RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies (original) (raw)
Abstract
Cajal bodies (CBs) are subnuclear organelles of animal and plant cells. A role of CBs in the assembly and maturation of small nuclear ribonucleoproteins (snRNP) has been proposed but is poorly understood. Here we have addressed the question where U4/U6.U5 tri-snRNP assembly occurs in the nucleus. The U4/U6.U5 tri-snRNP is a central unit of the spliceosome and must be re-formed from its components after each round of splicing. By combining RNAi and biochemical methods, we demonstrate that, after knockdown of the U4/U6-specific hPrp31 (61 K) or the U5-specific hPrp6 (102 K) protein in HeLa cells, tri-snRNP formation is inhibited and stable U5 mono-snRNPs and U4/U6 di-snRNPs containing U4/U6 proteins and the U4/U6 recycling factor p110 accumulate. Thus, hPrp31 and hPrp6 form an essential connection between the U4/U6 and U5 snRNPs in vivo. Using fluorescence microscopy, we show that, in the absence of either hPrp31 or hPrp6, U4/U6 di-snRNPs as well as p110 accumulate in Cajal bodies. In contrast, U5 snRNPs largely remain in nucleoplasmic speckles. Our data support the idea that CBs may play a role in tri-snRNP recycling.
Keywords: Cajal bodies, hPrp31, pre-mRNA splicing, retinitis pigmentosa, snRNP biogenesis
Introduction
Nuclear pre-mRNA splicing is catalysed by a large ribonucleoprotein complex, the spliceosome, which is assembled onto each intron in an ordered, multi-step process from the small nuclear RNPs (snRNPs) U1, U2, U4/U6, U5 and more than a hundred non-snRNP proteins (splicing factors; reviewed in Burge et al, 1999; Will and Lührmann, 2001; Jurica and Moore, 2003). During assembly of the spliceosome and catalysis of splicing, the U snRNPs, like the spliceosome as a whole, undergo major, ordered changes in composition and structure. This is particularly true of the U4/U6 and U5 snRNPs, which enter the spliceosome as a pre-assembled U4/U6.U5 tri-snRNP. During the transformation of the fully assembled spliceosome into a catalytically active machine, the intermolecular base pairing between U4 and U6 snRNAs is disrupted and the U4 snRNP is released. Next, the U6 snRNA base pairs with the U2 snRNA, a step leading to the formation of the catalytically active centre (Staley and Guthrie, 1998; Brow, 2002; Nilsen, 2003). After splicing, the spliceosome disintegrates and the released, individual U4, U6 and U5 snRNPs reassemble into a new U4/U6 di-snRNP and then into a U4/U6.U5 tri-snRNP, before entering the next splicing cycle.
The recycling of the tri-snRNP is essential for splicing. However, relatively little is known about the mechanism of this process and the nuclear location where the assembly of the tri-snRNP takes place. Assembly studies in vitro have provided certain clues as to the molecular organisation of the mammalian U4/U6 di-snRNP and have suggested a hierarchical assembly pathway for this particle. This process appears to be initiated by the binding of the 15.5K protein (also designated NHPX) to the 5′ stem–loop of the U4 snRNA, followed by binding of the hPrp31 (61K) protein. The subsequent integration of the proteins CypH (20K or USACyp), hPrp4 (60K) and hPrp3 (90K), which form a heteromeric complex (hereafter termed the CypH/hPrp4/hPrp3 complex), requires an intact U4/U6 duplex (Nottrott et al, 1999, 2002). Additional proteins associated with the U4 and U6 snRNAs are the Sm and LSm2–8 proteins, respectively (Will and Lührmann, 2001). For the formation of the U4/U6 di-snRNP from the individual U4 and U6 snRNPs, an additional factor is required: the p110 protein (also designated SART3 or hPrp24), which was recently identified as the human orthologue of the yeast U4/U6 di-snRNP recycling factor Prp24p (Raghunathan and Guthrie, 1998; Bell et al, 2002; Rader and Guthrie, 2002). p110 binds specifically to U6 snRNA and is present in both the U6 snRNP and the U4/U6 di-snRNP. However, since p110 has never been found as a constituent of the spliceosome or the tri-snRNP (Bell et al, 2002), it apparently leaves the tri-snRNP during a late assembly step, perhaps concomitantly with the incorporation of the 20S U5 snRNP.
An important protein for the interaction between the U4/U6 di-snRNP and the U5 snRNP is the U4/U6-specific hPrp31 protein, which is indispensable for the formation of the human tri-snRNP in vitro and is required for pre-mRNA splicing (Makarova et al, 2002). Two-hybrid analysis has shown that hPrp31 binds specifically to the U5-specific protein hPrp6, suggesting that these two proteins form a bridge in the tri-snRNP (Makarov et al, 2000; Makarova et al, 2002). The hPrp31 protein is also of medical interest, because mutations in its gene (PRPF31) are associated with autosomal dominant retinitis pigmentosa (Vithana et al, 2001), a disease that leads to the degeneration of the photoreceptors in the eye.
The site in the nucleus where the tri-snRNP is recycled is unknown. Candidates are subnuclear domains or regions such as the interchromatin granules (also termed ‘splice factor compartments' or ‘speckles') or the Cajal (coiled) bodies (characterised by the marker protein coilin) or the nucleoplasm, all of which are enriched in spliceosomal snRNPs (Misteli et al, 1997; Spector, 2001; Lamond and Spector, 2003). While the speckles very probably act as storage sites from which snRNPs and splicing factors can move to the sites of active splicing (Misteli et al, 1997), the function of the Cajal bodies has long been largely an object of speculation. While they contain all the spliceosomal snRNAs, they lack many essential non-snRNP splicing factors such as SR proteins and are therefore not likely to be sites of active splicing (Matera, 1999; Ogg and Lamond, 2002; Gall, 2003). Several recent studies, however, have indicated that the Cajal bodies may be involved in the assembly and maturation of snRNPs. For example, recently discovered RNAs (scaRNAs) that guide post-transcriptional modification of U1, U2, U4 and U5 snRNAs localise specifically to Cajal bodies (Darzacq et al, 2002), suggesting that snRNA base modifications may take place in CBs (Kiss et al, 2002; Jády et al, 2003). In addition, newly synthesised snRNPs, after their assembly in the cytoplasm and import into the nucleus (for a review, see Will and Lührmann, 2001), first accumulate in the Cajal bodies before they move on to other nuclear locations such as the speckles and nucleoplasm (Ferreira et al, 1994; Sleeman and Lamond, 1999; Sleeman et al, 2001; Ogg and Lamond, 2002). The Cajal bodies could therefore be the site where dismantled snRNPs are recycled into functional particles. Consistent with this, the U4/U6 di-snRNP recycling factor p110 has been found to be enriched in Cajal bodies and to play a role in targeting U6 snRNPs to Cajal bodies (Staněk et al, 2003).
In this study, we have addressed the question of the site of tri-snRNP assembly by combining RNAi depletion methods with fluorescence microscopy. The former was used to elucidate the functions of the putative bridging proteins hPrp31 and hPrp6 in tri-snRNP assembly in vivo, while the latter allowed the location of various components of the tri-snRNP particle in the cell nucleus. We show that both proteins are required for tri-snRNP stability in vivo. In the absence of either of these proteins, U4/U6 di-snRNPs accumulate in Cajal bodies, suggesting that the formation of U4/U6.U5 tri-snRNPs occurs in these subnuclear structures.
Results
RNAi knockdown of the hPrp31 and hPrp6 proteins
To investigate the role of the hPrp31 and hPrp6 proteins in tri-snRNP formation in intact cells, RNAi experiments were performed with HeLa SS6 cells and 21-nt siRNA duplexes (Elbashir et al, 2001, 2002). siRNA duplexes targeting either the coding region or 3′UTR of mRNAs encoding the hPrp31, hPrp6 and as controls the U2 protein SF3b14a and a non-human protein with no human homologues (firefly luciferase) were tested for their ability to inhibit cell growth. siRNA duplexes directed against all three snRNP proteins reduced cell growth to a significant extent after 72 h, ranging from 45% (hPrp31), 30% (hPrp6) and 20% (SF3b14a), as compared to the control cells treated with an siRNA duplex against luciferase (GL2). Thus, these proteins are essential for cell viability, consistent with studies in yeast demonstrating that disruption of genes encoding Prp31 or Prp6 results in lethality (Legrain et al, 1991; Weidenhammer et al, 1996). Cells treated with siRNAs against the spliceosomal proteins, but not cells treated with GL2, started to undergo apoptosis at this time point, as demonstrated by TUNEL analysis. However, no apoptotic cells were observed after 48 h. Therefore, all biochemical experiments were performed 48 h after transfection.
Taken together, these results indicate that the three snRNP proteins play essential roles in the cell, consistent with their functioning in pre-mRNA splicing. To demonstrate that the targeted proteins had indeed been depleted from the cells, we performed Western blotting with antibodies directed against each protein. As shown in Figure 1, the hPrp31 and hPrp6 proteins were reduced by about 95% (lanes 2 and 4, respectively) and SF3b14a by about 85% (lane 6) as compared to the control cells treated with an siRNA against luciferase (lanes 1, 3 and 5). hPrp4 and p110 levels were not affected by the loss of either protein, and the depletion of hPrp6 or hPrp31 did not affect the level of the other protein (lanes 2, 4 and 6).
Figure 1.
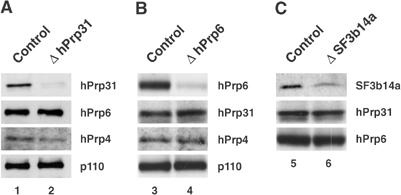
Suppression (knockdown) of spliceosomal proteins by siRNAs. (A) A Western blot showing levels of hPrp31, hPrp6, hPrp4 and p110 protein in the nuclear extract prepared from HeLa cells 48 h after transfection with hPrp31-specific siRNA (EA1) and a firefly luciferase siRNA control. (B) A similar blot made 48 h after transfection with hPrp6-specific siRNA (AL5). (C) A Western blot showing protein levels of hPrp31, hPrp6 and SF3b14a protein 48 h after transfection with the SF3b14a-specific siRNA (AS1). The same amounts of protein were loaded from nuclear extracts generated from control and knockdown cells.
For biochemical experiments, the transfection procedure was then scaled up from microlitre to the millilitre scale (details in Materials and methods), with transfection by electroporation instead of the Oligofectamine method usually employed. In this way, we obtained enough cells for a small-scale preparation of nuclear extract that was suitable for subsequent biochemical assays (Lee and Green, 1990).
hPrp31 and hPrp6 are required for tri-snRNP formation in vivo
To corroborate the function ascribed to hPrp31 and hPrp6 in vitro (see Introduction), we examined the effect of their depletion on the formation of the tri-snRNP in intact cells. Figure 2A illustrates this for the case of hPrp31 protein. Transfection with siRNA against hPrp31 does not affect the level of U4, U5 or U6 snRNAs in the cells (lane 3) as compared to cells treated with either siRNAs against the U2 protein SF3b14a (lane 2) or luciferase (lane 1). Immunoprecipitation with antibodies against the Sm proteins (α-Sm), which are common to U4/U6 and U5 snRNPs, shows that the amount of these snRNPs in the cells is not affected by knockdowns of the hPrp31 and SF3b14a proteins, when compared with the cells that had received an siRNA against luciferase (lanes 4–6). The critical effect is seen in immunoprecipitation with antibodies against protein 40K (α-40K), a protein that is a part of the U5 snRNP. While this antibody precipitates intact tri-snRNPs in extracts of cells transfected with an siRNA against a non-human or a U2 protein (lanes 7 and 8), in extracts of cells that have been depleted of protein hPrp31, the antibody precipitates primarily U5 snRNPs (lane 9). Therefore, the lack of protein hPrp31 has prevented interaction between the U5 snRNP and U4/U6 di-snRNP.
Figure 2.
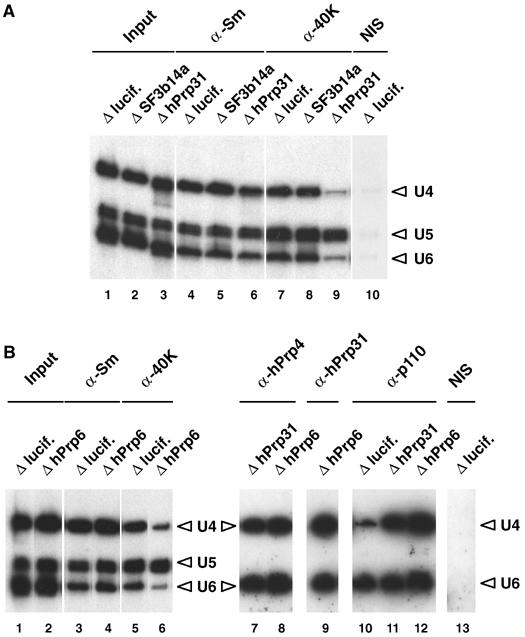
Disruption of the tri-snRNP in vivo by selective depletion of either the U4/U6 protein hPrp31 or the U5 protein hPrp6. (A) Anti-Sm and anti-40K antibodies (denoted α-Sm, α-40 K, etc.) were used to immunoprecipitate snRNPs from the nuclear extract prepared from HeLa cells 48 h after transfection with siRNAs targeting either luciferase, the U2 protein SF3b14a (AS1, ΔSF3b14a) or the U4/U6 protein hPrp31 (EA1, ΔhPrp31). NIS: nonimmune serum from α-40K. Separate controls with non-transfected cells (not shown) revealed patterns identical to those with luciferase-transfected cells. (B) Anti-Sm, anti-40K, anti-hPrp4, anti-hPrp31 and anti-p110 antibodies were used to immunoprecipitate snRNPs from the nuclear extract prepared from HeLa cells 48 h after transfection with siRNAs targeting luciferase, the U4/U6 protein hPrp31 (ΔhPrp31) or the U5 protein hPrp6 (AL5, ΔhPrp6). NIS: non-immune serum from α-hPrp4. In all experiments, RNA was isolated from the immunoprecipitated snRNPs and characterised by Northern blot analysis.
Similar results were obtained when the cells were depleted of the U5-specific protein hPrp6 (Figure 2B). The knockdown leaves unaffected the cells' complement of U4, U5 and U6 snRNAs (lane 2) and U4/U6 and U5 snRNPs (lane 4), as compared to cells transfected with an siRNA against luciferase (lanes 1 and 3, respectively). However, the absence of protein hPrp6 significantly suppresses the formation of tri-snRNPs (>80%), as revealed by the significant decrease in precipitation of intact tri-snRNPs by antibodies against the U5-specific 40K protein (lane 6, compare lane 5). These results show for the first time in vivo that intact tri-snRNP particles require the presence of the hPrp31 protein. Furthermore, these experiments provide for the first time direct evidence about the function of the U5 protein hPrp6 and show that, like hPrp31, it is essential for the stability of the tri-snRNP in vivo.
Accumulation of stable U4/U6 di-snRNPs upon depletion of hPrp31 or hPrp6
It is important to note that the suppression of tri-snRNP formation by depletion of protein hPrp31 or hPrp6 not only left unchanged the cells' complement of U4, U5 and U6 snRNA and snRNP, but also had no effect on the stability of the U4/U6 di-snRNP. That is, U4 and U6 snRNAs were always precipitated in the same ratio, and—more important—they were precipitated by antibodies against protein hPrp4, which is known (as part of a heterotrimeric complex CypH/hPrp4/hPrp3; see Introduction) to bind only to the base-paired U4/U6 di-snRNP but not the separate snRNPs U4 or U6 (Figure 2B, lanes 7 and 8). We also note that protein hPrp31 is stably bound in the U4/U6 snRNP in the absence of protein hPrp6 (Figure 2B, lane 9), although the stability of the U4/U6 di-snRNP even in the absence of protein hPrp31 shows that the latter is not a structural prerequisite for the di-snRNP.
Finally, our RNAi experiments provide strong evidence that the U4/U6 di-snRNPs, which accumulate upon removal of proteins hPrp31 or hPrp6, that is, under conditions where the tri-snRNP cannot form, are associated not only with the U4/U6-specific proteins but also with the U4/U6 di-snRNP recycling factor p110. That is, while under control transfection with an siRNA against luciferase mRNA, anti-p110 antibodies precipitate primarily free U6 snRNP and only small amounts of U4/U6 di-snRNP (Figure 2B, lane 10). The yield of U4/U6 precipitated by anti-p110 antibodies is much greater when the cells are depleted of proteins hPrp31 or hPrp6, that is, under conditions where the tri-snRNP cannot form (Figure 2B, lanes 11 and 12, respectively). The increased yield is consistent with accumulation of p110-bound U4/U6 di-snRNP in a stage that immediately precedes association with the U5 snRNP.
Targeting of U4/U6 di-snRNPs, but not U5 snRNPs, to Cajal bodies in the absence of hPrp31 or hPrp6
We proceeded to investigate the influence of the inhibition in vivo of tri-snRNP formation upon the distribution of the U4/U6 and U5 snRNPs in the nucleus. Proteins were located by fluorescence microscopy with appropriately labelled antibodies, and RNAs by fluorescence in situ hybridisation (FISH). The depletion by RNAi of hPrp31 or hPrp6 led to a pronounced accumulation of the U4/U6-specific protein hPrp4 in discrete nuclear foci at the expense of nucleoplasmic staining. The foci where the protein accumulates can be identified as Cajal bodies, as evidenced by co-immunofluorescence of hPrp4 protein and coilin (Figure 3B and C; compare Figure 3A). A similar accumulation in Cajal bodies was seen with hPrp3 (Figure 3D–F) and for the U6-specific protein hLSm4 (not shown). In all cases, the intensity of the fluorescent signals in the Cajal bodies increased significantly compared with cells that had undergone control transfections. Co-immunofluorescence of protein p110 and coilin (Figure 3G–I) also showed an accumulation of p110 in the Cajal bodies in the absence of protein hPrp31 or hPrp6. However, the decrease in nucleoplasmic staining was less pronounced than in the situation described for hPrp4 and hPrp3. The average intensity of fluorescence from proteins hPrp4, hPrp3 or p110 in the Cajal bodies and in the rest of the nucleus was measured (at low exposure, to avoid saturation effects) and, in each case, the ratio of intensity in the Cajal bodies to that in the rest of the nucleus was calculated. Results are shown in Table I; in each case, the intensity ratio increased by a factor of 3–5 when the cells were depleted of either hPrp31 or hPrp6.
Figure 3.
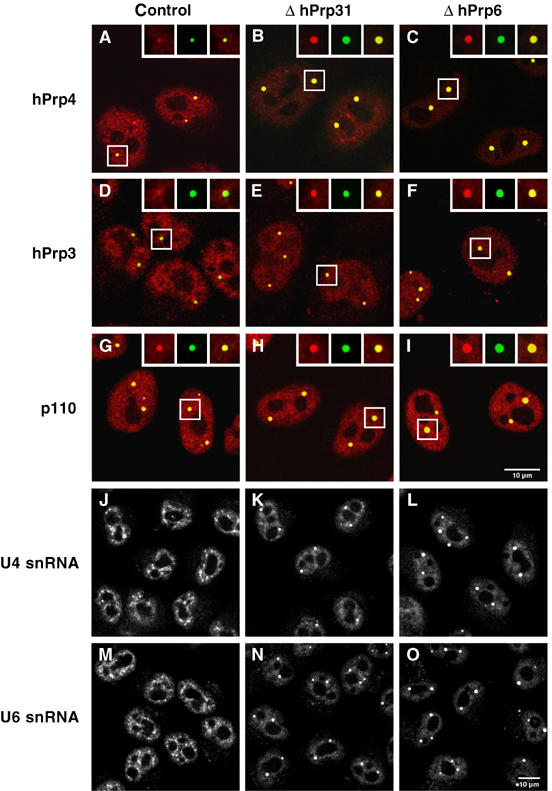
Inhibition of tri-snRNP formation leads to an accumulation of U4/U6-specific components in the Cajal bodies. HeLa cells transfected with siRNA targeting luciferase (control), the U4/U6 protein hPrp31 (EA1, ΔhPrp31) or the U5 protein hPrp6 (AL5, ΔhPrp6) were examined after 48 h using indirect immunofluorescence (A–I) or fluorescence in situ hybridisation (J–O). (A–I) Green, monoclonal antibodies against coilin (showing the Cajal bodies); red, antibodies against the protein in question. (A–C) Transfected HeLa cells were stained with antibodies against coilin (green) and against the U4/U6 protein hPrp4 (red). The accumulation of protein hPrp4 in the Cajal bodies is indicated by the superposition of these (yellow). (D–F) As (A–C), using antibodies against the protein hPrp3 instead of protein hPrp4. (G–I) As above, but using affinity-purified antibodies against protein p110. The red and green fluorescence signals were recorded independently and combined to give the overlay images shown; the insets show separate images for selected Cajal bodies. (J–O) For visualisation of snRNAs, cells were hybridised with Cy3-labelled oligonucleotides complementary to (J–L) U4 snRNA or (M–O) U6 snRNA. For optimal display, contrast and brightness of the images were adjusted.
Table 1.
Effect of inhibition of tri-snRNP formation on concentration of U4/U6-specific and U5-specific components in Cajal bodies
| | Cajal bodies/nucleoplasm ratioa±s.d.b (_n_c) | | | | | ------------------------------------------------------------------------------------------------------------------- | ------------ | ------------- | ------------- | | Protein or snRNA | Control | ΔhPrp31 | ΔhPrp6 | | hPrp4 | 4.1±0.4 (23) | 13.6±3.6 (23) | 19.8±7.3 (15) | | hPrp3 | 3.0±0.4 (23) | 12.4±3.8 (24) | 10.6±2.1 (23) | | p110 | 4.3±1.1 (23) | 11.6±3.5 (24) | 11.1±2.6 (23) | | U4 snRNA | 3.6±0.4 (24) | 14.0±5.8 (26) | 13.0±6.9 (25) | | U6 snRNA | 3.4±0.2 (23) | 19.0±5.4 (23) | 21.6±5.7 (24) | | hSnu114 | 1.4±0.2 (37) | 1.5±0.2 (31) | 1.2±0.1 (29) | | U5 snRNA | 3.6±0.7 (37) | 4.7±1.5 (26) | 5.1±2.1 (23) | | U1 snRNA | 2.7±0.3 (23) | 2.8±0.2 (23) | 2.8±0.2 (23) | | aRatio of average per-pixel fluorescence intensity in the Cajal bodies and nucleoplasm. | | | | | bStandard deviation of random sample of cells (all the photographs in the experiment were used, without selection). | | | | | c_n_=number of cells evaluated (with an average of 2–3 Cajal bodies per cell). | | | |
Next, we examined the distribution of U4 and U6 snRNAs in the nucleus by FISH. Knockdown of hPrp31 or hPrp6 resulted in a dramatic increase in the amount of U4 and U6 snRNA in the Cajal bodies and decrease of these RNAs in the speckles (Figure 3J–L and M–O). As described above, the enhancement of the signals from the snRNAs in the Cajal bodies was quantified (Table I) and revealed relative increases for U4 and U6 snRNAs of a magnitude similar to those seen for proteins hPrp4 and hPrp3. These results thus provide strong evidence that the absence of protein hPrp31 or hPrp6, which inhibits tri-snRNP formation, also results in an accumulation of U4 and U6 snRNAs and U4- and U6-specific proteins in the Cajal bodies.
To determine whether U4 snRNP, U6 snRNP and the protein complex CypH/hPrp4/hPrp3 accumulate in Cajal bodies separately, or as U4/U6 di-snRNP particles, we repeated the experiment shown in Figure 3, but subjected the cells to a short treatment with RNase A before staining with antibodies against protein hPrp4 or p110. This treatment destroys RNA and releases the protein associated with it (Spector et al, 1991). The results of such an experiment are shown in Figure 4. In the top row (A–C), a typical staining pattern for protein hPrp4 is seen, with accumulation resembling that observed in Figure 3. In the second row (D–F), in which cells had been treated with RNase, most of the hPrp4 protein in the cell (and all hPrp4 visible in the Cajal bodies) was released by RNase treatment. However, p110 was not affected (Figure 4G–I); that is, its localisation in the Cajal bodies was not influenced by the destruction of snRNAs. A control with U6 snRNA detected by FISH showed that RNase treatment conditions do indeed suffice to remove RNA (Figure 4M–R); furthermore, a control with U4 snRNA gave a similar result, while another with antibodies against coilin showed that the distribution of coilin and thus the Cajal bodies, was unaffected by the treatment with RNase A (not shown). The above observations, along with the fact that the complex CypH/hPrp4/hPrp3 binds in vitro and in vivo only to the complete U4/U6 snRNA duplex (and is not found in U4 or U6 mono-particles; Gonzalez-Santos et al, 2002; Nottrott et al, 2002), led us to conclude that the accumulation of the RNAs and proteins of the U4/U6 di-snRNP in the Cajal bodies, as shown above, is due to the accumulation of the assembled U4/U6 di-snRNP.
Figure 4.
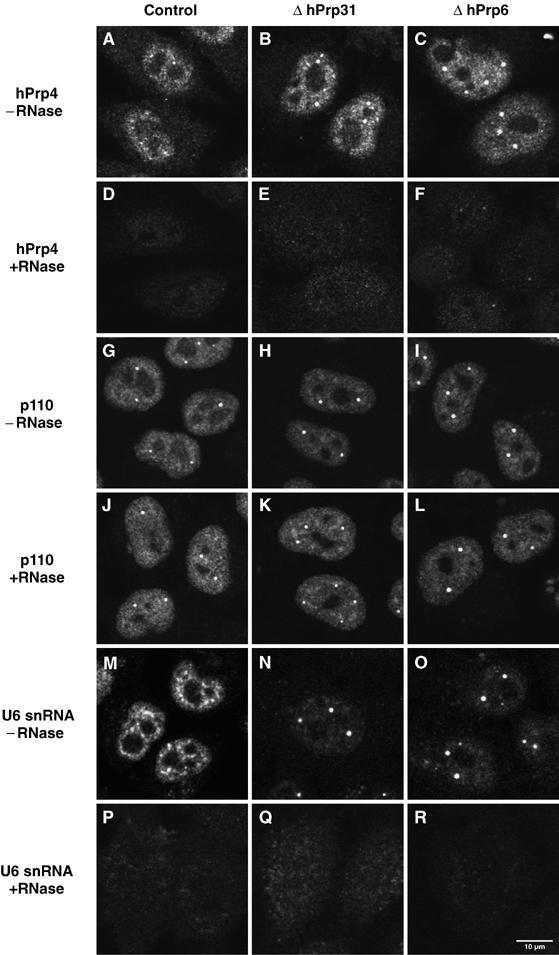
Sensitivity of the U4/U6 hPrp4 protein distribution pattern to treatment with RNase A. HeLa cells transfected with siRNA targeting the firefly luciferase protein (control), a U4/U6 hPrp31-specific (EA1, ΔhPrp31) or a U5 hPrp6-specific siRNA (AL5, ΔhPrp6) were treated either with (D–F, J–L, P–R) or without RNase A (A–C, G–I, M–O) 48 h after transfection before immunofluorescent investigation of hPrp4 (A–F), p110 distribution in the nucleus (G–L) or U6 snRNA as a control for RNase A digestion (M–R).
Next, we investigated the nuclear location of U5 snRNPs in HeLa cells under conditions where tri-snRNP formation is inhibited (Figure 2). In contrast to the situation observed for U4/U6 snRNPs, depletion by RNAi of hPrp31 or hPrp6 did not lead to a significant accumulation in Cajal bodies of the U5 snRNP-specific protein hSnu114 (116K) (Figure 5A–C, see also quantitative data in Table I). However, the speckled pattern appeared more coarse (Figure 5B–C). Consistent with the results obtained for the hSnu114 protein, U5 snRNA showed no significant accumulation in Cajal bodies when tri-snRNP formation was inhibited, but remained in speckles, which also appeared more coarsely grained (Figure 5D–F). Interestingly, as shown by FISH with anti-sense probes for U2 and U1 snRNAs, the distributions of these RNAs are identical (U2) or similar (U1) to the distribution of the U5 snRNA in the nucleus upon knockdown of protein hPrp31 or hPrp6 (Figure 5G–I and J–L and Table I).
Figure 5.
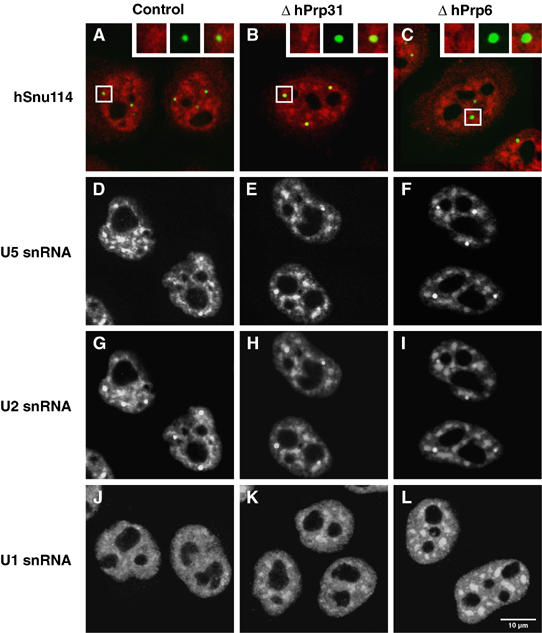
Inhibition of tri-snRNP formation does not lead to an accumulation of U5-specific components in Cajal bodies. HeLa cells transfected with an unspecific siRNA targeting the firefly luciferase protein (control), a U4/U6 hPrp31-specific (EA1, hPrp31) or a U5 hPrp6-specific siRNA (AL5, hPrp6) were analysed after 48 h using immunofluorescence (A–C) or fluorescence in situ hybridisation (D–L). (A–C) Transfected HeLa cells that were stained with antibodies against coilin (green) and against the U5 protein hSnu114 (red). The red and green fluorescence signals were recorded independently and combined to give the overlay images shown; the insets show separate images for selected Cajal bodies. For visualisation of snRNAs, cells were hybridised with fluorescent-labelled oligonucleotides complementary to (D–F) U5 snRNA, (G–I) U2 snRNA or (J–L) U1 snRNA. In rows 2 and 3, the same cells are shown, hybridised with both a Cy3-labelled U5 probe and an Alexa 488-labelled U2 probe to identify the Cajal bodies for quantitative measurements.
Discussion
This work combines (i) an siRNA-based biochemical analysis of RNA and protein constituents of the tri-snRNP in vivo with (ii) the use of fluorescence microscopy to locate these constituents in the nucleus. Our initial finding is that when the cells are depleted of hPrp31 or hPrp6 protein by RNAi knockdown, the formation of U4/U6.U5 tri-snRNP is inhibited, and stable U4/U6 di-snRNPs and U5 snRNPs accumulate in the cell (Figure 2). This demonstrates for the first time in vivo that the U4/U6 protein hPrp31 is essential for the interaction of the U4/U6 di-snRNP with the 20S U5 snRNP, and is consistent with previous studies performed in vitro (Makarova et al, 2002). A similar property must also now be ascribed to the U5 protein hPrp6 which, like the hPrp31 protein, is essential for the stability of the tri-snRNP in vivo. It is known from two-hybrid analyses that hPrp31 and hPrp6 bind strongly and specifically to one another (Makarova et al, 2002), and we therefore consider it likely that U4/U6 and U5 snRNPs are connected by a bridge comprised of at least these two proteins. Our results regarding the hPrp6 protein are also consistent with the independent observation (Galisson and Legrain, 1993) that when yeast cells are depleted of protein Prp6p, which is homologous to human protein hPrp6, tri-snRNP formation is inhibited and U4/U6 di-snRNPs and U5 snRNP mono-particles accumulate.
We examined cells that had been depleted of a given protein using fluorescence microscopy, and were thus able to draw conclusions from the combined biochemical and in situ localisation approaches. The most striking observation was that, under conditions where tri-snRNP formation was inhibited, both U4/U6-specific proteins (detected by specific antibodies) and U4 and U6 snRNAs (detected by FISH) accumulate in Cajal bodies (Figure 3 and Table I). Our further finding that RNase A treatment of these cells resulted in the loss of all of hPrp4 visible in the Cajal bodies (Figure 4), along with the fact that the CypH/hPrp4/hPrp3 protein complex requires the U4/U6 snRNA duplex for binding (Nottrott et al, 2002), allows us to conclude that U4/U6 di-snRNPs rather than individual U4 and U6 snRNP components accumulate in Cajal bodies under these conditions. Consistent with this conclusion, our biochemical results show that stable U4/U6 di-snRNPs can be immunoprecipitated from nuclear extracts of cells depleted of hPrp31 or hPrp6 by RNAi (Figure 2).
In contrast to the situation observed for the U4/U6 di-snRNPs, the U5 snRNPs did not accumulate in Cajal bodies when tri-snRNP formation was inhibited. However, the U5 snRNP speckle pattern appeared to be more coarsely grained. This coarsening of the speckle pattern has already been observed by others when splicing was blocked (Carmo-Fonseca et al, 1992; O'Keefe et al, 1994). Indeed, removal of hPrp31 or hPrp6 by RNAi also resulted in a coarsening of speckles containing U1 and U2 snRNPs (Figure 5) and the non-snRNP splicing factor SC35 (not shown), indicating that splicing is inhibited in the HeLa cells when tri-snRNP formation is inhibited.
Our combined results show that there is a major qualitative difference between the behaviour of the U4/U6 snRNPs and U5 snRNPs when the tri-snRNP cannot be formed: the U4/U6 snRNPs accumulate in the Cajal bodies, while the distribution of the U5 snRNPs remains largely unchanged. What could be the mechanism accounting for the selective accumulation of the U4/U6 di-snRNP in Cajal bodies? While this question cannot at present be answered completely, protein p110 is likely to play a role in the U4/U6 snRNP accumulation. It has been shown that a significant fraction of this protein is located in the Cajal bodies (Staněk et al, 2003), as also seen here for the ‘control' cells (Figure 3G). This accumulation is known to depend upon the so-called HAT domain located at the N-terminus of p110 (Staněk et al, 2003). Protein p110 is required for di-snRNP formation and binds to both the U6 (mono) snRNP and U4/U6 di-snRNPs, and yet it is not a part of the tri-snRNP (Bell et al, 2002). In our experiments, in the absence of correct bridging between the U4/U6 and U5 constituents of the tri-snRNP, an increased amount of U4/U6 di-snRNP is precipitated by anti-p110 (Figure 2B). Likewise, the accumulation of di-snRNP in the Cajal bodies is accompanied by a corresponding accumulation of p110 in these bodies (Figure 3 and Table I). It is therefore tempting to suggest that the U4/U6 di-snRNPs are retained in the Cajal bodies through the HAT domain of p110. At least two scenarios can be envisioned for the retention process of U4/U6 di-snRNPs in Cajal bodies. One possibility is that U4/U6 di-snRNP formation takes place in the Cajal bodies (as proposed by Staněk et al, 2003), with U6 mono-snRNPs being initially anchored in the Cajal bodies. Alternatively, and consistent with the relatively large amounts of p110 in the nucleoplasm, p110 may bind first to U6 mono-snRNPs followed by U4/U6 di-snRNP formation in the nucleoplasm (Bell et al, 2002). The resulting complexes could then move to the Cajal bodies.
All in all, the data presented here point towards a pivotal function of the Cajal bodies in the final step(s) of the biogenesis of the tri-snRNP before it enters the spliceosome, and strengthen the idea of a principal role of the Cajal bodies in the assembly and maturation of snRNPs (see Introduction). Our results fit best with a model in which the U4/U6 di-snRNP first becomes anchored, through p110, to an as yet unidentified structure within the Cajal bodies (by one of the scenarios discussed above). The U5 snRNP then binds to the di-snRNP, with subsequent or concomitant dissociation or displacement of p110 and liberation of the mature tri-snRNP from the Cajal body. This is also consistent with the known ‘recycling' function of p110 (Bell et al, 2002). However, this picture also raises new questions, such as why such a mechanism should be needed, when the binding reaction between the U4/U6 di-snRNP and the U5 snRNP can easily be pictured to occur by collision, making protein-mediated catalysis unnecessary; or why the di-snRNPs essentially require a special mechanism to anchor them in the Cajal bodies when their reaction partners, the U5 snRNPs, do not. Clearly, more experiments are needed to answer these points.
The ability to knock down hPrp31 and other tri-snRNP proteins is a first step towards the molecular dissection of their function in vivo. In future studies, depletion of hPrp31 by siRNA-mediated knockdown will be coupled with the simultaneous expression of mutant forms of hPrp31, by transfection of the corresponding cDNAs. In this way, the effects of hPrp31 mutation on tri-snRNP complex formation or snRNP subcellular localisation can be studied. Of particular medical interest will be those mutations in hPrp31 that are associated with retinitis pigmentosa (Vithana et al, 2001).
Materials and methods
siRNA transfection and cell culture
All siRNA duplexes were designed and synthesised in-house as 21-mers with 3′dTdT overhangs as described previously (Elbashir et al, 2002). A BLAST search against the human genome sequence (NCBI UniGene database) was used to confirm that only the gene of interest would be targeted. The sequences used to target each gene were as follows: The accession numbers given below are from GenBank and the positions of the targeting sequences are given relative to the first nucleotide of the start codon.
hPrp31(NM_015629): EA1 5′-AAGCCAAAGCUUCAGAAGUGAUG-3′ (220–242 in ORF), EA2 5′-CAGUAUGGGCUAGAGCAGGUCUU-3′ (1648–1706 in 3′ UTR),
EA3 5′-AAGGGACACAGAGGUCCAGUCCU-3′ (1533–1555 in 3′ UTR),
hPrp6 (NM_012469): AL5 5′-AAGCUACAAGUAGCUCGGAACCU-3′ (969–991 in ORF),
AL6 5′-GAGCGGUUGCCAUGGCCGGUCUC-3′ (2828–2850 in 3′ UTR),
AL8 5′-CAGGGUUGGGCCGCAUGUGGAAG-3′ (2858–2880 in 3′ UTR),
SF3b14a (NM_016047): AS1 5′-AAGAAUGCAUGUGAUCACCUAUC-3′ (210–232 in ORF),
AS2 5′-UAAAUCCCACGAAUGACAACUAC-3′ (403–425 in 3′ UTR),
AS4 5′-AAUCCCACGAAUGACAACUACCA-3′ (405–427 in 3′ UTR).
The GL2 siRNA, which targets the firefly (Photinus pyralis) luciferase gene, was used as a control (Elbashir et al, 2002).
Transient transfections of siRNA into cultured HeLa SS6 cells were performed at a cell confluence of ∼20% corresponding to 60 000 cells/ml, by using Oligofectamine (Invitrogen) as described by Elbashir et al (2002). Cells were assayed 48 h after transfection; at this point in time, no apoptosis was observed by the TUNEL assay performed according to the manufacturer's instruction (Roche) (Gorczyca et al, 1993). The effect of depletion of hPrp6, hPrp31 and SF3b14a on cell growth was monitored by determining the cell survival of the knockdowns in comparison with the control cells treated with an siRNA against GL2. After 72 h, the number of viable cells was determined by using a cell counter (CASYcounter TT, Schärfe Systems GmbH). The effect on cell growth was calculated by normalising against the number of viable cells after GL2 transfection.
For electroporation, 10 μl of 20 μM siRNA duplex and 300 μl of cold OptiMEM1 were combined in an electroporation cuvette with an electrode gap of 0.4 cm (Bio-Rad) and pre-chilled on ice. Then 500 μl of cell suspension (5 × 106 cells) was added into the cuvette, and the contents were mixed well and incubated on ice for a further 5 min. The cell suspension was pulsed once at 260 V, 950 μF with a Gene Pulser electroporator II (Bio-Rad). The cells were resuspended and transferred to 12 ml pre-warmed growth medium and incubated for 48 h at 37°C.
Immunoprecipitation, immunoblotting and indirect immunofluorescence
Anti-p110 antibodies were raised against a C-terminal peptide (amino-acid residues 945–963) in rabbits. The antibodies were affinity-purified on a SulfoLink column containing the immobilised peptide as described by the manufacturer (Pierce). In addition, antibodies raised against the following proteins were used: Sm-proteins (Y12, monoclonal; Lerner et al, 1981), U5-40K (Achsel et al, 1998), hPrp4 (Lauber et al, 1997), hPrp3 (Lauber et al, 1997), hPrp31 (Makarova et al, 2002), SF3b14a (Will et al, 2001), hPrp6 (Makarov et al, 2000) and p80-coilin (5P10 π; Almeida et al, 1998).
Immunoprecipitations were performed as described (Makarova et al, 2001) with a small-scale nuclear extract prepared according to the protocol of Lee and Green (1990) from cells that had been transfected with siRNA by electroporation as described above. Co-precipitated RNAs were separated on 10% polyacrylamide gels containing 8 M urea. Subsequently, snRNAs were detected by Northern hybridisation using probes generated from plasmids encoding U4, U5 or U6 snRNAs (Pikielny et al, 1989) by using a random primer labelling kit (Prime It II, Stratagene). For Western blot analysis, 10 μg of nuclear extract proteins was separated on an 8 or 13% SDS–polyacrylamide gel, transferred to a nitrocellulose membrane (Schleicher & Schuell) and subsequently immunostained with affinity-purified antibodies by using the ECL detection kit as described by the manufacturer (Amersham). Indirect immunofluorescence experiments with siRNA-transfected cells were performed as previously described (Ingelfinger et al, 2002).
Fluorescence in situ hybridisation
Fluorescence in situ hybridisation (FISH) was performed according to a protocol based on that of Taneja et al (1992). Coverslips were mounted on the slides with Mowiol (Calbiochem) and examined by fluorescence microscopy. The sequences of the oligonucleotides used for FISH were as follows: U1 snRNA, 5′-CCT TCG TGA TCA TGG TAT CTC CCC TGC CAG GTA AGT AT-3′; U2 snRNA, 5′-GAA CAG ATA CTA CAC TTG ATC TTA GCC AAA AGG CCG AGA AGC-3′; U4 snRNA, 5′-TCA CGG CGG GGT ATT GGG AAA AGT TTT CAA TTA GCA ATA ATC GCG CCT-3′; U5 snRNA, 5′-CTC TCC ACG GAA ATC TTT AGT AAA AGG CGA AAG ATT TAT ACG ATT TGA AGA G-3′; U6 snRNA, 5′-CAC GAA TTT GCG TGT CAT CCT TGC GCA GGG GCC ATG CTA ATC-3′ (all oligonucleotides were Cy3-labelled at their 5′ end, except for U2 snRNA that was Alexa 488-labelled at its 3′- and 5′-end). RNase A digestion of HeLa cells grown on coverslips was performed as described by Spector et al (1991). After several washes in PBS, cells were prepared for immunofluorescence or FISH as described before.
Fluorescence microscopy and quantification
Fluorescence samples were visualised with identical parameters for control and knockdown cells by using a Zeiss LSM 510 meta confocal microscope with a × 63/1.4 oil objective. All images shown here are single confocal sections (pinhole diameter: 106 μm, 1.12 Airy units ∼0.8 μm sections). After automated background subtraction (estimated from the background peak in the histogram), each cell nucleus was delineated by hand-defining a region of interest. For each immunostaining, the Cajal bodies were counterstained with antibodies against coilin bearing a different fluorescent label, and for hybridisation with probes against the different snRNAs, the CBs were counterstained with a probe against the U2 snRNA (U2 snRNA is very prominent in CBs also under control conditions). The Cajal body staining was used to generate a ‘mask' for the CBs. In this way, the average fluorescence intensity in the Cajal body region in the immunostaining and the U1, U4, U5 and U6 snRNA hybridisation could be defined precisely and could be quantified by superposing the independently generated mask upon the images. The average intensity in these Cajal body regions was compared with the average intensity in the remaining nucleoplasmic region of the same nucleus. The quantification was performed with the help of the Khoros software (Khoral Inc., NM).
Acknowledgments
We are grateful to M Carmo-Fonseca for kindly providing anti-coilin antibodies and CL Will for critically reading the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 526/A8), BMBF (031U215B), Ernst Jung Stiftung and Fonds der Chemischen Industrie to RL. NS was supported by a scholarship from the Boehringer Ingelheim Fonds.
References
- Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R (1998) The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol Cell Biol 18: 6756–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M (1998) Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J Cell Biol 142: 899–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A (2002) p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 21: 2724–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA (2002) Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, Gesteland RF, Cech TR, Atkins JF (eds), pp 525–560. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI (1992) Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 117: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 21: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213 [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI (1994) Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol 126: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisson F, Legrain P (1993) The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res 21: 1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG (2003) The centennial of the Cajal body. Nat Rev Mol Cell Biol 4: 975–980 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Santos JM, Wang A, Jones J, Ushida C, Liu J, Hu J (2002) Central region of the human splicing factor Hprp3p interacts with Hprp4p. J Biol Chem 277: 23764–23772 [DOI] [PubMed] [Google Scholar]
- Gorczyca W, Bigman K, Mittelman A, Ahmed T, Gong J, Melamed MR, Darzynkiewicz Z (1993) Induction of DNA strand breaks associated with apoptosis during treatment of leukemias. Leukemia 7: 659–670 [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T (2002) The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T (2003) Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 22: 1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ (2003) Pre-mRNA splicing: a wash in a sea of proteins. Mol Cell 12: 5–14 [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jády BE, Darzacq X, Verheggen C, Bertrand E, Kiss T (2002) A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res 30: 4643–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Lauber J, Plessel G, Prehn S, Will CL, Fabrizio P, Groning K, Lane WS, Lührmann R (1997) The human U4/U6 snRNP contains 60 and 90 kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA 3: 926–941 [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Green MR (1990) Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol 181: 20–30 [DOI] [PubMed] [Google Scholar]
- Legrain P, Chapon C, Schwob E, Martin R, Rosbash M, Dujon B (1991) Cloning of the two essential yeast genes, PRP6 and PRP9, and their rapid mapping, disruption and partial sequencing using a linker insertion strategy. Mol Gen Genet 225: 199–202 [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA (1981) Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA 78: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Achsel T, Lührmann R (2000) The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein–protein interactions. J Mol Biol 298: 567–575 [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Liu S, Vornlocher HP, Lührmann R (2002) Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J 21: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Lührmann R (2001) The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J 20: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol 9: 302–309 [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature 387: 523–527 [DOI] [PubMed] [Google Scholar]
- Nilsen TW (2003) The spliceosome: the most complex macromolecular machine in the cell? BioEssays 25: 1147–1149 [DOI] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R (1999) Functional interaction of a novel 15.5 kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J 18: 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Urlaub H, Lührmann R (2002) Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J 21: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Lamond AI (2002) Cajal bodies and coilin-moving towards function. J Cell Biol 159: 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL (1994) Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol 124: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny CW, Bindereif A, Green MR (1989) In vitro reconstitution of snRNPs: a reconstituted U4/U6 snRNP participates in splicing complex formation. Genes Dev 3: 479–487 [DOI] [PubMed] [Google Scholar]
- Rader SD, Guthrie C (2002) A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8: 1378–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C (1998) A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279: 857–860 [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Ajuh P, Lamond AI (2001) snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 114: 4407–4419 [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI (1999) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol 9: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Spector DL (2001) Nuclear domains. J Cell Sci 114: 2891–2893 [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T (1991) Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J 10: 3467–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Staněk D, Rader SD, Klingauf M, Neugebauer KM (2003) Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 160: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja KL, Lifshitz LM, Fay FS, Singer RH (1992) Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J Cell Biol 119: 1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, Bird AC, Hunt DM, Bhattacharya SS (2001) A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 8: 375–381 [DOI] [PubMed] [Google Scholar]
- Weidenhammer EM, Singh M, Ruiz-Noriega M, Woolford JL Jr (1996) The PRP31 gene encodes a novel protein required for pre-mRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res 24: 1164–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301 [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC (2001) A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]