated genes evolve new functions? Genetics 139**:** 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W., and Lawrence, C.E. 2000. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 26**:** 225–228. [DOI] [PubMed] [Google Scholar]
- Waterman, M.S. and Eggert, M. 1987. A new algorithm for best subsequence alignments with application to tRNA-rRNA comparisons. J. Mol. Biol. 197**:** 723–728. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. and Li, W.H. 2003. Molecular evolution meets the genomics revolution. Nat. Genet. 33**:** 255–265. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13**:** 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 2000. Phylogenetic Analysis by Maximum Likelihood (PAML), version 3.0, http://abacus.gene.ucl.ac.uk/software/paml.html
WEB SITE REFERENCES
- http://wormbase.org; Wormbase.
- http://www.oeb.harvard.edu/faculty/wakeley/; C source code of the SMM software (sharmot).