Asynchronous replication timing of telomeres at opposite arms of mammalian chromosomes (original) (raw)
Abstract
Telomeres are defining structural elements of all linear chromosomes, yet information concerning the timing of their replication in higher eukaryotes is surprisingly limited. We developed an approach that allowed a study of telomere replication patterns of specific mammalian chromosomes. In the Indian muntjac (Muntiacus muntjac), replication timing between respective telomeres of homologous chromosomes was highly coordinated, but no such synchrony was evident for p- and q-arm telomeres of the same chromosome. This finding contrasts with the coordinated timing of both ends of each chromosome in yeast. Also in contrast to yeast, where replication of all telomeres is confined to late S phase, we found specific telomeres in Indian muntjac chromosomes that replicated early in S and other telomeres that replicated later. Finally, replication timing of some but not all telomeres was influenced by telomere length. Knowledge of telomere replication timing represents a first step toward understanding the relationship between telomere replication and telomerase action. The approach, which we call replicative detargeting fluorescence in situ hybridization, is widely applicable to different species and genetic loci.
Keywords: DNA replication, Indian muntjac, telomerase
Located at the ends of all eukaryotic chromosomes, telomeres are essential for ensuring chromosomal integrity and maintaining overall genome stability (1–3). In vertebrates, each chromosome end is composed of a sequence of identical tandem TTAGGG sequences (telomeric repeats), which associate with proteins to form a specialized structure to protect chromosomes from end-to-end fusion, degradation, and inappropriate recombination (1–3). The accurate and timely duplication of the genome is a major task for eukaryotic cells and requires the cooperation of multiple factors to ensure the stability of the genetic information of each cell. Telomeres are essential for the complete replication of eukaryotic chromosomes. Replication of linear DNA results in what is commonly known as the end replication problem (4, 5), in which the DNA replication machinery is not able to completely replicate the very termini of chromosomes. As a result, 50–200 bp of telomeric DNA are lost during each S phase in cultured human cells. This loss can be prevented by telomerase (hTERT), a cellular reverse transcriptase. The catalytic subunit of telomerase contains an integral RNA molecule (hTR) with the sequence that is used as a template to add telomeric repeats onto the 3′ end of the telomere. Telomeres in yeast are short (≈250–350 nucleotides) and maintained at relatively constant length because of the constitutive expression of telomerase. Telomeres in both human and Indian muntjac (Muntiacus muntjac, a barking deer) are much longer, ranging from 4 to 20 kb (6–8). Heterogeneous telomere lengths have been observed at different chromosome ends and even at homologous telomere ends (9, 10). Most human and Indian muntjac fibroblasts do not express telomerase, and thus their telomeres shorten progressively during successive cellular divisions (6, 11, 12). Ectopic expression of the catalytic subunit of hTERT is sufficient to produce telomerase activity, elongate telomeres, and extend the life span of a variety of human cells and Indian muntjac fibroblasts (6, 13–15).
The telomeres of yeast are heterochromatic and replicate late in S phase, and the two telomeres at each end of the same chromosome replicate synchronously (16–18). However, there is limited information about the replication timing of telomeres in mammalian cells except that they replicate throughout S phase (19–22). Because these studies did not examine the replication of individual telomeres, there are a number of possible explanations. Telomeric replication could be a stochastic process where the time of replication of each telomere varies from one cell cycle to the next. Alternatively, there could be a specific interval in S phase during which any given telomere replicates, and this specific interval may differ from one telomere to the next. Finally, although it seems unlikely, it is formally possible that the replication of each telomere extends throughout S phase. To resolve questions about specific replication timing, we developed a protocol based on fluorescence in situ hybridization (FISH) that we call replicative detargeting FISH (ReDFISH), which allowed the study of mammalian telomere replication patterns for each chromosome end during defined stages of S phase. Indian muntjac fibroblasts were chosen for this study, as they have only a few chromosomes (23), and share similar characteristics of telomere biology with normal human cells (6). To validate the ReDFISH technique, we compared the timing of telomere replication in hydroxyurea (HU)-synchronized and unsynchronized Indian muntjac cells. We found that the HU treatment did not affect the temporal order of telomere replication. Each telomere of Indian muntjac cells had a characteristic and reproducible timing of replication that produced a collective replication of telomeric DNA throughout S phase. Whereas replication timing between the telomeres of homologous chromosomes was highly coordinated, no such synchrony was evident for p- and q-arm telomeres of the same chromosome, which contrasts with the pattern in yeast. Alteration of telomere length affected the timing of telomere replication for only a few chromosome ends. In summary, the timings and regulation of the replication of Indian muntjac telomeres are very different from those of yeast.
Materials and Methods
Cell Culture and Synchronization. Five Indian muntjac fibroblast cell lines were used in this study. Among these were the F4374 parental cell line and its telomerase-immortalized subclone, which was derived from a female Indian Muntjac with six chromosomes (a kind gift from Roger Shultz, University of Texas Southwestern Medical Center, Dallas). The others included the F4869 strain and its telomerase-immortalized clones, which are from a male Indian Muntjac with seven chromosomes (American Type Culture Collection no. F4869). All cells were grown in a 4:1 mixture of DMEM 199 (Life Technologies, Grand Island, NY) plus 20% Cosmic calf serum (HyClone) at 37°C in 5% CO2/95% air with split ratios of 1:4 and synchronized with serum starvation followed by HU as described in ref. 20. After release into complete growth medium, designated cultures were pulse–chase fed at hourly intervals with medium containing 10 μM BrdUrd and 3.3 μM 5′-bromo-2′-deoxycytidine (BrdC) (2-cm2 wells for anti-BrdUrd staining and 100-cm2 dishes for preparation of metaphase spread). For experiments with asynchronous cultures, cells were pulse-labeled with BrdUrd/BrdC at hourly intervals, then fed with regular medium until metaphase spreads were prepared.
Anti-BrdUrd Staining. Anti-BrdUrd staining was used to determine the length of G2 phase and to visualize the replication bands. Cells were denatured in a 70% formamide/2× SSC solution (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) at 70°C for 2 min and dehydrated by a 2-min serial incubation in 70%, 90%, and 100% ethanol then air-dried. The slides were blocked with 10% normal goat serum (Santa Cruz Biotechnology) for 1 h, stained with mouse anti-BrdUrd antibody (50 ng/ml in PBS) for 15 min at room temperature, washed with 1× PBS for 15 min, and incubated with FITC-conjugated anti-mouse antibody (Molecular Probes). The slides were washed and dehydrated as above, mounted with VECTASHIELD containing 4′,6-diamidino-2-phenylindole-dihydrochloride (DAPI, Vector Laboratories) at a 0.6 μg/ml final concentration, and imaged by using a Zeiss Axioplan 2 microscope (×63; 1.4 numerical aperture; Plan-Apochromat oil immersion objective) with precision FITC/DAPI bandpass filter sets.
Quantitative FISH Analysis. To get better quantitative analysis results, the slides were sequentially hybridized to both G- and C-rich telomere probes conjugated to the same fluorescent dye as described (24). The quantitative FISH analysis was performed by imagej software (www.nih.org). One clone of mouse NIH 3T3 cells was mixed with testing samples and used as an internal control to normalize results and to convert the fluorescence intensity measurements of human telomeres to kilobases (6, 9). The banding pattern from inverted DAPI images was used for chromosome identification.
Strand-Specific FISH (CO-FISH). CO-FISH (25, 26) was used with sequential hybridizations to C- and G-rich probes conjugated to different fluorescent dyes. Slides from cells grown in the presence of BrdUrd/dC were treated with 0.5 mg/ml ribonuclease A (Roche) for 10 min at 37°C, stained with 0.5 μg/ml Hoechst 33258 (Sigma) for 15 min at room temperature, mounted with McIlvaine's buffer at pH 8.0, and exposed to 365-nm UV light (Stratalinker 1800 UV irradiator) for 30 min at 55°C. The nicked BrdUrd/dC-substituted DNA was digested with 3 units/μl exonuclease III (Promega) in 50 mM Tris·HCl (pH 8.0), 5 mM MgCl2, and 5 mM DTT for 10 min at room temperature. After being air-dried, the slides were denatured in 70% formamide/2× SSC (pH 7.0) at 70°C for 2 min. The C-rich strands (templates for leading strand synthesis) were revealed by hybridizing (20 μl) in 70% formamide, 20 ng of Cy3-conjugated (TTAGGG)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate probe (27, 28), 0.25% (wt/vol) blocking reagent (Roche), and 5% MgCl2 in 10 mM Tris, pH 7.2, which was added to the slides containing single-stranded chromosomal target DNA. After a 2-h incubation at room temperature, the slides were washed twice with 70% formamide/0.1% BSA/10 mM Tris, pH 7.2, and washed twice with 0.15 M NaCl/0.05% Tween 20/0.05 M Tris. The slides were dehydrated through an ethanol series (70%, 85%, and 100%) and air-dried in the dark. The G-rich strands (templates for lagging strand synthesis) were then visualized by incubating with a second hybridization mixture (20 μl) containing 70% formamide, 20 ng of 3′-FITC-conjugated (CCCTAA)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate probe, 0.25% (wt/vol) blocking reagent (Roche), and 5% MgCl2 in 10 mM Tris, pH 7.2, for 2 h at room temperature. The slides were then washed, dehydrated, and air-dried as described above. Chromosomes were counterstained with DAPI (Vector Laboratories) and identified according to different banding patterns from inverted-DAPI images. The slides were digitally imaged on a Zeiss Axioplan 2 microscope (×63, 1.4 numerical aperture; Plan-Apochromatic oil immersion objective) with precision Cy3/FITC/DAPI cell bandpass filter sets. Cy3, FITC, and DAPI images were captured separately with a charge-coupled device camera (Hamamatsu, Bridgewater, NJ) and merged by using openlab software (Improvision, Lexington, MA).
Results and Discussion
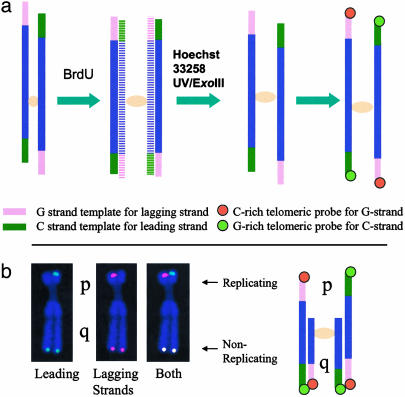
Principle of the ReDFISH Technique. ReDFISH is based on the CO-FISH technique (25, 26) (Fig. 1_a_). A requirement of CO-FISH is that a cell goes through a single complete round of DNA synthesis in the presence of a halogenated nucleotide precursor, such as BrdUrd. This produces unifilar (and complementary) labeling of the DNA in each sister chromatid. Treatment with Hoechst dye and exposure to UV radiation preferentially nicks BrdUrd-substituted nascent daughter strands, allowing them to become substrates for subsequent digestion with exonuclease III. Because the unsubstituted parental strands are left largely intact by this treatment, the end result, at least for the purposes of FISH, is a chromosome whose sister chromatids are rendered both single-stranded and complementary to one another. This means that a single-stranded probe, such as a synthetic oligonucleotide to a particular DNA sequence, can hybridize to only one of the two sister chromatids at the same locus.
Fig. 1.
Schema of ReDFISH technique. (a) ReDFISH of a chromosome that has replicated fully in the presence of BrdUrd/dC. Newly synthesized DNA incorporating BrdUrd/dC (horizontal stripes) is removed after nicking the DNA with Hoechst 33258 plus UV and digesting nicked DNA with exonuclease III, leaving only the parental strands. The G-rich telomeric strand is the template for lagging strand synthesis and anneals to a Cy3-conjugated C-rich telomeric probe, whereas the C-rich telomeric strand is the template for leading strand synthesis and anneals to an FITC-conjugated G-rich telomeric probe. This pattern defines which telomeric strands replicated at the time of BrdUrd/BrdC labeling. (b) ReDFISH of a partially replicated chromosome. In this example, only the p-arm telomere of the X chromosome was replicating during the 1-h BrdUrd/dC pulse; the q arm was not replicating. As a consequence, only the parental strands are available for hybridization in the p arm (schema, p arms), whereas both strands of unreplicated q-arm DNA survive digestion and hybridize to both probes (schema, q arms).
The (TTAGGG)n telomeric sequences of vertebrate chromosomes run 5′→3′ toward the termini of all chromatids. By using a C-rich probe complementary to this G-rich sequence, CO-FISH labels only the daughter telomeres replicated by lagging-strand synthesis (25, 26). A characteristic staining pattern is thus produced in which hybridization signals are confined to the p terminus of one chromatid and the q terminus of its sister. Conversely, G-rich probes complementary to the C-rich (CCCTAA)n strand identify telomeres replicated by leading-strand synthesis. When probes to G- and C-rich sequences are made distinguishable from one another through the use of different fluorochromes, sequential hybridization produces a characteristic mirror-image CO-FISH pattern, as indicated in Fig. 1_a_.
In the absence of BrdUrd incorporation, both the parental and daughter DNA strands of sister chromatids remain as targets for probe hybridization. In the case of telomeric sequences, differentially labeled probes to the G- and C-rich strands will both hybridize to the same locus, producing double-colored signals on each of the four chromosomal termini. This situation represents the normal FISH pattern observed under these conditions. Replication of a telomere during a short pulse in the presence of BrdUrd will cause a switch from a FISH pattern to a CO-FISH pattern upon passage of the replication fork. At this point, the telomere becomes detargeted, no longer able to serve as a target for normal FISH (Fig. 1_b_). In applying ReDFISH, we exploit this situation by introducing BrdUrd and BrdC at selective times during S phase, observing later in mitosis the appearance of CO-FISH in relation to FISH signals on different chromosomes, and on different arms of the same chromosome.
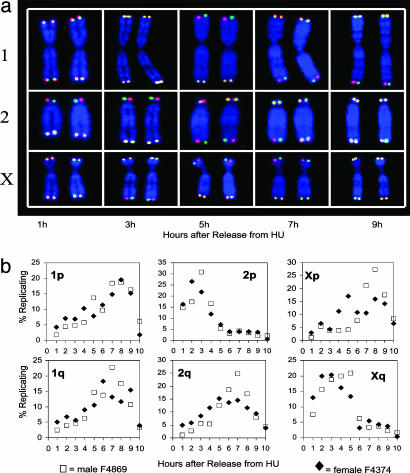
Each Telomere Has a Characteristic Replication Time During S Phase in HU-Synchronized Cell Cultures. Indian muntjac cells have a number of properties that make them useful for the present studies. They have the fewest number of chromosomes among mammalian species: 2_n_ = 6 in females (chromosomes 1, 2, and X); 2_n_ = 7 in males (chromosomes 1, 2, X, and Y1/Y2) (23). In both sexes, chromosomes are morphologically distinguishable from one another without resort to banding or chromosome painting, containing telomeres large enough to yield reproducibly bright FISH signals. Replication timing of telomeres was analyzed by ReDFISH in synchronized cell cultures. After a 2-day serum starvation and a 1-day HU treatment (20), cells were treated with BrdUrd/BrdC during hourly intervals following the release into S phase, and metaphase spreads were harvested from each time point when the synchronized cell cohort reached mitosis. A high degree of synchronization was confirmed by staining with anti-BrdUrd antibodies and flow cytometry of the different fractions (20) (data not shown).
Different telomeres of Indian muntjac cells were found to replicate at different times throughout S phase (Fig. 2_a_). Each telomere has a characteristic time of replication that may be caused either by a proterminal (subtelomeric) origin of replication in the adjacent chromosomal DNA or by a telomeric origin of replication, which initiates replication at different times from one telomere to the next. This results in the replication of total telomeric DNA throughout S phase as the sum of timing of specific telomere replication rather than the replication of each telomere throughout S phase. Telomere replication timing was similar in two different cell lines, one male and one female (Fig. 2_b_), thus validating the generality of our findings. Two ends (2p and Xq) replicated earlier than the other four telomeres. This is very different from what happens in yeast, in which all telomeres replicate late in S phase.
Fig. 2.
Telomere replication of male and female Indian muntjac chromosomes after G1/S synchronization with HU. (a) Representative chromosomes after release from HU with hourly BrdUrd/dC pulses. During the first 1-h pulse, only chromosome 2p replicated; thus, it is the only chromosome to show differential hybridization with the two probes (red for lagging strand template and green for leading strand template). Chromosomes were stained with DAPI. Different patterns of telomere replication timing were observed for each chromosome at various time points after release from HU. Each of the five columns illustrates ReDFISH staining for a single cell. (b) Timing patterns of telomere replication of each chromosome end. Male Indian muntjac cells F4869 (PD 13.8, □) and female Indian muntjac cells F4374 (PD 40, ♦) were growth-arrested in low serum and then synchronized at the G1/S interface by HU. Telomere replication patterns of a total of 64,044 chromosome ends were analyzed by ReDFISH. Note that telomeres replicate throughout S phase.
Yeast telomeres are tethered to the nuclear periphery, a nuclear compartment rich in chromatin-modifying factors that can establish a late-activation domain (29) and are constrained from chromatin movement in S phase, compared with other chromosome regions (30). Although late replication is not an obligatory feature of heterochromatin (31), yeast heterochromatic telomeres replicate in late S phase, and telomeric chromatin can confer late replication on telomere-proximal genes (32). Mammalian telomeres also form a large specialized protein–telomeric DNA chromatin structure, but they are tightly associated with the nuclear matrix components rather than the nuclear periphery (33). Whether and how this attachment to the nuclear matrix affects the temporal order of telomere replication remains unclear. Although the reason for this temporally programmed order of telomere replication is not understood, it might have functional significance. It is well established that there is a general correlation between replication timing and gene expression, with few exceptions. Most active genes replicate early in the first half of S phase, whereas many inactive gene sequences replicate late in S phase. The existence of some telomeres that replicate in early S phase raises the possibility that endogenous genes near these telomere ends could be actively expressed, which may have important biological functions.
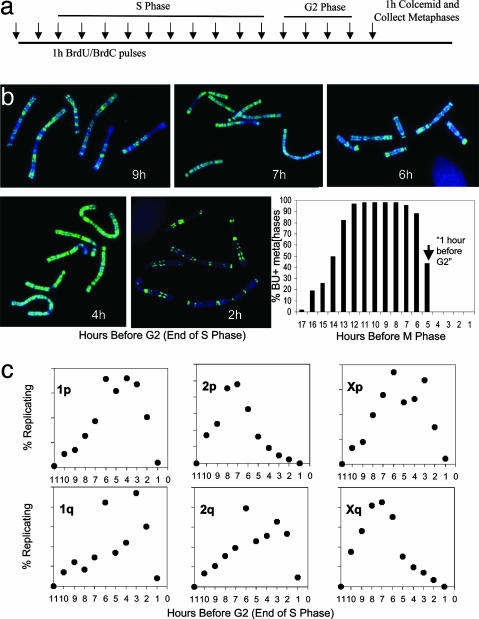
Similar Telomere Replication Pattern of Indian Muntjac in Unsynchronized Cell Cultures. We used asynchronous cultures to verify that telomere replication times were not affected by HU-induced chemical synchrony. Cultures of female muntjac cells in exponential growth were pulsed with BrdUrd/BrdC during hourly intervals, then fed fresh medium before collecting metaphases at the same end point for anti-BrdUrd staining and ReDFISH analysis (Fig. 3_a_). As determined by anti-BrdUrd staining, each chromosome of the Indian muntjac exhibited multiple replication bands, whose transverse pattern characterizes the particular substage of S phase (Fig. 3_b_) the cells were in at the time of BrdUrd/dC pulse (34). Note also the high fraction of BrdUrd-positive metaphases. The time course of the fraction of mitoses containing BrdUrd-positive chromosomes defines the transit times of G2 phase (before labeled chromosomes appear in metaphase) and of S phase (Fig. 3_b_). Fig. 3_c_ shows the fraction of telomeres replicating for each chromosome end during each hour before the start of G2 (end of S phase). In comparing Figs. 3 and 4, it is evident that both methods used for the analysis of replication timing yielded qualitatively similar results, namely that Indian muntjac telomeres replicate throughout S phase, and certain telomeres (e.g., 2p and Xq) showed a strong tendency to replicate earlier than others. Taken together; our results indicate that HU treatment does not affect the telomere replication timing of Indian muntjac cells within the 1-h resolution of this experiment.
Fig. 3.
Telomere replication of female Indian muntjac cells as measured during asynchronous growth. (a) Experimental design in which cells were pulse-labeled with BrdUrd/dC for 1 h to determine the length of G2 phase by anti-BrdUrd staining. Cells were treated with colcemid for 1 h before metaphase harvest. (b) Replication bands of metaphase spreads representing the labeling periods indicated (given as hours before the onset of G2 phase). Chromosomes were stained with anti-BrdUrd antibody (green) and counter-stained with DAPI (blue). Cells exhibited characteristic replication bands at each time point. (Lower Right and Lower Center) The absence of BrdUrd-positive metaphases after pulsing during the last 4 h before metaphase defines the G2 period. The 50% positive time point (vertical arrow) is defined as 1 h before G2/the last hour of S. (c) Time course of telomere replication in asynchronous cells. The telomere replication patterns representing a total of 52,632 chromosome ends of a female Indian muntjac cell (expressing hTERT) were analyzed. Each point represents the percentage of all replicating events of that chromosome end during the 11 h before the onset of G2 phase. The data from asynchronous cultures are very similar to that for HU-synchronized cells shown in Fig. 2.
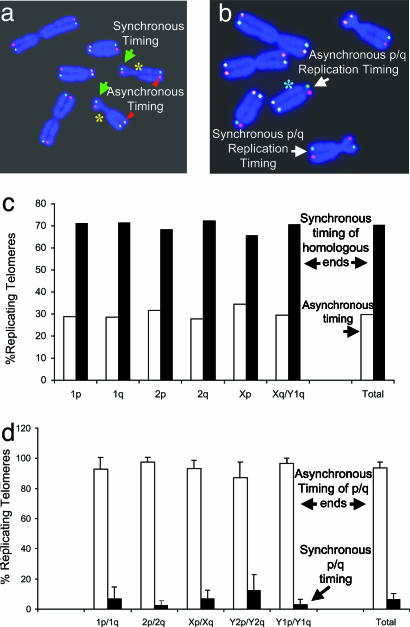
Fig. 4.
The timing of telomere replication between homologous chromosomes and between the p and q arms of the same chromosome. (a) Representative metaphase spread of an Indian muntjac cell (F4374) by ReDFISH. Yellow asterisks indicate homologous X chromosomes. Green arrows mark synchronous telomere replication. Red triangles mark homologous chromosome ends that replicated asynchronously. (b) Representative metaphase spread of Indian muntjac cell (F4374) illustrating replication timing of p versus q arms of the same chromosome. A green asterisk indicates a chromosome in which asynchronous timing of telomere replication for the long arm and the short arm is found. A chromosome with synchronous timing of p and q arms is also shown. (c) Timing of telomere replication between homologous chromosomes. About 70% of the homologous chromosome ends replicated within the same hour-long BrdUrd pulse. A total of 22,310 replicating homologous chromosome ends were analyzed. (d) Timing of p-versus q-arm telomere replication. For ≈90% of all chromosomes, the p- and q-arm telomeres did not replicate at the same time. A total of 50,148 replicating chromosome ends were analyzed.
Coordination of Telomere Replication of Homologues. Homologous regions on a pair of autosomes generally replicate at the same time in S phase; this has been shown by chromosome replication banding studies at a resolution of about several megabases (35). However, random monoallelic expression and asynchronous replication involving genes in the active and inactive X chromosome, autosomal imprinted genes, and randomly monoallelically transcribed autosomal genes have been reported (36–38). Replication timing of homologous alleles for a unique gene is easily assayed by FISH analysis of interphase nuclei, which is observed as a single spot for an unreplicated locus and a double signal after replication (39). However, this method cannot be used to study the replication timing of homologous telomere ends, as all chromosome ends share the identical telomeric DNA sequence, and each telomere will have a respective FISH signal in an interphase nucleus. Because there was no information about the replication behavior of homologue telomere ends in mammalian cells, we used the ReDFISH technique to address this question. We analyzed >20,000 replicating homologous telomere ends to determine whether replication timing was similar between homologues (Fig. 4 a and c). The analysis included five different Indian muntjac populations: the two parental cell lines and various clones thereof, including telomerase-expressing derivatives (6). Within a population of cells, the distribution of timing of replication is relatively broad (e.g., the half peak-width for 2q is 4–5 h in Fig. 2). Despite this, and irrespective of the fact that we had previously observed heterogeneous telomere lengths between homologues in these cells (6), more than 70% of homologous telomere ends within individual cells replicated synchronously during each hourly pulse (Fig. 4_b_). This suggests a genome-wide coordination of the replication timing of homologous telomere ends. The region located between chromosome-specific sequences and the array of telomeric repeats is called the subtelomere, which is ≈10–300 kb long in human. Although subtelomeres show commonalities of structure and function in organisms as diverse as human, yeast, and trypanosomes, they are extraordinarily dynamic and variable regions, and the same subtelomeric sequence can duplicate and disperse among many chromosome ends (40). Insertion/deletion polymorphisms caused by translocation, recombination, gene conversion, and/or duplication are common in subtelomeric alleles, and differences in size (up to 160 kb) between subtelomeric alleles have been shown to exist in the normal human population (40, 41). Interestingly, homologous telomeres replicate synchronously in the two different Indian muntjac cell strains that we studied, suggesting that the polymorphisms in the subtelomeric regions of homologues might not affect replication timing of homologous telomeres. Although the subtelomeric regions are less than 1% of the total human genome, ≈10% of idiopathic mental retardation patients have rearrangements or deletions of subtelomerically located genes (42). The analysis of replication of homologous telomeres present in normal human and patient cells might give insights into the chromatin structure and biological function of telomeres and subtelomere regions.
Asynchronous Telomere Replication of the Two Ends of the Same Chromosome. In budding yeast, both ends of each chromosome replicate at the same time, raising the possibility that intrachromosomal telomere interactions (perhaps involving the G-rich single-stranded telomeric DNA) might affect the time of origin activation (16, 43). To test for coordination in the timing of telomere replication between p and q arms of the same chromosome, we analyzed ≈25,000 replicating chromosomes, by using the same five different Indian muntjac populations as above (Fig. 4 b and d). Unlike yeast, 94% of telomere ends of the short and the long arm of the same chromosome did not replicate synchronously (Fig. 4_d_). These data suggest that intrachromosomal interactions do not influence the replication timing of mammalian telomeres.
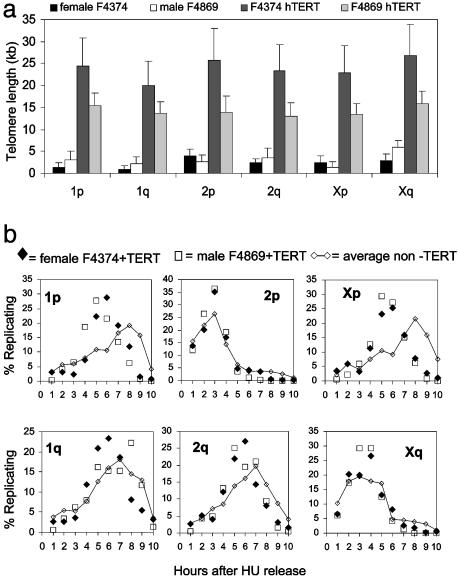
Telomere Length and Telomere Replication Timing. Epigenetic inactivation of genes introduced into telomere-proximal positions is called telomere position effect. Silencing of gene expression by telomere position effect has been demonstrated and conserved in several organisms from budding yeast (44, 45) to humans (46). Changes in histone acetylation correlate with both altered replication timing and telomere position effect (46, 47). However, the relationships among telomere replication timing, gene expression, and telomere position effect in higher eukaryotes are unknown. To examine the potential link between telomere length and replication timing, we elongated telomeres by expressing the catalytic subunit of human telomerase (hTERT). The telomere lengths among parental and hTERT-transfected cell lines were measured by Q-FISH. Whereas expression of hTERT led to rather dramatic increases in telomere lengths (Fig. 5_a_), the timing of telomere replication remained more or less unchanged (Fig. 5_b_). Exceptions included the p arms of chromosomes 1 and the X, for which replication appeared to shift toward earlier times in S phase compared to parental untransfected cells. Conceivably, the origins of replication normally used by these particular telomeres have become inactive as a result of telomeric elongation, perhaps through a telomere position effect-like mechanism, so that earlier-firing origins that are more centromeric are used instead.
Fig. 5.
The influence of telomere length on the timing of telomere replication. (a) The distribution of telomere length of individual chromosome ends. Telomere fluorescence signals from metaphase chromosomes of Indian muntjac F4374 and F4869, and their telomerase-expressing derivatives were measured by quantitative FISH. Note the significant lengthening in telomeres associated with hTERT expression. The F4374 cells were analyzed while still expressing telomerase (6); the F4869 cells had been infected with a lox-hTERT retrovirus and were analyzed after Cre-mediated excision of hTERT (50) to ensure that any differences were due to the elongation of telomeres rather than the presence of telomerase. (b) Telomere length and replication timing. The potential influence of telomere length on the timing of replication was examined by comparing the pattern of replication in the parental cells (average of both cell types from Fig. 2; diamonds connected by a line) with that after telomere elongation (individual points for each cell line). Only two of the telomeres (1p and Xp) shared a significant change in replication timing. At least 30,000 telomere ends of each cell line were analyzed.
Conclusions and Perspective. A unique strength of ReDFISH lies in its ability to distinguish, on a cell-by-cell basis, replicating and nonreplicating regions of individual chromosomes at defined stages of DNA synthesis. This makes an evaluation of intra- and interchromosome replication possible. We report here that each telomere of a mammalian cell has a characteristic timing for replication during S phase. Some specific telomeres replicate early in S phase, and p- and q-arm telomeres of the same chromosome replicate asynchronously. Thus, the timing and regulation of replication of Indian muntjac telomeres are very different from that found in yeast, in which all telomeres replicate late in S phase, and both arm telomeres of the same chromosome replicate synchronously (16–18). We also report a vertebrate cell showing a high coordination of replication timing between telomeres of homologous chromosomes, regardless of their heterogeneous telomere lengths.
In principle, the ReDFISH technique can be applied to any locus across the genomes of a variety of organisms, including human, where a wealth of gene sequence information is readily available. The main requirements are the availability of good quality metaphase spreads from cells in culture and strand-specific fluorescently labeled FISH probes for the desired target. The latter requirement is trivial for repetitive elements such as telomeric sequences, which can easily be synthesized as single-stranded oligomers. Whereas it should be simple to prepare single-stranded probes for unique-copy sequences from cloned libraries, to our knowledge this has not yet been attempted. Using single-stranded RNAs as probes to hybridize to the desired DNA target to perform ReDFISH is another option. ReDFISH should be able to provide qualitative and quantitative information about several additional aspects associated with replication timing, including the relationship between defects in replication timing and defects in chromosome condensation, sister chromatid cohesion, and genome stability (48, 49).
Acknowledgments
We thank Bill Walker and Jennifer Costley for technical assistance. This work was supported by National Institute on Aging Grant AG07992 (to W.E.W. and J.W.S.) and by National Cancer Institute Grant CA76260, U.S. Department of Energy Grant DE-FG03-02ER63442, and the National Aeronautical and Space Administration Office of Biological and Physical Research (to M.N.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FISH, fluorescence in situ hybridization; ReDFISH, replicative detargeting FISH; BrdC, 5′-bromo-2′-deoxycytidine; DAPI, 4′,6-diamidino-2-phenylindole-dihydrochloride; CO-FISH, strand-specific FISH; HU, hydroxyurea.
References
- 1.Blackburn, E. H. (2000) Nature 408**,** 53–56. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. (2001) Cell 106**,** 661–673. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira, M. G., Miller, K. M. & Cooper, J. P. (2004) Mol. Cell 13**,** 7–18. [DOI] [PubMed] [Google Scholar]
- 4.Watson, J. D. (1972) Nat. New Biol. 239**,** 197–201. [DOI] [PubMed] [Google Scholar]
- 5.Olovnikov, A. M. (1973) J. Theor. Biol. 41**,** 181–190. [DOI] [PubMed] [Google Scholar]
- 6.Zou, Y., Yi, X., Wright, W. E. & Shay, J. W. (2002) Exp. Cell Res. 281**,** 63–76. [DOI] [PubMed] [Google Scholar]
- 7.deLange, T., Shiue, L., Myers, R. M., Cox, D. R., Naylor, S. L., Killery, A. M. & Varmus, H. E. (1990) Mol. Cell. Biol. 10**,** 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harley, C. B., Futcher, A. B. & Greider, C. W. (1990) Nature 345**,** 458–460. [DOI] [PubMed] [Google Scholar]
- 9.Martens, U. M., Zijlmans, J. M., Poon, S. S., Dragowska, W., Yui, J., Chavez, E. A., Ward, R. K. & Lansdorp, P. M. (1998) Nat. Genet. 18**,** 76–80. [DOI] [PubMed] [Google Scholar]
- 10.Londono-Vallejo, J. A., DerSarkissian, H., Cazes, L. & Thomas, G. (2001) Nucleic Acids Res. 29**,** 3164–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastie, N. D., Dempster, M., Dunlop, M. G., Thompson, A. M., Green, D. K. & Allshire, R. C. (1990) Nature 346**,** 866–868. [DOI] [PubMed] [Google Scholar]
- 12.Lindsey, J., NMcGill, N. I., Lindsey, L. A., Green, D. K. & Cook, H. J. (1991) Mutat. Res. 256**,** 45–48. [DOI] [PubMed] [Google Scholar]
- 13.Bodnar, A. G., Ouellette, M. M., Frolkis, M., Holt, S. E., Chui, C.-P., Morin, G. B., Harley, C. B., Shay, J. W., Lichsteiner, S. & Wright, W. E. (1998) Science 279**,** 349–352. [DOI] [PubMed] [Google Scholar]
- 14.Vaziri, H. & Benchimol, S. (1998) Curr. Biol. 8**,** 279–282. [DOI] [PubMed] [Google Scholar]
- 15.Wang, J., Xie, L. Y., Allan, S., Beach, D. & Hannon, G. J. (1998) Genes Dev. 12**,** 1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J. & Fangman, W. L. (2001) Science 294**,** 115–212. [DOI] [PubMed] [Google Scholar]
- 17.McCarroll, R. M. & Fangman, W. L. (1988) Cell 88**,** 657–666. [DOI] [PubMed] [Google Scholar]
- 18.Wellinger, R. J., Wolf, A. J. & Zakian, V. A. (1993) Mol. Cell. Biol. 13**,** 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ten Hagen, K. G., Gilbert, D. M., Willard, H. F. & Cohen, S. N. (1990) Mol. Cell. Biol. 10**,** 6348–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright, W. E., Tesmer, V. M., Liao, M. L. & Shay, J. W. (1999) Exp. Cell Res. 251**,** 492–499. [DOI] [PubMed] [Google Scholar]
- 21.Hultdin, M., Gronlund, E., Norrback, K. F., Just, T., Taneja, K. & Roos, G. (2001) Exp. Cell Res. 271**,** 223–229. [DOI] [PubMed] [Google Scholar]
- 22.Hultdin, M., Gronlund, E., Norrback, K. F., Eriksson-Lindstrom, E., Just, T. & Roos, G. (1998) Nucleic Acids Res. 26**,** 3651–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurster, D. H. & Benirschke, K. (1970) Science 168**,** 1364–1366. [DOI] [PubMed] [Google Scholar]
- 24.Zou, Y., Sfeir, A., Gryaznov, S. M., Shay, J. W. & Wright, W. E. (2004) Mol. Biol. Cell. 15**,** 3709–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey, S. M., Cornforth, M. N., Kurimasa, A., Chen, D. J. & Goodwin, E. H. (2001) Science 293**,** 2462–2465. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin, E. & Meyne, J. (1993) Cytogenet. Cell Genet. 63**,** 126–127. [DOI] [PubMed] [Google Scholar]
- 27.Gryaznov, S. M. & Banait, N. S. (2002) Exp. Opin. Ther. Patents 12**,** 543–559. [Google Scholar]
- 28.Egli, M. & Gryaznov, S. M. (2000) Cell Mol. Life Sci. 57**,** 1440–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert, D. M. (2001) J. Cell Biol. 152**,** F11–F15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S. M. (2001) Science 294**,** 2181–2186. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S., Dubey, D. D. & Huberman, J. A. (2003) Genes Dev. 17**,** 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson, J. B. & Gottschling, D. E. (1999) Genes Dev. 13**,** 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.deLange, T. (1992) EMBO J. 11**,** 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aghamohammadi, S. Z. & Savage, J. R. (1990) Chromosoma 99**,** 76–82. [DOI] [PubMed] [Google Scholar]
- 35.Drouin, R., Lemieux, N. & Richer, C. (1990) Chromosoma 99**,** 273–280. [DOI] [PubMed] [Google Scholar]
- 36.Chess, A., Simon, I., Cedar, H. & Axel, R. (1994) Cell 78**,** 823–834. [DOI] [PubMed] [Google Scholar]
- 37.Efstratiadis, A. (1994) Curr. Opin. Genet. Dev. 4**,** 265–280. [DOI] [PubMed] [Google Scholar]
- 38.Lyon, M. F. (1986) Nature 320**,** 313. [DOI] [PubMed] [Google Scholar]
- 39.Selig, S., Okumura, K., Ward, D. C. & Cedar, H. (1992) EMBO J. 11**,** 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mefford, H. C. & Trask, B. J. (2002) Nat. Rev. Genet. 3**,** 91–102. [DOI] [PubMed] [Google Scholar]
- 41.Wilkie, A. O., Higgs, D. R., Rack, K. A., Buckle, V. J., Spurr, N. K., Fischel-Ghodsian, N., Ceccherini, I., Brown, W. R. & Harris, P. C. (1991) Cell 64**,** 595–606. [DOI] [PubMed] [Google Scholar]
- 42.Bocian, E., Helias-Rodzewicz, Z., Suchenek, K., Obersztyn, E., Kutkowska-Kazmierczak, A., Stankiewicz, P., Kostyk, E. & Mazurczak, T. (2004) Med. Sci. Monit. 10**,** 143–151. [PubMed] [Google Scholar]
- 43.Wellinger, R. J., Wolf, A. J. & Zakian, V. A. (1993) Cell 72**,** 51–60. [DOI] [PubMed] [Google Scholar]
- 44.Gottschling, D. E., Aparicio, O. M., Billington, B. L. & Zakian, V. A. (1990) Cell 63**,** 751–762. [DOI] [PubMed] [Google Scholar]
- 45.Tham, W. H. & Zakian, V. A. (2002) Oncogene 21**,** 512–521. [DOI] [PubMed] [Google Scholar]
- 46.Baur, J. A., Zou, Y., Shay, J. W. & Wright, W. E. (2001) Science 292**,** 2075–2077. [DOI] [PubMed] [Google Scholar]
- 47.Lin, C. M., Fu, H., Martinovsky, M., Bouhassira, E. & Aladjem, M. I. (2003) Curr. Biol. 13**,** 1019–1028. [DOI] [PubMed] [Google Scholar]
- 48.Loupart, M., Krause, S. & Heck, M. S. (2000) Curr. Biol. 10**,** 1547–1556. [DOI] [PubMed] [Google Scholar]
- 49.Pflumm, M. F. & Botchan, M. R. (2001) Development (Cambridge, U.K.) 128**,** 1697–1707. [DOI] [PubMed] [Google Scholar]
- 50.Steinert, S., Shay, J. W. & Wright, W. E. (2000) Biochem. Biophys. Res. Commun. 273**,** 1095–1098. [DOI] [PubMed] [Google Scholar]