Phylogenetic Footprint Analysis of IGF2 in Extant Mammals (original) (raw)
Abstract
Genomic imprinting results in monoallelic gene transcription that is directed by _cis_-acting regulatory elements epigenetically marked in a parent-of-origin-dependent manner. We performed phylogenetic sequence and epigenetic comparisons of IGF2 between the nonimprinted platypus (Ornithorhynchus anatinus) and imprinted opossum (Didelphis virginiana), mouse (Mus musculus), and human (Homo sapiens) to determine if their divergent imprint status would reflect differences in the conservation of genomic elements important in the regulation of imprinting. We report herein that IGF2 imprinting does not correlate evolutionarily with differential intragenic methylation, nor is it associated with motif 13, a reported _IGF2_-specific “imprint signature” located in the coding region. Instead, IGF2 imprinting is strongly associated with both the lack of short interspersed transposable elements (SINEs) and an intragenic conserved inverted repeat that contains candidate CTCF-binding sites, a role not previously ascribed to this particular sequence element. Our results are the first to demonstrate that comparative footprint analysis of species from evolutionarily distant mammalian clades, and exhibiting divergent imprint status is a powerful bioinformatics-based approach for identifying _cis_-acting elements potentially involved not only in the origins of genomic imprinting, but also in its maintenance in humans.
Genomic imprinting refers to epigenetic chromosomal modifications that result in the preferential expression of an allele in a parent-of-origin-dependant manner. The evolution of genomic imprinting more than 150 million years ago (Killian et al. 2000, 2001) is postulated to have resulted because of a parental conflict to control the amount of nutrients extracted from the mother by her offspring (Haig and Graham 1991; Murphy and Jirtle 2003). Since the discovery in the early 1990s that murine and human IGF2 predominantly exhibit expression from the paternally inherited chromosome (DeChiara et al. 1991), this gene has been a central focus of numerous studies involving genomic imprinting and its relationship to development and disease. IGF2 encodes for a powerful mitogenic growth factor that is frequently found to be overexpressed in cancer because of a loss of imprinting (Rainier et al. 1993; Falls et al. 1999; Cruz-Correa et al. 2004). This deregulation of IGF2 imprinting has been investigated in several human malignancies, including those that affect children such as Wilms' tumor and hepatoblastoma (Rainier et al. 1993, 1995; Cruz-Correa et al. 2004), and adult-onset cancers including colorectal carcinoma, bladder cancer, osteosarcoma, ovarian cancer, and breast cancer (Falls et al. 1999; Feinberg and Tycko 2004).
IGF2 is also implicated in the somatic overgrowth disorder referred to as Beckwith-Wiedemann Syndrome (BWS; Mannens et al. 1994; Reik et al. 1995). Moreover, individuals with BWS are at increased risk of developing childhood tumors, especially Wilms' tumors. Recent reports also implicate IGF2 deregulation in the increased incidence of BWS in individuals conceived through assisted reproductive technology (Gosden et al. 2003; Niemitz and Feinberg 2004). In addition, deregulation of IGF2 imprinting, with consequent overexpression following somatic nuclear transfer, contributes to large offspring syndrome, an impediment to successful reproductive cloning (Rideout III et al. 2001; Ogura et al. 2002; Ogawa et al. 2003). Thus, a greater understanding of the _cis_-acting elements necessary for the proper maintenance and regulation of IGF2 imprinting is required to prevent and treat developmental disorders and cancer.
Nevertheless, a comprehensive understanding of the sequence elements necessary for the proper maintenance of genomic imprinting at the IGF2 locus has remained elusive, despite years of intense investigation. IGF2 lies in a juxtapositioned reciprocally imprinted gene domain upstream of H19 in both mouse and human (Paulsen et al. 1998; Onyango et al. 2000). Imprinting at the IGF2/H19 locus at least partially relies on the presence of differential methylation at multiple binding sites for the zinc finger protein CCCTC-binding factor (CTCF), located upstream of the maternally expressed H19 (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000). Several additional important regulatory elements involved in imprinting at the Igf2 locus in mice have been identified (Lopes et al. 2003). These include a mesodermal silencing element located within Igf2 in differentially methylated region 1 (DMR1), located upstream of the fetal promoters of Igf2 (Constancia et al. 2000; Arney 2003), and a methylation-sensitive activating element in DMR2, located in the last exon of Igf2 (Feil et al. 1994; Murrell et al. 2001).
A major limitation to advances in these analyses is the poor understanding of the nonexonic regulatory sequences that are crucial to proper maintenance of IGF2 imprinting. One method for identifying such candidate elements is through comparative footprint analysis of orthologous gene domains across evolutionarily distanced species to distinguish between neutrally evolving regions and conserved motifs that correlate with imprinting. The IGF2/H19 domain and other independent imprinted regions within the genomes of Eutherian mammals have already been studied using such comparative analyses (Otte et al. 1998; Paulsen et al. 1998; Onyango et al. 2000; Wylie et al. 2000; Paulsen et al. 2001; Amarger et al. 2002; Takada et al. 2002). Although providing valuable information, these prior studies were limited in their ability to identify imprint-specific signals because high levels of interspecies sequence conservation typify animals that lie within the same Eutherian superordinal clade (Murphy et al. 2001).
In this study, we provide a novel expansion to these earlier IGF2 phylogenetic comparisons by including animals from both the Metatherian and Prototherian subclasses of mammals. We have cloned and sequenced IGF2 not only in the imprinted marsupial, American opossum (Didelphis virginiana), but also in the monotreme, Tasmanian platypus (Ornithorhynchus anatinus), because in contrast to marsupials, monotremes are not imprinted at the IGF2 locus (Killian et al. 2001). Therefore, marsupials are the most ancestral mammals in which this gene is imprinted (O'Neill et al. 2000; Killian et al. 2001). This comparative analysis of the IGF2 gene in imprinted and nonimprinted mammalian species allows for the identification of sequence features shared by all species excluding monotremes, thus providing valid candidates for mediating IGF2 imprinting. We analyzed orthologous regions for sequences and features that had previously been associated with IGF2 imprinting. We identified a novel imprinting signature that fully correlates with imprint status of IGF2 in the three extant mammalian subclasses. Our results show that phylogenetic comparison of IGF2 among Prototherian, Metatheria, and Eutherian mammals is a powerful method to elucidate conserved _cis_-acting elements potentially involved in regulating IGF2 imprinting.
RESULTS
Genomic Structure of Opossum and Platypus IGF2
Comparison of exonic sequences for both the opossum and platypus confirmed that the sequences were more similar to IGF2 of other species than other members of the insulin-like growth factor family. The coding nucleotide sequence of opossum IGF2 (o_IGF2_; accession no. AY552325) is 79% identical to that of human (AF517226), whereas the coding nucleotide sequence of platypus IGF2 (p_IGF2_; accession no. AY552324) is only 48% identical to that of human. The coding nucleotide sequences of o_IGF2_ and p_IGF2_ are 89% identical to one another. The genomic sequence of o_IGF2_ is 7143 bp, whereas that of p_IGF2_ is 30,584 bp. The overall exon/intron structure of IGF2 has been conserved in all species examined for well over 150 million years (Figs. 1 and 2). The coding sequences for IGF2 have also remained restricted to the three coding exons that we have designated as C1, C2, and C3.
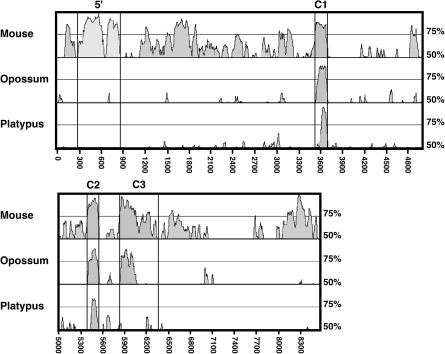
Figure 1.
mVISTA plot of mouse, opossum, and platypus IGF2 sequences with respect to that of human. Conserved segments are defined as regions in which every contiguous subsegment of 50 bp was at least 50% identical to its paired sequence. These segments were then merged to define the conserved regions (shaded peaks). The _x_-axis denotes the nucleotide positions for the human IGF2 domain sequence, where position 1 corresponds to position 2399 of GenBank accession no. AF517226. Exons are delineated by boxes.
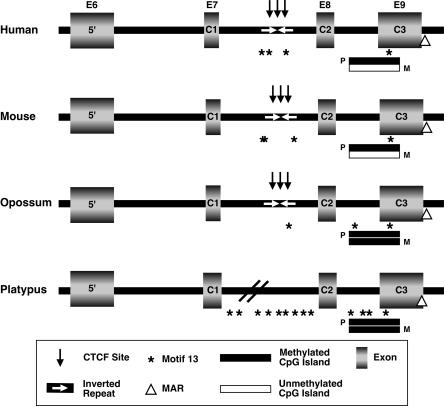
Figure 2.
Schematic representation of multispecies alignments of human, mouse, opossum, and platypus IGF2. (Rectangles) exons; (horizontal bars) CpG islands (filled, methylated; unfilled, unmethylated); (black arrows) consensus CTCF-binding sites; (triangle) putative matrix attachment region; (*) “imprint-specific” motif 13 sequences (Wang et al. 2004); (white arrows) inverted repeat regions; (E) exon; (C) coding exon.
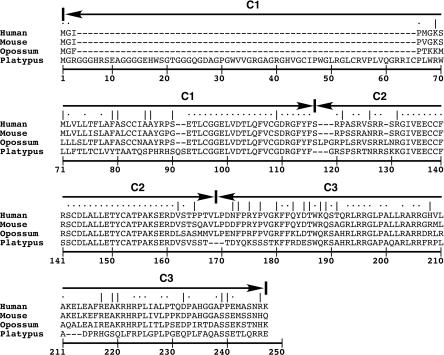
As expected, the 67 amino acids of the mature peptide are most highly conserved (Fig. 3). The opossum and platypus IGF2 mature peptides are 91% and 82% identical to that of human, respectively. The highest level of conservation between all species is found in exon C2, from which a large portion of the mature IGF2 peptide is produced. Exon C3, which encodes the E-domain and is clipped during processing of the peptide, displays a significant decrease in percent identity among these species (Fig. 3). The position of the o_IGF2_ and p_IGF2_ ATG start codon is consistent with the open reading frame of the contiguous coding exon sequences obtained through 5′-RACE, and matches the Kozak consensus sequence. Although both human and mouse IGF2 are under the control of several promoters preceding multiple 5′-noncoding exons, only one 5′-noncoding o_IGF2_ exon was identified by RACE in this study. Likewise, only one p_IGF2_ 5′-noncoding exon exists in the determined BAC sequence, as predicted by GrailExp (http://compbio.ornl.gov/grailexp/).
Figure 3.
Amino acid alignment of human, mouse, opossum, and platypus IGF2. (Solid arrows) Coding exons, C1, C2, and C3; (dots above sequence) perfect amino acid agreement; (vertical slash) conservative amino acid change. Mature IGF2 peptide, amino acids 87 to 160.
Comparative Analysis of Human, Mouse, Opossum, and Platypus IGF2 DMR2
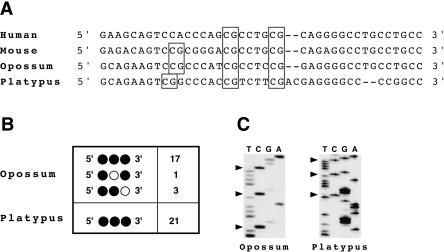
In the mouse and human IGF2 genes, a CpG island spans a portion of C3 and the intron preceding it (Fig. 2). This region is consistently differentially methylated in Eutherian mammals investigated, and is identified as DMR2 (Catchpoole et al. 2000; Murrell et al. 2001). The core differentially methylated region of this island in mouse includes CpGs 18-25 (Murrell et al. 2001); three of these CpG dinucleotides are also conserved in both the opossum and platypus (Fig. 4A). This suggests that differential methylation of these CpGs may also contribute to opossum IGF2 imprinting. In contrast, the nonimprinted platypus would not be expected to exhibit differential methylation of these CpGs. Bisulfite sequence analysis of 21 allele-specific clones containing the CpGs within this conserved core region of DMR2 in o_IGF2_ revealed 94% methylation of all CpGs (Fig. 4B,C). Similarly, bisulfite sequence analysis of 21 allele-specific clones from the platypus conserved core region orthologous to DMR2 exhibited 100% methylation (Fig. 4B,C).
Figure 4.
Epigenetic and sequence comparison of conserved core region of DMR2 in IGF2. (A) Multispecies alignment of DMR2 core; conserved CpGs are boxed. (B) Methylation profiles of opossum and platypus CG dinucleotides from individual cloned alleles following PCR amplification of bisulfite-modified genomic DNA. Filled circles represent methylated cytosines in the context of CpG dinucleotides. The number of individual clones with each methylation profile is listed. (C) Core CpG dinucleotide methylation sequencing analysis from the opossum and platypus orthologous DMR2 region following bisulfite modification. Methylated cytosines are labeled with arrowheads.
Matrix Attachment Regions
Matrix attachment regions (MARs) are AT-rich sequences involved in organization of chromatin structure and regulation of gene expression by localizing chromosomal domains to the nuclear envelope, a region rich in gene transcriptional activity. It has been postulated that genomic imprinting may be modulated by MARs (Greally et al. 1997, 1999; Burns et al. 2001). In human and mouse IGF2, an MAR adjacent to DMR2 has been identified. On the paternal allele of Igf2, this MAR falls under the control of genomic imprinting (Weber et al. 2003). We have identified conserved putative AT-rich MAR consensus sequences in orthologous regions of o_IGF2_ and p_IGF2_ genes using a bioinformatics-based algorithm (MAR-Wiz; http://www.futuresoft.org/MARWiz). Thus, these sequence elements are conserved in the imprinted opossum in addition to the platypus, where IGF2 is not imprinted (Fig. 2).
Human and Mouse Imprinting Motifs
In a recent comparative analysis of the genomic sequence of imprinted and nonimprinted genes in mouse and human, multiple conserved motifs were identified that are present only in imprinted genes (Wang et al. 2004). These motifs were labeled as “novel imprinting signatures,” and multiple copies of motif 13, consisting of the consensus sequence: 5′-GGCCTGCCCTCCAT CTTAG-3′, were identified in the human and mouse IGF2 but were not identified in any of the nonimprinted genes analyzed. Interestingly, these motif 13 sequences are present in both imprinted o_IGF2_ and nonimprinted p_IGF2_ genes (Fig. 2). Thus, this sequence element is not restricted to the IGF2 locus only when it is imprinted.
Short Interspersed Transposable Elements (SINEs)
Although the coding sequences for IGF2 have remained restricted to three coding exons (Fig. 2), the o_IGF2_ gene extends 7143 bp, whereas the p_IGF2_ encompasses 30,584 bp. This large difference in gene size mainly reflects the size of the intron between exons C1 and C2 of p_IGF2_ (25 kb), which contains several short interspersed transposable elements (MON1, MIR3). In fact, the p_IGF2_ genomic region contains a total of 88 SINE elements, occupying 9119 bp, or 28.59% of the sequence. In contrast, IGF2 sequences from orthologous regions in human, mouse, and opossum lack SINE elements.
Conserved Inverted Repeat and Putative CTCF Sites
MultiVISTA (http://gsd.lbl.gov/vista/index.shtml) allows for alignment of conserved regions from several related sequences on the same scale, adjusting for variable lengths of nonconserved sequence. In addition to conservation among coding sequences, an overall MultiVISTA plot of mouse, opossum, and platypus IGF2 gene sequences with respect to human reveals several conserved nonexonic regions that are present in species that are imprinted at IGF2 yet absent in p_IGF2_, which is not imprinted (Fig. 1).
One such region we identified is located between exons C1 and C2 (Fig. 2). It consists of a conserved inverted repeat in which protein binding in mouse is reduced upon methylation at CpG dinucleotides present within the inverted repeat (Otte et al. 1998). This region in humans also exhibits insulator activity and directly binds CTCF (Du et al. 2003). CTCF forms a chromatin insulator that functions in coordinating Igf2/H19 regulation (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000) as well as regulating an expanding list of other imprinted and nonimprinted genes throughout the genome (Awad et al. 1999; Bell et al. 1999; Arnold et al. 2000; Quitschke et al. 2000; Hikichi et al. 2003). Interspecies sequence comparisons identified 150 bp that are conserved among opossum (accession no. AY552325, beginning at 5622), mouse (accession no. U71085, beginning at nucleotide 23409), and human (accession no. AF517226, beginning at nucleotide 7258), but are lacking in the nonimprinted platypus. Three core recognition motifs for CTCF binding, 5′-CCCTC-3′ are present within this conserved region (Lobanenkov et al. 1990). The impact of CTCF binding within this inverted repeat on IGF2 imprinting in Eutherian mammals has yet to be evaluated.
DISCUSSION
Phylogenetic footprint analyses that are limited to imprinted Eutherian mammals lack the ability to effectively distinguish between neutrally evolving regions and conserved motifs that strongly correlate with imprinting because numerous “highly conserved elements” are identified even when very stringent selection criteria are used (Paulsen et al. 2001; Amarger et al. 2002). Thus, there is no simple way to distinguish between conservation due to function versus the possibility that these “conserved” sequences are merely a byproduct of evolutionary relatedness. In this report, we describe the first phylogenetic genomic sequence comparison of IGF2 between Eutherians and Metatherians, where this gene is imprinted, and Prototherians, where this gene is not imprinted, in order to identify conserved _cis_-acting elements that are precisely associated with the evolution of genomic imprinting. We show that IGF2 imprinting is strongly associated with both the lack of SINEs and a conserved inverted repeat that contains multiple candidate CTCF-binding sites, a role not previously ascribed to this element.
One of the most critical aspects of imprint regulation thus far identified is parent-of-origin-specific, differential methylation of regions rich in CpG dinucleotides (Reik and Walter 2001; Murphy and Jirtle 2003). Methylation of cytosines in this context regulates gene transcription by blocking access of DNA-binding proteins to methylated DNA or by recruiting methylation-specific DNA-binding proteins that lead to the induction of chromatin condensation and regional silencing. Although the majority of the CpGs within the human genome are methylated, CpG-rich regions are generally unmethylated. These CpG islands often are associated with gene promoters and are sometimes found intragenically. CpG islands associated with imprinting are differentially methylated; that is, one allele is relatively hypermethylated. This epigenetic characteristic is thought to contribute to the differential expression between the two alleles.
We have identified a conserved CpG island in both o_IGF2_ and p_IGF2_ that is orthologous to DMR2 in Eutherian mammals. Previous studies of DMR2 in mice and humans have implicated it as being a critical component of IGF2 imprinting. Studies in mice show that methylation on the paternal allele has an activator function (Murrell et al. 2001), whereas human studies have focused on the repression of transcription caused by the unmethylated status of the maternal allele (Catchpoole et al. 2000). Surprisingly, we found that the DMR2 core in both o_IGF2_ and p_IGF2_ is fully methylated. This methylation status is consistent with IGF2 not being imprinted in the monotremes (Killian et al. 2001); however, it is inconsistent with IGF2 being imprinted in marsupials (O'Neill et al. 2000) because full methylation of DMR2 would be expected to result in biallelic IGF2 expression. Although we cannot rule out a role for other potential DMRs, imprinting of o_IGF2_ is not dependent on differential methylation within this region.
Matrix attachment regions are important regulators of gene expression, and may have a role in controlling monoallelic expression of imprinted genes by differential tethering of the active and inactive alleles to the nuclear matrix. Previously, a MAR adjacent to DMR2 was found to be associated with imprinting (Weber et al. 2003). The DMR2/MAR region on the paternal allele is associated with the nuclear matrix in a DMR2-methylation-dependent manner. We found that the orthologous regions of both the opossum and platypus contained similar putative MAR sequences. Moreover, we have shown that in both opossum and platypus, the region orthologous to DMR2 is fully methylated. If these conserved putative MAR sequences normally function to attach the IGF2 locus to the nuclear matrix in combination with methylated DMR2, our findings suggest that either o_IGF2_ imprinting is independent of both DMR2 and the MAR or the ability of these sequences to modulate IGF2 imprinting in mouse and human is under the control of additional elements not yet identified.
In a comparative genomic sequence analysis of imprinted and nonimprinted genes in both mouse and human, specific sequence motifs were identified and proposed to comprise novel “imprinting signatures” (Wang et al. 2004). In their study, motif 13 is identified as the only sequence feature associated with imprinting of IGF2. In our phylogenetic comparisons, we found that motif 13 was present not only in the imprinted opossum, but also in the nonimprinted platypus. Given the presence of these motif 13 sequences, as well as their relative conservation in location within the IGF2 gene in both imprinted and nonimprinted states, our data indicate that these sequences did not play a fundamental role in the evolutionary origin of imprinting at the IGF2 locus.
Cross-species phylogenetic comparisons using MultiVISTA analysis illustrate the ability to selectively identify core elements of imprinted domains through inclusion of more distantly related species. For example, beginning at nucleotide 4800 in the human sequence in Figure 1, a wide peak of conservation is evident between human and mouse. In the opossum, however, this peak has tapered considerably, thus delineating putative essential sequences in a more precise manner. Within this defined region, an inverted repeat element ∼150 bp long was identified that is present in the imprinted human, mouse, and opossum, but not in the nonimprinted platypus. This inverted repeat is also present in the horse IGF2 region (Otte et al. 1998). Three core recognition motifs for CTCF binding are also located within this region in the imprinted species (Lobanenkov et al. 1990). Intriguingly, this inverted repeat does, indeed, bind CTCF protein in humans (Du et al. 2003), and exhibits protein-binding activity that is methylation-sensitive in horse and mouse (Otte et al. 1998). Given the conservation of the CTCF sites in the imprinted opossum, mouse, and human and the lack of a similarly positioned, CCCTC-containing inverted repeat in the platypus, these results indicate that this region may have provided an important function in the origin and maintenance of imprinting at this locus.
Recently, several groups have reported that SINEs tend to be excluded from imprinted domains in humans (Amarger et al. 2002; Greally 2002), and that the paucity of these repeats may be important in initiating and/or regulating imprinted gene expression in mammals. Our results are consistent with this postulate in that the nonimprinted platypus is replete with SINEs, whereas the imprinted human, mouse, and opossum IGF2 domain sequences are nearly devoid of these elements. This supports the hypothesis that the exclusion of SINE elements may be required for imprinted expression to occur.
Through comparisons of the IGF2 gene in imprinted human, mouse, opossum, and nonimprinted platypus, we have identified strong candidates for mechanistic involvement in the evolution of imprinting at this locus. Phylogenetic imprint analyses of species from evolutionarily distant mammalian clades with divergent imprint status are therefore a valid bioinformatics-based approach for the identification of _cis_-acting elements potentially involved in both the origins of genomic imprinting and its maintenance in humans.
METHODS
Tissue Samples
Tasmanian and mainland Australian platypus (Ornithorhynchus anatinus) visceral organs (i.e., spleen, liver, and kidney) and skin biopsies were obtained from wild animals that had either succumbed to dog attack or were under surveillance (kindly provided by Barry Munday, University of Tasmania). Kidney, liver, and brain tissues were taken from adult female American opossums (Didelphis virginiana; kindly provided by Michael Stoskopf from North Carolina State University) and their pouch young following euthanasia by North Carolina Wildlife Commission officials as part of a predator removal/disease epidemiology study in Hyde and Wilson Counties. Samples were transported in RNAlater (Ambion, Inc.) from either the University of Tasmania or Hyde and Wilson Counties, NC, to Duke University, where they were maintained at -80°C prior to nucleic acid extraction.
Opossum IGF2 Genomic Sequence Identification
Genomic DNA was isolated from adult opossum liver, using the Qiagen Genomic Tip protocol 100/G (QIAGEN Sciences, Inc.). To complete the sequencing of o_IGF2_, Genome Walker (Clontech) libraries were constructed as recommended by the manufacturer. These libraries consist of multiple independent restriction-enzyme-digested opossum genomic DNA pools that are ligated to adapters and subjected to PCR amplification using adapter-specific and gene-specific primers, followed by nucleotide sequence analysis. Amplification of o_IGF2_ gene fragments from these libraries was performed with nested PCR in the Expand Long Template PCR System (Roche), using buffer 3. IGF2_-specific primers (based on the opossum IGF2 cDNA sequence) were used in conjunction with the nested AP1 and AP2 primers provided with the Genome Walker Kit (Clontech). The primary PCR reaction mixture was diluted 10-fold and used as a template for the nested reaction. PCR products were resolved on agarose gels. The amplicons were recovered using GenElute spin columns (Sigma), and sequenced directly with an automated ABI 377 sequencer (PE Systems) at the Duke University DNA Sequencing Facility. From the sequence obtained, additional pairs of gene-specific primers were designed and used for further walking (primer sequences are available from the authors on request). The o_IGF2 (accession no. AY552325) gene sequence was assembled using GeneJockey (Biosoft), and the gene organization was confirmed by PCR amplification of genomic DNA from several individual opossums.
Opossum IGF2 cDNA Sequence Identification
Total RNA was isolated from opossum pouch young brain, liver, and kidney and from adult liver tissues by homogenization in RNA-Stat 60 (Tel-Test), and subsequent processing was performed as recommended by the manufacturer. First-strand cDNA was oligo(dT) primed and synthesized from DNase I-treated RNA using Superscript II as recommended by the manufacturer (Invitrogen). Nondegenerate cross-species IGF2 primers were used to amplify a conserved region of IGF2 from cDNA in subsequent combinations (forward primer, CS-_IGF2_F1, 5′-CGGCGGGGA GCTGGTGGACAC; or forward primer, CS-_IGF2_F2, 5′-TGGGGAC CGCGGCTTCTACTTCAG; and the reverse primers, CS-_IGF2_R1, 5′-GACTTGGCGGGGGTGGCACAG; or reverse primer, CS-IGF2_R2, 5′-GGGGTGGCACAGTACGTCTCC AG) using 1.5 U of Platinum Taq DNA polymerase (Invitrogen), 15 pmoles of primers, 1.5 mM MgCl2, and 10 mM dNTPs in a 30-μL PCR reaction volume (15 sec at 94°C, 5 sec at 55°C, and 45 sec at 72°C for 30-35 cycles). RT-PCR products were separated by electrophoresis on a 2.0% agarose gel, and appropriately sized fragments were excised and gel-extracted (GenElute; Sigma Chemical Co.). Upon nucleotide sequencing to confirm product identity (ABI 377 sequencer; PE Biosystems), the complete o_IGF2 sequence was determined with gene-specific primers using Rapid Amplification of cDNA Ends (RACE) as described by the manufacturer (Invitrogen).
Platypus _IGF2_-Containing BAC Identification and Sequencing
Our 5× coverage platypus genomic BAC library (Munday Platypus BAC Library) was generated by Amplicon Express using total DNA isolated from a platypus kidney in Qiagen buffer ATL and proteinase K (QIAGEN Sciences, Inc.) followed by phenol:chloroform:isoamyl alcohol extraction and ethanol precipitation. The genomic BAC library consists of 264 384-well plates for a total of 101,376 clones. Because the average insert size in the clones is 142 kb, this library provides ∼5× coverage of the platypus genome.
Owing to the high degree of homology between the mature peptides of known INS, IGF1, and IGF2 genes, the probe generated for hybridization of the platypus BAC genomic library was from the p_IGF2_ E-domain (Killian et al. 2001). A 379-bp region of the platypus IGF2 E-domain was successfully amplified from DNA with forward primer Probe1, 5′-CAAAAGCCATCCAGCA CAAAGTTC, and reverse primer Probe2, 5′-GGTAGAGGTCTGT GCCCACC, using 1.5 U of Platinum Taq DNA polymerase (Invitrogen), 15 pmoles of primers, 1.5 mM MgCl2, 100 μM dNTPs, and 1 M Betaine (Sigma Chemical Co.) in a 30-μL PCR reaction volume (30 sec at 94°C, 30 sec at 69°C, and 30 sec at 72°C for nine cycles, -1° per cycle, followed by 30 sec at 94°C, 30 sec at 59°C, and 30 sec at 72°C for 24 cycles). PCR products were resolved by electrophoresis on a 1.0% agarose gel, and the appropriately sized fragment was excised and gel-extracted (GenElute; Sigma Chemical Co.). Then 30 ng of this product was sequenced (ABI 377 sequencer; PE Biosystems) to confirm its identity, and 25 ng was subsequently labeled with the RTS RadPrime DNA Labeling System according to the manufacturer's instructions (Invitrogen).
The Munday Platypus BAC Library was screened according to a protocol adapted from Molecular Cloning: A Laboratory Manual. Each membrane was then exposed to autoradiographic film (Kodak X-OMAT MR) for 2-6 d. Positive clones were identified and cultured, and DNA was isolated using Gerard Transgene 500 mL Sequencing Grade Prep (Gerard Biotech) for subsequent screening by PCR (as above).
DNA from a single BAC containing a 100-kb insert (Field-Inversion Gel Electrophoresis; BioRad) was isolated by alkalinelysis/phenol:cholorform extraction, ethanol precipitation, and BAC DNA was mechanically fragmented to generate random fragments. These fragments were subcloned into appropriate library vectors and sequenced (ABI 3700 sequencer; PE Biosystems). Sequences were analyzed and assembled with the Pare/Phrased sequence package (Ewing and Green 1998; Ewing et al. 1998) and Consed (Gordon et al. 1998). Gap filling was accomplished using primer pairs specific to each end of the assembled contigs in various combinations using standard PCR amplification. Amplicons produced from these reactions were sequenced, and overlaps were joined with the contigs to complete assembly (primer sequences available upon request).
Platypus cDNA Sequence Identification
Total RNA was isolated from adult platypus kidney, lung, and liver tissues by homogenization in RNA-Stat 60 (Tel-Test), and subsequent processing was performed as recommended by the manufacturer. Platypus 5′-IGF2 sequence was obtained using the SMART RACE cDNA Amplification Kit, as described by the manufacturer (BD Biosciences). RACE products were sequenced with an ABI 377 sequencer (PE Biosystems), and compared with the platypus genomic BAC sequence (accession no. AY552324) using CONSED (Gordon et al. 1998) and GeneJockey (Biosoft) to identify coding exons and transcribed regions at the 3′- and 5′-ends of IGF2. The p_IGF2_ ATG start site was predicted by GrailEXP (http://compbio.ornl.gov/grailexp/), and is contiguous with the open reading frame of the coding exon sequences obtained through 5′-RACE experiments. This putative start codon meets criteria for a Kozak consensus sequence. The platypus IGF2 cDNA sequence has been deposited in GenBank (accession no. AY552324).
Global Comparison of IGF2 Sequences
The multiVISTA program (http://www-gsd.lbl.gov/vista/) was used to compare the IGF2 sequences of human (accession no. AF517226; used as the reference sequence), mouse (accession no. U71085), opossum (accession no. AY552325), and platypus (accession no. AY552324), with a criteria of 50% identity and a 50-bp window. Translational alignments of IGF2 sequences were performed with CLUSTALW (http://www.ebi.ac.uk/clustalw/). CpG islands were identified using WebGene (http://l25.itba.mi.cnr.it/cgi-bin/wwwcpg.pl) with a window length of 120 bp under default parameters. RepeatMasker (http:ftp.genome.washington.edu/cgi-bin/RepeatMasker) was used to identify simple and complex repeats. Potential matrix attachment regions were identified with MAR-Wiz software (http://www.futuresoft.org), with a window size of 300 bp stepped at 50-bp intervals. MEME and MAST programs (http://meme.sdsc.edu/meme/website/meme-download.html) were used for common motif search and consensus sequence identification. Consensus sequence searches were also performed using Gene Jockey (Biosoft) with a minimum 63% identity as criterion for a match (Wang et al. 2004).
Methylation Analysis
Sodium bisulfite modification of both opossum and platypus kidney and liver DNA was performed as previously described (Waterland and Jirtle 2003). Regions of interest were then amplified in nested PCR from platypus or opossum bisulfite-treated DNA using 1.5 U of Platinum Taq DNA polymerase (Invitrogen), 15 pmoles of primers, 1.5 mM MgCl2, 100 μM dNTPs, and 1 M Betaine (Sigma Chemical Co.) in a 30-μL PCR reaction volume (40 cycles; primer sequences available upon request). PCR products were resolved by electrophoresis on a 1.0% agarose gel, excised, and gel-extracted (GenElute; Sigma Chemical Co.). Amplicons were subcloned into the pGEMT-easy vector, transformed, and plated according to the manufacturer's instructions (Promega). DNA from single colony forming units was amplified by whole-cell PCR using standard T7 and SP6 promoter primers and sequenced manually (Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit; USB Corporation).
Acknowledgments
This work was supported by NIH grants CA25951, ES013053, and ES08823 and an Enterprise Ireland grant IC/2003/20 to C.M.N. The platypus genomic BAC library used in this study has been named the Munday Platypus BAC Library in memory of our collaborator, Dr. Barry L. Munday, who died in 2003.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[The sequence data from this study have been submitted to Entrez/NCBI under accession nos. AY552325 (opossum IGF2) and AY552325 (platypus IGF2). The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: M. Stoskopf and B. Munday.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2774804.
References
- Amarger, V., Nguyen, M., Van Laere, A.S., Braunschweig, M., Nezer, C., Georges, M., and Andersson, L. 2002. Comparative sequence analysis of the INS-IGF2-H19 gene cluster in pigs. Mamm. Genome 13**:** 388-398. [DOI] [PubMed] [Google Scholar]
- Arney, K.L. 2003. H19 and Igf2—Enhancing the confusion? Trends Genet. 19**:** 17-23. [DOI] [PubMed] [Google Scholar]
- Arnold, R., Maueler, W., Bassili, G., Lutz, M., Burke, L., Epplen, T.J., and Renkawitz, R. 2000. The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA-DRB1(*)0401 gene. Gene 253**:** 209-214. [DOI] [PubMed] [Google Scholar]
- Awad, T.A., Bigler, J., Ulmer, J.E., Hu, Y.J., Moore, J.M., Lutz, M., Neiman, P.E., Collins, S.J., Renkawitz, R., Lobanenkov, V.V., et al. 1999. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J. Biol. Chem. 274**:** 27092-27098. [DOI] [PubMed] [Google Scholar]
- Bell, A.C. and Felsenfeld, G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405**:** 482-485. [DOI] [PubMed] [Google Scholar]
- Bell, A.C., West, A.G., and Felsenfeld, G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98**:** 387-396. [DOI] [PubMed] [Google Scholar]
- Burns, J.L., Jackson, D.A., and Hassan, A.B. 2001. A view through the clouds of imprinting. FASEB J. 15**:** 1694-1703. [DOI] [PubMed] [Google Scholar]
- Catchpoole, D., Smallwood, A.V., Joyce, J.A., Murrell, A., Lam, W., Tang, T., Munroe, D., Reik, W., Schofield, P.N., and Maher, E.R. 2000. Mutation analysis of H19 and NAP1L4 (hNAP2) candidate genes and IGF2 DMR2 in Beckwith-Wiedemann syndrome. J. Med. Genet. 37**:** 212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia, M., Dean, W., Lopes, S., Moore, T., Kelsey, G., and Reik, W. 2000. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 26**:** 203-206. [DOI] [PubMed] [Google Scholar]
- Cruz-Correa, M., Cui, H., Giardiello, F.M., Powe, N.R., Hylind, L., Robinson, A., Hutcheon, D.F., Kafonek, D.R., Brandenburg, S., Wu, Y., et al. 2004. Loss of imprinting of insulin growth factor II gene: A potential heritable biomarker for colon neoplasia predisposition. Gastroenterology 126**:** 964-970. [DOI] [PubMed] [Google Scholar]
- DeChiara, T.M., Robertson, E.J., and Efstratiadis, A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64**:** 849-859. [DOI] [PubMed] [Google Scholar]
- Du, M., Beatty, L.G., Zhou, W., Lew, J., Schoenherr, C., Weksberg, R., and Sadowski, P.D. 2003. Insulator and silencer sequences in the imprinted region of human chromosome 11p15.5. Hum. Mol. Genet. 12**:** 1927-1939. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8**:** 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8**:** 175-185. [DOI] [PubMed] [Google Scholar]
- Falls, J.G., Pulford, D.J., Wylie, A.A., and Jirtle, R.L. 1999. Genomic imprinting: Implications for human disease. Am. J. Pathol. 154**:** 635-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil, R., Walter, J., Allen, N.D., and Reik, W. 1994. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 120**:** 2933-2943. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P. and Tycko, B. 2004. The history of cancer epigenetics. Nat. Rev. Cancer 4**:** 143-153. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8**:** 195-202. [DOI] [PubMed] [Google Scholar]
- Gosden, R., Trasler, J., Lucifero, D., and Faddy, M. 2003. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet 361**:** 1975-1977. [DOI] [PubMed] [Google Scholar]
- Greally, J.M. 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. 99**:** 327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally, J.M., Guinness, M.E., McGrath, J., and Zemel, S. 1997. Matrix-attachment regions in the mouse chromosome 7F imprinted domain. Mamm. Genome 8**:** 805-810. [DOI] [PubMed] [Google Scholar]
- Greally, J.M., Gray, T.A., Gabriel, J.M., Song, L., Zemel, S., and Nicholls, R.D. 1999. Conserved characteristics of heterochromatin-forming DNA at the 15q11-q13 imprinting center. Proc. Natl. Acad. Sci. 96**:** 14430-14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig, D. and Graham, C. 1991. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell 64**:** 1045-1046. [DOI] [PubMed] [Google Scholar]
- Hark, A.T., Schoenherr, C.J., Katz, D.J., Ingram, R.S., Levorse, J.M., and Tilghman, S.M. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405**:** 486-489. [DOI] [PubMed] [Google Scholar]
- Hikichi, T., Kohda, T., Kaneko-Ishino, T., and Ishino, F. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 31**:** 1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri, C., Pant, V., Loukinov, D., Pugacheva, E., Qi, C.F., Wolffe, A., Ohlsson, R., and Lobanenkov, V.V. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10**:** 853-856. [DOI] [PubMed] [Google Scholar]
- Killian, J.K., Byrd, J.C., Jirtle, J.V., Munday, B.L., Stoskopf, M.K., MacDonald, R.G., and Jirtle, R.L. 2000. M6P/IGF2R imprinting evolution in mammals. Mol. Cell 5**:** 707-716. [DOI] [PubMed] [Google Scholar]
- Killian, J.K., Nolan, C.M., Stewart, N., Munday, B.L., Andersen, N.A., Nicol, S., and Jirtle, R.L. 2001. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J. Exp. Zool. 291**:** 205-212. [DOI] [PubMed] [Google Scholar]
- Lobanenkov, V.V., Nicolas, R.H., Adler, V.V., Paterson, H., Klenova, E.M., Polotskaja, A.V., and Goodwin, G.H. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5**:** 1743-1753. [PubMed] [Google Scholar]
- Lopes, S., Lewis, A., Hajkova, P., Dean, W., Oswald, J., Forne, T., Murrell, A., Constancia, M., Bartolomei, M., Walter, J., et al. 2003. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 12**:** 295-305. [DOI] [PubMed] [Google Scholar]
- Mannens, M., Hoovers, J.M., Redeker, E., Verjaal, M., Feinberg, A.P., Little, P., Boavida, M., Coad, N., Steenman, M., Bliek, J., et al. 1994. Parental imprinting of human chromosome region 11p15.3-pter involved in the Beckwith-Wiedemann syndrome and various human neoplasia. Eur. J. Hum. Genet. 2**:** 3-23. [DOI] [PubMed] [Google Scholar]
- Murphy, S.K. and Jirtle, R.L. 2003. Imprinting evolution and the price of silence. BioEssays 25**:** 577-588. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Eizirik, E., Johnson, W.E., Zhang, Y.P., Ryder, O.A., and O'Brien, S.J. 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409**:** 614-618. [DOI] [PubMed] [Google Scholar]
- Murrell, A., Heeson, S., Bowden, L., Constancia, M., Dean, W., Kelsey, G., and Reik, W. 2001. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2**:** 1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemitz, E.L. and Feinberg, A.P. 2004. Epigenetics and assisted reproductive technology: A call for investigation. Am. J. Hum. Genet. 74**:** 599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, H., Ono, Y., Shimozawa, N., Sotomaru, Y., Katsuzawa, Y., Hiura, H., Ito, M., and Kono, T. 2003. Disruption of imprinting in cloned mouse fetuses from embryonic stem cells. Reproduction 126**:** 549-557. [DOI] [PubMed] [Google Scholar]
- Ogura, A., Inoue, K., Ogonuki, N., Lee, J., Kohda, T., and Ishino, F. 2002. Phenotypic effects of somatic cell cloning in the mouse. Cloning Stem Cells 4**:** 397-405. [DOI] [PubMed] [Google Scholar]
- O'Neill, M.J., Ingram, R.S., Vrana, P.B., and Tilghman, S.M. 2000. Allelic expression of IGF2 in marsupials and birds. Dev. Genes Evol. 210**:** 18-20. [DOI] [PubMed] [Google Scholar]
- Onyango, P., Miller, W., Lehoczky, J., Leung, C.T., Birren, B., Wheelan, S., Dewar, K., and Feinberg, A.P. 2000. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 10**:** 1697-1710. [DOI] [PubMed] [Google Scholar]
- Otte, K., Choudhury, D., Charalambous, M., Engstrom, W., and Rozell, B. 1998. A conserved structural element in horse and mouse IGF2 genes binds a methylation sensitive factor. Nucleic Acids Res. 26**:** 1605-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, M., Davies, K.R., Bowden, L.M., Villar, A.J., Franck, O., Fuermann, M., Dean, W.L., Moore, T.F., Rodrigues, N., Davies, K.E., et al. 1998. Syntenic organization of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5. Hum. Mol. Genet. 7**:** 1149-1159. [DOI] [PubMed] [Google Scholar]
- Paulsen, M., Takada, S., Youngson, N.A., Benchaib, M., Charlier, C., Segers, K., Georges, M., and Ferguson-Smith, A.C. 2001. Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res. 11**:** 2085-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke, W.W., Taheny, M.J., Fochtmann, L.J., and Vostrov, A.A. 2000. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 28**:** 3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier, S., Johnson, L.A., Dobry, C.J., Ping, A.J., Grundy, P.E., and Feinberg, A.P. 1993. Relaxation of imprinted genes in human cancer. Nature 362**:** 747-749. [DOI] [PubMed] [Google Scholar]
- Rainier, S., Dobry, C.J., and Feinberg, A.P. 1995. Loss of imprinting in hepatoblastoma. Cancer Res. 55**:** 1836-1838. [PubMed] [Google Scholar]
- Reik, W. and Walter, J. 2001. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2**:** 21-32. [DOI] [PubMed] [Google Scholar]
- Reik, W., Brown, K.W., Schneid, H., Le Bouc, Y., Bickmore, W., and Maher, E.R. 1995. Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum. Mol. Genet. 4**:** 2379-2385. [DOI] [PubMed] [Google Scholar]
- Rideout III, W.M., Eggan, K., and Jaenisch, R. 2001. Nuclear cloning and epigenetic reprogramming of the genome. Science 293**:** 1093-1098. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3rd ed., pp. 6050-6059. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Takada, S., Paulsen, M., Tevendale, M., Tsai, C.E., Kelsey, G., Cattanach, B.M., and Ferguson-Smith, A.C. 2002. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: Implications for imprinting control from comparison with Igf2-H19. Hum. Mol. Genet. 11**:** 77-86. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Fan, H., Yang, H.H., Hu, Y., Buetow, K.H., and Lee, M.P. 2004. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics 83**:** 395-401. [DOI] [PubMed] [Google Scholar]
- Waterland, R.A. and Jirtle, R.L. 2003. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23**:** 5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M., Hagege, H., Murrell, A., Brunel, C., Reik, W., Cathala, G., and Forne, T. 2003. Genomic imprinting controls matrix attachment regions in the Igf2 gene. Mol. Cell. Biol. 23**:** 8953-8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie, A.A., Murphy, S.K., Orton, T.C., and Jirtle, R.L. 2000. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 10**:** 1711-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://compbio.ornl.gov/grailexp/; GrailEXP.
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.
- http://l25.itba.mi.cnr.it/cgi-bin/wwwcpg.pl; WebGene.
- http://meme.sdsc.edu/meme/website/meme-download.html; MEME and MAST programs.
- http://www.ebi.ac.uk/clustalw/; CLUSTALW.
- http://www.futuresoft.org/MAR-Wiz; MAR-Wiz.
- http://www-gsd.lbl.gov/vista/; multiVISTA.