GATA4 is essential for formation of the proepicardium and regulates cardiogenesis (original) (raw)
Abstract
The role of GATA4 during the earliest stages of cardiogenesis has not been defined because Gata4 knockout embryos suffer an early developmental arrest caused by deficiencies in extraembryonic visceral endoderm function. We have used tetraploid embryo complementation to rescue these defects and generated clonal embryonic day 9.5 _Gata4_–/– embryos directly from embryonic stem cells. GATA4-null embryos display heart defects characterized by disrupted looping morphogenesis, septation, and a hypoplastic ventricular myocardium. We find that myocardial gene expression is relatively normal in GATA4-null hearts including expression of GATA6. Moreover, GATA4 expression in the endocardium is dispensable for trabeculae formation. Remarkably, the proepicardium is absent in GATA4-null embryos, blocking formation of the epicardium. Therefore, we propose that the observed myocardial defects may be a secondary consequence of loss of the proepicardium. These findings definitively demonstrate a requirement for GATA4 during early cardiac development and identify an essential factor for generation of the proepicardium.
Keywords: heart development, septum transversum mesenchyme, proepicardial organ
In the mouse, the cardiac lineage is specified from cells of the primitive streak at approximately embryonic day (E) 7.0. The nascent cardiac mesoderm migrates anterolaterally, where it fuses at the ventral aspect of the embryo to form a linear heart tube by E8.5. The linear tube subsequently undergoes complex looping and septation to form the characteristic four-chambered heart. Considerable effort has been made in defining the molecular networks controlling mammalian heart development, in part because this knowledge is applicable to human disease states such as congenital heart disease and cardiac hypertrophy. Central to these processes, cardiac-selective transcription factors such as MEF2c, NKX2–5, TBX5, and HAND1 have been shown to activate cardiac gene expression and are essential for the proper patterning of the developing heart (1, 2).
The zinc finger transcription factor GATA4 represents another such factor. It is expressed in the lateral plate mesoderm at the earliest stages of cardiac differentiation and subsequently maintained in the cardiac lineage throughout development (3). GATA4 regulates cardiac specific gene expression in vitro (4–6) and modulates cardiogenesis in Xenopus and in embryonic stem (ES) cell embryoid bodies (7, 8). In mice, an engineered point mutation in GATA4, predicted to disrupt the interaction with the cofactor friend-of-GATA 2 (FOG2), results in septation and coronary vasculature defects evident by E12.5 (9), a phenotype similar to two pedigrees of human congenital heart defects linked to mutations in GATA4 (10).
Despite these studies, the targeted disruption of GATA4 has failed to resolve a function for this factor at the earliest stages of heart development. GATA4-null embryos arrest at approximately E8.0 because of defects in ventral morphogenesis, including a failure of the cardiac mesoderm to form a linear heart tube (11, 12). However, these defects could be rescued by providing GATA4-null embryos with wild-type visceral endoderm, demonstrating that this phenotype is caused by a defect in extraembryonic tissue and not the heart, per se (13). Defects in the development of extraembryonic endoderm can be circumvented by producing embryos from ES cells by using tetraploid embryo complementation (14). Fetuses generated by this procedure are derived solely from ES cells, whereas the extraembryonic endoderm is tetraploid embryo-derived (15). We have used this procedure to generate _Gata4_–/– embryos. These embryos display defects in cardiogenesis and proepicardium development, demonstrating an essential function for GATA4 in the patterning of the ventral mesoderm.
Materials and Methods
Generation of Plasmids, Gata4–**/**– ES Cells, and Embryos. Targeting and Cre excision of the Gata4 locus was performed as described (16). A targeting vector pGATA4loxPDT was constructed to contain a _neo_-tk cassette flanked by two _lox_P sites from plasmid pHR-1 that were inserted into the first _Sma_I site upstream of Gata4 exon 3. A single _lox_P site was introduced into the _Bam_HI site between exons 5 and 6. Negative selection was provided by the diptheria toxin gene. Methodologies for the generation of ES cell-derived embryos by tetraploid embryo complementation have been described elsewhere (14). Pregnant mare serum gonadotropin (PMSG) used in superovulation was obtained from A. F. Parlow at the National Hormone and Peptide Program (Torrance, CA). Gata4loxP/+;Tie2Cre mice were generated by breeding Gata4loxP/+ mice (Gata4tm1Sad) with Tie2-Cre mice (17).
Histochemistry, Immunohistochemistry, and Electron Microscopy (EM). Immunohistochemistry was performed by using antigen retrieval as described elsewhere (16) by using antibodies against GATA4 (Santa Cruz Biotechnology), sarcomeric myosin (MF20, Developmental Studies Hybridoma Bank, Iowa City, IA), Wilms' tumor 1 (WT1) (Cell Marque, Hot Springs, AR), and platelet endothelial cell adhesion molecule (CD31, BD Pharmingen). For EM, tissues were fixed in 2% gluteraldehyde in cacodylate buffer and embedded in epoxy resin. Semithin (2-μm) sections were stained with toluidine blue, and 80- to 100-nm sections were contrasted with uranyl acetate and lead citrate for EM.
RT-PCR. RT–PCR was carried out as described (14). Primer sequences are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
We used a two-step _Cre-lox_P strategy to delete exons 3–5 of the Gata4 gene in ES cells (Fig. 1_A_). These exons contain both zinc finger DNA-binding domains and the nuclear localization signal that are essential for GATA4 function (12). Both genomic copies of Gata4 were targeted sequentially to produce _Gata4_–/– ES cells whose genotype was confirmed by Southern blot (Fig. 1_B_). ES cells harboring a single Gata4 “floxed” allele were also produced and used to generate a line of mice whose deletion of Gata4 depended on the presence of Cre recombinase. In agreement with previous studies (18), _Gata4_–/– ES cells were unable to differentiate into visceral endoderm in vitro confirming that they are functionally null for GATA4 (Fig. 6, which is published as supporting information on the PNAS web site). To address the role of GATA4 in cardiogenesis, we generated embryos from Gata4+/– and _Gata4_–/– ES cells by aggregating them with wild-type tetraploid embryos. As has been shown (13), _Gata4_–/– embryos complemented with wild-type visceral endoderm survive to E9.5, rescuing the developmental defects, including cardia bifida, associated with conventionally produced Gata4 knockout embryos (Fig. 1 C and D). Southern blot analyses demonstrated that these embryos were genetically identical to the _Gata4_–/– ES cells (Fig. 7, which is published as supporting information on the PNAS web site), and immunostaining experiments confirmed that they contained no functional GATA4 protein (Fig. 1 E and F). Control Gata4+/– ES cell-derived embryos were indistinguishable from wild-type embryos and were viable until at least E12. By comparison, _Gata4_–/– ES cell-derived embryos arrested between E9.5 and E10 (data not shown). It is worth noting that embryos harboring a mutation affecting the interaction of GATA4 with friend-of-GATA 2 (FOG2) survived until E12.5 (9). The earlier embryonic lethality of GATA4-null embryos, therefore, demonstrates a FOG2-independent function for GATA4 during development.
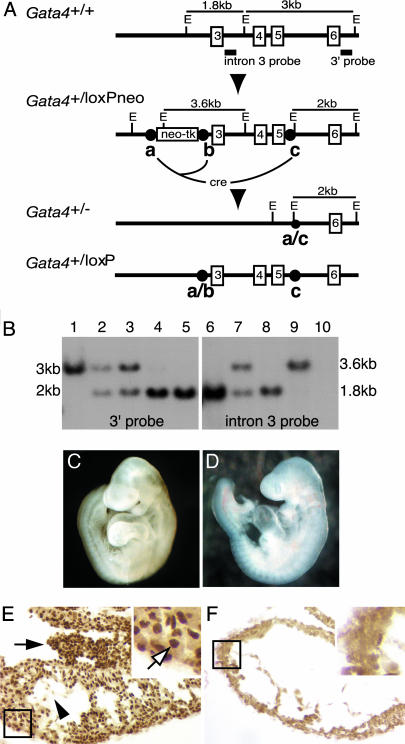
Fig. 1.
Generation of Gata4_–/– embryos by tetraploid embryo complementation. (A) Schematic of the targeting strategy and Eco_RI (E) restriction enzyme map of Gata4+/+, Gata4+/loxPneo, Gata4+/loxP, and Gata4+/– genomic loci. Gata4 exons 3–6 (numbered boxes) and Southern blot probes (solid bars) are shown. _Lox_P sites are shown as circles labeled a, b, and c. Cre-mediated recombination between sites a and b results in a “floxed” allele, and Cremediated recombination between sites a and c results in a null allele. (B) Genomic Southern blot using 3′ (lanes 1–5) and intron 3 (lanes 6–10) probes on _Eco_RI-digested DNA from Gata4+/+ (lanes 1 and 6), Gata4+/loxPneo (lanes 2 and 7), Gata4+/– (lanes 3 and 8), _Gata4_loxPneo/– (lanes 4 and 9), and _Gata4_–/– (lanes 5 and 10). (C and D) Micrographs of E9.5 embryos generated by tetraploid embryo complementation from Gata4+/– (C) and _Gata4_–/– (D) ES cells. (E and F) Immunohistochemistry using an α-GATA4 antibody on transverse sections of Gata4+/– (E) and _Gata4_–/– (F) embryos identifies nuclear GATA4 in the myocardium (boxed region, white arrow), proepicardium (black arrow), and endocardium (black arrowhead) only in control embryos.
To determine the cause of the developmental arrest, we compared somite-matched Gata4+/– and _Gata4_–/– embryos ranging from 18 to 22 somites. The mutant phenotype reported below was confirmed in embryos from two independent _Gata4_–/– ES cell lines. The most striking anatomical abnormality associated with _Gata4_–/– embryos was a failure of the heart tube to undergo correct looping morphogenesis, characterized by the atrial/inflow tract region failing to migrate rostrally (Fig. 2 A_–_D). In addition, the atrioventricular canal and bulboventricular groove, structures representing early events in chamber formation, are absent (Fig. 2 A_–_D). These defects are evident on sections of _Gata4_–/– embryos as a dilated heart tube that has failed to form distinct chambers (Fig. 2 E_–_H). Histology also reveals that the inflow tract region, which bifurcates at the level of the liver bud into the left and right sinus venosus in Gata4+/– embryos, is present as a single vessel in _Gata4_–/– embryos (Fig. 2 I and J). Interestingly, vitamin A-deficient (VAD) quail embryos display a similar fused inflow tract (19). However, unlike the VAD model, GATA4-null embryos do not appear to have a blind-ended heart tube (data not shown). The failure of GATA4-null hearts to initiate chamber formation is particularly provocative given the recent finding that septation defects are associated with GATA4 mutations in human congenital heart disease (10).
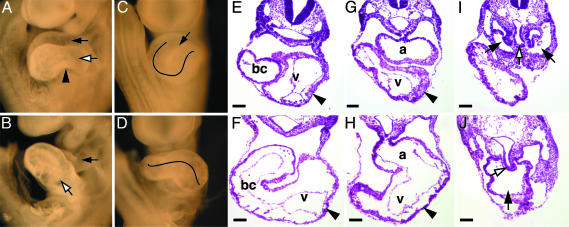
Fig. 2.
Heart defects in _Gata4_–/– embryos. Whole-mount E9.5 Gata4+/– (A and C) and _Gata4_–/– (B and D) embryos are shown. (A) Left lateral view of Gata4+/– heart shows the atrial chamber (white arrow) positioned immediately caudal to the outflow tract (black arrow) and the constriction of the atrioventricular canal (arrowhead). (B) Same view of _Gata4_–/– heart shows a caudal displacement of the presumptive atrial region (white arrow) relative to the outflow tract (black arrow). No constriction corresponding to the atrioventricular canal is detected. (C) Ventral view of Gata4+/– heart shows the bulboventricular groove (black arrowhead) and a characteristic U-shaped loop, whereas the bulboventricular groove is absent in _Gata4_–/– hearts (D). Hematoxylin and eosin-stained rostral to caudal transverse sections of Gata4+/– (E, G, and I) and _Gata4_–/– (F, H, and J) embryos. Myocardium (black arrowheads), liver bud (white arrows), and inflow tract (black arrows) are all indicated. bc, Bulbus cordis; v, left ventricle; a, common atrial chamber.
Close examination of sections through _Gata4_–/– hearts revealed a thinning of the myocardium in the presumptive ventricular region compared to control hearts (Fig. 2 E_–_H). In addition, there was no evidence that the endocardial cushions had formed in the null embryos, although they were readily detectable in the controls. Immunohistochemical staining for sarcomeric myosin was performed to define whether there was a defect in the generation of the myocardium in the absence of GATA4. In Gata4+/– hearts (Fig. 3_A_), the ventricular region is characterized by the presence of cardiomyocyte projections, called trabeculae, that extend into the lumen of the heart tube. In contrast, the degree of ventricular trabeculation in _Gata4_–/– hearts is drastically reduced (Fig. 3_B_). The cellular architecture of these regions was examined in toluidine blue-stained semithin sections (Fig. 3 C and D). The ventricular cardiomyocytes of Gata4+/– hearts form a well defined, tightly packed outer (future subepicardial) cell layer and a distinctive trabecular organization to the inner surface (Fig. 3_C_). The individual cardiomyocytes have a globular morphology with prominent nuclei. In _Gata4_–/– hearts, the ventricular cardiomyocytes have an elongated appearance with clear intercellular gaps. Consequently, there is less integrity to the outer surface of the myocardium (Fig. 3_D_). Based on the disrupted cytoarchitecture and reduced trabeculation of _Gata4_–/– hearts, we believe that a deficit in cardiac output is the most likely cause of lethality of _Gata4_–/– embryos. Defects in trabeculation are a feature of other mouse mutants, including Nkx2–5, Raldh2, and Mef2c, which result in developmental arrest between E9.5 and E10.5 (20–22). This may reflect the role of trabeculae in adding contractile power to the ventricles at this stage in development (23).
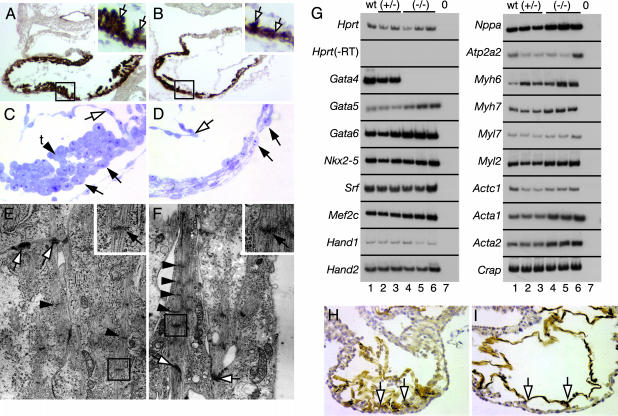
Fig. 3.
Myocardial defects in _Gata4_–/– embryos. (A and B) Immunohistochemistry for sarcomeric myosin (brown staining) on transverse sections of Gata4+/– (A) and _Gata4_–/– (B) embryos. Trabeculation (boxed region, white arrows) seen in Gata4+/– hearts is reduced in _Gata4_–/– hearts. (C and D) Micrographs of semithin toluidine blue-stained sections of the ventricular myocardium from Gata4+/– (C) and _Gata4_–/– (D) embryos. Tightly packed Gata4+/– cardiomyocytes (black arrows) form trabeculae (t) on the inner mural myocardial surface (arrowhead), and the myocardium closely associates with endocardium (white arrow). _Gata4_–/– cardiomyocytes have a squamous morphology with distinct spaces between cells (black arrows), no trabeculae are evident, and the endocardium (white arrow) does not appose the myocardium. (E and F) Electron microscopic analysis of Gata4+/– (E) and _Gata4_–/– (F) cardiomyocytes identifies myofibrils, associated Z lines (Insets, black arrows), which are more abundant in _Gata4_–/– cardiomyocytes, and intercalated disks (white arrows). (G) Expression analysis of _Gata4_–/– hearts. RT-PCR was performed on RNA isolated from individual heart tubes of E9.5 somite-matched wild-type (lane 1), Gata4+/– (lanes 2 and 3), and _Gata4_–/– (lanes 4–6) embryos by using primers against cardiac transcripts. Lane 7 is a no-cDNA control. Hprt controlled for loading and Hprt in the absence of reverse transcriptase (-RT) confirmed absence of genomic DNA. (H and I) Platelet endothelial cell adhesion molecule staining (brown) of transverse sections through Gata4+/– (H) and _Gata4_–/– (I) embryos showing Gata4+/– ventricles with an intricate endocardial network (white arrows) in contrast to that in _Gata4_–/– embryos, which remains underdeveloped (white arrows).
The flattened morphology of GATA4-null cardiomyocytes is suggestive of a more differentiated phenotype. We therefore examined the extent of differentiation in the future subepicardial cardiomyocytes by using electron microscopy. We were able to detect bundles of myofibrils and associated Z lines, structures indicative of sarcomere assembly in control and _Gata4_–/– cardiomyocytes, indicating that differentiation had occurred in both (Fig. 3 E and F). However, these structures were more predominant in _Gata4_–/– cardiomyocytes, suggesting that differentiation may be more advanced in these cells. Intercalated disks, intercellular junctions connecting adjacent cardiomyocytes, could also be identified in both control and mutant embryos (Fig. 3 E and F).
Although GATA4 has been shown to regulate cardiac gene expression, our finding that cardiomyocyte differentiation may be enhanced in _Gata4_–/– hearts seemed to contradict this. Therefore, we examined the consequence of loss of GATA4 on myocardial gene expression. The steady-state levels of 17 characteristic cardiomyocyte mRNAs were compared between wild-type and _Gata4_–/– hearts by RT-PCR. Fig. 3_G_ shows that no overt disruption of gene expression was associated with loss of GATA4, even though several of these genes have been shown to contain GATA4-responsive regulatory elements (4, 24). In addition, expression of Raldh2 (Aldh1a2R) and Vcam, mutations in which defects in trabeculae formation occur at later developmental stages, was normal, as was expression of the GATA-binding proteins Fog1 (Zfpm1) and Fog2 (Zfpm2) (Fig. 8, which is published as supporting information on the PNAS web site). Interestingly, mRNA levels of some genes appeared to be slightly, but reproducibly, increased in _Gata4_–/– hearts, including Gata5, Gata6, Nppa, Myh6, Myl2, Acta1, and Acta2 (Fig. 3_G_). The relatively benign affect that loss of GATA4 function has on cardiac gene expression may be a consequence of redundancy between GATA factors. GATA6 has been implicated in rescuing GATA4 function in other gene disruption studies (11, 12, 25), raising the possibility that the observed increase in Gata5/6 expression (Fig. 3_G_) may be sufficient to compensate for loss of GATA4 function.
The absence of changes in GATA-target gene expression in _Gata4_–/– embryos suggests that the observed disruption to myocardial development is non-cell autonomous. Indeed, chimera analysis has shown that GATA4-null cardiomyocytes could contribute to a functional myocardium up to at least E18 (25). In addition to the cardiomyocytes, GATA4 is expressed in two tissues that influence myocardial differentiation, the endocardium and proepicardium (Figs. 1_E_ and 4_C_). Therefore, we examined the development of these tissues to determine whether they may be contributing to the observed cardiogenic defects.
Fig. 4.
Expression of GATA4 in the endocardium is dispensable for trabecular formation at E10.5. Embryos were generated that lack GATA4 specifically in the endocardium by breeding Gata4loxP/loxP mice to Gata4 loxP/+;Tie2Cre mice. Immunostaining using an anti-GATA4 antibody identified GATA4 in the nuclei of both the myocardium (white arrow) and endocardium (black arrow) in control Gata4 loxP/+;Tie2Cre embryos (A and C) at E10.5. In contrast, GATA4 is detected only in the myocardium and is absent from the endocardium of Gata4loxP/loxP;Tie2Cre embryos (B and D). Boxes in A and B represent high-magnification images shown in C and D. (E and F) Hematoxylin and eosin-stained nearby sections identify trabeculae (line) regardless of the presence (E) or absence (F) of GATA4 in the endocardium.
The endocardium develops from the precardiac mesoderm in parallel with the myocardium, forming an endothelial cell layer which lines the inside of the primitive heart tube (26). Later in development, the endocardium, through a process dependent on interaction with the myocardium, contributes to the formation of the valvuloseptal tissues via intermediate cardiac cushion tissue (27, 28). In Fig. 3 H and I, immunohistochemical staining for platelet endothelial cell adhesion molecule clearly identifies endocardial formation in the ventricular region of Gata4+/– and _Gata4_–/– hearts. As trabeculae form, the endocardium infiltrates the myocardium to line these structures (20). This network of endothelial cells can be seen in the ventricular region of transverse sections of Gata4+/– hearts (Figs. 3_H_ and 4_C_). However, in the _Gata4_–/– hearts, the structure of the endocardial network is less elaborate, with clear gaps observed between the myocardium and endocardium (Fig. 3_I_). The cellular origin of this defect is unclear. Deletion of Nkx2–5, which is expressed only in the myocardium, results in a similar phenotype (20). Consequently, this defect was attributed to the attenuated development of the myocardium rather than a specific defect in endocardial development because reduced contact between the myocardium and endocardium is a feature of a more primitive pretrabecular ventricle (29). In support of this interpretation for _Gata4_–/– hearts, ultrastructural analysis found no differences between cells of control and mutant endocardium (data not shown). Disruption of Nrg1, which is expressed in the endocardium, or its receptors ErbB2 or ErbB4 results in a disruption to development of trabeculae (30–32). To test directly whether GATA4 expression in the endocardium was required for trabeculae formation, we produced embryos in which Gata4 was disrupted specifically in the endocardium by generating Gata4loxP/loxP;Tie2Cre mice. At E10.5, all embryos, regardless of the presence or absence of GATA4 in the endocardium, formed trabeculae (Fig. 4). Moreover, RT-PCR analyses of _Gata4_–/– ES cell-derived embryos identified no difference from controls in the steady-state mRNA levels encoding Nrg1, ErbB2, ErbB3, or ErbB4, or in expression of Tie2, VegfA, VegfB, VegfRI, VegfRII, and Flt1 (Fig. 8).
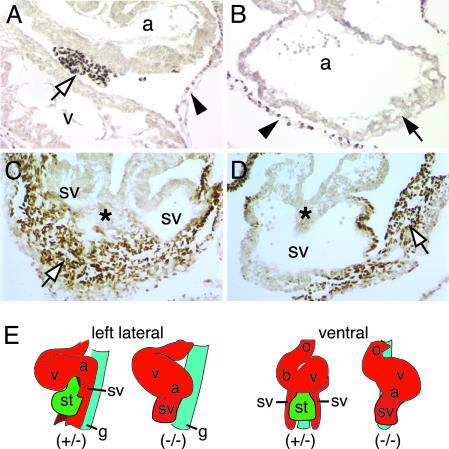
GATA4 is also robustly expressed in the proepicardium (Fig. 1_E_), which is an outgrowth of the anterior surface of the septum transversum mesenchyme (STM) that migrates over the surface of the heart to form the epicardium. Cells in this epicardial layer are subsequently capable of migrating into the myocardium, where they form cardiac fibroblasts and the smooth muscle and endothelium of the coronary vessels (33). Studies in chickens have clearly described the morphogenetic movements made by cells of the proepicardium and have identified the cell lineages to which the proepicardium gives rise (33). However, the molecular mechanisms that control proepicardium development are virtually unknown, which is surprising given that cells derived from the proepicardium have been shown to directly impact myocardial differentiation including valvuloseptal development (34–36). The tumor suppressor WT1 protein is one of the few characterized markers of the proepicardium, so we used it to trace development of this tissue in GATA4-null embryos (37). As shown in Fig. 5_A_, the proepicardium can be identified as a cluster of WT1-positive cells migrating between the atrial and ventricular regions of the myocardium. In contrast to control embryos, sections through the entire heart tube of _Gata4_–/– embryos showed no indication of any WT1-positive cells in contact with the myocardium (Fig. 5_B_). Caudal to the heart in Gata4+/– embryos, WT1-positive STM, the source of the proepicardium, is located ventrally between the bifurcated arms of the sinus venosus (Fig. 5_C_). At an equivalent level in _Gata4_–/– embryos, the sinus venosus exists as a single vessel with no WT1-positive mesenchyme present ventral to this structure. There does exist a small outgrowth of WT1-positive mesenchyme continuous with the peritoneum in a more lateral position, which may represent a remnant of the STM (Fig. 5_D_). In an effort to determine which were primary versus secondary effects, we compared the heart tubes of Gata4+/– and _Gata4_–/– embryos containing 10–11 somites (E8.5). Although not as pronounced as a day later, looping was slightly perturbed in GATA4-null embryos, consistent with a caudal displacement of the atrial region (data not shown). Otherwise, the myocardium was indistinguishable from Gata4+/– control embryos (Fig. 9, which is published as supporting information on the PNAS web site). At this same stage in development, the STM can be identified in control embryos, but, as is the case at later developmental stages, it is undetectable in _Gata4_–/– embryos (Fig. 9). These data identify GATA4 as a transcription factor that is essential for the genesis of the proepicardium and for correct STM development. It is noteworthy that, as in cardiomyocytes, GATA6 is also expressed in the STM and proepicardium (data not shown). However, in contrast to cardiomyocytes, the complete absence of the proepicardium in _Gata4_–/– embryos demonstrates that the role of GATA4 in the formation of this tissue is indispensable and cannot be compensated by GATA6. These data are supported by defects in the proepicardium derived coronary vasculature of E12.5 embryos with a point mutation in GATA4 (9).
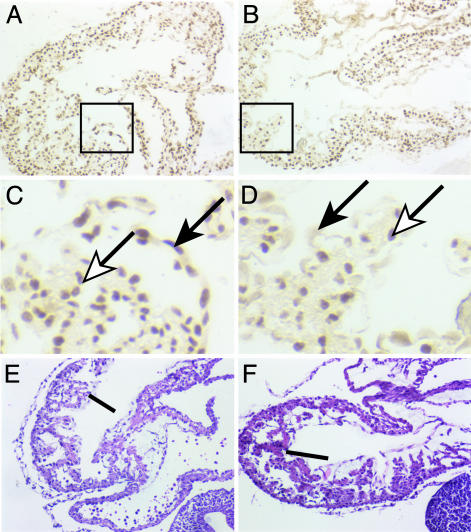
Fig. 5.
_Gata4_–/– embryos lack a proepicardium. (A_–_D) Micrographs show WT1 staining of Gata4+/– (A and C) and _Gata4_–/– (B and D) embryos. WT1 staining identifies the proepicardium as an outgrowth of the STM between atrial (a) and ventricular (v) regions in Gata4+/– embryos (A; white arrow) that is absent in _Gata4_–/– embryos (B; black arrow). The parietal pericardium stains positively for WT1 in both Gata4+/– and _Gata4_–/– embryos (A and B; black arrowhead). At the level of the liver bud (C and D; asterisk), WT1-positive cells are present ventral to the sinus venosus (sv) in Gata4+/– embryos (C; white arrow). In contrast, mutant embryos only have WT1-positive cells in an irregular outgrowth of the peritoneum lateral to the single lumen of the sinus venosus (D; white arrow). (E) Schematic representation of GATA4 mutant phenotype. In Gata4+/– embryos the STM (st) is positioned immediately caudal to the heart. The absence of STM in _Gata4_–/– embryos results in a displaced atrioventricular region and a single sinus venosus. a, Atrial region; b, bulbus cordis; g, gut endoderm; o, outflow tract; st, STM/proepicardium; sv, sinus venosus; v, ventricular region.
Although we cannot, at this time, rule out a cell-autonomous requirement for GATA4 in the myocardium, we propose that the absence of the STM and the proepicardium is a primary cause of the cardiac phenotype associated with loss of GATA4. This proposal seems particularly plausible given the spatial association between the heart, STM, and proepicardium during development and the specific myocardial defects observed in _Gata4_–/– embryos. The STM and, by association, proepicardium are derived from splanchnic mesoderm, and development of these tissues is closely associated with the cardiac mesoderm (38). By E9.5, the STM and proepicardium are positioned immediately caudal to the atrioventricular region of the heart, bisecting the sinus venosus (Fig. 5_E_). The absence of the STM/proepicardium may, therefore, disrupt the development of the atrioventricular region and the bifurcation of the sinus venosus, as is observed in _Gata4_–/– embryos (Fig. 5_E_). Our data support a role for the STM/proepicardium in myocardial development from E8.5. Although no data exist for the role of the STM/proepicardium in myocardial development at such an early developmental stage, several studies have identified a role for the epicardium in myocardial development at later stages. In quail embryos, ablation of the proepicardium at the Hamburger/Hamilton (HH) 15 stage (E9.5 in mouse) results in myocardial defects, including a thin compact zone and aberrant septation, which are manifest at HH29 (E13.5 in mouse) (39). Similar myocardial defects are observed when migration of the proepicardium to the myocardium is impaired in gene knockout studies that block the interaction between vascular endothelial adhesion molecule 1 and α4-integrin (40–42). Recent analyses have also begun to address the molecular basis of epicardial function in myocardial development. Retinoic acid (RA) and erythropoietin (Epo), produced by the epicardium, are thought to act as autocrine signals mediating the secretion of trophic factors that influence myocardial development (34–36). Interestingly, reduced trabeculation, enhanced cardiomyocyte differentiation, and defective outf low tract separation, as seen in _Gata4_–/– embryos, are features of RA deficiency and inactivation of genes in the RA signaling pathway (43). Although none of the above studies explicitly address the development of the endocardium, embryos null for the RA metabolizing enzyme RALDH2 appear to have a less developed endocardium, similar to _Gata4_–/– embr yos (22). The STM/proepicardium also expresses a number of growth factors, including bone morphogenetic proteins, implicated in heart development (26, 44), and is essential for the outgrowth of the hepatic primordia, highlighting the role of this tissue as an inductive mesenchyme (45).
In summary, we have uncovered a role for GATA4 in the generation of the STM and the related proepicardium, thereby identifying GATA4 as the only known factor to be essential for the genesis of this inductive tissue. We postulate that, because of redundancy between GATA factors, the additional cardiogenic defects are likely caused by the absence of the proepicardium rather than a cell-autonomous defect in the myocardium. This model may also suggest a nonmyocardial component to the human congenital heart defects associated with mutations in GATA4.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. J. Lough, R. Misra, and T. Nelson for advice, and Jim Bain and Clive Wells for electron microscopy. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK55743 and DK60064 and American Heart Association Postdoctoral Fellowship 0120668Z (to A.J.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: E_n_, embryonic day n; ES, embryonic stem; WT1, Wilms' tumor 1; STM, septum transversum mesenchyme.
References
- 1.Cripps, R. M. & Olson, E. N. (2002) Dev. Biol. 246**,** 14–28. [DOI] [PubMed] [Google Scholar]
- 2.Bruneau, B. G. (2002) Circ. Res. 90**,** 509–519. [DOI] [PubMed] [Google Scholar]
- 3.Parmacek, M. S. & Leiden, J. M. (1999) in Heart Development, eds. Harvey, R. P. & Rosenthal, N. (Academic, San Diego), pp. 291–306.
- 4.Molkentin, J. D. (2000) J. Biol. Chem. 275**,** 38949–38952. [DOI] [PubMed] [Google Scholar]
- 5.Stennard, F. A., Costa, M. W., Elliott, D. A., Rankin, S., Haast, S. J., Lai, D., McDonald, L. P., Niederreither, K., Dolle, P., Bruneau, B. G., et al. (2003) Dev. Biol. 262**,** 206–224. [DOI] [PubMed] [Google Scholar]
- 6.Sepulveda, J. L., Vlahopoulos, S., Iyer, D., Belaguli, N. & Schwartz, R. J. (2002) J. Biol. Chem. 277**,** 25775–25782. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, Y. & Evans, T. (1996) Dev. Biol. 174**,** 258–270. [DOI] [PubMed] [Google Scholar]
- 8.Grepin, C., Nemer, G. & Nemer, M. (1997) Development (Cambridge, U.K.) 124**,** 2387–2395. [DOI] [PubMed] [Google Scholar]
- 9.Crispino, J. D., Lodish, M. B., Thurberg, B. L., Litovsky, S. H., Collins, T., Molkentin, J. D. & Orkin, S. H. (2001) Genes Dev. 15**,** 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garg, V., Kathiriya, I. S., Barnes, R., Schluterman, M. K., King, I. N., Butler, C. A., Rothrock, C. R., Eapen, R. S., Hirayama-Yamada, K., Joo, K., et al. (2003) Nature 424**,** 443–447. [DOI] [PubMed] [Google Scholar]
- 11.Kuo, C. T., Morrisey, E. E., Anandappa, R., Sigrist, K., Lu, M. M., Parmacek, M. S., Soudais, C. & Leiden, J. M. (1997) Genes Dev. 11**,** 1048–1060. [DOI] [PubMed] [Google Scholar]
- 12.Molkentin, J. D., Lin, Q., Duncan, S. A. & Olson, E. N. (1997) Genes Dev. 11**,** 1061–1072. [DOI] [PubMed] [Google Scholar]
- 13.Narita, N., Bielinska, M. & Wilson, D. B. (1997) Dev. Biol. 189**,** 270–274. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, S. A., Nagy, A. & Chan, W. (1997) Development (Cambridge, U.K.) 124**,** 279–287. [DOI] [PubMed] [Google Scholar]
- 15.Nagy, A. & Rossant, J. (1993) in Gene Targeting: A Practical Approach, ed. Joyner, A. (Oxford Univ. Press, Oxford), pp. 147–179.
- 16.Parviz, F., Matullo, C., Garrison, W. D., Savatski, L., Adamson, J. W., Ning, G., Kaestner, K. H., Rossi, J. M., Zaret, K. S. & Duncan, S. A. (2003) Nat. Genet. 34**,** 292–296. [DOI] [PubMed] [Google Scholar]
- 17.Kisanuki, Y. Y., Hammer, R. E., Miyazaki, J., Williams, S. C., Richardson, J. A. & Yanagisawa, M. (2001) Dev. Biol. 230**,** 230–242. [DOI] [PubMed] [Google Scholar]
- 18.Soudais, C., Bielinska, M., Heikinheimo, M., MacArthur, C. A., Narita, N., Saffitz, J. E., Simon, M. C., Leiden, J. M. & Wilson, D. B. (1995) Development (Cambridge, U.K.) 121**,** 3877–3888. [DOI] [PubMed] [Google Scholar]
- 19.Romeih, M., Cui, J., Michaille, J. J., Jiang, W. & Zile, M. H. (2003) Dev. Dyn. 228**,** 697–708. [DOI] [PubMed] [Google Scholar]
- 20.Lyons, I., Parsons, L. M., Hartley, L., Li, R., Andrews, J. E., Robb, L. & Harvey, R. P. (1995) Genes Dev. 9**,** 1654–1666. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Q., Schwarz, J., Bucana, C. & Olson, E. N. (1997) Science 276**,** 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niederreither, K., Vermot, J., Messaddeq, N., Schuhbaur, B., Chambon, P. & Dolle, P. (2001) Development (Cambridge, U.K.) 128**,** 1019–1031. [DOI] [PubMed] [Google Scholar]
- 23.Sedmera, D. & Thomas, P. S. (1996) BioEssays 18**,** 607. [DOI] [PubMed] [Google Scholar]
- 24.McFadden, D. G., Charite, J., Richardson, J. A., Srivastava, D., Firulli, A. B. & Olson, E. N. (2000) Development (Cambridge, U.K.) 127**,** 5331–5341. [DOI] [PubMed] [Google Scholar]
- 25.Narita, N., Bielinska, M. & Wilson, D. B. (1997) Development (Cambridge, U.K.) 124**,** 3755–3764. [DOI] [PubMed] [Google Scholar]
- 26.Lough, J. & Sugi, Y. (2000) Dev. Dyn. 217**,** 327–342. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg, L. M. & Markwald, R. R. (1995) Circ. Res. 77**,** 1–6. [DOI] [PubMed] [Google Scholar]
- 28.Mjaatvedt, C. H., Yamamura, H., Wessels, A., Ramsdell, A., Turner, D. & Markwald, R. R. (1999) in Heart Development, eds. Harvey, R. P. & Rosenthal, N. (Academic, San Diego), pp. 159–177.
- 29.Viragh, S. & Challice, C. E. (1973) J. Ultrastruct. Res. 42**,** 1–24. [DOI] [PubMed] [Google Scholar]
- 30.Gassmann, M., Casagranda, F., Orioli, D., Simon, H., Lai, C., Klein, R. & Lemke, G. (1995) Nature 378**,** 390–394. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, D. & Birchmeier, C. (1995) Nature 378**,** 386–390. [DOI] [PubMed] [Google Scholar]
- 32.Lee, K. F., Simon, H., Chen, H., Bates, B., Hung, M. C. & Hauser, C. (1995) Nature 378**,** 394–398. [DOI] [PubMed] [Google Scholar]
- 33.Mikawa, T. (1999) in Heart Development, eds. Harvey, R. P. & Rosenthal, N. (Academic, San Diego), pp. 19–33.
- 34.Perez-Pomares, J. M., Phelps, A., Sedmerova, M., Carmona, R., Gonzalez-Iriarte, M., Munoz-Chapuli, R. & Wessels, A. (2002) Dev. Biol. 247**,** 307–326. [DOI] [PubMed] [Google Scholar]
- 35.Chen, T. H., Chang, T. C., Kang, J. O., Choudhary, B., Makita, T., Tran, C. M., Burch, J. B., Eid, H. & Sucov, H. M. (2002) Dev. Biol. 250**,** 198–207. [DOI] [PubMed] [Google Scholar]
- 36.Stuckmann, I., Evans, S. & Lassar, A. B. (2003) Dev. Biol. 255**,** 334–349. [DOI] [PubMed] [Google Scholar]
- 37.Moore, A. W., Schedl, A., McInnes, L., Doyle, M., Hecksher-Sorensen, J. & Hastie, N. D. (1998) Mech. Dev. 79**,** 169–184. [DOI] [PubMed] [Google Scholar]
- 38.Dunwoodie, S. L., Rodriguez, T. A. & Beddington, R. S. (1998) Mech. Dev. 72**,** 27–40. [DOI] [PubMed] [Google Scholar]
- 39.Gittenberger-de Groot, A. C., Vrancken Peeters, M. P., Bergwerff, M., Mentink, M. M. & Poelmann, R. E. (2000) Circ. Res. 87**,** 969–971. [DOI] [PubMed] [Google Scholar]
- 40.Kwee, L., Baldwin, H. S., Shen, H. M., Stewart, C. L., Buck, C., Buck, C. A. & Labow, M. A. (1995) Development (Cambridge, U.K.) 121**,** 489–503. [DOI] [PubMed] [Google Scholar]
- 41.Sengbusch, J. K., He, W., Pinco, K. A. & Yang, J. T. (2002) J. Cell Biol. 157**,** 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, J. T., Rayburn, H. & Hynes, R. O. (1995) Development (Cambridge, U.K.) 121**,** 549–560. [DOI] [PubMed] [Google Scholar]
- 43.Kubalak, S. W. & Sucov, H. M. (1999) in Heart Development, eds. Harvey, R. P. & Rosenthal, N. (Academic, San Diego), pp. 209–216.
- 44.Winnier, G., Blessing, M., Labosky, P. A. & Hogan, B. L. (1995) Genes Dev. 9**,** 2105–2116. [DOI] [PubMed] [Google Scholar]
- 45.Rossi, J. M., Dunn, R. N., Hogan, B. L. M. & Zaret, K. S. (2001) Genes Dev. 15**,** 1998–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information