Down-Regulation of Rad51 and Decreased Homologous Recombination in Hypoxic Cancer Cells (original) (raw)
Abstract
There is an emerging concept that acquired genetic instability in cancer cells can arise from the dysregulation of critical DNA repair pathways due to cell stresses such as inflammation and hypoxia. Here we report that hypoxia specifically down-regulates the expression of RAD51, a key mediator of homologous recombination in mammalian cells. Decreased levels of Rad51 were observed in multiple cancer cell types during hypoxic exposure and were not associated with the cell cycle profile or with expression of hypoxia-inducible factor. Analyses of RAD51 gene promoter activity, as well as mRNA and protein stability, indicate that the hypoxia-mediated regulation of this gene occurs via transcriptional repression. Decreased expression of Rad51 was also observed to persist in posthypoxic cells for as long as 48 h following reoxygenation. Correspondingly, we found reduced levels of homologous recombination in both hypoxic and posthypoxic cells, suggesting that the hypoxia-associated reduction in Rad51 expression has functional consequences for DNA repair. In addition, hypoxia-mediated down-regulation of Rad51 was confirmed in vivo via immunofluorescent image analysis of experimental tumors in mice. Based on these findings, we propose a novel mechanism of genetic instability in the tumor microenvironment mediated by hypoxia-induced suppression of the homologous recombination pathway in cancer cells. The aberrant regulation of Rad51 expression may also create heterogeneity in the DNA damage response among cells within tumors, with implications for the response to cancer therapies.
Solid tumors constitute a unique tissue type, characterized by hypoxia, low pH, and nutrient deprivation (45). Although decreased oxygen tension is potentially toxic to normal human cells, cancer cells acquire genetic and adaptive changes allowing them to survive and proliferate in a hypoxic microenvironment. Intratumoral hypoxia induces profound alterations in numerous physiological processes, including altered glucose metabolism, up-regulated angiogenesis, increased invasive capacity, and dysregulation of apoptotic programs (37).
From a clinical standpoint, many studies have established hypoxia as an independent and adverse prognostic variable in patients with head and neck, cervical, or soft tissue (sarcoma) tumors (3, 26). With regard to the extent of hypoxia observed in tumors, it has been proposed that cells within hypoxic regions of solid tumors often derive almost all metabolic energy requirements from up-regulated glycolytic pathways. This phenomenon has been referred to as the Pasteur effect (34) and provides a partial physiologic explanation for the viability of tumor cells exposed to severe hypoxia within the tumor microenvironment. Polarographic needle electrode studies used to measure oxygen tension directly in cancer patients have revealed that a significant proportion of breast carcinomas (up to 40%) contain regions of severely decreased oxygen tension (0 to 2.5 mm Hg, compared to the normal tissue range of 24 to 66 mm Hg) while still supporting viable tumor cells (40). In addition, hypoxic cells are more resistant to radiotherapy and chemotherapy; this resistance represents a significant challenge in achieving maximal treatment efficacy (32). Collectively, these studies underscore the importance of elucidating the effects of hypoxia at the molecular level and the mechanisms by which such conditions can lead to a more aggressive phenotype and tumor progression.
Tumor progression has been specifically correlated with genetic instability (27). Furthermore, it has long been argued that the large number of mutations found in malignant cells cannot be accounted for by the low rate of mutation observed in somatic cells, leading to the suggestion that cancer cells assume a mutator phenotype during tumorigenesis (23). We and others have proposed that the tumor microenvironment contributes to such genetic instability (31). Indeed, several studies using both reporter genes and endogenous loci have demonstrated increased mutation rates in cells grown in tumors relative to those in identical cells grown in culture (22, 29, 31). Hypoxia appears to be a key microenvironmental factor involved in the development of genetic instability. Studies have suggested that it is associated with increased DNA damage, enhanced mutagenesis, and functional impairment in DNA repair pathways. With regard to DNA damage, hypoxia and subsequent reoxygenation induce DNA strand breaks and oxidative base damage such as 8-oxoguanine and thymine glycols (47). Exposure of cells in culture to hypoxic conditions yields increased frequencies of point mutations at reporter gene loci (31). Hypoxia-reoxygenation cycles are also associated with other genetic aberrations, including gene amplification and DNA overreplication, although the mechanism by which they occur has not been fully elucidated (7, 46). Studies in our laboratory have also established that hypoxia induces functional decreases in the nucleotide excision repair (NER) pathway (48). Additionally, it has been reported previously that severe hypoxia (0.01% O2) specifically down-regulates the expression of the DNA mismatch repair gene MLH1, contributing to increased mutation rates (25). Collectively, these phenomena constitute a source of genetic instability induced by hypoxia, thus potentially accelerating the multistep process of tumor progression.
Given the dynamic and complex gene expression changes that have been observed under hypoxia, we have been investigating whether alterations in the expression of other DNA repair genes could also occur in response to hypoxia and thereby play a role in hypoxia-induced genetic instability. In the present study, we utilized transcriptome profiling to identify DNA repair pathways which may be regulated by hypoxia. We report that the expression of RAD51, a critical mediator of homologous recombination (HR) in mammalian cells, is specifically down-regulated by hypoxia at the mRNA level, resulting in marked decreases in the protein expression of this gene. Decreased Rad51 protein expression under hypoxia was observed in numerous cell lines from a wide range of tissues, and importantly, these decreases persisted even during the posthypoxic phase following reoxygenation. This finding is especially relevant to solid tumors that typically experience fluctuating vascular perfusion, resulting in oxygen tensions that vary spatially and temporally. Analyses of protein stability, mRNA stability, and promoter activity indicate that hypoxia regulates this gene via a mechanism involving repression of the RAD51 gene promoter. Rad51 down-regulation also appears to be independent of both the cell cycle and hypoxia-inducible factor (HIF), since no correlations were found between Rad51 expression and either the cell cycle profile or HIF induction. We also detected significantly decreased HR in both hypoxic and posthypoxic cells, indicating that hypoxia-mediated reductions in RAD51 gene expression have functional consequences. Hypoxia-mediated Rad51 down-regulation was also confirmed in vivo within the tumor microenvironment: we observed a consistent inverse association between staining with a hypoxia marker (EF5) and Rad51 protein expression in vivo by use of immunofluorescent image analysis of cervical and prostate cancer xenografts. We propose the existence of a hypoxic and posthypoxic phenotype in solid tumors characterized by decreased expression of critical DNA repair genes, representing a novel mechanism of acquired genetic instability within the tumor microenvironment.
MATERIALS AND METHODS
Cells.
MCF-7 (HTB-22), A549, RKO, SW-480, HeLa (CCL-2), ME180, SiHa, PC3, DU145, and A431 cells were obtained from the American Type Culture Collection (Manassas, Va.) and were grown according to supplier instructions and as previously described (4). 786-0 cell lines were a gift from W. G. Kaelin and were cultured as described previously (24).
Plasmids and transfections.
The HIF-1α expression vector pCEP4-HIF-1α was a generous gift from G. Semenza and has been described previously (12). The 5X-HRE luciferase reporter plasmid was a gift from Zhong Yun and has been described previously (35). Transfections were performed by using the Fugene 6 reagent (Roche Diagnostics Corporation, Indianapolis, Ind.) according to the manufacturer's recommendations.
Hypoxia exposure.
Cells were exposed to hypoxia in vitro as described previously (6). Briefly, for 0.2 to 0.5% oxygen experiments, a NAPCO incubator (Precision Scientific, Winchester, Va.) was equipped with a PROOX 710 sensor (BioSpherix, Redfield, N.Y.) to regulate the flow of 100% N2 at low pressure (<25 lb/in2) in order to achieve a constant oxygen concentration within the entire incubator for the indicated times. The CO2 level was maintained at 5% by using a regulated internal CO2 regulation system. Severe hypoxia (0.01% O2) was established as described previously (31) by using a continuous flow of a humidified mixture of 95% N2 and 5% CO2 gas certified to contain less than 10 ppm O2 (Airgas Northeast, Cheshire, Conn.). Desferrioxamine mesylate (DFX) (Sigma, St. Louis, Mo.) treatment was carried out by supplementation of the culture medium at a concentration of 250 μM under normoxic conditions for 24 h.
Transcriptome profiling.
Microarray analysis was performed with the GenCompass universal microarray system (Agilix Corporation, New Haven, Conn.). The specific details of this technology have been described recently (33). The data reference the UniGene database Build 161 (National Center for Biotechnology Information, Bethesda, Md.). Gene regulation was calculated by averaging the ratios of the signal intensities of the tags of each hypoxia-exposed condition to the signal intensities of the tags from the control (normoxic) cells.
Western blot analysis.
Radioimmunoprecipitation assay protein lysates were prepared, and Western blot analyses were performed, as described previously (4, 25). The following antibodies were used for analysis: anti-VHL (clone IG32) and anti-Rad51 (BD Pharmingen, Franklin Lakes, N.J.), anti-tubulin (clone B-512; Sigma), anti-β-actin (Research Diagnostics, Flanders, N.J.), anti-HIF-1α (clone 54; BD Transduction Laboratories, Franklin Lakes, N.J.), and anti-Glut1 (Alpha Diagnostic International, San Antonio, Tex.). Bands were quantified by using NIH Image (version 1.63; National Institutes of Health [NIH], Bethesda, Md.). Protein bands were visualized with horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G and an enhanced chemiluminescence detection system (Amersham Biosciences, Little Chalfont, Buckinghamshire, England).
Semiquantitative RT-PCR and qPCR analysis.
Semiquantitative reverse transcriptase PCR (RT-PCR) analyses were performed using the SuperScript One-Step RT-PCR kit (Life Technologies, Carlsbad, Calif.). Primer sequences for each gene used in all analyses are listed in the next section. Linear amplification ranges were determined for each gene by using 100-ng samples of normoxic and hypoxic RNA previously analyzed by Northern blotting. RT-PCR amplification was performed according to the manufacturer's recommendations. Additional information regarding the optimized RT-PCR protocol is available upon request. Bands were quantified by using NIH Image (version 1.63) analysis of SYBR Gold (Molecular Probes, Eugene, Oreg.)- or ethidium bromide (Sigma)-stained gels. For quantitative real-time PCRs (qPCRs), primers and probes were designed with Beacon Designer 2.06 (Premier Biosoft International, Palo Alto, Calif.), and analyses were performed using the MX400 Multiplex Quantitative PCR system (Stratagene, La Jolla, Calif.) as previously described (33). Primers and probes for all analyses are available upon request.
Northern blot analysis.
Total RNA was isolated by using the TRIzol RNA isolation system (Life Technologies) followed by phenol-chloroform extraction. Northern blot analyses were performed as described previously (25). The following primer pairs were used to construct the probes: for RAD51, 5′-TGGCCCACAACCCATTTCAC-3′ (sense) and 5′-TCAATGTACATGGCCTTTCCTTCAC-3′ (antisense); for the vascular endothelial growth factor gene (VEGF), 5′-CTTCACTGGATGTATTTGACTGCTGTGG-3′ (sense) and 5′-GCTAGTGACTGTCACCGATCAGGGAG-3′ (antisense). β_-Actin_ primers were obtained from Ambion. Blots were visualized by autoradiography and quantified either by phosphorimager analysis or by use of NIH Image software.
Protein and mRNA stability analyses.
Cells were plated in 10-cm-diameter dishes and allowed to attach for 24 h, followed by a 24-h incubation in the presence or absence of DFX (250 μm). Cycloheximide (CHX) (10 μg/ml; Sigma), as an inhibitor of protein synthesis, or actinomycin D (ActD) (5 μg/ml; Sigma), as an inhibitor of transcription, was then added to the cell cultures. Cells were harvested at the indicated times after the addition of inhibitor, and Rad51 protein and mRNA expression levels were determined by Western and Northern blotting, respectively. RAD51 mRNA half-lives were deduced from the regression line based on mRNA degradation plots.
Luciferase reporter gene assays.
For luciferase reporter gene analyses, a 1.8-kb fragment from the 5′-flanking region of the RAD51 gene was identified by using the UCSC genome browser (University of California—Santa Cruz), isolated by genomic PCR, and subsequently subcloned into the pGL3-basic vector (Promega, Madison, Wis.). Primers are available upon request. The subcloned fragment contains the core promoter region(s) described in the Eukaryotic Promoter Database (30) and that identified by in silico analysis using the Genomatix promoter identification algorithm PromoterInspector (44). For transfection, 5 × 105 RKO cells were seeded in duplicate into 6-well culture plates and transfected with 1 μg of each reporter construct by using the Fugene 6 reagent (Roche Diagnostics Corporation). Firefly and Renilla luciferase activities were measured by using the Dual-Luciferase Reporter Assay System kit (Promega) according to the manufacturer's instructions. Renilla luciferase activity from a cotransfected pRL-SV40 control vector (5 ng/well) was used for normalization.
Cell cycle and fluorescence-activated cell sorter (FACS) analysis.
Cell cycle analyses were performed as described previously (2). Stained cells were analyzed on a Becton Dickinson FACS-Calibur flow cytometer. Data capture, density plots, and histogram construction were performed by using BD CellQuest Pro software (Becton Dickinson), and cell cycle distribution profiles were determined by using ModFit LT software (Verity House Software, Topsham, Maine).
For FACS experiments, normoxic and hypoxic cells were incubated with 10 μg of Hoechst 33342 (Molecular Probes)/ml for 1 h at 37°C in the dark. Verapamil (100 mM; Sigma) was also added to the culture medium to inhibit dye efflux during the incubation. Cells were then trypsinized and resuspended in the culture medium supplemented with 0.1% fetal bovine serum, 10 μg of Hoechst 33342 dye/ml, and 100 mM verapamil to further enhance dye uptake. Equal numbers of specific G1- and S-phase cell populations were sorted on the basis of DNA content by using a BD FACSvantage flow cytometer fitted with a UV laser, and cells were collected in 0.5 ml of culture medium supplemented with 10% fetal bovine serum. RNA was then extracted from the isolated cells by using a modified TRIzol protocol. This technique has recently been reported to facilitate the recovery of intact mRNA from viable cells in distinct phases of the cell cycle (20).
HR assay.
The shuttle vector plasmid pSupFG1/G144C, containing a supFG1 gene with an inactivating G:C-to-C:G point mutation at position 144, has been described previously (5). For all shuttle assays, 5 μg of pSupFG1/G144C plasmid DNA was mixed with 10 μg of a PCR-generated, 1-kb homologous donor fragment containing the wild-type supFG1 gene in 50 mM Tris (pH 6.8). After transfection of the plasmid-donor mixture into cells for the indicated times, plasmid DNA was recovered by using a modified Hirt lysate procedure, as described previously (5). The purified plasmid was then used to transform Escherichia coli SY302 cells by electroporation, followed by growth of the cells on indicator plates for genetic analysis of supFG1 gene function as described previously (5).
Hypoxia immunostaining within human xenografts.
For each cell line, tumor cell suspensions (50 μl) containing 5 × 105 cells were injected into the gastrocnemius muscle of SCID mice. When a tumor diameter of 4 to 5 mm was reached, the nitroimidazole hypoxic marker EF5 (Ben Venue Laboratories, Bedford, Ohio) was injected (200 μl of a 10 mM stock solution) intravenously, and tumors were excised 3 h later, placed in Tissue-TekOCT compound (Sakura Finetek, Torrance, Calif.), and snap-frozen in liquid nitrogen. Immunofluorescence analyses were performed as described previously (42). Frozen sections were incubated overnight at 4°C with a monoclonal anti-Rad51 antibody (Affinity Bioreagents, Inc., Golden, Colo.) before incubation with a donkey anti-mouse Cy3-conjugated secondary antibody (diluted 1:400; Jackson ImmunoResearch Laboratories, West Grove, Pa.). Tissue-bound EF5 was labeled by using a 1:30 dilution of the monoclonal antibody ELK3-51 (provided by Cameron Koch, University of Pennsylvania, Philadelphia) directly labeled with Cy5. Sections were then imaged by using a cooled charge-coupled device camera (Quantix, Photometrics, Tucson, Ariz.) mounted on an epifluorescence microscope (Olympus, Melville, N.Y.) fitted with a computer-controlled motorized stage (Ludl Electronic Products, Hawthorne, N.Y.).
RESULTS
Transcriptome response to hypoxia.
Several reports have been published with the primary goal of assessing gene expression patterns under hypoxic conditions (37). Most of these studies have focused on induced expression patterns during brief periods of exposure to mild or moderate hypoxia. In this work, we sought to further characterize gene expression patterns under more prolonged exposure to hypoxia (24 h at 0.5% O2) by using a transcriptome profiling-based approach to screen for both known and novel genes which are regulated by hypoxia.
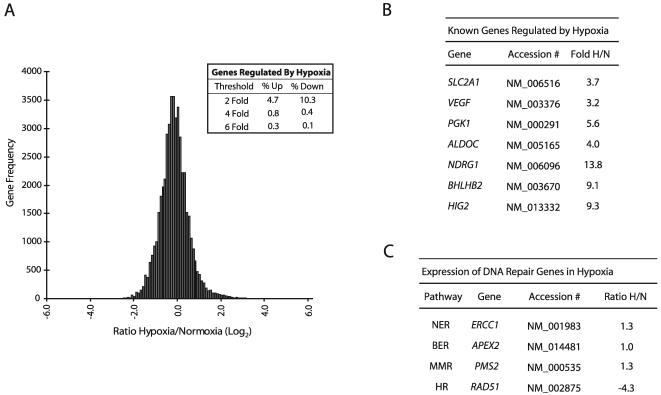
Analysis of hypoxia/normoxia (H/N) expression ratios for approximately 48,000 transcripts (16,000 unique transcripts in three biological replicates) detected in human MCF-7 breast cancer cells by transcriptome profiling revealed that the expression levels of the great majority of genes (∼85%) are not altered by a 24-h exposure to hypoxia (0.5% O2) (Fig. 1A); at a threshold of ≥2-fold regulation, approximately 5% of the genes detected demonstrated up-regulation, while 10% were down-regulated, by hypoxia. These findings were derived from pooled RNA samples analyzed in triplicate and obtained in two independent hypoxia-normoxia experiments. Interestingly, when the threshold for analysis is increased to a ≥6-fold change, more genes appear to be up-regulated than down-regulated by hypoxia (0.3 versus 0.1%, respectively). These results show that specific patterns of both up- and down-regulation occur in response to hypoxia, and they suggest that decreased expression of certain genes may be as important as increased expression in determining the phenotype of hypoxic cells.
FIG. 1.
Transcriptome response to hypoxia after 24-h exposure of MCF-7 cells to 0.5% O2. (A) Histogram analysis of the approximately 48,000 transcripts (16,000 unique transcripts for each of three biological replicates) detected by the GenCompass microarray, based on H/N expression ratios (log2). (Inset) Percentages of genes up-regulated and down-regulated at two-, four-, and sixfold thresholds. (B) Selection of genes previously identified as regulated by hypoxia which were detected by the GenCompass microarray. Accession numbers are given for reference. H/N ratios are averaged from three independent experiments. NDRG1, N-myc downstream-regulated gene 1. (C) GenCompass H/N expression ratios of selected DNA repair genes, with the respective pathways listed. H/N ratios are averaged from two experiments. BER, base excision repair; MMR, mismatch repair. ERCC1, excision repair cross-complementing group 1; APEX2, apurinic/apyrimidinic endonuclease/redox factor 2; PMS2, postmeiotic segregation increased 2; RAD51, RAD51 homolog.
As expected, hypoxic exposure led to increased expression of several known HIF target genes (Fig. 1B), such as glucose transporter 1 (SLCA21, or Glut1) and VEGF (17). Glycolysis-associated genes previously shown to be up-regulated by hypoxia, such as phosphoglycerate kinase I (PGK1) and aldolase C (ALDOC), were also significantly up-regulated. In addition, several other genes recently reported to be hypoxia inducible, such as DEC1 (BHLHB2) and hypoxia-inducible gene 2 (HIG2), displayed marked elevation, with 9.1- and 9.3-fold changes in H/N ratios, respectively. The H/N fold changes for the genes shown in Fig. 1B were verified by qPCR (data not shown).
Expression of DNA repair genes under hypoxia.
Given that a comprehensive analysis of DNA repair gene expression under prolonged hypoxia had not been performed previously, we sought to determine whether there are specific genes in this category that exhibit novel regulation by hypoxia. The microarray detected the expression of more than 50 transcripts from genes that directly play a role in DNA repair, and representative genes from several repair pathways are shown in Fig. 1C. The expression levels of the majority of DNA repair genes detected were not substantially altered by hypoxia, including genes in pathways such as NER and base excision repair, consistent with previous work in our laboratory examining expression patterns of factors in these pathways by Western blot analyses (48). A more comprehensive list of DNA repair genes from these pathways, including qPCR expression validation for several of these genes, is available upon request. We did detect a moderate decrease in the level of MLH1 expression under hypoxia (1.3-fold [data not shown]). It had been demonstrated previously that MLH1 expression is down-regulated by hypoxia, especially after prolonged periods (>24 h) of severe hypoxia (<0.1% O2), and thus the finding that MLH1 mRNA is only slightly decreased at 24 h of moderate hypoxia (0.5% O2) is consistent with previous data (25).
Intriguingly, we detected substantial decreases in the expression of RAD51 under hypoxia (4.3-fold). This decrease was verified by qPCR and was specific to the RAD51 gene; we did not detect hypoxia-induced decreases in the expression of other HR genes in the RAD52 epistasis group (38), including RAD51B, RAD54B, RAD52, and RAD50 (data not shown). Consequently, we reasoned that such decreases in a critical HR-associated gene could compromise recombinational repair in cells exposed to hypoxia.
Decreased expression of Rad51 protein in response to hypoxia or the Fe2+ chelator DFX.
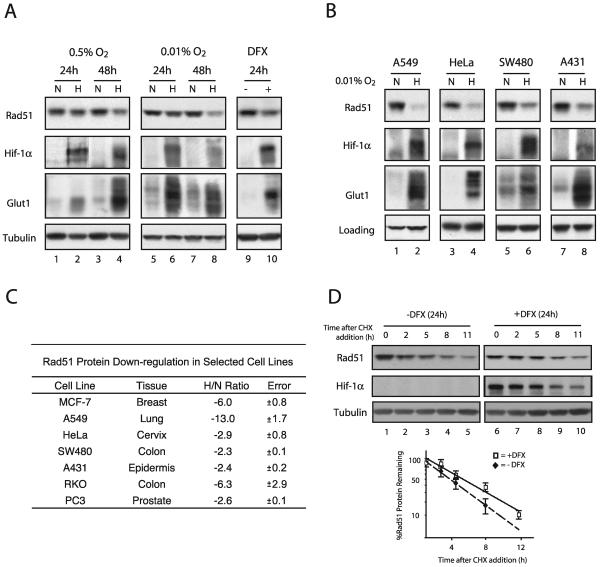
We sought to determine whether the changes observed in the microarray and qPCR experiments were also manifested at the protein level. In order to account for the effects of protein stability, we examined Rad51 protein expression levels not only at the 24-h time point used in the microarray experiments but also after 48 h of hypoxia. As shown in Fig. 2A, Western blot analysis revealed that Rad51 protein levels were substantially decreased after 48 h of hypoxia (approximately threefold) (lane 4), with minimal decreases observed after 24 h of hypoxia (lane 2). The expression of HIF-1α and its downstream target Glut1 (which appears as multiple isoforms between 37 and 75 kDa) is shown for comparison, to confirm physiologically relevant levels of hypoxia. In addition, levels of tubulin were unchanged and served as standards to confirm equal loading of cellular protein samples.
FIG. 2.
Decreased levels of Rad51 protein in cell lines in response to hypoxia or the Fe2+ chelator DFX, and determination of Rad51 protein stability in the presence of DFX. (A) Western blot analyses were performed to determine the expression of the HR-associated protein Rad51 in MCF-7 cells after exposure to normoxia (lanes N), hypoxia (0.5 or 0.01% O2) (lanes H), or DFX (250 μM). The time for which the cells were maintained under each condition (24 or 48 h) is shown. Expression of HIF-1α and Glut1 is shown for comparison, to verify that physiologically relevant levels of hypoxia were present in the treated cells. Note that Glut1 appears as several isoforms which are detected by our antibody. Tubulin expression is also presented to confirm equal sample loading. (B) Western blot analyses of Rad51 protein expression in A549, HeLa, SW480, and A431 cells after a 48-h exposure to normoxia or hypoxia (0.01% O2). Tubulin expression is presented to confirm equal loading of samples for HeLa and SW480 cells, while β-actin was used as a loading control for A549 and A431 cells. (C) Average reduction in Rad51 protein levels after a 48-h exposure to hypoxia (0.01% O2), as determined by densitometry analysis of Western blots generated from duplicate (RKO, A431, SW480, and PC3 cells) or triplicate (MCF-7, A549, and HeLa cells) hypoxia experiments. The tissue of origin for each cell line is shown. Reductions are expressed as H/N ratios normalized to the expression of β-actin and tubulin, and standard errors for each ratio are given. (D) To assess Rad51 protein stability, MCF-7 cells were either exposed to DFX (250 μM) or left untreated for 24 h, followed by coincubation with CHX (10 μg/ml) to block new protein synthesis. (Upper panels) Cells were harvested at the indicated times after addition of CHX, and Rad51 protein expression was determined by Western blotting. Expression of HIF-1α is shown to confirm both the induction of chemical hypoxia and successful abolition of new protein synthesis. In addition, tubulin protein levels were unchanged and served as standards to confirm equal loading of cellular protein samples. (Lower panel) Analysis of Rad51 protein expression at each time point after addition of CHX in cells exposed to DFX or left untreated, quantified as described in the legend to panel C. Each data point is the percentage of Rad51 protein remaining (after normalization to tubulin levels) in either DFX-treated or untreated cells at the indicated time.
As discussed earlier, we had previously reported the specific down-regulation of the MLH1 gene after prolonged exposure to more severe hypoxia. Based on this finding and on the relevance of such conditions to chronically hypoxic regions in tumors, we subjected MCF-7 cells to more severe hypoxia (0.01% O2) for prolonged periods in order to determine whether there might be even greater reductions in Rad51 protein expression. As shown, a 48-h exposure to 0.01% O2 resulted in a larger decrease (approximately sixfold) in Rad51 protein expression (Fig. 2A, lane 8). As expected, decreases were also observed in Mlh1 protein expression, but no reductions were observed in the expression of several other DNA repair proteins, including Msh2 and Msh6 (data not shown). Thus, both moderate hypoxia and severe hypoxia are associated with profound and specific decreases in the expression of the RAD51 gene in MCF-7 cells.
The hypoxic state can be mimicked in cell culture by using the iron chelator DFX, which has been proposed to disrupt normal oxygen-sensing pathways in mammalian cells by inhibiting heme-Fe2+ interactions (43). As shown in Fig. 2A, a 24-h exposure of MCF-7 cells to DFX also resulted in decreased Rad51 protein expression (lane 10). In addition, HIF-1α and Glut1 levels were increased in DFX-treated MCF7 cells, confirming that this chemical treatment mimicked aspects of gas-induced hypoxia. These results show that Rad51 protein expression is reduced not only in truly hypoxic cells but also in cells in which hypoxia is simulated by interference with normal cellular oxygen sensing.
Given that such profound decreases in Rad51 protein expression after hypoxic stress could have broad implications for genetic instability in the tumor microenvironment, we sought to determine whether this phenomenon could also be detected in tumor cell lines derived from other tissues. A selection of tumor cell lines from various tissues were cultured under hypoxic and normoxic conditions, followed by Western blot analysis to assess Rad51 protein levels. As shown in Fig. 2B, significant decreases in Rad51 protein expression were observed after 48 h of hypoxia in a wide range of human cell lines, including the A549 epithelial lung carcinoma cell line and the HeLa cervical cancer cell line. Densitometry analysis was used to approximate H/N protein expression ratios for several cell lines evaluated in this study, based on triplicate hypoxia experiments and normalization to either β-actin or tubulin expression. As shown in Fig. 2C, A549 and MCF-7 cells displayed the highest levels of down-regulation, with H/N ratios of −13 and −6, respectively. Taken together, significant hypoxia-mediated down-regulation of Rad51 protein is observed in numerous human cell lines derived from a wide range of tissues.
To determine whether the observed decreases in Rad51 protein expression could be accounted for by an increase in protein degradation, MCF-7 cells were either left untreated or exposed to DFX, followed by coincubation with CHX to block new protein synthesis. Cells were then harvested at various times after the addition of CHX, and Rad51 protein expression was determined by Western blotting. As shown in Fig. 2D, while a 24-h DFX exposure resulted in a threefold decrease in Rad51 protein expression (compare lanes 1 and 6), no significant differences in Rad51 protein stability between untreated cells and cells exposed to DFX were observed after the addition of CHX. In contrast, HIF-1α expression was substantially increased in the presence of DFX, and HIF-1α degraded rapidly after CHX addition, confirming both the induction of chemical hypoxia and successful abolition of new protein synthesis. In addition, levels of tubulin protein were unchanged and served as standards to confirm equal loading of cellular protein samples. These findings suggest that the hypoxia-mediated down-regulation of the RAD51 gene does not occur at the posttranslational level.
Prolonged hypoxia leads to the down-regulation of RAD51 mRNA expression.
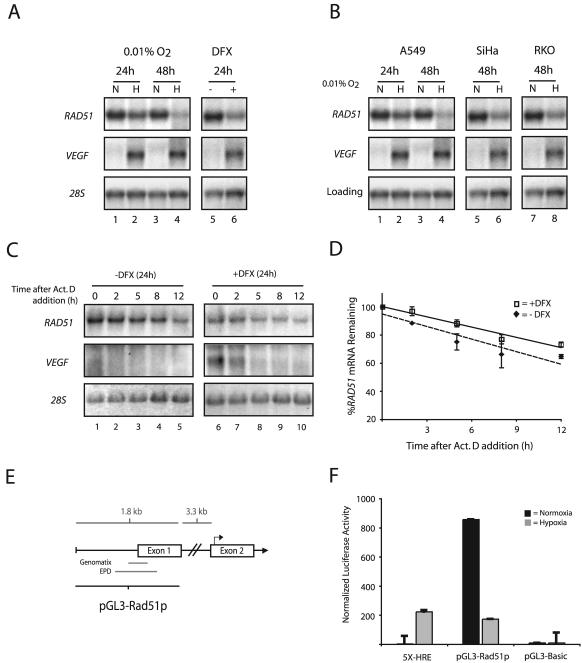
To determine if the decreases in Rad51 protein expression observed after severe hypoxia are also associated with decreased mRNA levels, Northern blot analyses were performed on total RNAs extracted from MCF-7 and A549 cells after 24- and 48-h exposures to 0.01% O2. RAD51 mRNA levels were substantially decreased at the 48-h time point, with H/N ratios of approximately −8 and −9 in MCF-7 and A549 cells, respectively (Fig. 3A and B, respectively). Ratios were averaged from multiple experiments and normalized to either 28S rRNA or β_-actin_ mRNA levels. Significant decreases were also observed at the 24-h time point in both cell lines. The expression of VEGF is shown, to verify the induction of physiological levels of hypoxia, and β_-actin_ and 28S rRNA are presented as loading controls. Consistent with the protein expression levels, a 24-h exposure of MCF-7 cells to DFX also resulted in decreased expression of RAD51 mRNA (Fig. 3A, lane 6), and this finding was also verified by qPCR analysis (data not shown). Additionally, hypoxia-induced decreases in RAD51 mRNA expression were observed in a number of other human cell lines, including SiHa, RKO, and DU145 (Fig. 3B; also data not shown). These findings provide consistent evidence that hypoxia regulates the expression of the RAD51 gene at the mRNA level.
FIG. 3.
Transcriptional repression of the RAD51 gene by hypoxia. (A) Northern blot analyses were performed on total RNA extracted from MCF-7 cells after exposure to normoxia (lanes N), hypoxia (0.01% O2) (lanes H), or DFX (250 μM). The time for which cells were maintained under each condition (24 or 48 h) is given. VEGF expression is shown for comparison, to verify that physiologically relevant levels of hypoxia were present in the treated cells, and expression of 28S rRNA is presented to confirm equal sample loading. (B) Northern blot analysis of RAD51 mRNA expression in A549, SiHa, and RKO cells after a 24- or 48-h exposure to hypoxia (0.01% O2). VEGF expression is shown for comparison, to verify that physiologically relevant levels of hypoxia were present in the treated cells. Expression of 28S rRNA (MCF-7, SiHa, and RKO cells) and β_-actin_ mRNA (A549 cells) is presented to confirm equal sample loading. (C) To assess the stability of RAD51 mRNA, MCF-7 cells were either left untreated or exposed to DFX (250 μM) for 24 h, followed by coincubation with ActD (5 μg/ml) to block transcription. Cells were harvested at the indicated times after the addition of ActD, and RAD51 mRNA expression was determined by Northern blotting. Expression of VEGF is shown to confirm both the induction of chemical hypoxia and successful abolition of transcription. In addition, 28S rRNA levels were unchanged and served as standards to confirm equal sample loading. (D) Analysis of RAD51 mRNA expression at each time point after ActD addition in cells exposed to DFX or left untreated, as determined by phosphorimager analysis of Northern blots. Values are the percentage of RAD51 mRNA remaining in either DFX-treated or untreated cells at each time point, and error bars are based on standard errors calculated from duplicate experiments. (E) Schematic of the 5′-flanking region of the RAD51 gene, with delineation of the promoter fragment used for luciferase reporter gene assays (pGL3-Rad51p). Approximate locations of the core promoter regions, as described in the Eukaryotic Promoter Database (EPD) and as identified by in silico analysis using the Genomatix promoter identification algorithm PromoterInspector, are shown for reference. Bent arrow above exon 2 indicates the ATG translation start codon. (F) To determine the effect of hypoxia on RAD51 gene promoter activity, the pGL3-Rad51p luciferase (firefly) reporter plasmid was transiently transfected into RKO cells 4 h prior to normoxic or hypoxic exposure (for 48 h), immediately followed by measurement of luciferase activity. Firefly luciferase values were normalized to Renilla luciferase activity from a cotransfected pRL-SV40 control vector, and error bars are based on standard errors calculated from duplicate experiments. The activity of the luciferase reporter plasmid 5X-HRE, which contains five HREs tandemly ligated to a human cytomegalovirus minimal promoter, is shown as a control to confirm physiologically relevant levels of hypoxia. The activity of the promoterless luciferase reporter gene construct pGL3-Basic is also shown as a control.
Transcriptional repression of the RAD51 gene by hypoxia.
We next sought to determine the mechanism by which hypoxia down-regulates steady-state levels of RAD51 mRNA. To test a possible effect on RAD51 mRNA stability, MCF-7 cells were exposed to DFX, followed by coincubation with ActD to block transcription. Cells were then harvested at various times after the addition of ActD, and RAD51 mRNA expression was determined by Northern blotting. As shown in Fig. 3C, while a 24-h DFX exposure resulted in a twofold decrease in RAD51 mRNA expression (compare lanes 1 and 6), no significant differences in mRNA stability after ActD addition were observed between cells exposed to DFX and cells left untreated (Fig. 3D). RAD51 mRNA half-lives were determined to be approximately 20 and 15 h in DFX-exposed and untreated cells, respectively, and these values were based on duplicate ActD experiments. In contrast, VEGF expression is substantially increased in the presence of DFX, and VEGF mRNA degrades rapidly after ActD addition, confirming both the induction of chemical hypoxia and the successful inhibition of transcription. In addition, 28S rRNA levels were unchanged and served as standards to confirm equal sample loading. These data indicate that the hypoxia-induced down-regulation of RAD51 mRNA expression cannot be accounted for by decreases in the stability of the mRNA; thus, they suggest that regulation occurs at the level of transcription.
To test whether the reduction in steady-state levels of RAD51 mRNA under hypoxia is dependent on RAD51 promoter regulation, the effect of hypoxia on RAD51 promoter activity was examined by using a promoter-luciferase reporter system. As shown in Fig. 3E, a 1.8-kb fragment from the 5′-flanking region of the RAD51 gene, containing the core promoter region(s) described in the Eukaryotic Promoter Database (30) and that identified by in silico analysis using the Genomatix promoter identification algorithm PromoterInspector (44), was isolated. A firefly luciferase reporter plasmid containing this fragment (pGL3-Rad51p) was transiently transfected into RKO cells 4 h prior to normoxic or hypoxic exposure (48 h), immediately followed by measurement of luciferase activity. Renilla luciferase activity from a cotransfected pRL-SV40 control vector was used for normalization. The luciferase reporter plasmid 5X-HRE, which contains five hypoxia response elements (HREs) tandemly ligated to a human cytomegalovirus minimal promoter, was used as a control to confirm physiologically relevant levels of hypoxia. As shown in Fig. 3F, exposure to hypoxia resulted in a fivefold repression of pGL3-Rad51p reporter activity. In contrast, 5X-HRE reporter activity increased approximately 50-fold, and no change in activity was observed in the promoterless control plasmid pGL3-Basic, in hypoxic cells. RKO cells were used in these studies due to the ease with which they are transfected and because of their ability to support high levels of RAD51 promoter activities. RAD51 promoter activity was also repressed by hypoxia in MCF-7 and A549 cells, with H/N ratios of −1.8- and −1.5-fold, respectively (data not shown). Taken together, these findings suggest that hypoxia down-regulates the expression of the RAD51 gene via a mechanism involving transcriptional repression.
Persistent down-regulation of Rad51 expression posthypoxia.
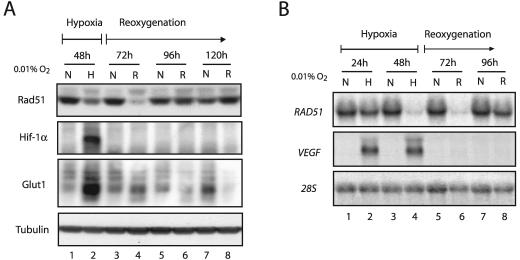
Given the dramatic decreases that we observed in RAD51 mRNA and protein levels after 48 h of hypoxia, we sought to determine the extent to which these alterations persisted after reoxygenation. MCF-7 cells were exposed to 48 h of hypoxia, followed by a return to normoxic conditions for several days thereafter. Cell lysates were prepared at 24-h intervals throughout the time course, and Western blot analyses were performed to assess Rad51 expression levels during and after hypoxic exposure. Intriguingly, we detected the lowest levels of Rad51 protein in the period following hypoxia. As shown in Fig. 4A, Rad51 levels were lowest at 24 h after reoxygenation (referred to as the 72-h time point) (lane 4), and these levels did not return to normal, prehypoxia expression levels until after the 96-h time point (lane 6), which corresponds to 48 h posthypoxia. The HIF-1α protein is rapidly degraded under normoxic conditions, and thus it was not detected upon reoxygenation at any of the posthypoxia time points shown. Furthermore, we also observed a rapid decrease in Glut1 expression following reoxygenation, a response that is expected for this gene in the transition from hypoxia to normoxia. Rad51 protein expression levels at the 48- and 72-h time points were reduced approximately 6- and 12-fold, respectively, and these values were based on triplicate experiments. Reductions in Rad51 protein expression were also observed 20 h after a shorter, 24-h hypoxia exposure, although these decreases were not as pronounced (data not shown). In addition, similar trends were also observed at the protein level in A549 cells, with a slightly more rapid return of protein levels following reoxygenation (Fig. 5D).
FIG. 4.
Persistent down-regulation of Rad51 expression after hypoxia. (A) Western blot analyses were performed to determine the expression pattern of Rad51 protein in MCF-7 cells cultured under hypoxia for 48 h (0.01% O2) (lane H) and then following reoxygenation (R), as indicated by the timeline shown. Samples were obtained at 24-h intervals posthypoxia (lanes R), with the 72-, 96-, and 120-h time points representing 24, 48, and 72 h following reoxygenation, respectively. As a control to show that Rad51 levels do not change over the same period under normoxia, Rad51 protein expression in cells grown in parallel under consistently normoxic conditions is shown for each time point (lanes N). Expression of HIF-1α and Glut1 is shown for comparison, to verify that physiologically relevant states of hypoxia and reoxygenation were observed in the treated cells. Tubulin expression is also presented to confirm equal sample loading. (B) Northern blot analysis was performed to determine the expression of RAD51 mRNA in MCF-7 cells during the same time course and under the same conditions described for panel A. As indicated by the timeline shown, RAD51 mRNA expression was assessed at 24 and 48 h of hypoxia (lanes H). RNA samples were also analyzed at 24-h intervals posthypoxia (lanes R), with the 72- and 96-h time points representing 24- and 48-h periods of reoxygenation, respectively. As a control to show that RAD51 expression does not change over the same period under normoxia, RAD51 mRNA expression in normoxic cells grown in parallel is shown for each time point (lanes N). VEGF expression is shown for comparison, to verify that physiologically relevant states of hypoxia and reoxygenation were obtained in the treated cells. Expression of 28S rRNA is also presented to confirm equal sample loading.
FIG. 5.
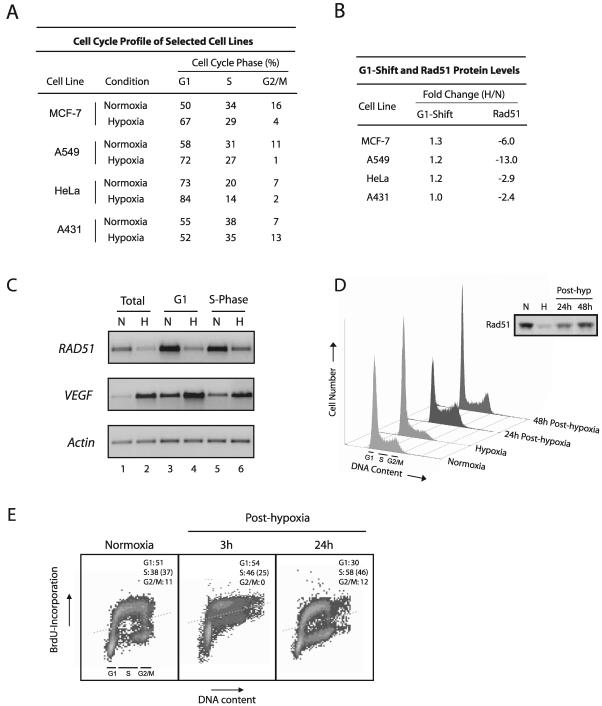
Decreased Rad51 expression is not associated with the cell cycle profile. (A) Quantitative assessment of cell cycle profiles of four cell lines exposed to hypoxia (0.01% O2) or normoxia for 48 h. Calculated proportions are expressed as percentages based on triplicate hypoxia experiments using flow cytometric analysis of PI-stained cells and histogram analysis software. (B) Fold changes in both the percentage of cells in the G1 phase and the Rad51 protein level in hypoxia compared to normoxia, as calculated from panel A and Fig. 2C, respectively. (C) Semiquantitative RT-PCR analysis of RAD51 mRNA expression in isolated G1- and S-phase populations of normoxic and hypoxic cells. Equal numbers of cell cycle-specific populations were obtained by DNA staining of cells with the vital fluorochrome Hoechst 33342, followed by flow cytometric analysis and cell sorting. Unsorted cells processed in parallel are also shown for reference (lanes 1 and 2). VEGF expression is shown for comparison, to verify physiologically relevant states of hypoxia in normoxic and hypoxic cell populations, and the expression of β_-actin_ is presented to confirm equal sample loading. (D) Cell cycle profiles of A549 cells exposed to either normoxia or hypoxia for 48 h and of A549 cells reoxygenated immediately following hypoxia for the indicated times, shown as histograms based on PI staining for DNA content. Approximate ranges of G1-, S-, and G2/M-phase populations are shown for reference. (Inset) Rad51 protein expression in the A549 cells at the corresponding time points. (E) S-phase proliferation in normoxic A549 cells or in A549 cells reoxygenated for the indicated times, as assessed by BrdU incorporation. Dotted lines represent the threshold for positive BrdU incorporation, based on cells assayed in parallel without BrdU incubation at each time point. Quantitative assessments of cell cycle profiles at each time point are shown in each panel and were calculated as described in the legend to panel A. The percentage of total cells in each sample that incorporated BrdU is given in parentheses.
Northern blot analysis revealed that RAD51 mRNA levels were also maximally decreased at the 72-h time point, representing 24 h posthypoxia (Fig. 4B, lane 6). Furthermore, mRNA and protein expression data from three independent experiments revealed that the hypoxia-induced decrease in Rad51 protein levels was consistently preceded by decreased RAD51 mRNA levels (data not shown). As expected, the expression of VEGF decreased rapidly to undetectable levels after reoxygenation and thus serves as a control for the physiological transition from hypoxia to normoxia. Taken together, these results indicate that hypoxia induces substantial decreases in RAD51 mRNA levels, which are followed by corresponding reductions in Rad51 protein levels. Importantly, these reductions are most pronounced in the posthypoxia reoxygenation phase and persist for a significant period thereafter.
Hypoxia-mediated down-regulation of the RAD51 gene is independent of the cell cycle profile.
RAD51 mRNA expression levels have been shown to be highest in the S and G2 phases of the cell cycle, and lowest during G0 and G1, in mammalian cells (11). We thus sought to determine whether the observed decreases in Rad51 expression could be accounted for by specific changes in the cell cycle profile induced by hypoxia. To this end, we used flow cytometric analyses to determine the cell cycle profiles of several of the human cell lines listed in Fig. 2C upon exposure to hypoxia. As shown in Fig. 5A, a range of cell cycle profile changes were observed under hypoxia among the four cell lines studied. We reasoned that if Rad51 expression decreases could be accounted for by increases in the proportion of cells in the G1 phase under hypoxia, for example, then there should be direct correlations between hypoxia-induced G1 shifts and H/N ratios of Rad51 expression in the cell lines evaluated. As shown in Fig. 5B, no such correlations were observed; MCF7, A549, and HeLa cells all displayed small increases in the proportion of G1-phase cells upon hypoxia exposure (1.3-, 1.2-, and 1.2-fold, respectively) yet exhibited substantial (but variable) H/N ratios of Rad51 expression (−6.0, −13.0, and −2.9, respectively). A431 cells, by contrast, displayed no detectable G1 shift under hypoxia yet displayed H/N expression ratios similar to those seen in HeLa cells (−2.4-fold).
In order to further confirm the cell cycle independence of the down-regulation of RAD51 gene expression by hypoxia, specific populations of G1- and S-phase cells were isolated from both normoxic and hypoxic A549 cells, followed by analysis of RAD51 mRNA expression by semiquantitative RT-PCR. Cell cycle phase-specific cell populations were obtained by DNA staining of cells with the vital fluorochrome Hoechst 33342, followed by flow cytometric analysis and cell sorting. This technique has recently been reported to facilitate the recovery of intact mRNA from viable cells in distinct phases of the cell cycle (20). As shown in Fig. 5C, substantial decreases in RAD51 mRNA expression were observed in both G1- and S-phase cells after a 48-h exposure to hypoxia (lanes 4 and 6, respectively), with H/N ratios of −3.2 and −2.7, respectively. These H/N ratios were similar in magnitude to those observed in unsorted hypoxic cells processed in parallel (Fig. 5C, lanes 1 and 2), which displayed H/N ratios of approximately −3.4. VEGF expression is shown for comparison, to verify physiologically relevant states of hypoxia in the individual normoxic and hypoxic G1- and S-phase cell populations, and the expression of β_-actin_ is also presented to confirm equal sample loading. These data provide direct evidence that the down-regulation of RAD51 gene expression by hypoxia occurs in both the G1 and S phases of the cell cycle and thus cannot be attributed to changes in the cell cycle profile.
A lack of correlation between the cell cycle phase and decreased Rad51 expression was also observed during the posthypoxia reoxygenation period, based on propidium iodide (PI) analyses of DNA content and on the technique of bromodeoxyuridine (BrdU) incorporation to assess rates of S-phase proliferation. As shown in Fig. 5D, marked increases in the proportions of A549 cells in the S and G2/M phases were observed 24 h posthypoxia (58 and 12%, respectively), yet Rad51 levels in these cells were persistently decreased. In addition, cells were clearly entering S phase and undergoing DNA replication in the early posthypoxic period, when Rad51 expression remained at its lowest levels. For example, S-phase proliferation rates at 3 and 24 h posthypoxia were 25 and 46%, respectively (Fig. 5E). Collectively, these data demonstrate that posthypoxic A549 cells resume S-phase replication and enter the G2/M phase of the cell cycle yet still show substantial decreases in Rad51 protein expression, indicating further that the down-regulation of Rad51 is not governed by the proportion of cells in G1 or S phase and demonstrating an uncoupling between cell proliferation and DNA repair gene expression. Taken together, these findings provide strong evidence that decreased RAD51 gene expression is more hypoxia specific than cell cycle phase dependent.
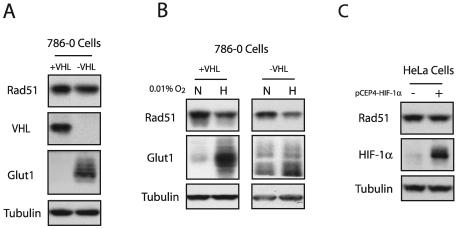
Reduced expression of Rad51 is not associated with HIF expression.
The HIF family of proteins has been shown to regulate the expression of numerous genes that play roles in angiogenesis, glycolysis, invasion, and metastasis in response to hypoxia (17). It has also been demonstrated recently that HIF-1α can induce the expression of transcriptional repressors such as DEC1, a gene revealed by our microarray analysis to be induced by hypoxia (Fig. 1B). Hence, we tested whether the observed decreases in RAD51 expression induced by hypoxia might be mediated by HIF-1α or HIF-2α. HIF proteins are highly unstable under normal oxygen tensions, because both HIF-1α and HIF-2α are ubiquitinated through the interaction with the von Hippel-Lindau tumor suppressor protein (pVHL) and subsequently degraded by the 26S proteasome under normoxic conditions (19). The pVHL-deficient cell line 786-0 specifically overexpresses HIF-2α and consequently overexpresses HIF-2α downstream target genes, including Glut1 and VEGF (24). Expression of the VHL cDNA in these cells restores the normoxic regulation of HIF-2α. We thus examined the expression of Rad51 protein in 786-0 cells complemented either with the VHL cDNA or with an empty vector. As shown in Fig. 6A, the expression of Rad51 was not affected by VHL status or HIF-2α expression. The expression of VHL and Glut-1 proteins are shown to confirm VHL status and a constitutively hypoxic phenotype, respectively, in these two cell lines. Northern blotting also revealed no differences in steady-state RAD51 mRNA levels between the VHL mutant and wild-type cells (data not shown). Interestingly, exposure of 786-0 cells expressing either wild-type or mutant VHL to hypoxia resulted in similar decreases in Rad51 expression (Fig. 6B). Taken together, these data suggest that the hypoxia-mediated down-regulation of Rad51 expression occurs independently of HIF-2α. It also suggests that the down-regulation does not require the expression of HIF-1α, since 786-0 cells lack HIF-1α expression (19). As an alternative approach to assessing the role of HIF-1α in the regulation of Rad51 expression by hypoxia, we transiently overexpressed a full-length HIF-1α cDNA in normoxic HeLa cells. Figure 6C demonstrates that exogenous overexpression of HIF-1α was not associated with decreased expression of Rad51. Collectively, these data suggest that hypoxia-induced decreases in RAD51 gene expression are not mediated by HIF-dependent pathways.
FIG. 6.
Decreased Rad51 expression is not associated with HIF expression. (A) Western blot analyses were performed to determine the expression of Rad51 protein in log-phase 786-0 cells expressing a wild-type (+VHL) or mutant (−VHL) VHL gene. Expression of VHL and Glut1 is shown for comparison to verify the status of these cells. Tubulin expression is also presented to confirm equal sample loading. (B) Western blot analysis of Rad51 protein expression in 786-0 cells expressing either wild-type or mutant VHL following exposure to hypoxia (0.01% O2) for 48 h. (C) Western blot analysis of Rad51 and HIF-1α expression levels in HeLa cells 48 h after transfection with the HIF-1α expression vector pCEP4-HIF-1α.
Decreased HR in hypoxic and posthypoxic cells.
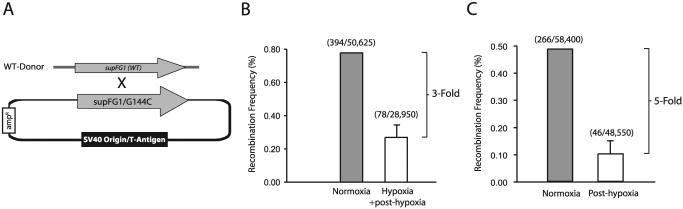
We next sought to determine whether hypoxia-induced reductions in Rad51 protein expression were associated with functional decreases in HR. We utilized a shuttle vector recombination assay involving the transient transfection of a plasmid containing a mutated reporter gene along with a wild-type donor fragment into MCF-7 cells exposed to normoxia or hypoxia in order to assess frequencies of HR. Plasmid pSupFG1/G144C, containing a mutated version of the supFG1 amber suppressor tRNA gene, _supFG1_-144, was used as the recombination substrate. A homologous fragment containing the wild-type supFG1 gene was used as a donor for recombination. The function of the supFG1 gene can be assayed by recovery of the episomal shuttle vector plasmid from the MCF-7 cells, with subsequent transformation into indicator bacteria carrying an amber stop codon in the lacZ gene. In this manner, _supFG1_-144 reports recombination events that cause the gene to revert to the functional sequence as detected in a blue/white colony screen (5). A schematic of this reporter system is presented in Fig. 7A for reference. Important features of this assay are the facts that vector replication and recombination are independent of the cell cycle and that previous studies in our lab have provided evidence that recombination events in this assay are dependent on Rad51 (8).
FIG. 7.
Decreased HR in hypoxic and posthypoxic cells. (A) Schematic of the shuttle vector plasmid and donor DNA fragment used to assess HR. Plasmid pSupFG1/G144C contains a supFG1 gene with an inactivating G:C-to-C:G point mutation at position 144. The donor is a homologous DNA fragment containing a portion of the wild-type supFG1 gene. Abbreviations: ampr, ampicillin resistance gene; WT, wild type. (B) Recombination frequencies in MCF-7 cells cotransfected with the shuttle vector and donor fragment, followed by culture either under normoxic conditions alone (72 h) or under hypoxia (0.01% O2) for 48 h, immediately followed by reoxygenation and 24 h of normoxia (Hypoxia + post-hypoxia). The total number of blue colonies/ total number of colonies in each sample is given in parentheses. Error bars are based on standard errors calculated from duplicate experiments. (C) Recombination frequencies in MCF-7 cells cotransfected with the shuttle vector and donor fragment under normoxic conditions immediately following a 48-h exposure either to normoxia or to hypoxia (0.01% O2) (Post-hypoxia).
As shown in Fig. 7B, cotransfection of the shuttle vector and wild-type fragment into MCF-7 cells immediately prior to hypoxic exposure (for 48 h), followed by plasmid recovery 24 h posthypoxia, revealed a ∼3-fold decrease in recombination frequency in hypoxic cells relative to that observed in MCF-7 cells incubated under normoxia for the same total time. Intriguingly, we observed the most substantial decreases in HR frequencies when the cells were transfected during the posthypoxic period. Figure 7C shows that transfection of the shuttle vector and donor fragment into MCF-7 cells immediately following the hypoxic (48-h) period resulted in an almost fivefold decrease in HR, again relative to the frequency observed in cells maintained under normoxic conditions throughout. Thus, the observed decreases in HR parallel the changes in expression of Rad51 protein in hypoxia and posthypoxia. Taken together, these data provide evidence that the decreased expression of Rad51 caused by hypoxia is associated with a substantial and prolonged reduction in the capacity of cells to carry out HR both during and after hypoxia, for as long as 48 h following hypoxic exposure.
Inverse association between hypoxia and Rad51 expression in cervical and prostate cancer xenografts.
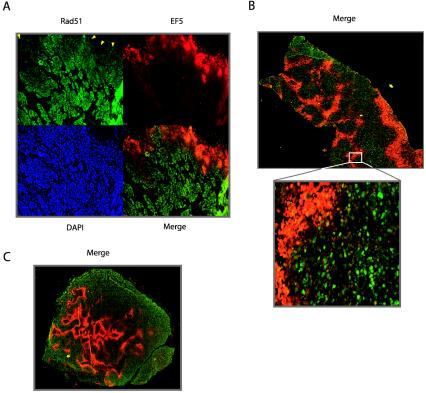
We next sought to determine whether hypoxia-induced decreases in Rad51 expression could be detected in vivo in the tumor microenvironment. To this end, human prostate and cervical cancer xenografts in mice were generated from the PC3, Me180, and SiHa6 cell lines. These cell lines had all exhibited hypoxia-induced decreases in Rad51 expression in culture (Fig. 2C and 3B; also data not shown). Histological sections from the resulting tumors were analyzed by immunofluorescence for Rad51 expression and staining with the hypoxia marker EF5. The binding of EF5 to cellular macromolecules occurs as a result of hypoxia-dependent bioreduction by cellular nitroreductases and thus can be used to detect hypoxia in solid tumors (10). As shown in Fig. 8, we detected an inverse association between Rad51 protein expression and EF5 staining in xenografts from all three cell lines. All tumors were found to contain hypoxic areas, as measured by an Eppendorf pO2 probe prior to removal for immunostaining (data not shown) (9). This inverse association is particularly striking along the upper border of the section from a Me180 cervical xenograft shown in Fig. 8A, in which Rad51 expression is substantially decreased within a region of strong EF5 staining. In the merged image, the overlay of Rad51 expression and EF5 binding reveals minimal overlap in the majority of the section (Fig. 8A). Figure 8B and C also demonstrate consistent inverse associations between Rad51 expression and EF5 staining in PC3 and SiHa6 xenografts, respectively. Collectively, these findings demonstrate that hypoxia within the tumor microenvironment is associated with decreased Rad51 expression; thus, they extend our in vitro observations to the in vivo situation in tumors.
FIG. 8.
Inverse association between hypoxia marker staining and Rad51 expression in cervical and prostate cancer xenografts. Shown are immunofluorescence analyses of staining with the hypoxia marker EF5 (red) and Rad51 expression (green) in tumor xenografts derived from Me180 cervical cancer cells (A), PC3 prostate cancer cells (B), and SiHa6 cervical cancer cells (C). Individual EF5, Rad51, and DAPI staining (blue), and Rad51-EF5 merged staining (yellow), are shown in panel A. Arrowheads in panel A indicate substantially decreased Rad51 expression within a region of strong EF5 staining.
DISCUSSION
In the present study, we have demonstrated that hypoxia specifically down-regulates the expression of RAD51, a critical mediator of HR, both in vitro in cell culture and in vivo within the tumor microenvironment. Substantial decreases in Rad51 expression both during and after hypoxic exposure were observed in a wide range of cell types, and the mechanism of regulation appears to be independent of cell cycle profile and HIF expression. Analyses of protein stability, mRNA stability, and promoter activity indicate that hypoxia regulates RAD51 gene expression via a mechanism involving transcriptional repression. We detected decreased HR in cells both during and after hypoxic stress, demonstrating that the down-regulation of Rad51 expression by hypoxia has functional consequences for DNA repair. These findings were extended to the tumor microenvironment; we detected decreased expression of Rad51 in hypoxic regions of cervical and prostate tumor xenografts. Taken together, we propose a novel mechanism of genetic instability in the tumor microenvironment mediated by hypoxia-induced suppression of the HR pathway.
In a recent review regarding genetic instability and tumorigenesis, Loeb and colleagues proposed a paradigm shift in the way in which DNA repair pathways are thought to be regulated in mammalian cells (23). In the traditional view, it was thought that DNA repair genes were expressed constitutively in cells, such that they may be readily available as new DNA damage lesions arise from exogenous insults or endogenous processes such as replication errors or oxidative metabolism. Inactivation of these pathways in cancer was thought to occur primarily through genetic mutation or silencing by promoter methylation, thus resulting in an irreversible mutator phenotype. Recent studies, however, have supported the concept that genetic instability can arise from dysregulation, rather than complete inactivation, of DNA repair pathways (13). Our results are in accord with such a paradigm shift, as we detected substantial but reversible hypoxia-induced decreases in Rad51 expression that were associated with significant functional consequences with respect to recombinational repair.
The HR pathway in the maintenance of genetic stability.
Despite extensive studies, there have been few reports describing mutations in the RAD51 gene in human tumors (18). However, it was recently shown that overexpression of a dominant-negative form of the RAD51 gene (dnRAD51), but not a wild-type form, was associated with increased tumorigenicity in Chinese hamster ovary (CHO) cells (1), suggesting that RAD51 indeed acts as a tumor suppressor. Furthermore, recent studies have suggested that BRCA1, a well-documented tumor suppressor (41), may function specifically to promote high-fidelity HR while simultaneously suppressing the error-prone, nonhomologous end-joining (NHEJ) pathway (49). Along these lines, several reports have demonstrated up-regulated NHEJ repair activity in the context of impaired HR, and vice versa (39). Thus, the BRCA1-RAD51 pathway likely represents an axis of tumor suppression, in which genome integrity is maintained by high-fidelity HR-mediated repair. As proposed for the case of inherited BRCA1 deficency, hypoxia-induced acquired decreases in RAD51 expression, and consequently diminished HR frequencies, thus may lead to genetic instability by shifting the balance between HR and NHEJ. We are currently investigating this possibility in further detail.
Hypoxia, DNA damage, and genetic instability.
As discussed earlier, hypoxia is associated with a diverse spectrum of DNA damage and genetic aberrations. In particular, hypoxia-reoxygenation cycles are associated with oxidative stress and the production of reactive oxygen species, which are thought to induce high levels of both single-strand breaks and double-strand breaks. Studies have demonstrated that reoxygenation after a brief period of hypoxia can induce a high level of DNA damage (in the form of double-strand breaks) comparable to that observed after exposure to 4 to 5 Gy of ionizing radiation (IR) (15). In spite of this potentially large number of DNA strand breaks following hypoxic exposure, it is striking that we observed such profound decreases in Rad51 expression in the same time period, as Rad51 is a central protein in the pathway responsible for the error-free repair of such DNA lesions. In addition, studies have demonstrated that Rad51 is required for normal S-phase progression in mammalian cells, because of the role of Rad51 in resolving stalled and collapsed replication forks (36). Thus, the finding that posthypoxic cells resume DNA replication in the setting of decreased Rad51 expression suggests that hypoxia-induced down-regulation of Rad51 may have a major impact on genome integrity in the tumor microenvironment. This uncoupling of proliferation and Rad51 expression may be particularly important in regions of tumors undergoing fluctuating perfusion and consequently repeated cycles of hypoxia followed by reoxygenation. Hypoxia itself has been shown to induce an S-phase arrest which is reversible upon reoxygenation and may be associated with stalled replication forks (14, 16). In this situation, hypoxia-induced reductions in Rad51 expression would again have a major impact on the ability of tumor cells to maintain genomic integrity.
Regulation of DNA repair gene expression.
Microarray data from this study have provided further evidence that the down-regulation of specific genes by hypoxia is as important as up-regulation in accurately characterizing gene expression profiles associated with hypoxia. The data presented here demonstrate that the hypoxia-induced decreases in Rad51 expression occur through a HIF-independent pathway involving transcriptional repression of the RAD51 promoter. Initial analyses of promoters from genes involved in the HR pathway have revealed a complex picture of regulatory elements, and many of these elements appear to be conserved in mice and humans (18). Interestingly, the RAD51 transcript contains an untranslated first exon (Fig. 3E), suggestive of gene regulation at the mRNA level. While the 5′ regulatory region of the RAD51 gene contains a CpG-rich region and lacks a TATA box (a typical arrangement found in many housekeeping genes), recent analyses have demonstrated numerous potential regulatory elements in the core promoter region of this gene (18). In one study by Levy-Lahad et al., a single-nucleotide polymorphism at nucleotide 135 of the untranslated first exon of the RAD51 gene was associated with increased breast cancer risk in BRCA2 mutation carriers (21). These analyses strongly suggest that the expression of RAD51 may be regulated by specific promoter elements in response to various stimuli, and they represent potential sites of dysregulation in the context of tumorigenesis and the tumor microenvironment.
Implications for cancer therapy.
It has been established that hypoxic cells are more resistant to IR than their well-oxygenated counterparts due to decreased potentiation of free radical damage mediated by oxygen, and numerous studies have quantitatively associated tumor oxygenation with response to radiotherapy (32). Interestingly, however, it has also been reported that cells irradiated under normoxic conditions in the period immediately following hypoxia are actually more radiosensitive than cells irradiated without such hypoxic pretreatment (50). At the time, the underlying mechanism for this sensitivity was not clear. This phenomenon was observed in numerous cell lines, including two used in the present study, HeLa and A431 cells. Interestingly, cells with inactivated HR components exhibit hypersensitivity to DNA-damaging agents, including IR (38). Specifically, several studies have demonstrated an association between down-regulation of Rad51 expression and increased radiosensitivity in a number of cell types (28). We propose that the persistent posthypoxic decreases in Rad51 expression reported here may partially account for the phenomenon of posthypoxia-associated radiosensitivity. These findings suggest that gene expression changes that persist in the posthypoxic period may significantly impact the response of cancer cells to therapy. Daily fractionated radiotherapy is thought to preferentially kill oxygenated cells and promote the reoxygenation of previously hypoxic cells. Hence, the extra sensitivity of immediately posthypoxic, reoxygenated cells (perhaps due to the dynamics of RAD51 expression and dysregulation of other DNA repair genes) may provide insight into the basis for the efficacy of fractionated radiotherapy.
Substantial evidence now exists implicating tumor hypoxia in the development of aggressive tumor phenotypes. Recent studies have further clarified this association, leading to the finding that hypoxia up-regulates numerous genes involved in invasion and metastasis (37). We and others have demonstrated that the tumor microenvironment contributes to genetic instability and tumor progression, and the data presented in this study provide a mechanistic basis for this phenomenon. Furthermore, Rad51 dysregulation may also create heterogeneity in the DNA damage response among cells within tumors, with implications for the response to cancer therapies.
Acknowledgments
We thank Tom Taylor for technical expertise in the FACS analyses, Geoffrey Lyon for assistance with tools for flow cytometry analysis, the Department of Microbial Pathogenesis for use of its BD FACscan flow cytometer, Anthony Valerio for assistance with real-time PCR assays, B. Kuba for excellent technical assistance, Greg Semenza for the pCEP-HIF-1α expression vector, William Kaelin for the 786-0 VHL cell lines, and Melissa Knauert for technical expertise in the HR shuttle assays. Finally, we thank Jianling Yuan, Shannon Gibson, Ryan Jensen, Denise Hegan, other members of the Glazer laboratory, and Jasjit, Ranjna, and Kavitha Bindra for insightful discussions regarding the manuscript.
This work was supported by a grant from the NIH (ES05775) to P.M.G. and by NCIC Project Program and U.S. Army DOD Prostate Program grants to D.W.H. and R.G.B. R.S.B. was supported by NIH/National Institute of General Medical Sciences Medical Scientist Training Grant GM07205.
REFERENCES
- 1.Bertrand, P., S. Lambert, C. Joubert, and B. S. Lopez. 2003. Overexpression of mammalian Rad51 does not stimulate tumorigenesis while a dominant-negative Rad51 affects centrosome fragmentation, ploidy and stimulates tumorigenesis, in p53-defective CHO cells. Oncogene 22**:**7587-7592. [DOI] [PubMed] [Google Scholar]
- 2.Bindra, R. S., J. R. Vasselli, R. Stearman, W. M. Linehan, and R. D. Klausner. 2002. VHL-mediated hypoxia regulation of cyclin D1 in renal carcinoma cells. Cancer Res. 62**:**3014-3019. [PubMed] [Google Scholar]
- 3.Brizel, D. M., S. P. Scully, J. M. Harrelson, L. J. Layfield, R. K. Dodge, H. C. Charles, T. V. Samulski, L. R. Prosnitz, and M. W. Dewhirst. 1996. Radiation therapy and hyperthermia improve the oxygenation of human soft tissue sarcomas. Cancer Res. 56**:**5347-5350. [PubMed] [Google Scholar]
- 4.Bromfield, G. P., A. Meng, P. Warde, and R. G. Bristow. 2003. Cell death in irradiated prostate epithelial cells: role of apoptotic and clonogenic cell kill. Prostate Cancer Prostatic Dis. 6**:**73-85. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. P., M. Lin, A. F. Faruqi, J. Powell, M. M. Seidman, and P. M. Glazer. 1999. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J. Biol. Chem. 274**:**11541-11548. [DOI] [PubMed] [Google Scholar]
- 6.Chiarotto, J. A., and R. P. Hill. 1999. A quantitative analysis of the reduction in oxygen levels required to induce up-regulation of vascular endothelial growth factor (VEGF) mRNA in cervical cancer cell lines. Br. J. Cancer 80**:**1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coquelle, A., F. Toledo, S. Stern, A. Bieth, and M. Debatisse. 1998. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol. Cell 2**:**259-265. [DOI] [PubMed] [Google Scholar]
- 8.Datta, H. J., P. P. Chan, K. M. Vasquez, R. C. Gupta, and P. M. Glazer. 2001. Triplex-induced recombination in human cell-free extracts. Dependence on XPA and HsRad51. J. Biol. Chem. 276**:**18018-18023. [DOI] [PubMed] [Google Scholar]
- 9.De Jaeger, K., M. C. Kavanagh, and R. P. Hill. 2001. Relationship of hypoxia to metastatic ability in rodent tumours. Br. J. Cancer 84**:**1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, S. M., B. Joiner, W. T. Jenkins, K. M. Laughlin, E. M. Lord, and C. J. Koch. 1995. Identification of hypoxia in cells and tissues of epigastric 9L rat glioma using EF5 [2-(2-nitro-1H-imidazol-1-yl)-_N_-(2,2,3,3,3-pentafluoropropyl) acetamide]. Br. J. Cancer 72**:**875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flygare, J., F. Benson, and D. Hellgren. 1996. Expression of the human RAD51 gene during the cell cycle in primary human peripheral blood lymphocytes. Biochim. Biophys. Acta 1312**:**231-236. [DOI] [PubMed] [Google Scholar]
- 12.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16**:**4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, H. H., and L. A. Loeb. 2003. Tumbling down a different pathway to genetic instability. J. Clin. Investig. 112**:**1793-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond, E. M., N. C. Denko, M. J. Dorie, R. T. Abraham, and A. J. Giaccia. 2002. Hypoxia links ATR and p53 through replication arrest. Mol. Cell. Biol. 22**:**1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond, E. M., M. J. Dorie, and A. J. Giaccia. 2003. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J. Biol. Chem. 278**:**12207-12213. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, E. M., S. L. Green, and A. J. Giaccia. 2003. Comparison of hypoxia-induced replication arrest with hydroxyurea and aphidicolin-induced arrest. Mutat. Res. 532**:**205-213. [DOI] [PubMed] [Google Scholar]
- 17.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2**:**38-47. [DOI] [PubMed] [Google Scholar]
- 18.Henning, W., and H. W. Sturzbecher. 2003. Homologous recombination and cell cycle checkpoints: Rad51 in tumour progression and therapy resistance. Toxicology 193**:**91-109. [DOI] [PubMed] [Google Scholar]
- 19.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292**:**464-468. [DOI] [PubMed] [Google Scholar]
- 20.Juan, G., E. Hernando, and C. Cordon-Cardo. 2002. Separation of live cells in different phases of the cell cycle for gene expression analysis. Cytometry 49**:**170-175. [DOI] [PubMed] [Google Scholar]
- 21.Levy-Lahad, E., A. Lahad, S. Eisenberg, E. Dagan, T. Paperna, L. Kasinetz, R. Catane, B. Kaufman, U. Beller, P. Renbaum, and R. Gershoni-Baruch. 2001. A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc. Natl. Acad. Sci. USA 98**:**3232-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, C. Y., J. B. Little, K. Hu, W. Zhang, L. Zhang, M. W. Dewhirst, and Q. Huang. 2001. Persistent genetic instability in cancer cells induced by non-DNA-damaging stress exposures. Cancer Res. 61**:**428-432. [PubMed] [Google Scholar]
- 23.Loeb, L. A., K. R. Loeb, and J. P. Anderson. 2003. Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA 100**:**776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonergan, K. M., O. Iliopoulos, M. Ohh, T. Kamura, R. C. Conaway, J. W. Conaway, and W. G. Kaelin, Jr. 1998. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 18**:**732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihaylova, V. T., R. S. Bindra, J. Yuan, D. Campisi, L. Narayanan, R. Jensen, F. Giordano, R. S. Johnson, S. Rockwell, and P. M. Glazer. 2003. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell. Biol. 23**:**3265-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordsmark, M., M. Overgaard, and J. Overgaard. 1996. Pretreatment oxygenation predicts radiation response in advanced squamous cell carcinoma of the head and neck. Radiother. Oncol. 41**:**31-39. [DOI] [PubMed] [Google Scholar]
- 27.Nowell, P. C. 1976. The clonal evolution of tumor cell populations. Science 194**:**23-28. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi, T., T. Taki, S. Hiraga, N. Arita, and T. Morita. 1998. In vitro and in vivo potentiation of radiosensitivity of malignant gliomas by antisense inhibition of the RAD51 gene. Biochem. Biophys. Res. Commun. 245**:**319-324. [DOI] [PubMed] [Google Scholar]
- 29.Paquette, B., and J. B. Little. 1994. In vivo enhancement of genomic instability in minisatellite sequences of mouse C3H/10T1/2 cells transformed in vitro by X-rays. Cancer Res. 54**:**3173-3178. [PubMed] [Google Scholar]
- 30.Perier, R. C., V. Praz, T. Junier, C. Bonnard, and P. Bucher. 2000. The eukaryotic promoter database (EPD). Nucleic Acids Res. 28**:**302-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds, T. Y., S. Rockwell, and P. M. Glazer. 1996. Genetic instability induced by the tumor microenvironment. Cancer Res. 56**:**5754-5757. [PubMed] [Google Scholar]
- 32.Rockwell, S. 1997. Oxygen delivery: implications for the biology and therapy of solid tumors. Oncol. Res. 9**:**383-390. [PubMed] [Google Scholar]
- 33.Roth, M. E., L. Feng, K. J. McConnell, P. J. Schaffer, C. E. Guerra, J. P. Affourtit, K. R. Piper, L. Guccione, J. Hariharan, M. J. Ford, S. W. Powell, H. Krishnaswamy, J. Lane, G. Intrieri, J. S. Merkel, C. Perbost, A. Valerio, B. Zolla, C. D. Graham, J. Hnath, C. Michaelson, R. Wang, B. Ying, C. Halling, C. E. Parman, D. Raha, B. Orr, B. Jedrzkiewicz, J. Liao, A. Tevelev, M. J. Mattessich, D. M. Kranz, M. Lacey, J. C. Kaufman, J. Kim, D. R. Latimer, and P. M. Lizardi. 2004. Expression profiling using a hexamer-based universal microarray. Nat. Biotechnol. 22**:**418-426. [DOI] [PubMed] [Google Scholar]
- 34.Seagroves, T. N., H. E. Ryan, H. Lu, B. G. Wouters, M. Knapp, P. Thibault, K. Laderoute, and R. S. Johnson. 2001. Transcription factor HIF-1 is a necessary mediator of the Pasteur effect in mammalian cells. Mol. Cell. Biol. 21**:**3436-3444.11313469 [Google Scholar]
- 35.Shibata, T., A. J. Giaccia, and J. M. Brown. 2000. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 7**:**493-498. [DOI] [PubMed] [Google Scholar]
- 36.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17**:**598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subarsky, P., and R. P. Hill. 2003. The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis 20**:**237-250. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509**:**49-78. [DOI] [PubMed] [Google Scholar]
- 39.Valerie, K., and L. F. Povirk. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22**:**5792-5812. [DOI] [PubMed] [Google Scholar]
- 40.Vaupel, P., K. Schlenger, C. Knoop, and M. Hockel. 1991. Oxygenation of human tumors: evaluation of tissue oxygen distribution in breast cancers by computerized O2 tension measurements. Cancer Res. 51**:**3316-3322. [PubMed] [Google Scholar]
- 41.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108**:**171-182. [DOI] [PubMed] [Google Scholar]
- 42.Vukovic, V., H. K. Haugland, T. Nicklee, A. J. Morrison, and D. W. Hedley. 2001. Hypoxia-inducible factor-1α is an intrinsic marker for hypoxia in cervical cancer xenografts. Cancer Res. 61**:**7394-7398. [PubMed] [Google Scholar]
- 43.Wang, G. L., and G. L. Semenza. 1993. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood 82**:**3610-3615. [PubMed] [Google Scholar]
- 44.Werner, T. 2003. The state of the art of mammalian promoter recognition. Brief Bioinform. 4**:**22-30. [DOI] [PubMed] [Google Scholar]
- 45.Williams, K. J., R. L. Cowen, and I. J. Stratford. 2001. Hypoxia and oxidative stress. Tumour hypoxia—therapeutic considerations. Breast Cancer Res. 3**:**328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, S. D., R. S. Marshall, and R. P. Hill. 1988. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc. Natl. Acad. Sci. USA 85**:**9533-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan, J., and P. M. Glazer. 1998. Mutagenesis induced by the tumor microenvironment. Mutat. Res. 400**:**439-446. [DOI] [PubMed] [Google Scholar]
- 48.Yuan, J., L. Narayanan, S. Rockwell, and P. M. Glazer. 2000. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 60**:**4372-4376. [PubMed] [Google Scholar]
- 49.Zhang, J., H. Willers, Z. Feng, J. C. Ghosh, S. Kim, D. T. Weaver, J. H. Chung, S. N. Powell, and F. Xia. 2004. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 24**:**708-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zolzer, F., and C. Streffer. 2002. Increased radiosensitivity with chronic hypoxia in four human tumor cell lines. Int. J. Radiat. Oncol. Biol. Phys. 54**:**910-920. [DOI] [PubMed] [Google Scholar]