Evidence for Gradients of Human Genetic Diversity Within and Among Continents (original) (raw)
Abstract
Genetic variation in humans is sometimes described as being discontinuous among continents or among groups of individuals, and by some this has been interpreted as genetic support for “races.” A recent study in which >350 microsatellites were studied in a global sample of humans showed that they could be grouped according to their continental origin, and this was widely interpreted as evidence for a discrete distribution of human genetic diversity. Here, we investigate how study design can influence such conclusions. Our results show that when individuals are sampled homogeneously from around the globe, the pattern seen is one of gradients of allele frequencies that extend over the entire world, rather than discrete clusters. Therefore, there is no reason to assume that major genetic discontinuities exist between different continents or “races.”
Early studies on human diversity showed that most genetic diversity was found between individuals rather than between populations or continents (e.g., Boyd 1950; Lewontin 1972) and that variation in human diversity is best described by geographic clines (Livingstone 1962; Cavalli-Sforza et al. 1994). In spite of this, many recent studies using DNA polymorphisms have suggested that human genetic diversity is organized in continental clades (Cavalli-Sforza et al. 1988; Bowcock and Cavalli-Sforza 1991; Bowcock et al. 1994; Jorde et al. 1995; Nei and Takezaki 1996; Tishkoff et al. 1996; Mountain and Cavalli-Sforza 1997; Perez-Lezaun et al. 1997; Calafell et al. 1998; Stephens et al. 2001; Bamshad et al. 2003). An understanding of how genetic diversity is structured in the human species is not only of anthropological and political importance, but also of medical relevance. For example, if major differences in allele frequencies exist between populations, individuals from different origins may often be expected to respond differently to medical treatments (Wilson et al. 2001). In agreement with this, it was recently suggested that “race” represents a useful proxy for genetic susceptibility in clinical practice and medical interventions (Risch et al. 2002). Furthermore, an understanding of population structure is of crucial importance for efforts to identify disease genes by association with marker loci (Lander and Schork 1994; Cardon and Bell 2001). However, it is not obvious how best to approach the question of how human genetic diversity is structured.
One issue of central importance is how samples are collected. For historical and practical reasons, the approach most commonly used is to collect individuals from “populations” (such as “Norwegians,” “Yorubas,” “Ashkenazi Jews”) defined by cultural traits such as a shared language, shared religion, or shared myths of origins. Under such sampling schemes, populations considered to be “admixed,” as well as individuals of “mixed ancestry,” are often excluded from sampling (Bowcock and Cavalli-Sforza 1991). One problem with this approach may be that the cultural traits used to define “populations” are at the most a few thousand years old and often substantially younger. In addition, it is often not known if these populations represent any relevant reproductive units in humans even a few generations back in time. An alternative approach is to sample humans according to geography without regard to cultural traits. Although this approach has been advocated (e.g., King and Motulsky 2002; Kittles and Weiss 2003), it has not been widely implemented because of logistical difficulties. However, studies of genetic diversity from restricted geographical areas, where large numbers of individuals are sampled and a reasonable geographic coverage of the variation is achieved, generally reveal spatial gradients of allele frequencies (Barbujani et al. 1995; Krings et al. 1999; Ding et al. 2000; Rosser et al. 2000; Karafet et al. 2001) that are only occasionally disrupted by local discontinuities corresponding to linguistic or geographical barriers (Barbujani and Sokal 1990; Sokal et al. 1990). This suggests that isolation by distance (i.e., decreasing gene flow with increasing geographical distances) may be the most appropriate description of human genetic diversity (Cavalli-Sforza et al. 1994). In contrast, worldwide studies of human diversity based on “populations” generally find that individuals cluster discretely depending on their continents of origin (Cavalli-Sforza et al. 1988; Bowcock and Cavalli-Sforza 1991; Bowcock et al. 1994; Jorde et al. 1995; Nei and Takezaki 1996; Tishkoff et al. 1996; Mountain and Cavalli-Sforza 1997; Perez-Lezaun et al. 1997; Calafell et al. 1998; Stephens et al. 2001; Bamshad et al. 2003), and this is sometimes taken to mean that human genetic diversity is structured according to “race” (Risch et al. 2002; Burchard et al. 2003). The discrepancy in results between regional and global surveys of human genetic diversity could suggest that gradients in allele frequencies are restricted to smaller geographic regions, whereas the continents are distinguished by discontinuities in genetic diversity. Alternatively, they may result from differences in study design (suggested, e.g., by Kittles and Weiss 2003).
To understand if worldwide genetic diversity is best described as discrete units (an “island model”; Wright 1969) or by continuous variation in allele frequencies (“isolation by distance”; Wright 1969), we have explored the influence of study design on investigations of human diversity. We find that if sampling is based on individuals and geography rather than on “populations,” gradual variation and isolation by distance on a worldwide scale are better representations of global genetic diversity than are discontinuities among continents or “races.”
RESULTS
Influence of Sampling Strategy
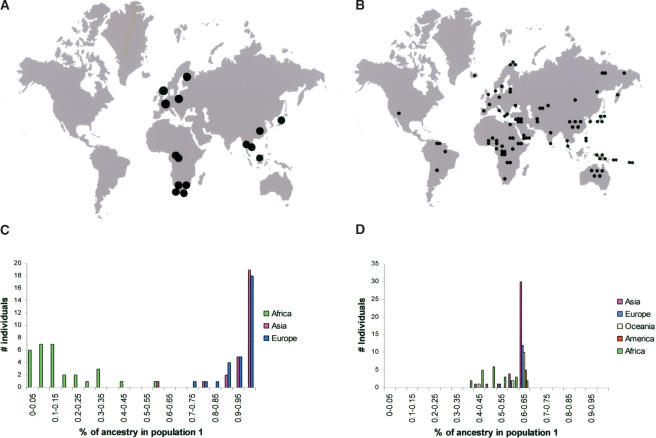
To determine the influence of the sampling strategy on the view of genetic diversity in humans, we first conducted a small study in which we compared two sampling schemes. The first data set (Fig. 1A) is published and based on populations. It consists of 89 individuals sampled from 15 populations (Jorde et al. 1995, 1997). In contrast, the second data set (Fig. 1B) is based on geography and consists of 90 individuals from 52 different populations, selected such that their geographic distribution around the world approximates the distribution of the human population as a whole and includes areas where Africa, Asia, and Europe meet.
Figure 1.
Geographic distribution of the samples used in the (A) population-based and (B) geography-based data sets. In A, each circle represents five to eight individuals and in B a single individual. The assignment of individuals into two inferred populations from the population-based (C) and the geography-based (D) samples. See text for details.
Genotype data from 20 unlinked autosomal microsatellites were collected for the individuals of the second data set and were used together with genotype data for the same microsatellite loci in the first data set to infer the extent to which substructure can be detected in the two data sets by the program Structure (Pritchard et al. 2000). This program takes genotype data from individuals without considering their origin and infers populations (called “inferred populations” below) to which the individuals are assigned in such a way that linkage disequilibrium as well as Hardy-Weinberg disequilibrium are minimized within each inferred population. Each analyzed individual can belong to one or several inferred populations according to its “coefficients of ancestry.” When two inferred populations are used to analyze the population-based data set (Fig. 1A), most individuals (83%) are estimated to belong to either one or the other of the two inferred populations with high coefficients of ancestry (100%-85%; Fig. 1C). Moreover, one of the inferred populations is made up of African individuals and the other of non-African individuals, suggesting a division in the human gene pool between Africans and non-Africans. In contrast, in the geography-based data set in which the sampling is based on individuals rather than populations, all individuals are estimated to be 40%-50% admixed between two inferred populations, and no qualitative difference between Africans and non-Africans can be detected (Fig. 1D).
Thus, the population-based sampling scheme results in a view of human diversity that suggests two discrete continental units of diversity. In contrast, when individuals that cover the geographic distribution of humans across the continents better (albeit still imperfectly) are used, no such subdivision is observed. Given the small number of markers and individuals analyzed, this result does not mean that no genetic subdivision exists among humans. However, it demonstrates that the difference seen between individuals from different continents when relatively few individuals and few markers are analyzed can be due to the discontinuous sampling scheme that results from basing the sampling on “populations,” which often come from the extremes of continental land masses (Fig. 1A).
A Robust Representation of the Human Genetic Diversity?
Recently, 1066 individuals from the CEPH human genome diversity cell line panel (referred to here as the CEPH diversity panel; Cann et al. 2002) were genotyped for 377 autosomal microsatellites (http://research.marshfieldclinic.org/). This is the largest study of human genetic diversity to date. Rosenberg et al. (2002) used the program Structure to describe different levels of substructure in these data. Although the authors note that they see admixture between inferred populations, they found strong evidence of continental clustering, with individuals grouping in inferred populations according to their geographical origin with high coefficients of ancestry. For example, when they used three inferred populations, they could assign individuals from Africa to one population, individuals from Europe and central Asia to another, and individuals from East Asia and America to a third population. With four inferred populations, Native Americans separated from the East Asian group. However, it is worth noting that the assignments of individuals to inferred populations were not stable when the number of inferred populations was changed: Some individuals drastically changed their coefficients of ancestry between inferred populations when the number of inferred populations was increased. Moreover, the assignment of the individuals in different inferred populations cannot be easily interpreted in terms of population history because Structure does not give any indication of the relevance of these units nor of their differentiation with regard to the other inferred populations. For example, in Rosenberg et al. (2002), the Kalash population from Pakistan splits from the Eurasian populations when six inferred populations are used (K = 6), whereas the Kalash can hardly be interpreted as a major subdivision of the human species. It is thus difficult to evaluate whether human genetic diversity is best represented by the results given by Structure for two, five, 10, or any other number of inferred populations.
We tested if it is possible to find an assignment of the individuals of the CEPH diversity panel that remains stable when the number of inferred populations is increased above any particular number. The analyses performed by Rosenberg et al. (2002) use a model in which the allele frequencies in the inferred populations at each locus are correlated with each other. The choice of this model is presumably based on the assumption that all human populations originated from a single ancestral source population quite recently. We tested if it is possible to find a stable assignment of the individuals to inferred populations using a model in which allele frequencies in the inferred populations are allowed to be independent of each other. This would best represent a situation in which colonizations of various parts of the world originated from ancestral populations in which genetic drift would have been strong enough to allow microsatellite allele frequencies to become independent from each other (see, e.g., Harpending and Rogers 2000). This would also represent a situation in which archaic human populations (e.g., the Neandertals) contributed at some locations to the gene pool of early modern humans. When the 1066 individuals of the CEPH diversity panel are analyzed using a model of uncorrelated allele frequencies, the results are similar to those found by Rosenberg et al. (2002), with slightly lower coefficients of ancestry. Additionally, as in Rosenberg et al. (2002), some individuals change their assignment drastically when inferred populations are added (data not shown). Thus, when the whole data set is analyzed, the choice of the model does not change the results, but neither does it allow finding a stable representation of human genetic diversity.
However, as indicated by the results described in Figure 1, a sampling strategy that maximizes the geographic distribution of samples and keeps similar sample size for each geographical area may be essential to avoid the creation of apparent substructures. We therefore analyzed subsamples of the individuals of the CEPH diversity panel that equalized the number of individuals per population. Three different such subsamples were constructed by sampling, where possible, five different individuals from each of the 52 populations present in the CEPH diversity panel. Using a model of uncorrelated allele frequencies, the results are very different from those when the entire CEPH diversity panel is analyzed. For all three subsamples, the assignment of the individuals to inferred populations differs if we consider two, three, or four inferred populations. However, when more than four inferred populations are considered, the assignment of the individuals is identical to that of the four inferred populations, and no individual was assigned >1% ancestry from the additional inferred populations. Interestingly, the assignments of the individuals of all three subsamples in four inferred populations are very similar to each other despite the fact that they are mostly composed of different individuals. When the first subsample is analyzed using a model of correlated allele frequencies (i.e., the model used in Rosenberg et al. 2002), the results are very similar to those found by Rosenberg et al. (2002), despite the reduced number of individuals analyzed (261 individuals instead of 1061). Thus, the assignment of individuals to inferred populations changes when the numbers of inferred populations are changed, and individuals tend to cluster according to continents. For example, for K = 4, individuals from Africa, Europe, America, and Asia/Oceania group together with high coefficients of ancestry (Supplemental Fig. 1). It is, therefore, unlikely that the results obtained by analyzing three subsamples of the CEPH diversity panel with a model of uncorrelated allele frequencies is the result of a lack of power caused by a reduced number of individuals while it leads to a robust solution.
Thus, a geographically homogeneous worldwide sampling of humans and analysis of the data with a model in which the allele frequencies in the inferred populations are allowed to be uncorrelated allows us to obtain a stable and reproducible assignment of the individuals into four inferred populations.
Worldwide Patterns of Admixture
Figure 2 shows the proportion of admixture from each of the four inferred populations (termed A-D) for each of the 261 individuals of the first subsample given by Structure using a model of uncorrelated allele frequencies. In contrast to the results obtained by Rosenberg et al. (2002), most individuals are found to be highly admixed between two or three of the inferred populations. In Africa, individuals show admixture from the inferred populations A, B, and C. Eurasian and Oceanian individuals show admixture of the inferred populations A and B, whereas Native Americans exhibit admixture from the inferred populations B and D.
Figure 2.
Assignment of 261 individuals from the CEPH diversity panel (Cann et al. 2002) according to their coefficients of ancestry of four inferred populations (A, B, C, and D). Green circles represent African individuals, blue circles European individuals, pink circles Asians individuals, yellow circles Oceanian individuals, and red circles Native American individuals. The appropriate representation would be a pyramid. The two faces of the pyramid represented here included all individuals at least once.
Notably, and in contrast to the finding of discrete continental clustering often reported in the literature, three major geographical gradients become apparent. In Africa, a north-south gradient is seen. Thus, individuals from Kenya, as well as the only individual from the Nile Valley in the panel, have a lower contribution from inferred population C than Pygmy individuals in the Democratic Republic of Congo and the Central African Republic. The latter, in turn, have a lower contribution of the inferred population C than the San individuals from Namibia (data not shown). Sub-Saharans and the only North Africans in the panel (Mozabites from Algeria) appear to be distinct in this analysis. The latter have a 5%-10% admixture from inferred population C but fall closer to Eurasians than to other African individuals. However, it is likely that the lack of individuals from other regions of North Africa in the diversity panel is responsible for this apparent separation between Sub-Saharan Africans and North Africans because gene frequencies in Egypt and Ethiopia are known to be intermediate between Europe and sub-Saharan Africa (Passarino et al. 1998; Krings et al. 1999; Chen et al. 2000; Manni et al. 2002).
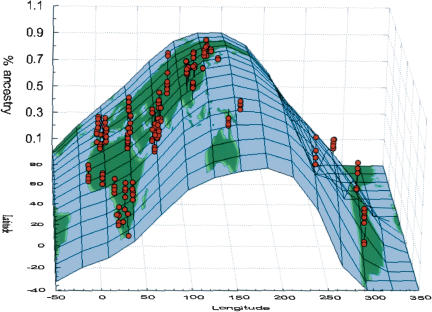
In Eurasia, genetic affiliation of individuals changes gradually with longitude (Fig. 3) such that western European individuals are at one end of a gradient and Southeast Asian individuals at the other. Oceanian individuals tend to fall on the eastern extremity of the Eurasian gradient, with similar patterns of admixture in Melanesians as in Southeast Asians. Papuans have a 1%-10% proportion of admixture from inferred population D that is otherwise represented only in Native American individuals.
Figure 3.
Geographic representation of the proportion of ancestry in inferred population B obtained by Structure. Red dots represent individuals defined by their longitude (_x_-axis), latitude (_y_-axis), and coefficient of ancestry (_z_-axis) in the inferred population B. The surface of the world was fitted to the xyz coordinates using a distance weighted least-squares smoothing method.
In the Americas, the genetic variation is similarly organized according to a geographical gradient. All individuals show mixed ancestry composed of inferred population B (the Asian/Oceanian end of the Eurasian gradient) and inferred population D, found only in Native Americans. Mexican individuals are closer to Asians, whereas Native Americans from Brazil are at the other extreme of the gradient, with Colombian individuals in an intermediate position. A discontinuity is apparent between Native Americans and East Asians on Figure 2. However, it is worth noting that it corresponds to a “sampling gap,” with no native individuals from North America sampled in the CEPH diversity panel.
DISCUSSION
Populations Versus Individuals
Our results show that population-based sampling schemes such as the one depicted in Figure 1A produce views of the human genetic diversity characterized by discrete units of diversity that tend to correspond to continents. This is especially true for samples in which few geographically disconnected populations from different continents are sampled. In contrast, when individuals that better cover the geographic distribution of humans across continents (Fig. 1B) are sampled, the human gene pool no longer appears to be composed of discrete clusters. It is noteworthy that the discrete clusters described by Rosenberg et al. (2002) from analyzing more than one thousand individuals of the CEPH diversity panel might be caused by discontinuities in the sampling, because when samples that have equal numbers of individuals of each population are analyzed (Fig. 2), the inferred populations yielded by Structure do not match continents or geographical regions but represent theoretical “populations” in which all individuals show admixture to at least two such “populations.” Therefore, when the aim is to investigate genetic diversity on a worldwide scale, we recommend an approach in which individuals from as many localities as possible are sampled. Sampling schemes based on populations should only be used if the aim of the study is to unravel the history of these specific populations or their relationship with surrounding populations (e.g., the origin and relationships of the different Polynesian populations; Kayser et al. 2000).
Representations of Worldwide Human Genetic Diversity
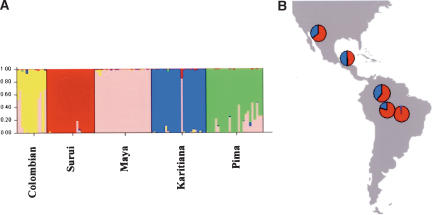
The high degree of clustering of genetic diversity according to populations described by Rosenberg et al. (2002) for the CEPH diversity panel is in disagreement with our results obtained by analyzing subsamples of the CEPH diversity panel that equalize population sizes, and a model of uncorrelated allele frequencies. It should not be overlooked, however, that this disagreement is largely a result of how the data are depicted and discussed, because the genetic units described by Rosenberg et al. (2002) often correspond to a splitting of the gradients we observe into several smaller inferred populations. For example, in America, we find evidence of an allele frequency cline whereas Rosenberg et al. (2002) assign all individuals to five inferred populations that correspond to their populations of origin (Fig. 4). Thus, whereas Rosenberg's group investigates whether individuals can be assigned to culturally predefined populations on the basis of their genotypes, we investigate the patterns of relatedness across the human gene pool. The goals of the two approaches are both valid but clearly distinct. However, it is important to stress that when the goal of a study is to identify the geographical origin of one individual (e.g., in forensics) by his/her genotype, the results will be very dependent on the populations used as references and to their genetic relatedness with the sample investigated.
Figure 4.
Assignment of Native American individuals using (A) correlated allele frequencies as used in Rosenberg et al. (2002) and (B) uncorrelated allele frequencies. In B, the assignment of the individuals has been determined using all individuals from the first subsample and four inferred populations as in Figure 2. In A, each bar represents a single individual, and the colors correspond to the coefficients of ancestry. In B, the coefficients of ancestry were averaged over the five individuals for each population. Blue and red segments represent the proportion of ancestry in inferred population B and D, respectively.
Clines, Not “Races”
Using a homogeneous sampling strategy and a model in which allele frequencies in the different inferred populations are allowed to be independent, we find a stable and reproducible representation of human genetic diversity in which the extent of admixture between individuals in Eurasia and the Americas changes continuously with geographical distance without any major discontinuities (Figs. 2 and 3). Between Eurasia and Africa, the Mediterranean Sea does not seem to act as a major barrier because people from Algeria are similar to people from western Europe except for an ∼10% admixture from inferred population C. Between Eurasia and the Americas, the lack of individuals from North America in the CEPH diversity panel limits the ability to draw conclusions. However, it is interesting that Mayan individuals from Mexico are more similar to East Asian individuals than are individuals in South America. In the light of these results, and in agreement with extensive studies of classical genetic markers (Cavalli-Sforza et al. 1994), it seems that gradual variation and isolation by distance rather than major genetic discontinuities is typical of global human genetic diversity. Obviously, this does not imply that genetic discontinuities do not exist on a more local scale, for example, between people from different linguistic groups (e.g., Barbujani and Sokal 1990; Sokal et al. 1990). It also does not mean that no differences whatsoever exist between continental groups. In fact, what Rosenberg et al. (2002) have shown is that given enough markers and the extraordinary power of Structure, the tiny amounts of genetic differences that exist between continents can also be discerned. However, this should not obscure the fact that on a worldwide scale, clines are a better representation of the human diversity than clades, and that continents do not represent more substantial discontinuities in such clines than many other geographical and cultural barriers.
That clines are a more adequate representation of human genetic diversity than clades is not unexpected in view of earlier works that show that most genetic variation is found among individuals rather than among continents (e.g., Boyd 1950; Livingstone 1962; Lewontin 1972; Cavalli-Sforza et al. 1994). In fact, also in the current data set, 87.6% percent of the total diversity is found among individuals and only 9.2% among continents (Excoffier and Hamilton 2003), in agreement with many previous studies (e.g. Lewontin 1972; Owens and King 1999; but see also Edwards 2003). The current results are also not unexpected in view of the fact that identical DNA sequences of several kilobases are found on different continents (Kaessmann et al. 1999; Gabriel et al. 2002). In fact, as much as a third of the entire human diversity of common haplotypes may be contained within single individuals (Pääbo 2003). However, in spite of this, there is a great tendency in the literature to use a few populations from the extremes of continental landmasses (such as in Fig. 1A) to make worldwide inferences about substructures in the human gene pool. In fact, because human genetic diversity tends to be distributed clinally, it is especially problematic to sample the extremes of continents because this will create the impression of sharp discontinuities in the distribution of genetic variants.
In this regard, it is worth noting that the colonization history of the United States has resulted in a “sampling” of the human population made up largely of people from western Europe, western Africa, and Southeast Asia. Thus, studies in which individuals from Europe, sub-Saharan Africa, and Southeast Asia are used (e.g., Jorde et al. 1997) might be an adequate description of the major components of the U.S. population (e.g., see Kayser et al. 2003). However, it would be incorrect to conclude that such studies necessarily generalize to subdivisions of the human gene pool on a worldwide scale.
The absence of strong continental clustering in the human gene pool is of practical importance. It has recently been claimed that “the greatest genetic structure that exists in the human population occurs at the racial level” (Risch et al. 2002). Our results show that this is not the case, and we see no reason to assume that “races” represent any units of relevance for understanding human genetic history. In clinical practice, the “classification” of people into “races,” as recently suggested (Risch et al. 2002; Burchard et al. 2003), could perhaps have some justification as a proxy for differences in environmental and other factors of relevance for public health or to help identify rare disease alleles (Phimister 2003). However, in the absence of other knowledge, most alleles influencing susceptibility to disease or outcome of medical interventions cannot be expected to show significantly different frequencies between “races.” An exception may be genes where different selection regimes have acted in different geographical regions. However, even in those cases, the genetic discontinuities seen are generally not “racial” or continental in nature but depend on historical and cultural factors that are more local in nature. For example, the hemoglobin S allele that causes resistance to malaria occurs not only in sub-Saharan Africa but also in southern Europe, the middle East, and India (Cavalli-Sforza et al. 1994). Lactose tolerance occurs both in Europe and in Africa (Sahi 1994), and the deleted allele of CCR5 that confers resistance to human immunodeficiency virus occurs in Europe as well as in Asia (Martinson et al. 1997). Thus, even for a rapid and rough evaluation of genetic risk factors, “racial” background is of limited use, and direct analysis of the relevant gene is the only reliable way to evaluate genetic risk in an individual (Cooper et al. 2003). Fortunately, this will become increasingly possible as the genetic components of more diseases become elucidated.
METHODS
Influence of Sampling Strategy
First, we constructed a population-based data set (Fig. 1A) by selecting five to eight individuals from each of 15 populations studied by Jorde et al. (1995, 1997). Individuals were chosen to minimize the extent of missing data at 20 microsatellite loci genotyped in the geography-based data set (see below). In all, 30 individuals came from sub-Saharan Africa, 29 individuals from Southeast Asia, and 30 individuals from Western Europe (see Supplemental Table S1 for more details).
For comparison to this data set, we produced a geography-based data set (Fig. 1B). We analyzed DNA samples from 90 individuals widely distributed across the globe to roughly reflect the distribution of humans. In all, 20 individuals came from Africa, 36 individuals from Asia, 16 individuals from Europe, 13 individuals from Oceania, and five individuals from Native American groups (Supplemental Table S2). These individuals were genotyped for 20 unlinked autosomal microsatellite loci that had been genotyped in the population-based sample (Fig. 1A) by Jorde et al. (1997). These were chosen to be widely distributed across different chromosomes. Genomic DNA sequences were amplified using 20 ng of genomic DNA from each individual and fluorescence-labeled primers. PCR products were analyzed on an ABI Prism 3100 Genetic Analyser (Applied Bio-system). DNA from two individuals analyzed in Jorde et al. (1997), kindly provided by L.B. Jorde (Dept. of Human Genetics, Univ. of Utah), were used to calibrate the scoring of allele lengths between the two studies. The geography-based data set has <1.4% missing data, and no individual is missing information at more than two loci.
Reanalyses of the CEPH Diversity Panel Data Set
We reanalyzed a data set of 1066 individuals from the HGDPCEPH Human Genome Diversity Cell Line Panel (CEPH diversity panel; Cann et al. 2002), which have been genotyped for 377 autosomal microsatellites at the Center for Medical Genetics at the Marshfield Medical Research Foundation (http://research.marshfieldclinic.org/) and analyzed by Rosenberg et al. (2002). These individuals were sampled across all five continents and were assigned to 52 different populations. To date, they represent one of the most extensive geographical samplings of human diversity. In our analyses, we considered Bantu speakers from Kenya and from South Africa as two different populations. For some analyses, we equalized the sample size of the different populations by creating three subsamples of 261 individuals from the 1066 individual data set, such that five individuals were sampled from each of the 52 populations present in the diversity panel (plus a single Nilotic individual). Whenever possible, five different individuals from each population were taken for each subsample.
Data Analyses
Individuals were assigned to a prespecified number (K) of “inferred populations” according to their genotype using a Bayesian approach in the program Structure (Pritchard et al. 2000). The inferred populations are theoretical entities whose allele frequencies are estimated so that the ancestry of the observed individual genotypes can be explained by admixture of one or more inferred population(s). The term “admixture” as used in this study thus refers to the assignment of the individuals into inferred populations by Structure. It does not necessarily imply any actual admixture between any actual historical populations. We used a version of Structure that allows for such admixture and uses non-correlated allele frequencies among inferred populations (the choice of model is discussed in Results). Every run consisted of 200,000 burn-in steps followed by 100,000 Markov Chain Monte Carlo steps. We ran five independent replicates for each value of K, allowing K to vary between 1 and 6. The five runs always converged to a single representation with similar estimated likelihoods but for one of the three subsamples of the CEPH diversity panel for which two of the five replicates at K = 6 converged to a second solution of similar likelihood but with another assignment of individuals (data not shown).
Acknowledgments
This work is supported by the Max Planck Society and the Bundesministerium für Bildung und Forschung. We thank Lynn Jorde for providing the genotypes of the individuals and DNA samples and Aravinda Chakravarti, David Hughes, Michi Hofreiter, Jonathan Pritchard, Susan Ptak, Noah Rosenberg, Mark Stoneking, Linda Vigilant, and especially Molly Przeworski, David Cutler, and two anonymous reviewers for constructive discussions, advice, and help.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: L.B. Jorde.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2529604.
References
- Bamshad, M.J., Wooding, S., Watkins, W.S., Ostler, C.T., Batzer, M.A., and Jorde, L.B. 2003. Human population genetic structure and inference of group membership. Am. J. Hum. Genet. 72**:** 578-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbujani, G. and Sokal, R.R. 1990. Zones of sharp genetic change in Europe are also linguistic boundaries. Proc. Natl. Acad. Sci. 87**:** 1816-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbujani, G., Bertorelle, G., Capitani, G., and Scozzari, R. 1995. Geographical structuring in the mtDNA of Italians. Proc. Natl. Acad. Sci. 92**:** 9171-9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock, A. and Cavalli-Sforza, L. 1991. The study of variation in the human genome. Genomics 11**:** 491-498. [DOI] [PubMed] [Google Scholar]
- Bowcock, A.M., Ruiz-Linares, A., Tomfohrde, J., Minch, E., Kidd, J.R., and Cavalli-Sforza, L.L. 1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368**:** 455-457. [DOI] [PubMed] [Google Scholar]
- Boyd, W.C. 1950. Genetics and the races of man. Blackwell, Oxford, UK.
- Burchard, E.G., Ziv, E., Coyle, N., Gomez, S.L., Tang, H., Karter, A.J., Mountain, J.L., Perez-Stable, E.J., Sheppard, D., and Risch, N. 2003. The importance of race and ethnic background in biomedical research and clinical practice. N. Engl. J. Med. 348**:** 1170-1175. [DOI] [PubMed] [Google Scholar]
- Calafell, F., Shuster, A., Speed, W.C., Kidd, J.R., and Kidd, K.K. 1998. Short tandem repeat polymorphism evolution in humans. Eur. J. Hum. Genet. 6**:** 38-49. [DOI] [PubMed] [Google Scholar]
- Cann, H.M., de Toma, C., Cazes, L., Legrand, M.F., Morel, V., Piouffre, L., Bodmer, J., Bodmer, W.F., Bonne-Tamir, B., Cambon-Thomsen, A., et al. 2002. A human genome diversity cell line panel. Science 296**:** 261-262. [DOI] [PubMed] [Google Scholar]
- Cardon, L.R. and Bell, J.I. 2001. Association study designs for complex diseases. Nat. Rev. Genet. 2**:** 91-99. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza, L.L., Piazza, A., Menozzi, P., and Mountain, J. 1988. Reconstruction of human evolution: Bringing together genetic, archaeological, and linguistic data. Proc. Natl. Acad. Sci. 85**:** 6002-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza, L.L., Menozzi, P., and Piazza, A. 1994. The history and geography of human genes. Princeton University Press, Princeton, NJ.
- Chen, Y.S., Olckers, A., Schurr, T.G., Kogelnik, A.M., Huoponen, K., and Wallace, D.C. 2000. mtDNA variation in the South African Kung and Khwe—and their genetic relationships to other African populations. Am. J. Hum. Genet. 66**:** 1362-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, R.S., Kaufman, J.S., and Ward, R. 2003. Race and genomics. N. Engl. J. Med. 348**:** 1166-1170. [DOI] [PubMed] [Google Scholar]
- Ding, Y.C., Wooding, S., Harpending, H.C., Chi, H.C., Li, H.P., Fu, Y.X., Pang, J.F., Yao, Y.G., Yu, J.G., Moyzis, R., et al. 2000. Population structure and history in east Asia. Proc. Natl. Acad. Sci. 97**:** 14003-14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, A.W. 2003. Human genetic diversity: Lewontin's fallacy. Bioessays 25**:** 798-801. [DOI] [PubMed] [Google Scholar]
- Excoffier, L. and Hamilton, G. 2003. Comment on “Genetic structure of human populations.” Science 300**:** 1877. [DOI] [PubMed] [Google Scholar]
- Gabriel, S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296**:** 2225-2229. [DOI] [PubMed] [Google Scholar]
- Harpending, H. and Rogers, A. 2000. Genetic perspectives on human origins and differentiation. Annu. Rev. Genomics Hum. Genet. 1**:** 361-385. [DOI] [PubMed] [Google Scholar]
- Jorde, L.B., Bamshad, M.J., Watkins, W.S., Zenger, R., Fraley, A.E., Krakowiak, P.A., Carpenter, K.D., Soodyall, H., Jenkins, T., and Rogers, A.R. 1995. Origins and affinities of modern humans: A comparison of mitochondrial and nuclear genetic data. Am. J. Hum. Genet. 57**:** 523-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde, L.B., Rogers, A.R., Bamshad, M., Watkins, W.S., Krakowiak, P., Sung, S., Kere, J., and Harpending, H.C. 1997. Microsatellite diversity and the demographic history of modern humans. Proc. Natl. Acad. Sci. 94**:** 3100-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann, H., Heissig, F., von Haeseler, A., and Pääbo, S. 1999. DNA sequence variation in a noncoding region of low recombination on the human X chromosome. Nat. Genet. 22**:** 78-81. [DOI] [PubMed] [Google Scholar]
- Karafet, T., Xu, L., Du, R., Wang, W., Feng, S., Wells, R.S., Redd, A.J., Zegura, S.L., and Hammer, M.F. 2001. Paternal population history of East Asia: Sources, patterns, and microevolutionary processes. Am. J. Hum. Genet. 69**:** 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser, M., Brauer, S., Weiss, G., Underhill, P.A., Roewer, L., Schiefenhovel, W., and Stoneking, M. 2000. Melanesian origin of Polynesian Y chromosomes. Curr. Biol. 10**:** 1237-1246. [DOI] [PubMed] [Google Scholar]
- Kayser, M., Brauer, S., Schadlich, H., Prinz, M., Batzer, M.A., Zimmerman, P.A., Boatin, B.A., and Stoneking, M. 2003. Y chromosome STR haplotypes and the genetic structure of U.S. populations of African, European, and Hispanic ancestry. Genome Res. 13**:** 624-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, M.C. and Motulsky, A.G. 2002. Human genetics. Mapping human history. Science 298**:** 2342-2343. [DOI] [PubMed] [Google Scholar]
- Kittles, R.A. and Weiss, K.M. 2003. Race, ancestry, and genes: Implications for defining disease risk. Annu. Rev. Genomics Hum. Genet. 4**:** 33-67. [DOI] [PubMed] [Google Scholar]
- Krings, M., Salem, A., Bauer, K., Geisert, H., Malek, A.K., Chaix, L., Simon, C., Welsby, D., Di Rienzo, A., Utermann, G., et al. 1999. mtDNA analysis of Nile River Valley populations: A genetic corridor or a barrier to migration? Am. J. Hum. Genet. 64**:** 1166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S. and Schork, N.J. 1994. Genetic dissection of complex traits. Science 265**:** 2037-2048. [DOI] [PubMed] [Google Scholar]
- Lewontin, R.C. 1972. The apportionment of human diversity. Evol. Biol. 6**:** 381-398. [Google Scholar]
- Livingstone, F.B. 1962. On the non-existence of human races. Curr. Anthropology 3**:** 279-281. [Google Scholar]
- Manni, F., Leonardi, P., Barakat, A., Rouba, H., Heyer, E., Klintschar, M., McElreavey, K., and Quintana-Murci, L. 2002. Y-Chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa. Hum. Biol. 74**:** 645-658. [DOI] [PubMed] [Google Scholar]
- Martinson, J.J., Chapman, N.H., Rees, D.C., Liu, Y.T., and Clegg, J.B. 1997. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet. 16**:** 100-103. [DOI] [PubMed] [Google Scholar]
- Mountain, J.L. and Cavalli-Sforza, L.L. 1997. Multilocus genotypes, a tree of individuals, and human evolutionary history. Am. J. Hum. Genet. 61**:** 705-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. and Takezaki, N. 1996. The root of the phylogenetic tree of human populations. Mol. Biol. Evol. 13**:** 170-177. [DOI] [PubMed] [Google Scholar]
- Owens, K. and King, M.C. 1999. Genomic views of human history. Science 286**:** 451-453. [DOI] [PubMed] [Google Scholar]
- Pääbo, S. 2003. The mosaic that is our genome. Nature 421**:** 409-412. [DOI] [PubMed] [Google Scholar]
- Passarino, G., Semino, O., Quintana-Murci, L., Excoffier, L., Hammer, M., and Santachiara-Benerecetti, A.S. 1998. Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am. J. Hum. Genet. 62**:** 420-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lezaun, A., Calafell, F., Mateu, E., Comas, D., Ruiz-Pacheco, R., and Bertranpetit, J. 1997. Microsatellite variation and the differentiation of modern humans. Hum. Genet. 99**:** 1-7. [DOI] [PubMed] [Google Scholar]
- Phimister, E.G. 2003. Medicine and the racial divide. N. Engl. J. Med. 348**:** 1081-1082. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K., Stephens, M., and Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics 155**:** 945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch, N., Burchard, E., Ziv, E., and Tang, H. 2002. Categorization of humans in biomedical research: Genes, race and disease. Genome Biol. 3**:** comment 2007. [DOI] [PMC free article] [PubMed]
- Rosenberg, N.A., Pritchard, J.K., Weber, J.L., Cann, H.M., Kidd, K.K., Zhivotovsky, L.A., and Feldman, M.W. 2002. Genetic structure of human populations. Science 298**:** 2381-2385. [DOI] [PubMed] [Google Scholar]
- Rosser, Z.H., Zerjal, T., Hurles, M.E., Adojaan, M., Alavantic, D., Amorim, A., Amos, W., Armenteros, M., Arroyo, E., Barbujani, G., et al. 2000. Y-Chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 67**:** 1526-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahi, T. 1994. Genetics and epidemiology of adult-type hypolactasia. Scand. J. Gastroenterol. Suppl 202: 7-20. [DOI] [PubMed]
- Sokal, R.R., Oden, N.L., Legendre, P., Fortin, M.J., Kim, J.Y., Thomson, B.A., Vaudor, A., Harding, R.M., and Barbujani, G. 1990. Genetics and language in European populations. American Naturalist 135**:** 157-175. [Google Scholar]
- Stephens, J.C., Schneider, J.A., Tanguay, D.A., Choi, J., Acharya, T., Stanley, S.E., Jiang, R., Messer, C.J., Chew, A., Han, J.H., et al. 2001. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293**:** 489-493. [DOI] [PubMed] [Google Scholar]
- Tishkoff, S.A., Dietzsch, E., Speed, W., Pakstis, A.J., Kidd, J.R., Cheung, K., Bonne-Tamir, B., Santachiara-Benerecetti, A.S., Moral, P., and Krings, M. 1996. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271**:** 1380-1387. [DOI] [PubMed] [Google Scholar]
- Wilson, J.F., Weale, M.E., Smith, A.C., Gratrix, F., Fletcher, B., Thomas, M.G., Bradman, N., and Goldstein, D.B. 2001. Population genetic structure of variable drug response. Nat. Genet. 29**:** 265-269. [DOI] [PubMed] [Google Scholar]
- Wright, S. 1969. Evolution and genetics of populations. The theory of gene frequencies. Vol. 2. University of Chicago Press, Chicago, IL.
WEB SITE REFERENCES
- http://research.marshfieldclinic.org/; Marshfield Clinic Research Foundation.