Mapping the Golgi Targeting and Retention Signal of Bunyamwera Virus Glycoproteins (original) (raw)
Abstract
The membrane glycoproteins (Gn and Gc) of Bunyamwera virus (BUN; family Bunyaviridae) accumulate in the Golgi complex, where virion maturation occurs. The Golgi targeting and retention signal has previously been shown to reside within the Gn protein. A series of truncated Gn and glycoprotein precursor cDNAs were constructed by progressively deleting the coding region of the transmembrane domain (TMD) and the cytoplasmic tail. We also constructed chimeric proteins of BUN Gc, enhanced green fluorescent protein (EGFP), and human respiratory syncytial virus (HRSV) fusion (F) protein that contain the Gn TMD with various lengths of its adjacent cytoplasmic tails. The subcellular localization of mutated BUN glycoproteins and chimeric proteins was investigated by double-staining immunofluorescence with antibodies against BUN glycoproteins or the HRSV F protein and with antibodies specific for the Golgi complex. The results revealed that Gn and all truncated Gn proteins that contained the intact TMD (residues 206 to 224) were able to translocate to the Golgi complex and also rescued the Gc protein, which is retained in the endoplasmic reticulum when expressed alone, to this organelle. The rescued Gc proteins acquired endo-β-_N_-acetylglucosaminidase H resistance. The Gn TMD could also target chimeric EGFP to the Golgi and retain the F protein, which is characteristically expressed on the surface of HRSV-infected cells, in the Golgi. However, chimeric BUN Gc did not translocate to the Golgi, suggesting that an interaction with Gn is involved in Golgi retention of the Gc protein. Collectively, these data demonstrate that the Golgi targeting and retention signal of BUN glycoproteins resides in the TMD of the Gn protein.
Enveloped viruses acquire their lipid bilayers by budding through a cellular membrane into which the viral glycoproteins have been inserted (reviewed in reference 10). Whereas most enveloped viruses are assembled at the plasma membrane, a few viruses mature at intracellular membranes (31). Viruses of the Bunyaviridae family are characterized by maturation and budding at the Golgi complex (17, 18, 27) due to the accumulation of the two viral glycoproteins, Gn and Gc, in this organelle (31, 32).
The Bunyaviridae family contains more than 300 viruses, classified into five genera, which share biochemical and morphological characteristics. All of the viruses have a tripartite negative-sense RNA genome that encodes four structural proteins. The largest segment (L) codes for an RNA-dependent RNA polymerase (L protein), the medium segment (M) codes for a precursor containing the two glycoproteins (Gn and Gc), and the smallest segment codes for the nucleoprotein (N). Nonstructural proteins are encoded by some viruses on the M (called NSm) and S (called NSs) segments. The pattern of sizes of the genomic RNAs and of the structural proteins varies between the different genera. Bunyamwera virus (BUN) is the prototype of both the Bunyaviridae family and the Orthobunyavirus genus (6-8).
For most viruses in the family that have been studied to date, the glycoprotein located at the amino terminus of the precursor, Gn, contains the Golgi retention and targeting signal (32). When BUN Gn is expressed on its own, it localizes to this organelle; in contrast, the glycoprotein located at the carboxy terminus of the precursor, Gc, remains in the endoplasmic reticulum (ER) when it is expressed alone and is only transported to the Golgi after an interaction with Gn (19). Similar findings were reported for the glycoproteins of the plant-infecting Tomato spotted wilt virus (Tospovirus genus) when they were expressed in BHK cells (15). Golgi retention of Uukuniemi virus (Phlebovirus genus) glycoproteins is mediated by a short region in the cytoplasmic tail of the Gn glycoprotein (1, 2), but for two other phleboviruses, Punta Tora fever virus and Rift Valley fever virus, the Golgi retention signal has been mapped to a region encompassing the transmembrane domain (TMD) and part of the adjacent cytoplasmic tail (11, 23, 24). However, for members of the Hantavirus genus, including the Old World Hantaan virus and the New World Sin Nombre virus, Golgi localization requires heterodimerization of both the Gn and Gc proteins (34, 37, 38). So far, no consensus sequence for retention in the Golgi complex has been identified in the glycoproteins of viruses in the family, and it appears that different viruses, even within a single genus, have their own strategies for Golgi targeting.
For this report, we have mapped the Golgi retention signal for BUN glycoproteins to the TMD of the Gn protein. We also investigated the role of the retention signal in rescuing Gc to the Golgi complex.
MATERIALS AND METHODS
Cells and viruses.
HeLa T4+ cells (22) and Vero E6 (ATCC C1008) cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. A recombinant vaccinia virus, vTF7-3, which synthesizes bacteriophage T7 RNA polymerase (9), was a gift from Bernard Moss (National Institutes of Health, Bethesda, Md.).
Antibodies.
A rabbit antiserum against purified BUN virions (anti-BUN) and the Gc-specific mouse monoclonal antibody (MAb) 742 have been described previously (19, 42). MAb 19, specific for the fusion protein (F) of human respiratory syncytial virus (HRSV) (39), was provided by Richard J. Sugrue (this department) and was originally a gift from Geraldine Taylor (Institute of Animal Health, Compton, United Kingdom). A rabbit polyclonal antibody against GM130, a _cis_-Golgi matrix protein (28), was provided by Martin Lowe (School of Biological Sciences, University of Manchester, Manchester, United Kingdom). A MAb against human golgin 97 (12) was purchased from Molecular Probes Inc. (Leiden, The Netherlands**)**. A goat anti-rabbit antibody conjugated with fluorescein isothiocyanate was purchased from Sigma, and a goat anti-mouse antibody conjugated with Cy5 was purchased from Amersham Pharmacia Biotech (Buckingham, United Kingdom).
Construction of recombinant cDNA.
The complete open reading frame of the BUN M segment cDNA was removed from pTF7-5BUNM (29) and cloned into pTM1 (25) under the control of the bacteriophage T7 promoter and the encephalomyocarditis virus internal ribosome entry site (IRES) (F. Weber and R. M. Elliott, unpublished data) to give pTM-BUNM. Truncated versions of the BUN Gn cDNA were derived from pTF7-5G2 (19) by digestion with the BAL-31 exonuclease (Fig. 1A). The BAL-31-treated DNA was made blunt ended and then ligated to a synthetic linker, Xba-STOP (5′CTAGTCTAGACTAG3′), which contains translational stop codons in all three reading frames (underlined) and an internal XbaI site. BUN Gc and BUN M cDNA mutants with internal in-frame deletions within the Gn and NSm coding regions were made by site-directed PCR mutagenesis with Turbo-Pfu DNA polymerase (Stratagene) (see Fig. 3A). Briefly, primer pairs in outward orientations were annealed to pTM-BUNM, and the amplified DNA was purified, digested with DpnI, and recircularized with T4 DNA ligase. All junctions across the deleted regions were confirmed by sequencing. The primers used and details of PCRs are available upon request.
FIG. 1.
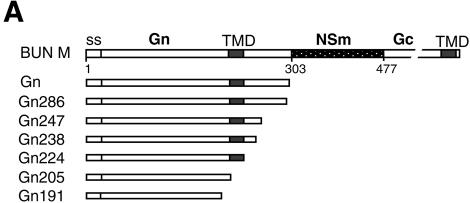
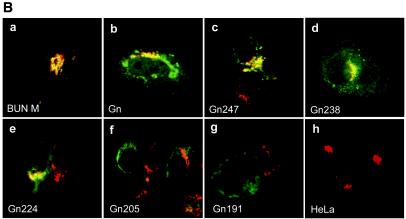
Intracellular localization of BUN Gn mutants. (A) Diagram of BUN glycoprotein precursor (BUN M) and Gn mutants. Amino acid positions are indicated beneath the top bar. ss, signal sequence. Gn mutants are designated according to the numbers of residues that they contain. (B) HeLa T4+ cells were infected with vTF7-3, followed by transfection with BUN Gn mutant cDNAs as indicated. The cells were then doubly stained with a mixture of anti-BUN antiserum and a MAb specific for the Golgi marker golgin 97. Merged confocal microscopic images are shown, with the Gn glycoprotein stained green and the Golgi stained red.
FIG. 3.
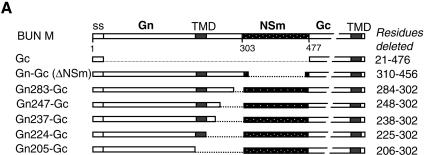
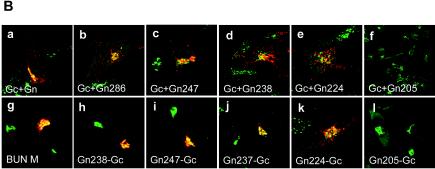
Rescue of BUN Gc from ER to the Golgi complex by truncated Gn proteins. (A) Diagram of BUN M cDNA constructs containing internal deletions. The amino acid residues deleted from the precursor cDNA are listed on the right. (B) Confocal images of transfected cells. HeLa T4+ cells were transfected with either separate Gc and mutant Gn cDNAs (a to f) or M segment cDNAs with internal deletions (g to l). The cells were doubly stained with a mixture of anti-Gc MAb 742 and anti-GM130 antibodies. Merged confocal microscopic images are shown, with the Gc glycoprotein stained red and the Golgi stained green.
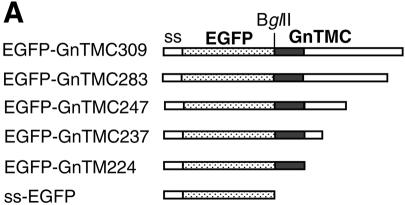
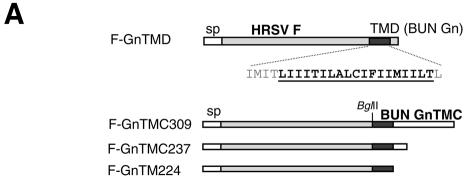
Twelve plasmids expressing chimeric proteins were constructed by either PCR or DNA cloning techniques. For the construction of plasmids expressing chimeric BUN Gc proteins that contain the TMD and cytoplasmic tail of the Gn protein (GnTMC), a SacI restriction enzyme site was introduced just upstream of the Gc TMD (at nucleotides 4218 to 4223, coding for glutamine and leucine). PCR-amplified DNAs encoding the TMD with various lengths of cytoplasmic tail from the Gn protein were ligated at this site without changing the amino acid sequences of either protein (see Fig. 5A). For the construction of plasmids expressing chimeric enhanced green fluorescent protein (EGFP), PCR-amplified DNAs encompassing the coding region for the TMD and different lengths of the cytoplasmic tail of the Gn protein were cloned downstream of the EGFP coding region in plasmid pEGFP-C3 (Clontech Laboratories), between BglII and SmaI sites (see Fig. 6A). The plasmid was further modified by inserting the signal sequence for the Hantaan virus Gn protein (coding for amino acids 1 to 18) (36) upstream of the EGFP coding region. pTM-F, containing the coding region of the full-length HRSV F protein, was provided by Ping Li (this department) and was used as a parental vector for the construction of plasmids expressing chimeric F proteins that contain the TMD of the BUN Gn protein (see Fig. 7A). In F-GnTM, the inner 19 residues of the F protein TMD (residues 529 to 547) were replaced with 19 residues of the BUN Gn TMD. For construction of the other three chimeric F proteins (F-GnTMC309, -TMC237, and -TM224), a BglII restriction enzyme site was introduced just upstream of the F TMD, and the cytoplasmic tail coding region was replaced with that of the Gn protein. The TMDs for BUN glycoproteins and the HRSV F protein were predicted with the Web-based EMBnet program Tmpred (www.ch.embnet.org). This algorithm is based on the statistical analysis of Tmbase, a database of naturally occurring transmembrane proteins, and the prediction is made by use of a combination of several weight matrices for scoring (13). The membrane-spanning regions were defined as residues 206 to 224 (19 residues) for BUN Gn and residues 525 to 548 (24 residues) for the HRSV F protein.
FIG. 5.
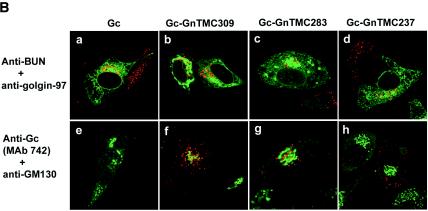
Intracellular localization of chimeric Gc containing the Gn TMD and cytoplasmic tail. (A) Diagram of chimeric Gc constructs containing the Gn TMD and parts of the cytoplasmic tail (Gn-TMC). (B) HeLa T4+ cells were infected with vTF7-3, followed by transfection with full-length Gc (a and e)- or chimeric Gc (b to d and f to h)-expressing plasmids. The cells in panels f to h were cotransfected with a Gn-expressing plasmid. The cells were stained with anti-BUN and anti-golgin 97 antibodies (a to d) or with anti-Gc MAb 742 and anti-GM130 antibodies (e to h) and were examined by confocal microscopy. Merged images are shown in which BUN Gc is stained green and the Golgi is stained red in panels a to d, whereas in panels e to h the Golgi is stained green and Gc is stained red.
FIG. 6.
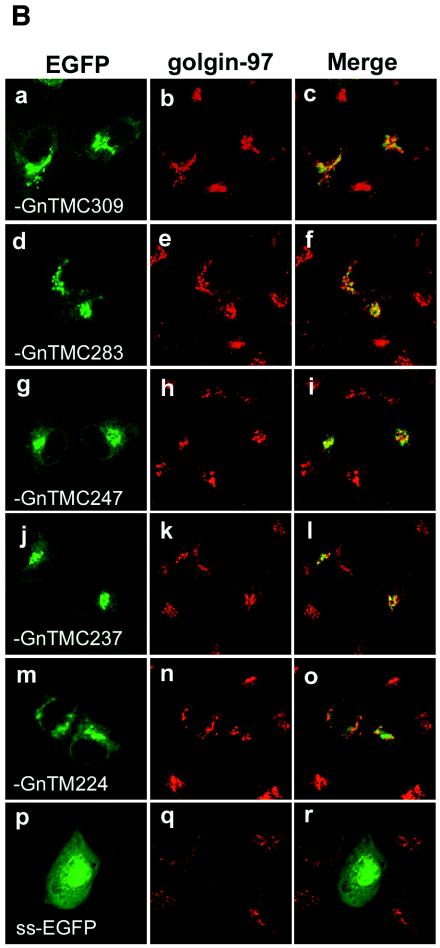
Intracellular localization of chimeric EGFP containing BUN Gn TMD and cytoplasmic tail. (A) Diagram of chimeric EGFP constructs containing the signal sequence (ss) of Hantaan virus Gn, the BUN Gn TMD, and different lengths of the cytoplasmic tail (Gn-TMC). EGFP-GnTM224 has no cytoplasmic tail at its C terminus. (B) Vero E6 cells were transfected with chimeric EGFP cDNAs as indicated and then incubated for 24 h. The cells were stained with an anti-golgin 97 MAb and Cy5-conjugated anti-mouse IgG and then examined by confocal microscopy. The images show GFP fluorescence (green), golgin 97 staining (red), and merged images.
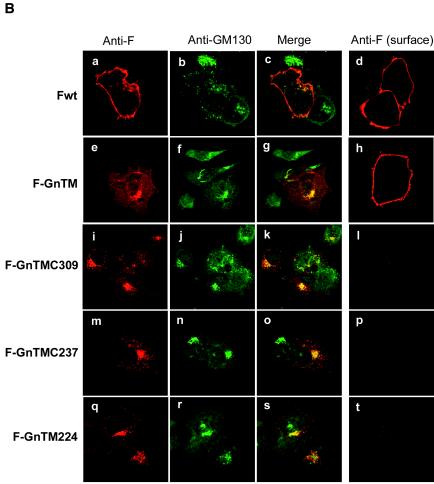
FIG. 7.
Intracellular and cell surface expression of parental HRSV F protein (Fwt) and chimeric F proteins containing the TMD and cytoplasmic tail of BUN Gn protein. (A) Diagram of chimeric HRSV F containing BUN Gn TMD. Gn TMD amino acids are underlined. The remaining original F TMD residues that flank the Gn TMD are shown in gray. (B) HeLa T4+ cells were infected with vTF7-3, followed by transfection with the wild-type F or chimeric F cDNAs. For the examination of cell surface expression, the cells were not permeabilized with Triton X-100 before incubation with antibodies. The cells were then stained with anti-F MAb 19 and anti-GM130 antibody for permeabilized samples or with only MAb 19 for nonpermeabilized samples (cell surface staining). The F protein and chimeric F proteins are stained red and GM130 is stained green.
Infection and transfection of cells.
Subconfluent monolayers of Vero E6 cells were grown in 35-mm-diameter petri dishes for immunoprecipitation experiments, and HeLa T4+ or Vero E6 cells were grown on 13-mm-diameter glass coverslips for immunofluorescence assays. The cells were infected with vTF7-3 at 5 PFU/cell for 60 min and then transfected with plasmid DNA as described previously (19). Non-vTF7-3-infected Vero E6 cells were used to express the chimeric EGFP gene, which was under the control of the cytomegalovirus promoter. Briefly, for cells grown in 35-mm-diameter dishes, 2 μg of DNA and 15 μl of cationic liposomes were diluted in 500 μl of OptiMEM (BRL-Life Technologies, Paisley, United Kingdom), and for cells grown on coverslips, 0.5 μg of plasmid DNA and 6 μl of liposomes were diluted in 250 μl of OptiMEM. The DNA-liposome mixtures were incubated for 10 min at room temperature before being added to cells that had been washed previously with OptiMEM. At 3 h posttransfection, 0.5 ml of Dulbecco's modified Eagle's medium containing 10% fetal bovine serum was added, and incubation was continued at 37°C.
Metabolic radiolabeling and immunoprecipitation.
At 7 h posttransfection, the cells were incubated for 1 h in starvation medium lacking methionine, washed, and then labeled with [35S]methionine (50 μCi/ml; Amersham Pharmacia Biotech) for 15 h. The cells were then lysed on ice with 300 μl of nondenaturing RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% Triton X-100, 300 mM NaCl, 5 mM EDTA) containing a cocktail of protease inhibitors (Roche). BUN glycoproteins were immunoprecipitated with an anti-BUN serum that had been conjugated to protein A-agarose (Sigma). The beads were washed five times with RIPA buffer containing 0.1% Triton X-100 and once with ice-cold phosphate-buffered saline (PBS), and the bound proteins were either analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) under reducing conditions or subjected to digestion with endo-β-_N_-acetylglucosaminidase (endo H) or _N_-glycosidase F (PNGase F).
Endo H and PNGase F treatment.
Immunoprecipitates were denatured in 30 μl of denaturing buffer (0.5% SDS and 1% β-mercaptoethanol) at 100°C for 10 min and then were cooled to room temperature. The denatured samples were then subjected to digestion with 150 mU of endo H (New England Biolabs) in 40 μl of reaction buffer (50 mM sodium citrate [pH 5.5], 0.5% SDS, 1% β-mercaptoethanol) or 4 mU of PNGase F (New England Biolabs) in 40 μl of reaction buffer (50 mM sodium phosphate [pH 7.5], 0.5% SDS, 1% β-mercaptoethanol) for 20 h at 37°C. The treated samples were analyzed by SDS-10% PAGE under reducing conditions.
Indirect immunofluorescence staining.
Immunofluorescence assays were performed as previously described (37). Briefly, at 5 h posttransfection of vTF7-3-infected HeLa T4+ cells or at 20 h posttransfection of noninfected Vero E6 cells, cycloheximide was added to the culture medium to a final concentration of 50 μg/ml and cells were incubated for a further 4 h. The cells were then fixed for 20 min with 4% paraformaldehyde and permeabilized by incubation in PBS containing 0.1% Triton X-100 for 30 min. The cells were incubated for 30 min with antibodies against BUN glycoproteins (anti-BUN serum or anti-Gc MAb 742) and Golgi markers (GM130 antiserum or anti-golgin 97 MAb). After being thoroughly washed with PBS, the cells were stained for 30 min with Cy5-conjugated anti-mouse immunoglobulin G (IgG) and fluorescein isothiocyanate-conjugated anti-rabbit IgG. The localization of fluorescently labeled proteins was examined under a Zeiss LSM confocal microscope.
RESULTS
Intracellular localization of truncated Gn proteins.
It was shown previously that the BUN glycoproteins Gn and Gc, as well as NSm, accumulate in the Golgi complex when they are expressed from a full-length M segment cDNA (29) and that when the Gn protein is expressed on its own, it localizes to this organelle (19), indicating that the Golgi retention signal resides in Gn. As illustrated in Fig. 1A, the complete BUN Gn coding region encompasses 302 amino acids, comprising the cleavable signal sequence (residues 1 to 17), the ectodomain (residues 18 to 205), the hydrophobic TMD (residues 206 to 224), and the cytoplasmic tail (residues 225 to 302) (20). To map the domain responsible for Golgi targeting and retention, we constructed a series of BUN Gn mutants by progressively deleting the coding region of the cytoplasmic tail and TMD from the 3′ end of pTF7-5G2 by BAL-31 digestion (Fig. 1A). The intracellular localization of the mutated Gn proteins was analyzed by indirect double-staining immunofluorescence with a rabbit anti-BUN serum that recognizes both Gn and Gc (42) and with a MAb against a Golgi marker, human golgin 97 (Fig. 1B). The results revealed that all Gn constructs expressing 224 or more amino acids, i.e., with an intact TMD, gave a predominately Golgi staining pattern for Gn that was confirmed by their colocalization with the Golgi marker (Fig. 1B, panels a to e). However, truncated Gn proteins containing 205 or fewer residues (Gn205 and Gn191) showed a more generalized cytoplasmic staining pattern (suggestive of an ER distribution), and no colocalization with golgin 97 was observed (Fig. 1B, panels f and g). In Gn205 and Gn191, both the TMD and cytoplasmic tail residues had been deleted. No fluorescent staining of the cell surface with the anti-BUN serum was observed for any of the mutated Gn proteins (data not shown). These results suggest that the Gn TMD is crucial for Golgi targeting and retention of the protein.
Rescue of BUN Gc from ER to Golgi complex by truncated Gn proteins.
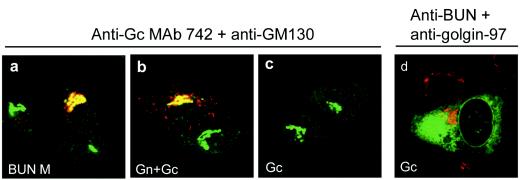
As reported previously (19), the BUN Gc protein was retained in the ER when expressed on its own (Fig. 2d), and Golgi targeting of Gc required the coexpression of Gn (Fig. 2a and b). We observed that individually expressed Gc did not react with the anti-Gc MAb 742 (Fig. 2c) but was recognized by the polyclonal anti-BUN rabbit serum, showing a typical ER staining pattern (Fig. 2d). MAb 742 only reacted with Gc that was expressed either from a full-length M segment cDNA (Fig. 2a) or after the coexpression of Gn and Gc from separate cDNAs (Fig. 2b). Under these conditions, Gc was transported to the Golgi complex and colocalized with the Golgi marker GM130 (Fig. 2a and b). These results suggest that the BUN Gc protein is in a different conformation when it is expressed alone and that it undergoes a conformational change when associated with Gn before exiting the ER or en route to the Golgi complex. Thus, it seems a feature of all viruses in the Bunyaviridae family that have been studied so far that the C-terminal glycoprotein requires the other viral glycoprotein (Gn) for trafficking to the Golgi complex (32).
FIG. 2.
Intracellular localization of coexpressed and separately expressed BUN Gc proteins. Gc proteins were coexpressed with Gn from either the whole M segment cDNA (a) or separate Gn and Gc cDNAs (b). (c and d) Gc expressed alone from pTM-BUNGc. The cells were doubly stained with the anti-Gc MAb 742 and an anti-GM130 serum (a to c) or with anti-BUN serum and an anti-golgin 97 antibody (d). Merged confocal microscopic images are shown. BUN glycoproteins are stained red and the Golgi is stained green in panels a to c, whereas Gc is stained green and the Golgi is stained red in panel d.
To investigate whether the Golgi targeting and retention domain of the Gn protein was also sufficient to rescue Gc to the Golgi complex, we observed the intracellular localization of Gc when it was coexpressed with individual truncated Gn constructs or from a single BUN M segment cDNA mutant containing internal deletions (Fig. 3A). As shown in Fig. 3B, the results for coexpressing Gc and truncated Gn from separate plasmids (Fig. 3B, panels a to f) were consistent with those when single BUN M cDNAs containing internal deletions were expressed (Fig. 3B, panels g to l). In both cases, the cells were stained with the conformation-sensitive anti-Gc MAb 742, and we noted that an appreciable amount of Gc protein was rescued to the Golgi complex by the truncated Gn proteins that contained 224 or more residues, i.e., they had an intact TMD (Fig. 3B, panels a to e and g to k). However, when the Gn TMD was deleted, as in the case of Gn constructs Gn205 and Gn205-Gc (panels f and l), Gc was not detected, suggesting that these Gc proteins were unable to undergo conformational maturation and were thus retained in the ER.
The results for the Golgi localization of Gn and Gc from immunofluorescence experiments were corroborated by an analysis of their resistance to endo H, which cleaves N-linked oligosaccharides in the high-mannose form but not those that have been converted, in the medial Golgi, to the complex form (16). The BUN glycoproteins contain three potential sites for the attachment of N-linked oligosaccharides, one site on Gn and two sites on Gc, and the electrophoretic mobility difference between glycosylated Gc and nonglycosylated Gc, for example, that synthesized in the presence of tunicamycin (20), is very small. However, members of our laboratory were able to show previously that the Gc protein expressed from a recombinant vaccinia virus containing the full-length BUN M cDNA acquired endo H resistance, although the expression levels of Gn were too weak to detect adequately (29).
Vero E6 cells were infected with vTF7-3, followed by transfection with the full-length M segment cDNA or constructs containing internal deletions. Radiolabeled proteins were immunoprecipitated, treated with endo H or PNGase F, and separated by SDS-PAGE, and the dried gel was exposed to a phosphorimager screen. Consistent with our previous results, nearly all of the Gc protein expressed from the full-length cDNA acquired endo H resistance, in contrast with Gc that was deglycosylated by PNGase F treatment (Fig. 4, lanes 1 and 2), whereas no endo H-resistant Gc was found when Gc was expressed alone (lane 15). A significant amount of Gc protein (between 38 and 48%, as estimated by phosphorimaging) became endo H resistant when it was expressed with truncated Gn proteins that contained more than 224 residues, i.e., with an intact TMD (Fig. 4, lanes 3 to 12). Notably, about half of the Gc expressed from the construct Gn-Gc, which contains the intact coding regions for Gn and Gc but has a deletion of the NSm coding region, became endo H sensitive (lane 3), suggesting that NSm is required for the efficient maturation of BUN glycoproteins. No endo H-resistant Gc was detected from expressed Gn205-Gc, in which both the TMD and the cytoplasmic tail were removed (lane 13). Although the truncated versions of Gn could be detected on the gel, it was not possible, as found previously, to determine electrophoretic mobility differences between glycosylated and deglycosylated forms.
FIG. 4.
Endo H resistance of BUN glycoproteins. Vero E6 cells were infected with vTF7-3 and subsequently transfected with the parental BUN M segment cDNA (pTM-BUNM) and mutants carrying internal deletions as indicated. The cells were radiolabeled with [35S]methionine, and radiolabeled proteins were immunoprecipitated with an anti-BUN antiserum. The resulting precipitates were subjected to digestion with endo H (H) or PNGase F (F) as shown and were analyzed by SDS-10% PAGE under reducing conditions. The truncated Gn proteins expressed from M cDNA mutants are marked with asterisks. The gel was exposed to a phosphorimager (Molecular Imager FX; Bio-Rad Laboratories), and the relative intensities of the two forms of Gc were estimated from a computer-enlarged image by the use of Quantity One (v. 4.2.2) software.
Replacement of Gc TMD with Gn TMD.
The above results show (i) that the truncated Gn proteins (either Gn224 or Gn224-Gc) that lacked the cytoplasmic tail were able to target themselves to the Golgi complex and rescue the Gc protein to this organelle, although less efficiently than full-length Gn, and (ii) that truncated Gn lacking both the TMD and the cytoplasmic tail failed to do so. This suggests that the Golgi targeting and retention signal resides in the Gn transmembrane region and is also required for Golgi targeting and retention of the Gc protein. To investigate the role of the Gn TMD and its cytoplasmic tail in the Golgi targeting and retention of Gc, we made three chimeric Gc proteins (Fig. 5A) in which the TMD and the short cytoplasmic tail of the Gc protein were replaced with either the complete TMD and cytoplasmic tail (Gc-GnTMC309) or the TMD and a truncated cytoplasmic tail of the Gn protein (Gc-GnTMC283 and -237). These proteins were either transfected alone into vTF7-3-infected HeLa T4+ cells or cotransfected with the plasmid expressing wild-type Gn (Fig. 5B); native Gc was expressed as a control. When stained with the anti-BUN polyclonal antibody, all Gc constructs showed an ER staining pattern and no localization to the Golgi was observed (Fig. 5B, panels a to d). None of the chimeric Gc proteins was recognized by the anti-Gc MAb 742 (data not shown). For cotransfection experiments, the cells were stained with the anti-Gc MAb 742; as noted previously, this antibody failed to recognize native Gc when it was expressed on its own (Fig. 5B, panel e). Interestingly, the chimeric Gc proteins were recognized by the antibody when they were coexpressed with native Gn, indicating a conformational change in Gc, but no Golgi localization was observed (panels f to h). In fact, a speckled cytoplasmic staining pattern was seen, perhaps indicative of aggregation of the immature protein.
Intracellular localization of chimeric EGFP-Gn TMC.
GFP is a useful marker for monitoring intracellular gene expression (5). To confirm that the Golgi target and retention signal mapped to the Gn TMD and to examine the possible role of the Gn cytoplasmic tail in Golgi trafficking, we constructed a panel of plasmids expressing EGFP chimeras by fusing the BUN Gn TMD with different lengths of cytoplasmic tail to the C terminus of EGFP (Fig. 6A). The signal sequence from the Hantaan virus Gn protein was placed at the N terminus of EGFP to allow entry into the secretory pathway. After transfection into Vero E6 cells, the cells were stained with an anti-golgin 97 antibody, specific for the Golgi marker, and were examined for the colocalization of EGFP fluorescence.
As shown in Fig. 6B, all of the chimeric EGFP proteins that contained the Gn TMD, with or without cytoplasmic tail residues, had a perinuclear distribution and colocalized with the Golgi marker (Fig. 6B, panels a to o), behaving like the analogous Gn constructs (Fig. 1). EGFP containing just the signal sequence was used as a control and showed diffuse fluorescence in both the cytoplasm and the nucleus (Fig. 6B, panels p and r). Chimeric EGFP proteins containing the intact Gn TMD plus some length of cytoplasmic tail (EGFP-GnTMC309, -GnTMC283, -GnTMC247, and -GnTMC237) showed more obvious colocalization with golgin 97 (Fig. 6B, panels c, f, i, and l) than did EGFP-GnTM224, which contained only the Gn TMD. However, even EGFP-GnTMC224 was able to target itself to the Golgi complex (Fig. 6B, panel o), indicating that the Gn TMD possesses all of the signals that are required for Golgi targeting and retention. Therefore, these results indicate that a signal for the Golgi trafficking of BUN glycoproteins is located in the transmembrane region, rather than the cytoplasmic tail or ectodomain, of the Gn protein.
Subcellular localization of chimeric HRSV fusion proteins containing BUN Gn TMD and cytoplasmic tail sequences.
To further study the role of the Gn TMD and its cytoplasmic tail in Golgi trafficking, we constructed plasmids expressing chimeric HRSV F proteins (Fig. 7A). The HRSV F protein is characterized by cell surface expression in infected cells, where it induces fusion between the plasma membranes of adjacent cells, leading to syncytium formation (40). The wild-type F protein showed typical cell surface expression in both permeabilized and nonpermeabilized HeLa T4+ cells (Fig. 7B, panels a, c, and d). Construct F-GnTM, in which only the inner 19 residues of the F TMD were replaced with the Gn TMD, showed evidence of both Golgi and cell surface staining (Fig. 7B, panels e, g, and h). We suggest that the presence of the Gn TMD resulted in delayed transport through, or some retention in, the Golgi of the chimeric F protein, although clearly it did not completely block passage to the plasma membrane.
However, when the complete F TMD and cytoplasmic tail were replaced with the TMD of the BUN Gn protein, with or without its cytoplasmic tail (Fig. 7A), not only was the chimeric F protein efficiently targeted to the Golgi region, but also no obvious cell surface staining was detected. These data are consistent with the results presented in Fig. 1 and 6 showing that the Golgi targeting and retention signal resides in the TMD of Gn. We also noted that the Golgi structures of the transfected cells appeared to be different: whereas the Golgi complex of HeLa T4+ cells expressing the chimeric F proteins appeared to be intact, that of cells expressing wild-type F appeared to be disrupted (Fig. 7B, panels b, f, j, n, and r).
DISCUSSION
Viruses of the Bunyaviridae family are characterized by intracellular maturation and budding at the Golgi complex, which is thought to be mediated by the retention and accumulation of the viral glycoproteins in this cellular compartment (32). Previous work has shown that the BUN Gn glycoprotein is required for the localization of the Gc protein to the Golgi complex (19). Similar results have also been obtained for viruses in other genera of the family whose glycoproteins are unrelated in sequence and have quite different sizes, indicating that the signal for Golgi targeting and retention is usually located in the protein (Gn) located at the N terminus of the glycoprotein precursor. Transport of the other viral glycoprotein (Gc) to the Golgi requires an association with the Gn protein (reviewed in reference 32).
In this report, we demonstrated that the TMD of BUN Gn harbors the signal for Golgi targeting and retention and may also be important for heterodimerization of the glycoproteins. This conclusion was drawn from the following observations. Firstly, a separately expressed truncated Gn (BUN Gn224) protein from which the complete cytoplasmic tail was deleted was transported and retained in the Golgi (Fig. 1B). Secondly, the same Gn224 or other truncated Gn proteins containing more than 224 residues were able to rescue Gc to the Golgi complex (Fig. 3). When in association with these truncated Gn proteins, a significant amount of the Gc proteins acquired endo H resistance. Thirdly, the TMD of BUN Gn was able to retain chimeric EGFP and HRSV F proteins in the Golgi complex (Fig. 6 and 7). Lastly, although chimeric Gc containing the Gn TMD and cytoplasmic tail changed its conformation when it was coexpressed with native Gn, it was not transported to the Golgi (Fig. 3). For all of these observations, Golgi localization of the BUN glycoproteins and the chimeric EGFP and HRSV F proteins was confirmed by their colocalization with the Golgi marker GM130 or golgin 97.
As assessed by the acquisition of endo H resistance, Golgi trafficking of Gc was impaired when NSm or the Gn cytoplasmic tail (part or whole) was not expressed. A recent study of the structural maturation of BUN showed that the Golgi complex, in particular the trans subcompartment, is necessary for maturation of the virus (35). This suggests that NSm and the Gn cytoplasmic tail, though unnecessary for Golgi targeting and retention, are required for efficient maturation and Golgi trafficking of the glycoproteins, especially for retaining glycoproteins in the medial and/or _trans_-Golgi, where the acquisition of endo H resistance occurs.
The mapping of targeting and retention signals has been done for several viruses in this family, but no consensus amino acid sequences or motifs have been delineated (1, 2, 11, 23, 24). The signal for Golgi targeting and retention was mapped either to the cytoplasmic tail of the Gn protein for Uukuniemi virus (1, 2) or to a region including the TMD and part of the cytoplasmic tail for the Punta Tora phlebovirus (24) and the Rift Valley fever phlebovirus (11). No specific Golgi targeting and retention signal has been found in hantavirus glycoproteins. In this case, Golgi trafficking of the glycoproteins requires the coexpression of both Gn and Gc; either protein is retained in the ER when it is expressed separately (37, 38). Here we have defined the Golgi retention signal of BUN glycoproteins as being located in the TDM of the Gn protein, as confirmed by the Golgi localization of the truncated Gn224 (Fig. 1B, panel e), chimeric EGFP-GnTMC224 (Fig. 6B, panel o), and HRSV F-GnTMC224 (Fig. 7B, panel s) proteins. All of these constructs contain only the intact Gn TMD and no cytoplasmic tail residues. Therefore, for BUN, the signal for Golgi targeting and retention resides in the TMD of Gn. It seems that different viruses in this diverse family have developed their own specific strategies for Golgi trafficking rather than displaying a consensus Golgi retention motif.
Like the native parental HRSV F protein, the chimeric F-GnTM protein, in which only the inner 19 residues within the TMD were replaced with the Gn TMD, was readily transported to the cell surface of the transfected cells, indicating that it retained the ability of being transported through the secretory pathway to the plasma membrane. This result suggests that there is more than one factor that contributes to Golgi targeting and retention of the BUN glycoproteins, such as folding or oligomerization. Indeed, this chimeric protein also showed a typical Golgi staining pattern, and the Golgi structure of the transfected cells remained intact, in contrast to that of wild-type F-transfected cells. In this case, the Golgi targeting motif, i.e., the internal 19 residues of the Gn TMD, seemed partially functional in retaining or delaying the cell surface trafficking of the chimeric F protein, but it was not sufficient to retain this larger and more complex protein completely in the Golgi.
Several mechanisms for the retention of membrane proteins in the Golgi complex have been suggested, including specific Golgi targeting and retention signals, membrane thickness in particular organelles (26, 30), and oligomerization of proteins within the lipid bilayer (21). In the case of the Golgi trafficking of BUN glycoproteins, it seems that more than one factor may be involved. The predicted TMD of the Gn protein contains 19 amino acids, and that of HRSV F has 24 residues. It is relevant to the lipid bilayer sorting model proposed by Bretscher and Munro that Golgi-localized proteins tend to have shorter TMDs than plasma membrane proteins (4, 26). The TMD length of chimeric F-GnTM was in fact unchanged, as we only replaced the inner 19 residues with the BUN Gn TMD (Fig. 7A). This could be why the chimeric protein was still detected on the cell surface. When the entire TMD and the cytoplasmic tail of F were swapped with the Gn TMD (F-GnTMC224) or the Gn-TMD with a full-length (F-GnTMC309) or shortened (F-GnTMC237) cytoplasmic tail region, the chimeric F protein showed a typical Golgi staining pattern (Fig. 7B). The data imply that the thickness of the TMD, rather than the primary amino acid sequence, is one of the crucial factors contributing to the cell surface trafficking of the HRSV F protein and to the Golgi targeting of the Gn protein. While it will be interesting to know what other properties or functions of HRSV F are affected when its TMD plus cytoplasmic tail are replaced with those of the BUN Gn protein, it was not a priority for this study.
For many membrane proteins, oligomerization seems to be necessary for exit from the ER (14), and the same appears to pertain to the bunyavirus glycoproteins studied so far (11, 32, 37, 38). Our results indicate that oligomerization or heterodimerization between Gn and Gc proteins is crucial for the exit of the Gc protein from the ER to the Golgi. We observed that Gc undergoes a conformational change when it is in association with the Gn protein, as monitored by its reactivity with MAb 742. Different conformations of Hantaan virus Gn when it is expressed separately or coexpressed with Gc have also been reported (41; X. Shi and R. M. Elliott, Abstr. 10th Int. Conf. Negative Strand Viruses, abstr. 271, 1997). For BUN glycoproteins, the presence of the Gn TMD seems crucial not only for Golgi retention of the Gn protein itself, but also for Golgi targeting and retention of Gc. We presume, therefore, that Gn and Gc heterodimerize through an interaction of their TMDs. Most likely, such an interaction occurs by the side-to-side assembly of Gn and Gc TMD regions, as in many other bitopic membrane proteins whose oligomerization occurs due to interactions between their transmembrane α-helices (3, 33).
In summary, our work has shed new light on the understanding of the Golgi trafficking and maturation of the glycoproteins of BUN. The signal for Golgi targeting and retention of the virus was mapped to the Gn TMD, and the TMD also seems to be crucial for the interaction between Gn and Gc. Our future studies will focus on the role of TMD interactions on the mechanism of Golgi targeting and retention of bunyavirus glycoproteins.
Acknowledgments
We thank Ping Li, Martin Lowe, Bernard Moss, Richard J. Sugrue, and Geraldine Taylor for providing reagents used for this work.
This work was supported by grants 046745 and 048378 from the Wellcome Trust and grant G9604479 from the Medical Research Council to R.M.E.
REFERENCES
- 1.Andersson, A. M., L. Melin, A. Bean, and R. F. Pettersson. 1997. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J. Virol. 71**:**4717-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, A. M., and R. F. Pettersson. 1998. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus (Bunyaviridae) to the Golgi complex. J. Virol. 72**:**9585-9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkin, I. T. 2002. Structural aspects of oligomerization taking place between the transmembrane alpha-helices of bitopic membrane proteins. Biochim. Biophys. Acta 1565**:**347-363. [DOI] [PubMed] [Google Scholar]
- 4.Bretscher, M. S., and S. Munro. 1993. Cholesterol and the Golgi apparatus. Science 261**:**1280-1281. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263**:**802-805. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, R., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Pettersson, A. Plyusnin, and C. S. Schmaljohn. 2000. Bunyaviridae, p. 599-621. In C. M. Fauquet, M. H. V. van Regenmortel, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 7.Elliott, R. M. 1996. The Bunyaviridae: concluding remarks and future prospects, p. 295-332. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 8.Elliott, R. M. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3**:**572-577. [PMC free article] [PubMed] [Google Scholar]
- 9.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83**:**8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62**:**1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerrard, S. R., and S. T. Nichol. 2002. Characterization of the Golgi retention motif of Rift Valley fever virus GN glycoprotein. J. Virol. 76**:**12200-12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffith, K. J., E. K. Chan, C. C. Lung, J. C. Hamel, X. Guo, K. Miyachi, and M. J. Fritzler. 1997. Molecular cloning of a novel 97-kd Golgi complex autoantigen associated with Sjogren's syndrome. Arthritis Rheum. 40**:**1693-1702. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Hoppe-Seyler's Biol. Chem. 374**:**166. [Google Scholar]
- 14.Hurtley, S. M., and A. Helenius. 1989. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 5**:**277-307. [DOI] [PubMed] [Google Scholar]
- 15.Kikkert, M., A. Verschoor, R. Kormelink, P. Rottier, and R. Goldbach. 2001. Tomato spotted wilt virus glycoproteins exhibit trafficking and localization signals that are functional in mammalian cells. J. Virol. 75**:**1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54**:**631-664. [DOI] [PubMed] [Google Scholar]
- 17.Kuismanen, E., B. Bang, M. Hurme, and R. F. Pettersson. 1984. Uukuniemi virus maturation: immunofluorescence microscopy with monoclonal glycoprotein-specific antibodies. J. Virol. 51**:**137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuismanen, E., K. Hedman, J. Saraste, and R. F. Pettersson. 1982. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol. Cell. Biol. 2**:**1444-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappin, D. F., G. W. Nakitare, J. W. Palfreyman, and R. M. Elliott. 1994. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J. Gen. Virol. 75**:**3441-3451. [DOI] [PubMed] [Google Scholar]
- 20.Lees, J. F., C. R. Pringle, and R. M. Elliott. 1986. Nucleotide sequence of the Bunyamwera virus M RNA segment: conservation of structural features in the bunyavirus glycoprotein gene product. Virology 148**:**1-14. [DOI] [PubMed] [Google Scholar]
- 21.Machamer, C. E. 1993. Targeting and retention of Golgi membrane proteins. Curr. Opin. Cell Biol. 5**:**606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47**:**333-348. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka, Y., S. Y. Chen, and R. W. Compans. 1994. A signal for Golgi retention in the bunyavirus G1 glycoprotein. J. Biol. Chem. 269**:**22565-22573. [PubMed] [Google Scholar]
- 24.Matsuoka, Y., S. Y. Chen, C. E. Holland, and R. W. Compans. 1996. Molecular determinants of Golgi retention in the Punta Toro virus G1 protein. Arch. Biochem. Biophys. 336**:**184-189. [DOI] [PubMed] [Google Scholar]
- 25.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348**:**91-92. [DOI] [PubMed] [Google Scholar]
- 26.Munro, S. 1998. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8**:**11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, F. A., A. K. Harrison, and S. G. Whitfield. 1973. Bunyaviridae: morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other arthropod-borne viruses. Intervirology 1**:**297-316. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a _cis_-Golgi matrix protein, GM130. J. Cell Biol. 131**:**1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakitare, G. W., and R. M. Elliott. 1993. Expression of the Bunyamwera virus M genome segment and intracellular localization of NSm. Virology 195**:**511-520. [DOI] [PubMed] [Google Scholar]
- 30.Opat, A. S., C. van Vliet, and P. A. Gleeson. 2001. Trafficking and localisation of resident Golgi glycosylation enzymes. Biochimie 83**:**763-773. [DOI] [PubMed] [Google Scholar]
- 31.Pettersson, R. F. 1991. Protein localization and virus assembly at intracellular membranes. Curr. Top. Microbiol. Immunol. 170**:**67-106. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson, R. F., and L. Melin. 1996. Synthesis, assembly, and intracellular transport of Bunyaviridae membrane proteins, p. 159-188. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 33.Popot, J. L., and D. M. Engelman. 2000. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69**:**881-922. [DOI] [PubMed] [Google Scholar]
- 34.Ruusala, A., R. Persson, C. S. Schmaljohn, and R. F. Pettersson. 1992. Coexpression of the membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi complex. Virology 186**:**53-64. [DOI] [PubMed] [Google Scholar]
- 35.Salanueva, I. J., R. R. Novoa, P. Cabezas, C. Lopez-Iglesias, J. L. Carrascosa, R. M. Elliott, and C. Risco. 2003. Polymorphism and structural maturation of Bunyamwera virus in Golgi and post-Golgi compartments. J. Virol. 77**:**1368-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmaljohn, C. S., A. L. Schmaljohn, and J. M. Dalrymple. 1987. Hantaan virus M RNA: coding strategy, nucleotide sequence, and gene order. Virology 157**:**31-39. [DOI] [PubMed] [Google Scholar]
- 37.Shi, X., and R. M. Elliott. 2002. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology 300**:**31-38. [DOI] [PubMed] [Google Scholar]
- 38.Spiropoulou, C. F., C. S. Goldsmith, T. R. Shoemaker, C. J. Peters, and R. W. Compans. 2003. Sin nombre virus glycoprotein trafficking. Virology 308**:**48-63. [DOI] [PubMed] [Google Scholar]
- 39.Taylor, G., E. J. Stott, J. Furze, J. Ford, and P. Sopp. 1992. Protective epitopes on the fusion protein of respiratory syncytial virus recognized by murine and bovine monoclonal antibodies. J. Gen. Virol. 73**:**2217-2223. [DOI] [PubMed] [Google Scholar]
- 40.Walsh, E. E., and J. Hruska. 1983. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 47**:**171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, M., D. G. Pennock, K. W. Spik, and C. S. Schmaljohn. 1993. Epitope mapping studies with neutralizing and non-neutralizing monoclonal antibodies to the G1 and G2 envelope glycoproteins of Hantaan virus. Virology 197**:**757-766. [DOI] [PubMed] [Google Scholar]
- 42.Watret, G. E., C. R. Pringle, and R. M. Elliott. 1985. Synthesis of bunyavirus-specific proteins in a continuous cell line (XTC-2) derived from Xenopus laevis. J. Gen. Virol. 66**:**473-482. [DOI] [PubMed] [Google Scholar]