Severe Global DNA Hypomethylation Blocks Differentiation and Induces Histone Hyperacetylation in Embryonic Stem Cells (original) (raw)
Abstract
It has been reported that DNA methyltransferase 1-deficient (_Dnmt1_−/−) embryonic stem (ES) cells are hypomethylated (20% CpG methylation) and die through apoptosis when induced to differentiate. Here, we show that _Dnmt_[_3a_−/−,_3b_−/−] ES cells with just 0.6% of their CpG dinucleotides behave differently: the majority of cells within the culture are partially or completely blocked in their ability to initiate differentiation, remaining viable while retaining the stem cell characteristics of alkaline phosphatase and Oct4 expression. Restoration of DNA methylation levels rescues these defects. Severely hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] ES cells have increased histone acetylation levels, and those cells that can differentiate aberrantly express extraembryonic markers of differentiation. _Dnmt_[_3a_−/−,_3b_−/−] ES cells with >10% CpG methylation are able to terminally differentiate, whereas _Dnmt1_−/− ES cells with 20% of the CpG methylated cannot differentiate. This demonstrates that successful terminal differentiation is not dependent simply on adequate methylation levels. There is an absolute requirement that the methylation be delivered by the maintenance enzyme Dnmt1.
Modifications in chromatin, including DNA methylation and histone modification, are known to be important epigenetic determinants of gene transcription. DNA methylation levels fluctuate markedly in early mouse development. In preimplantation development, the mouse embryo undergoes active and passive genomic demethylation (3, 15). This is restored at the time of implantation by the combined action of de novo and maintenance DNA methyltransferases (Dnmts). Studies of DNA methyltransferase 1-deficient (_Dnmt1_−/−) and Dnmt3a/Dnmt3b-deficient (_Dnmt_[_3a_−/−,_3b_−/−]) mouse embryos have demonstrated that restoring DNA methylation is essential for development (13, 19). _Dnmt1_−/− and _Dnmt_[_3a_−/−,_3b_−/−] embryos exhibit an early-lethal phenotype. At day 9.5 postcoitus, the embryos appear to have gastrulated but exhibit marked growth delay, having failed to turn or develop somites. In the presence of Dnmt1 deficiency, development is thought to fail because of cell death. Dnmt1-deficient embryoid bodies (EBs) aberrantly express Xist, down-regulate X-linked genes, and apoptose when induced to differentiate (20). Late-passage hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] embryonic stem (ES) cells are unable to form teratomas in vivo, but the cause of their differentiation failure has not been studied (4).
Early embryonic development is characterized by high levels of Dnmt3a and Dnmt3b expression. These enzymes clearly have roles in initiating remethylation of the genome following preimplantation demethylation, but it is not known whether continued de novo methyltransferase activity is required for development once global remethylation has taken place. This was our reason for studying the differentiation of _Dnmt_[_3a_−/−,_3b_−/−] ES cells in vitro. These mutant ES cells were derived from fully methylated wild-type ES cells and would have been predicted to have retained most of their methylation because of the continued presence of the maintenance methyltransferase Dnmt1. In fact, while early-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells are well methylated, DNA methylation levels fall progressively in culture (4). However, the rate of loss and the precise levels of methylation remaining have not been quantified. We have used a quantitative assay of DNA methylation to examine the effects of progressively decreasing genomic methylation levels on differentiation in vitro. Our studies reveal a clear but unexpected difference between the behaviors of hypomethylated _Dnmt1_−/− and _Dnmt_[_3a_−/−,_3b_−/−] ES cells in in vitro assays of differentiation. At very low levels of DNA methylation, _Dnmt_[_3a_−/−,_3b_−/−] ES cells demonstrate an inability to initiate differentiation upon leukemia inhibitory factor (LIF) withdrawal, remaining viable and retaining markers characteristic of undifferentiated ES cells.
MATERIALS AND METHODS
ES cell culture.
ES cells were maintained on gelatin in a Glasgow modification of Eagle medium (Invitrogen) supplemented with 10% fetal calf serum, 100 μM 2-mercaptoethanol, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 2 mM l-glutamine, and 100 U of leukemia inhibitory factor (LIF)/ml (24).
Alkaline phosphatase assay.
To determine the number of self-renewing ES cells in monolayer or EB culture, single-cell suspensions were prepared by trypsinization of monolayers or dispase treatment of EBs. Viable cells (103 per well; trypan blue exclusion) were plated for ES cell suspensions, and 5 × 103 cells per well were plated for the EB suspensions on gelatin-coated six-well dishes in Glasgow minimum essential medium (GMEM) supplemented with LIF (100 U/ml) or unsupplemented. After 5 days of culture, the cells were stained for alkaline phosphatase (86-R; Sigma).
Hematopoietic differentiation.
Differentiation of ES cells was performed as previously described (30). Briefly, 600 to 1,200 cells were plated in Iscove’s modified Dulbecco’s medium (Invitrogen); 1% methylcellulose (Stem Cell Technologies); 10% fetal calf serum; erythropoietin (1 U/ml; Roche); insulin (10 μg/ml; Sigma); murine interleukin-3 (2 U/ml; Roche); 2 mM l-glutamine, penicillin, and streptomycin; and 340 μM monothioglycerol (Sigma). The percentage of hemoglobinized EBs was scored on day 10, and the percentage of colonies showing myeloid differentiation was assessed on day 20 of culture.
Cardiomyocyte differentiation.
Embryoid bodies were generated by hanging drop (300 ES cells per 10 μl of GMEM plus LIF) for 2 days. EBs were cultured in suspension in GMEM without LIF for a further 5 days before being plated on gelatin-coated 24-well plates (1 EB/well). The percentage of EBs with beating cardiomyocytes was scored for 10 further days of culture, and a cumulative score was recorded.
Methylation rescue of demethylated ES cells with Dnmt3a or Dnmt3b transgenes.
Full-length cDNAs for Dnmt3a and Dnmt3b were generated by reverse transcription (RT)-PCR and subcloned into pCAGASIZ (a kind gift from A. Smith), bringing the Dnmt3 cDNAs under the control of the CAG promoter (18) with an internal ribosomal entry site (IRES) linking zeocin resistance with methyltransferase expression. Late-passage highly demethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells were electroporated with the Dnmt rescue construct. Three Dnmt3α-rescued and three Dnmt3β-rescued zeocin (15 μg/ml)-resistant ES cell clones were isolated. The controls were wild-type J1 ES cells and highly demethylated late-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells transfected with pCAGASIZ (vector control). Three J1 vector control ES cell clones and six _Dnmt_[_3a_−/−,_3b_−/−] vector-only zeocin-resistant ES cell clones were isolated.
Western blots.
ES cells (2 × 105) were lysed in 2× Laemmli sample buffer and incubated at 95°C for 5 min before being separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose; 10 μl/lane was loaded on a sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis gel and blotted onto nitrocellulose. The blots were blocked overnight in 5% Marvel-TBST (Tris-buffered saline-0.1% Tween 20) before incubation with primary antibody diluted in 5% bovine serum albumin-TBST. The blots were washed in TBST and incubated in horseradish peroxidase (HRP) secondary antibody before detection in enhanced-chemiluminescence reagent. The primary antibodies were anti-Oct4 (C10; Santa Cruz), rabbit anti-[acetyl-K5]H4 antibody (R41; B. Turner); and rabbit anti-H3 (loading control; A. Verault).
Immunocytochemistry.
ES cells cultured in GMEM plus LIF were fixed in 4% paraformaldehyde before incubation with rat antilaminin monoclonal antibody (LAMB; Developmental Studies Hybridoma Bank, University of Iowa) or mouse anti-Oct4 monoclonal antibody (C10). The cells were washed and incubated with secondary antibodies (anti-mouse antibody-HRP and anti-rat antibody-HRP [Santa Cruz]) before being developed in True-Blue peroxidase substrate (KPL Laboratories).
RT-PCR.
cDNA was synthesized from total RNA using oligo(dT) and Moloney murine leukemia virus reverse transcriptase (Roche). The PCR primers were as follows: Oct4, 5′GGCGTTCTCTTTGGAAAGGTGTTC3′ and 5′CTCGAACCACATCCTTCTCT3′; β_H1-globin_, 5′AGTCCCCATGGAGTCAAAGA3′ and 5′CTCAAGGAGACCTTTGCTCA3′; α_-globin_, 5′CTCTCTGGGGAAGACAAAAGCAAC3′ and 5′GGTGGCTAGCCAAGGTCACCAGCA3′; Xist, 5′GTAACTCATCCCAGTGCAGG3′ and 5′CTGTATAGGCTGCAGG3′; Hnf4a, 5′ACACGTCCCCATCTGAAG3′ and 5′CTTCCTTCTTCATGCCAG3′; PL-1, 5′AGCTGACTTTGAATCTTTCAGGCTCCG3′ and 5′TTATGGATGTCCCTTTTAATGCAGCGGC3′; Tpbpa, 5′AATCTTCCTAGTCATCCTATGCC3′ and 5′CGCCACTCTCTGTGTAATCC3′; Albumin, 5′GATGAAACATATGTCCCCAAAGA3′ and 5′TGTGTTCCTAGGGTGTTGATTTTA3′; Hprt, 5′GCCTGTATCCAACACTTCG3′ and 5′AGCGTCGTGATTAGCGATG3′; β-tubulin, 5′GGAACATAGCCGTAAACTGC3′ and 5′TCACTGTGCCTGAACTTACC3′; Brachyury, 5′ATGCCAAAGAAAGAAACGAC3′ and 5′AGAGGCTGTAGAACATGATT3′; and Gapdh, 5′GGGTGGAGCCAAACGGGTC3′ and 5′GGAGTTGCTGTTGAAGTCGCA3′.
5Methylcytosine quantification.
5Methylcytosine was quantified by nearest-neighbor analysis (22).
RESULTS
_Dnmt_[_3a_−/−,_3b_−/−] ES cells progressively lose methylation after prolonged passage in vitro.
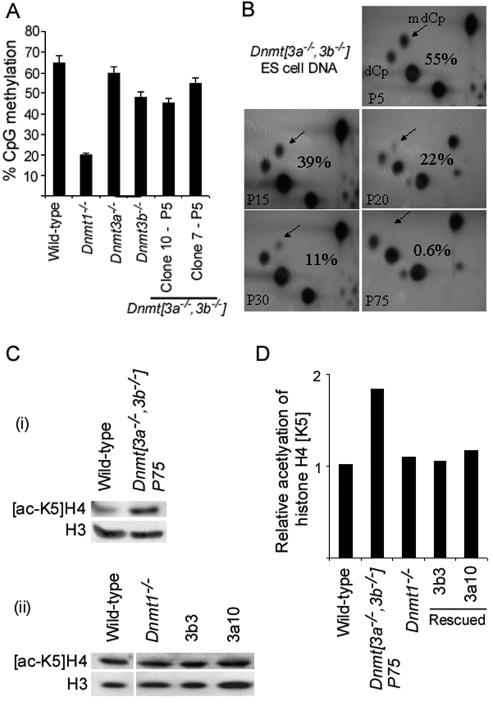
Wild-type (J1); _Dnmt1_−/−, null s allele (13); and _Dnmt_[_3a_−/−,_3b_−/−] ES cells (clone 7) (19) were cultured in GMEM plus 100 U of LIF/ml. At various passage numbers, samples were taken for DNA extraction and quantitative methylation analysis. The methylation levels of wild-type and _Dnmt1_−/− ES cells remained constant at 65 and 20%, respectively, throughout (Fig. 1A). Single-mutant _Dnmt3a_−/− and _Dnmt3b_−/− ES cells had methylation levels just less than those of wild-type ES cells, and a second, independently derived early-passage clone of _Dnmt_[_3a_−/−,_3b_−/−] ES cells (clone 10; passage 5 [P5]) had 45% of the CpG methylated (Fig. 1A). The fraction of CpG dinucleotides methylated was found to fall continuously in the _Dnmt_[_3a_−/−,_3b_−/−] ES cells, with the lowest level of 0.6% seen in P75 cells (Fig. 1B).
FIG. 1.
(A) Percent CpG methylation in the indicated ES cell lines as determined by nearest-neighbor analysis. The error bars indicate standard deviations. (B) Percent CpG methylation levels fall in _Dnmt_[_3a_−/−,_3b_−/−] ES cells with continued culture. The relative intensities of the dCp and mdCP spots reflect the frequencies CpG and mCpG at MboI sites. (C) Histone H4 lysine-5 acetylation ([acetyl-K5]H4) levels are increased in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells and restored in Dnmt3a and Dnmt3b cDNA-rescued _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cell lines. Acetylation levels are not appreciably increased in _Dnmt1_−/− ES cells. Separate experiments showing (i) [acetyl-K5]H4 ([ac-K5]H4) levels in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells compared with wild-type cells (total H3 serves as a loading control) and (ii) [acetyl-K5]H4 levels in _Dnmt1_−/− ES cells and the rescued _Dnmt_[_3a_−/−,_3b_−/−] cell lines 3b3 and 3a10 compared with wild-type ES cells. (D) Relative [acetyl-K5]H4 calculated by densitometry after normalization to a total histone H3 loading control.
Dnmt1 has a global maintenance fidelity of ∼98% in ES cells.
The tendency for _Dnmt_[_3a_−/−,_3b_−/−] ES cells to lose methylation in culture indicates that Dnmt1 alone is unable to maintain methylation levels in these cells. Pulse-labeling of exponentially growing _Dnmt_[_3a_−/−,_3b_−/−] ES cells with tritiated thymidine indicated a doubling time of 18.6 h compared with 14 h in wild-type J1 ES cells (data not shown). As the late-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells were grown for 165 days, the rate of loss of DNA methylation from 22 to 0.6% of CpG between passage 20 and passage 75 accords with ∼216 population doublings and a maintenance methylation fidelity of 98.3% [χ216 = 0.6/22; ∴ χ = 0.0272731/216 = 0.983]. This estimate assumes that DNA methylation falls exponentially, with the fraction lost every cell division being proportionate to the amount of methylation that remains. If one assumes that the number of population doublings (i.e., 216) could be incorrect by a factor of 30% either way, this would indicate a maintenance fidelity range of 97.7 to 98.7% (based on 151 to 281 population doublings). This estimate of the fidelity of maintenance methylation is considerably lower than previous estimates based on the maintenance of methylation at a specific genomic segment (21). In wild-type ES cells, the de novo methyltransferases Dnmt3a and Dnmt3b are clearly necessary for countering methylation losses due to imperfect maintenance of methylation by Dnmt1.
Stable integration of a Dnmt3 cDNA transgene into the hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] ES cell genome restores DNA methylation.
We restored DNA methylation to the demethylated cells by random stable integration of a Dnmt3a or Dnmt3b cDNA transgene under the control of the CAG promoter (18) using an IRES-zeocin-selectable marker. To assist in the interpretation of subsequent differentiation experiments, a single subclone of _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells was used for integration of the cDNA transgenes. Three Dnmt3a-rescued clones (3a4a, 3a10, and 3a16), three Dnmt3b-rescued clones (3b3, 3b17, and 3b23), and six mock-rescued clones (no Dnmt transgene; mock 1 to 6) were derived, and the methylation levels of the rescued clones were determined by nearest-neighbor analysis. The methylation levels approached those of wild-type ES cells (60 to 70% of the CpG methylated) in all but one clone, 3a4a, in which 26% of the CpG was methylated. Western blotting indicated that this clone had the lowest expression of the transgene (data not shown). Methylation levels were very low (<1% of CpG) in all mock-rescued clones.
Severe DNA hypomethylation causes global histone H4 lysine-5 hyperacetylation, which is reversed when DNA methylation is restored.
As previous studies have shown that there is a biochemical association among DNA methylation, proteins that interact with methylated DNA (methyl-CpG binding proteins), and histone deacetylase (HDAC), we investigated whether histone acetylation levels were affected by a near absence of DNA methylation. Western blots of ES cell lysates using an antibody specific for the acetylated lysine-5 residue of histone H4 ([acetyl-K5]H4) demonstrated that acetylation is increased in hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] ES cells (Fig. 1C). However, _Dnmt1_−/− ES cells and Dnmt3a- and Dnmt3b-rescued ES cells showed wild-type levels of [acetyl-K5]H4 (Fig. 1C). Quantification of the Western blotting results from independent experiments showed that _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells had approximately twice the level of [acetyl-K5]H4 as wild-type ES cells, suggesting that very low levels of DNA methylation result in less histone deacetylase activity being recruited to the chromatin.
Maintenance but not de novo methylation is required for efficient ES cell differentiation in vitro.
We studied the differentiation of _Dnmt_[_3a_−/−,_3b_−/−] ES cells with specific levels of overall DNA methylation (at different passage numbers) and compared this with passage-matched wild-type and _Dnmt1_−/− ES cells, both of which had a constant level of DNA methylation. This enabled us to determine the importance of Dnmt3-related de novo methylation compared with Dnmt1-related maintenance methylation in ES cell differentiation.
Methylcellulose hematopoietic progenitor assay.
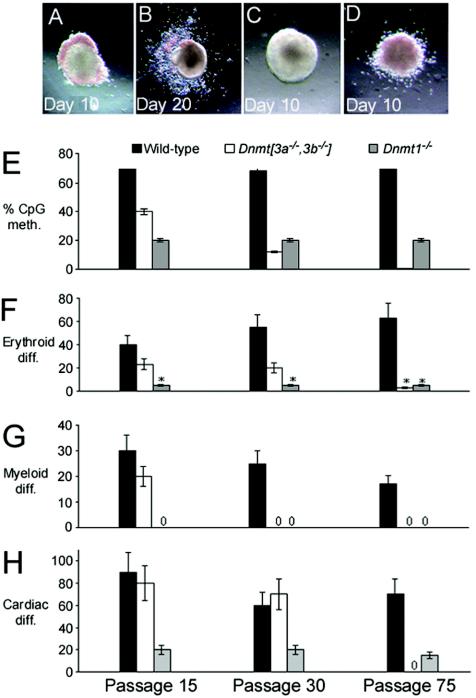
The methylcellulose hematopoietic progenitor assay determines the ability of ES cells to differentiate into hematopoietic lineages in the presence of interleukin-3 and erythropoietin. In wild-type ES cells, erythroid colonies typically have a localized accumulation of hemoglobin at the periphery of the colony and were scored on day 10 of the assay (Fig. 2A). Myeloid cells emerge and spread outward from the central core and were scored on day 20 (Fig. 2B). Cell lines that failed to differentiate typically showed no obvious hemoglobinization by eye (Fig. 2C), but occasionally low-level diffuse hemoglobinization was observed (Fig. 2D).
FIG. 2.
_Dnmt_[_3a_−/−,_3b_−/−] ES cells lose differentiation capacities with prolonged passage and progressive demethylation. Photomicrographs of normal erythroid (day 10) (A) and myeloid (day 20) (B) differentiation of wild-type ES cell colonies. Examples of absent (C) and atypical (diffuse low-level) (D) erythroid differentiation are also shown. Bar charts depicting the extent of DNA methylation (meth.) (E), percentage of colonies showing erythroid differentiation (diff.) (F), myeloid differentiation (G), and EBs showing cardiomyocyte differentiation (H) are shown for wild-type, _Dnmt1_−/−, and _Dnmt_[_3a_−/−,_3b_−/−] ES cells of different passages. The x axis labels shown in panel H are the same for all graphs. The error bars indicate standard deviations. Asterisks indicate a low level of diffuse hemoglobinization.
_Dnmt1_−/− ES cells (20% of CpG methylated) showed markedly reduced differentiation into erythroid colonies, and no myeloid colonies were observed (Fig. 2F and G). The single-knockout _Dnmt3a_−/− and _Dnmt3b_−/− ES cell lines showed both erythroid and myeloid differentiation (data not shown). However, ES cells with combined deletions of Dnmt3a and Dnmt3b exhibited erythroid and myeloid differentiation that was apparently dependent on the level of genomic methylation. Thus, early-passage _Dnmt_[_3a_−/−,_3b_−/−] P15 ES cells from clone 7 (39% of CpG methylated) and P5 cells from clone 10 (45% of CpG methylated) (data not shown) were able to differentiate into erythroid and myeloid colonies that were indistinguishable from wild-type colonies. However with increasing passage number and declining DNA methylation, the _Dnmt_[_3a_−/−,_3b_−/−] ES cells lost the ability to differentiate. By passage 30, the _Dnmt_[_3a_−/−,_3b_−/−] ES cells had lost the ability to form myeloid colonies, and by passage 75, no colonies could be scored as having erythroid differentiation typical of that seen in wild-type ES cells, though an atypical pattern of hemoglobinization could be seen in 5% of the colonies (Fig. 2D and F).
Cardiomyocyte differentiation.
We determined whether methylation loss would result in a defect in cardiomyocyte differentiation. The cardiomyocyte assay measures the proportion of EBs that beat (spontaneous contractions) within a 5- to 15-day period after LIF withdrawal. Very low-level differentiation was observed in _Dnmt1_−/− EBs independent of the passage number. Early-passage _Dnmt_[_3a_−/−,_3b_−/−] P15 ES cells with 39% CpG methylation differentiated well (Fig. 2H). Two single-knockout ES cell lines (_Dnmt3a_−/− and _Dnmt3b_−/−) were both shown to have near-normal levels of DNA methylation and were also able to differentiate efficiently (data not shown). _Dnmt_[_3a_−/−,_3b_−/−] P30 EBs differentiated well, but highly demethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs failed to differentiate within 15 days of LIF withdrawal (Fig. 2H). Beyond 15 days of differentiation, occasional EBs (<5%) showed intermittent beating in a small number of cells.
Differentiation is rescued by remethylation of the _Dnmt_[_3a_−/−,_3b_−/−] genome.
All Dnmt3-rescued ES cell lines except 3a4a showed wild-type levels of DNA methylation (Fig. 3A), and with the exception of 3b17, all showed levels of cardiomyocyte and erythroid differentiation, whereas mock-rescued clones failed to differentiate (Fig. 3B and D). Indeed, cardiomyocyte differentiation was absent in all six mock-rescued clones within 15 days of LIF withdrawal. Very low-level cardiomyocyte differentiation was observed in 3b17 EBs, and heme production was observed only after benzidine staining (data not shown). Some level of myeloid differentiation was restored in the rescued cell lines, but the level of restoration was less marked than that observed with cardiomyocyte and erythroid differentiation (Fig. 3C). Taken together, the success of the rescue experiments indicated that differentiation failure in the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cell line is the result of methylation loss rather than prolonged passage.
FIG. 3.
Differentiation is restored by random integration of Dnmt3a and Dnmt3b expression vectors into the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES genome. Nearest-neighbor analysis shows the extent of CpG methylation (meth.) in Dnmt3a-rescued (3a4, 3a10, and 3a16) and Dnmt3b-rescued (3b3, 3b17, and 3b23) clones and a mock-rescued clone 1 (A). The percentages of colonies showing erythroid differentiation (diff.) (B) and myeloid differentiation (C) and the percentage of EBs showing cardiomyocyte differentiation (D) are shown. The x axis labels shown in Fig. 3D are the same for all graphs. The error bars indicate standard deviations. ND, not determined. *, occasional beating seen after 15 days.
Hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs retain large numbers of undifferentiated cells following differentiation induction and fail to cavitate.
To investigate the cause of differentiation failure, we analyzed EB morphology. Cavitation is an early structural feature of EB differentiation (14). We analyzed the morphology of EBs to determine whether these cystic structures were present in the hypomethylated EBs. In fact, all EBs that failed to differentiate were small and lacked cavities (unpublished data). EBs derived from late-passage _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells were the smallest of all the cell lines studied. Alkaline phosphatase staining showed that these EBs, though small, consisted almost entirely of alkaline phosphatase-positive cells. To confirm that viable undifferentiated ES cells were retained in the _Dnmt_[_3a_−/−,_3b−/−_] P75 EBs, we disaggregated them and replated the cells at cloning density in 100 U of LIF/ml. Large numbers of undifferentiated colonies per 5 × 103 viable cells plated were recovered from the _Dnmt_[_3a_−/−,_3b_−/−] P75 and 3b17 EBs, but not from the wild-type _Dnmt1_−/− EBs or Dnmt3-rescued EBs (Fig. 4).
FIG. 4.
Quantification of alkaline phosphatase-positive undifferentiated ES cells remaining in day 15 EBs by colony assay in 100 U of LIF/ml. The error bars indicate standard deviations.
Hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES monolayers fail to initiate differentiation on LIF withdrawal.
Since large numbers of undifferentiated stem cells remain in the _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs 18 days after LIF withdrawal, we questioned whether this was an inherent feature of the hypomethylated cells or whether the close cell contact with an EB was in some way exacerbating the failure to differentiate. We therefore studied differentiation in monolayer cultures at low density (cloning density) and at standard densities for culturing ES cells.
Wild-type, Dnmt mutant, and Dnmt3-rescued ES cell lines were cultured either with LIF or without LIF for 7 days. On day 7, 103 cells were seeded into six-well plates in triplicate in either 0 or 100 U of LIF/ml. After a further 5 days of culture, the colonies were stained for alkaline phosphatase and scored as either stem cell (totally undifferentiated and alkaline phosphatase-positive), mixed (retaining alkaline phosphatase in part of the colony), or differentiated (with little or no evidence of alkaline phosphatase staining) colonies (17).
All cell lines showed a tendency to plate less efficiently in 0 U of LIF than in 100 U of LIF/ml (data not shown). In general, ∼20 to 30% of ES cells plated produced colonies in 100 U of LIF/ml at cloning density. However, in 0 U of LIF/ml, the plating efficiency could be as low as 10%. The reduction in plating efficiency upon LIF withdrawal indicates that a proportion of ES cells die when induced to differentiate. There was no correlation between plating efficiency in 0 U of LIF/ml and the ability of the various cell lines to differentiate as embryoid bodies.
We examined the morphology of ES colonies plated in 0 U of LIF/ml (Fig. 5A). Quantification of these morphologies is shown in Fig. 5B. The results demonstrate that while wild-type _Dnmt1_−/− ES cells and early-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells form predominantly differentiated colonies when plated in 0 U of LIF/ml, a considerable impediment to differentiation is observed in the hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells and the mock-rescued _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. In these cell lines, ∼30% of the colonies remained undifferentiated, 30% were partially differentiated, and 40% were differentiated. The observation of differentiated _Dnmt1_−/− and _Dnmt_[_3a_−/−,_3b_−/−] colonies in 0 U of LIF/ml demonstrates that, despite the reported death by apoptosis upon differentiation induction in _Dnmt1_−/− ES cells (20), a significant fraction of these cells do not die. We have also shown by pulse-labeling with tritiated thymidine that _Dnmt1_−/− ES cells continue to grow at a reduced rate 7 days after LIF withdrawal (unpublished data). The growth of _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells is less affected by LIF withdrawal because of the number of undifferentiated cells remaining in the culture.
FIG. 5.
Hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells fail to initiate differentiation upon LIF withdrawal. ES cells were cultured either with LIF (100 U/ml) or without LIF before being replated at low density into 100 or 0 U of LIF/ml. (A) Alkaline phosphatase staining of colonies previously cultured in LIF (100 U/ml) and replated into 0 U of LIF/ml. (B) Percentages of stem cell (black), mixed (grey), and differentiated (white) colonies observed in panel A. (C) ES cell colonies recovered (100 U of LIF/ml; alkaline phosphatase stain) after growth with and without LIF for 7 days. (D) Total numbers of stem cell (black), mixed (grey), and differentiated (white) colonies recovered after 7 days without LIF. (E) Oct4 and SHP2 Western blot of ES cells grown in 100 or 0 U of LIF/ml for 7 days.
ES cells that have been grown without LIF for 7 days would be predicted to differentiate and lose their capacity to form undifferentiated colonies or colonies of any kind when plated back into LIF. When LIF was withdrawn from wild-type ES cells, this was indeed the case. Only 23 colonies were recovered per 103 wild-type cells plated, and just 5 of these colonies were stem cell colonies (Fig. 5C and D). Thus, as expected, the wild-type ES cells differentiate upon LIF withdrawal and lose their potential to form colonies. In contrast, in the case of the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells, ∼300 colonies could still be derived from the cultures after 7 days without LIF (Fig. 5C and D). This indicates that a large number of cells within the cultures were incapable of differentiating in the absence of LIF. While the number of ES cell colonies recovered from the _Dnmt1_−/− ES cell cultures and from the early-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cell cultures was also a little greater than that from the wild-type cultures (50 to 100 versus 25 per 103 cells plated), the degree of differentiation inhibition was far less than that observed with the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. A Western blot analysis using protein lysates from cultures before and after LIF withdrawal confirmed that Oct4, a marker of undifferentiated ES cells, was down regulated in the wild-type cultures but not in the _Dnmt_[_3a_−/−,_3b_−/−] P75 or _Dnmt_[_3a_−/−,_3b_−/−] P75 mock-rescued cultures (Fig. 5E).
Five out of six Dnmt3a- and Dnmt3b-rescued cell lines recovered the ability to differentiate in the absence of LIF to various extents (Fig. 5A, B, C, and D). The differentiation defect was most efficiently rescued in the Dnmt3b-rescued cell lines 3b3 and 3b23 and was less efficiently rescued in the Dnmt3a-rescued cell lines. When the 3b3 and 3b23 cell lines were cultured in LIF and subsequently replated in 0 U of LIF/ml, ∼80% of the resulting colonies were completely differentiated, almost 20% were partially differentiated, and virtually none were completely undifferentiated (Fig. 5B). However, not all rescued clones differentiated well. Under identical conditions, 8% of the 3b17 colonies were completely undifferentiated and a further 80% were partially differentiated. A substantial proportion of the Dnmt3a-rescued colonies (80 to 95%) were also scored as partially differentiated.
LIF signaling through Stat3 is normal in demethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells.
We sought a mechanism that might explain the differentiation inhibition observed in the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. First, conditioned medium derived from these cells was diluted 1 in 2 with fresh medium and tested for its ability to support and maintain the growth of undifferentiated wild-type ES cells. No evidence for a _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cell-derived inhibiting factor was observed (data not shown). We next asked whether Stat3, a downstream target of the LIF signaling pathway (5), might be phosphorylated (activated) in the absence of exogenous LIF in the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. However, Western blots using an antibody specific for phosphorylated tyrosine-705 revealed that there was no constitutive tyrosine-705 phosphorylation and that Stat3 was phosphorylated normally in response to LIF in the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells (unpublished data).
Expression analysis of lineage-specific markers in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells.
Having established that differentiation into cardiomyocytes and hematopoietic progenitors was defective in the _Dnmt1_−/− and _Dnmt_[_3a_−/−,_3b_−/−] P75 cells, we were interested to see whether other markers of lineage commitment were affected in the differentiating EBs.
Hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells exhibit low-level spontaneous extraembryonic differentiation into parietal endoderm in the presence of LIF.
Hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 and mock-rescued subclones have a sporadic tendency to differentiate in the presence of LIF into cells that have a cobblestone appearance suggestive of parietal endoderm. This was not a feature of the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells that had been rescued with either Dnmt3a or Dnmt3b transgenes. We confirmed by immunocytochemistry that these differentiated cells were of extraembryonic lineage, because they failed to express Oct4 and alkaline phosphatase but expressed high levels of laminin B1, a protein specific for yolk sac parietal endoderm (29) (Fig. 6A). As laminin B1-positive cells were not conspicuous in the wild-type or Dnmt-rescued ES cells, we conclude that their presence was a direct result of hypomethylation.
FIG. 6.
Deregulated mRNA and protein expression is evident in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. (A) _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells and wild-type and 3b3 rescued ES cells cultured in 100 U of LIF/ml and stained for alkaline phosphatase (Alk. Phos.), laminin B1, and Oct4 by immunocytochemistry. Ab, antibody. (B) RT-PCR analysis of Oct4, Xist, and Gapdh in wild-type, _Dnmt_[_3a_−/−,_3b_−/−] P75, and _Dnmt1_−/− EBs after induction to differentiate by LIF withdrawal. diff., differentiation. (C) RT-PCR analysis of trophoblast (Tpbp; Pl-1), endoderm (Hnf4a; albumin), mesoderm (brachyury; βh1 globin and α-globin), and housekeeping (Hprt and β-tubulin) genes in differentiating EBs generated from wild-type, _Dnmt_[_3a_−/−,_3b_−/−] P75, and _Dnmt1_−/− ES cells.
Oct4 mRNA levels are maintained after differentiation induction in _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs.
Consistent with the complete inability of 30% of the hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells to initiate differentiation, we also found a delay in the down regulation of Oct4 mRNA levels after LIF withdrawal (Fig. 6B). Interestingly, while _Dnmt1_−/− EBs down regulated Oct4 at the expected time, the gene was reexpressed at day 20 post-LIF withdrawal (Fig. 6B). Also, and as expected, Xist mRNA expression was markedly deregulated by DNA hypomethylation, with expression increasing on differentiation induction in both the _Dnmt_[_3a_−/−,_3b_−/−] P75 and _Dnmt1_−/− EBs. This is in agreement with previous findings for _Dnmt1_−/− EBs (20).
_Dnmt_[_3a_−/−,_3b_−/−] P75 EBs aberrantly express trophoblast-specific mRNA transcripts on induction to differentiate.
We investigated whether other transcripts characteristic of extraembryonic differentiation might be expressed on LIF withdrawal. We analyzed placental lactogen 1 (PL-1α) and trophoblast-specific protein α (Tpbp) (26). These are specific for trophoblast giant cells, and their expression is not normally induced upon differentiation of wild-type ES cells. We found that that PL-1 and Tpbp mRNA transcripts are induced in the _Dnmt_[_3a_−/−, _3b_−/−] P75 EBs and to a lesser extent in the _Dnmt1_−/− EBs but are not induced significantly in wild-type EBs (Fig. 6C).
Markers of mesodermal and endodermal differentiation are deregulated in _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs after differentiation induction.
Brachyury, a marker of early mesodermal differentiation (8), was induced at day 3 as expected in the _Dnmt1_−/− EBs but was markedly diminished in the _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs. The expression profiles of later markers of mesodermal differentiation (α-globin and βH1-globin) indicated that these transcripts were being induced at the expected time (28) but that their expression was not being maintained in either the _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs or the _Dnmt1_−/− EBs (Fig. 6C).
Hnf4 (a marker of visceral endoderm and liver) (6), is not usually expressed significantly in undifferentiated ES cells (1) but is induced shortly after LIF withdrawal. Both the _Dnmt_[_3a_−/−,_3b_−/−] P75 and the _Dnmt1_−/− EBs expressed higher levels of Hnf4 than wild-type EBs (Fig. 6C). However, albumin, a later marker of endodermal differentiation (expressed in fetal liver and yolk sac and in day 10 EBs) (1), was not induced to the high levels that normally accompany wild-type EB differentiation. These data, in addition to the expression of laminin B1, support the notion that hypomethylated ES cells have an enhanced propensity for differentiation into primitive and visceral endoderm in both the presence and absence of LIF. The relatively weak expression of albumin mRNA suggests that later endodermal differentiation is compromised.
Histone deacetylase inhibitors are toxic to differentiated hypomethylated ES cells.
To investigate whether this secondary effect of DNA hypomethylation on histone acetylation might actually account for the failure of hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] ES cells to initiate differentiation spontaneously on LIF withdrawal, we attempted to exacerbate the phenotype by further increasing acetylation using agents that inhibit HDAC. Prolonged treatment (2 to 5 days) with HDAC inhibitors was found to be toxic to ES cells, but an 8-h pulse with either 50 mM sodium butyrate or 160 nM trichostatin A (TSA) was well tolerated by wild-type ES cells. Experiments to assess the effects of HDAC inhibition on histone acetylation levels in both wild-type and _Dnmt_[_3a_−/−,_3b_−/−] ES cells showed that the overall effect of HDAC inhibition was marked but was short lasting in both cell lines (Fig. 7A).
FIG. 7.
DNA hypomethylation exacerbates the toxic effects of histone deacetylase inhibition upon differentiation induction. (A) Western blot of ES cell lysates probed with anti-[acetyl-K5]H4. Time course to show the return of acetylation levels to baseline 0, 1.5, and 3.5 h after an 8-h treatment with 50 mM sodium butyrate. (B) Effect on ES cell differentiation of pretreatment with either 50 mM sodium butyrate (pBut) or 160 nM trichostatin A (pTSA). (C) Effect of pretreatment with TSA on the differentiation of _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cell colonies after LIF withdrawal (i) without TSA pretreatment and (ii) with TSA (160 nM) pretreatment. ES colonies were stained for alkaline phosphatase (red).
To assess the effect of HDAC inhibition on the cell lines, we first looked at the effect on cloning efficiency in 0 U of LIF/ml. In the absence of treatment, there is usually a modest loss in colony number when ES cells are plated in 0 compared with 100 U of LIF/ml. Prior treatment with the HDAC inhibitor exacerbated this loss markedly in the early-passage _Dnmt_[_3a_−/−,_3b_−/−] P18 and _Dnmt1_−/− ES cell lines, but colony numbers were well maintained in the more severely hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 and wild-type ES cells (Fig. 7B). Maintaining the ES cells in LIF after exposure to the HDAC inhibitor prevented this effect, indicating that HDAC inhibitor treatment is most toxic to hypomethylated differentiating ES cells. This was confirmed when the extent of differentiation in the colonies was scored. LIF withdrawal allows the differentiation of early-passage _Dnmt_[_3a_−/−,_3b_−/−] P18 and _Dnmt1_−/− colonies. The overall effect of prior HDAC inhibitor treatment was to dramatically reduce the number of differentiated and partially differentiated colonies in these cell lines. However as the more severely hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells are resistant to differentiation on LIF withdrawal, there was a less dramatic effect on total colony numbers, but close examination of the colonies revealed that they had lost their differentiated component (Fig. 7C). Together, these findings indicate that HDAC inhibitor treatment is toxic to differentiating ES cells that are deficient in DNA methylation.
DISCUSSION
We have investigated the effect of DNA methyltransferase deletion on DNA hypomethylation upon ES cell differentiation in vitro using ES cells with targeted deletions of the DNA methyltransferases Dnmt1, Dnmt3a, and Dnmt3b. The gradual loss of DNA methylation from _Dnmt_[_3a_−/−,_3b_−/−] ES cells indicates that the only remaining DNA methyltransferase within these cells, Dnmt1, is not able to maintain methylation with 100% efficiency. The published estimate of the efficiency of maintenance of methylation is 99.9% (21). This figure is based on the maintenance fidelity within a specific sequence of DNA. Our estimate is a global average and is considerably less than this, lying between 97.7 and 98.7%.
To understand the effect of DNA methyltransferase deletion and DNA hypomethylation on ES cell differentiation, it is useful to think of the process in two stages. First, the cell must initiate differentiation. Successful initiation results in the loss of stem cell characteristics, such as alkaline phosphatase expression. Second, the initiated cells must proceed through a series of stages (most of which are unknown) that lead finally to the production of a terminally differentiated cell. Differentiation might fail completely because of a failure to initiate differentiation successfully. Alternatively, it might fail because, following the successful initiation of differentiation, there might be failure to progress to a terminally differentiated cell.
_Dnmt1_−/− ES cells with 20% of the CpG methylated are able to initiate differentiation in response to LIF withdrawal but fail to differentiate efficiently into terminally differentiated cardiomyocytes or hematopoietic cells. In contrast, differentiation in _Dnmt_[_3a_−/−,_3b_−/−] ES cells depends on their residual level of DNA methylation catalyzed and maintained by Dnmt1. Early-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells are well methylated and are able to initiate differentiation and proceed to terminally differentiated cardiomyocytes and hematopoietic cells. The presence of Dnmt1 in these cells may be crucial for the regulation of certain genes that are essential for differentiation. This observation also demonstrates that neither de novo methylation catalyzed by the Dnmt3 enzymes nor any protein-protein interactions that these enzymes may be involved in is required for ES cell differentiation in vitro. These findings are in agreement with the observation that early-passage but not late-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells can form teratomas in vivo (4). Furthermore, in the present study, because we were able to accurately quantify DNA methylation, we have been able to show that _Dnmt_[_3a_−/−,_3b_−/−] P30 ES cells have less methylation than _Dnmt1_−/− cells (11 versus 20% CpG methylation) but are able to differentiate considerably more efficiently. This indicates that maintenance of methylation by Dnmt1 is a specific requirement for terminal differentiation. The preservation of similar or even higher levels of global DNA methylation by continuous reiterative Dnmt3-related de novo methylation cannot support terminal differentiation. Maintenance of methylation by Dnmt1, which occurs at the time of DNA replication, presumably results in a more stable chromatin structure than continuous reiterative de novo methylation, which occurs in _Dnmt1_−/− ES cells. In the latter case, global methylation levels appear to be reduced but stable, but the methylation at any particular site may not be preserved from one cell generation to the next. Maintenance of methylation may be fundamental in propagating the stepwise changes in transcription that are necessary for differentiation.
Severely hypomethylated late-passage _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells fail to differentiate into hematopoietic cells and cardiomyocytes. Since these cells retain alkaline phosphatase and Oct4 expression upon LIF withdrawal, it is clear that differentiation failure is not a simple consequence of cell death upon differentiation induction, as has been shown for _Dnmt1_−/− ES cells (20). _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells have a defect in the initiation of differentiation upon LIF withdrawal, so that ∼30% of these cells retain alkaline phosphatase, a marker of undifferentiated stem cells, upon LIF withdrawal. Restoration of DNA methylation to the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells by random integration of Dnmt3a or Dnmt3b cDNA transgenes rescued this differentiation initiation phenotype and enabled terminal differentiation into hematopoietic cells and cardiomyocytes, although some rescued cell lines, such as 3b3 and 3b23, were better differentiators than others.
The Dnmt3-rescued clones were generated after we first selected and expanded a single clone from the _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cell culture. This was done to minimize underlying differences between the rescued clones. However, there were still appreciable differences in the abilities of the Dnmt3b-rescued cell lines to differentiate. Failure to differentiate despite apparent remethylation might have been caused by an unfavorable integration event. Alternatively, as a result of hypomethylation, secondary changes in chromatin structure (such as histone modification) might have occurred, making particular loci more or less amenable to remethylation. For example, histone methylation has been shown to influence DNA methylation (12, 25). Apparent resistance to remethylation has been described for imprinted genes that become biallelically expressed in Dnmt mutant ES cells (4, 27), possibly for similar reasons.
We were interested to see if the transcriptional repertoire of the severely hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells was in any way deregulated. Hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells tended to differentiate sporadically into laminin B1-positive parietal endoderm cells (extraembryonic lineage) even in the presence of LIF. Removing LIF from hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 EBs resulted in a spectrum of mRNA expression defects. Some genes, such as that for α-globin, appeared to be expressed as expected for wild-type differentiating EBs. In other cases, the timing of transcript induction appeared to be normal, but the transcripts were found at reduced levels compared with wild-type EBs (brachyury, Bh1 globin, and albumin). However, Hnf4 transcripts, which are associated with primitive endoderm specification in the early embryo (6), were markedly increased in hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells and embryoid bodies. The trophoblast-specific transcripts Pl1 (specific for trophoblast secondary giant cells) and Tpbp (ectoplacental cone and spongiotrophoblast) were also expressed in differentiating severely hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] EBs, but not in wild-type ES cells. These data indicate that hypomethylation leads to a general loss of embryonic in favor of extraembryonic specification, and this is likely to contribute to the failure of embryonic mesodermal differentiation that we have observed in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. It is interesting that trophoblast specification occurs at a time of extreme global DNA hypomethylation in the preimplantation embryo (10), whereas the initiation of embryonic differentiation correlates with embryonic remethylation (23).
To investigate the cause of the differentiation defect in hypomethylated cell lines, we investigated whether there might be a secondary effect of DNA hypomethylation on histone modification. A number of studies have demonstrated biochemical interactions between methyl-CpG binding proteins and HDACs (7, 16), and a number of other proteins located in the chromatin are also known to associate with and potentially recruit HDAC (9). DNA hypomethylation might therefore have been predicted to cause histone hyperacetylation. Western blot analysis using antibodies specific for the acetylated lysine-5 residue of histone H4 (anti-[acetyl-K5]H4) demonstrated a definite increase in the amount of acetylation in late-passage _Dnmt_[_3a_−/−,_3b_−/−] ES cells in five independent experiments. A similar effect could not be reliably demonstrated in the _Dnmt1_−/− ES cells.
We reasoned that histone hyperacetylation might in some way be responsible for the differentiation failure of hypomethylated ES cells, either by inhibiting differentiation or by promoting death upon differentiation induction. To gain further insight into these possibilities, we tried to augment any affect that histone hyperacetylation might have by further increasing histone acetylation levels using HDAC inhibitors. The effects (on acetylation) of an 8-h treatment with either TSA or sodium butyrate were short lasting, indicating that most chromatin-associated HDAC activity is not DNA methylation dependent. Pretreatment of the early-passage _Dnmt_[_3a_−/−,_3b_−/−] P18 ES cell and the _Dnmt1_−/− ES cells with an HDAC inhibitor resulted in a marked reduction in the number of differentiated colonies, but there was no gain in the combined number of undifferentiated or partially differentiated colonies. The mechanism of this effect is not known, but normally methylated wild-type ES cell colonies were apparently not susceptible to this toxic effect. In the late-passage _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells, the loss of differentiated cells from partially differentiated colonies was striking, but again, there was no net gain in the number of undifferentiated or partially differentiated colonies. The toxicity observed toward differentiated cells is likely to be a specific effect of HDAC inhibition, because it was observed using two chemically dissimilar inhibitors of HDAC activity. It is tempting to speculate that, as histone deacetylation and DNA methylation have additive effects on gene silencing (2), the transcriptional deregulatory effect of pharmacologically inducing histone hyperacetylation are more profound in hypomethylated ES cells than they are in normally methylated cells. The degree of transcriptional deregulation caused by HDAC inhibition may correlate with the level of toxicity.
Though histone hyperacetylation was the presumed mechanism of toxicity in the experiment with HDAC inhibitors, the level of hyperacetylation induced was far higher than that observed in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells in the absence of HDAC inhibitor. Therefore, it may not be safe to conclude that histone hyperacetylation causes death when untreated _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells are induced to differentiate. Furthermore, as histone hyperacetylation could not be sustained (because prolonged treatment with HDAC inhibitors was toxic), we cannot exclude the possibility that moderate histone hyperacetylation causes the differentiation inhibition in _Dnmt_[_3a_−/−,_3b_−/−] P75 ES cells. One group has recently shown that histone deacetylation occurs during the first 24 h of ES cell differentiation induction and that treatment with much lower doses of trichostatin A than used here prevents this deacetylation and inhibits differentiation (11).
In summary, our results show that at equivalent levels of global DNA methylation, Dnmt1-related maintenance methylation is qualitatively superior to reiterative Dnmt3-related de novo methylation in terms of its ability to support terminal embryonic differentiation in vitro. In vitro, the Dnmt3 enzymes are not required for differentiation per se but are necessary for correcting methylation losses due to maintenance failures by Dnmt1. However, at very low levels of DNA methylation, ES cells that lack Dnmt3a and Dnmt3b but retain Dnmt1 do fail to terminally differentiate in vitro. The mechanisms underlying differentiation failure are complex. We show that the initiation of differentiation is defective in very hypomethylated _Dnmt_[_3a_−/−,_3b_−/−] ES cells. The chromatin of these cells is hyperacetylated, and transcription is grossly deregulated upon induction to differentiate. The cells aberrantly express Xist and tend to express extraembryonic rather than embryonic markers of differentiation. These features are all likely to be detrimental to terminal embryonic differentiation.
Acknowledgments
We thank En Li for providing Dnmt mutant ES cells and Austin Smith for providing the CAG-IRES-zeocin expression vector.
This work was funded by the Leukemia Research Fund (M.J.) and the Medical Research Council (B.R.).
REFERENCES
- 1.Abe, K., H. Niwa, K. Iwase, M. Takiguchi, M. Mori, S. I. Abe, and K. I. Yamamura. 1996. Endoderm-specific gene expression in embryonic stem cells differentiated to embryoid bodies. Exp. Cell Res. 229**:**27-34. [DOI] [PubMed] [Google Scholar]
- 2.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21**:**103-107. [DOI] [PubMed] [Google Scholar]
- 3.Carlson, L. L., A. W. Page, and T. H. Bestor. 1992. Properties and localisation of DNA methyltransferase in pre-implantation mouse embryos: implications for genomic imprinting. Genes Dev. 6**:**2536-2541. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T., Y. Ueda, J. E. Dodge, J. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23**:**5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277**:**1630-1635. [DOI] [PubMed] [Google Scholar]
- 6.Duncan, S. A., K. Manova, W. S. Chen, P. Hoodless, D. C. Weinstein, R. F. Bachvarova, and J. E. Darnell, Jr. 1994. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. USA 91**:**7598-7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuks, F., P. J. Hurd, D. Wolf, X. Nan, A. P. Bird, and T. Kouzarides. 2002. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278**:**4035-4040. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann, B. G., S. Labeit, A. Poustka, T. R. King, and H. Lehrach. 1990. Cloning of the T gene required in mesoderm formation in the mouse. Nature 343**:**617-622. [DOI] [PubMed] [Google Scholar]
- 9.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115**:**689-698. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, M. H., and C. A. Ziomek. 1981. The foundation of two distinct cell lineages within the mouse morula. Cell 24**:**71-80. [DOI] [PubMed] [Google Scholar]
- 11.Lee, J. H., S. R. Hart, and D. G. Skalnik. 2004. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis 38**:**32-38. [DOI] [PubMed] [Google Scholar]
- 12.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone h3 lysine-9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13**:**1192-1200. [DOI] [PubMed] [Google Scholar]
- 13.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122**:**3195-3205. [DOI] [PubMed] [Google Scholar]
- 14.Martin, G. R., L. M. Wiley, and I. Damjanov. 1977. The development of cystic embryoid bodies in vitro from clonal teratocarcinoma stem cells. Dev. Biol. 61**:**230-244. [DOI] [PubMed] [Google Scholar]
- 15.Mayer, W., A. Niveleau, J. Walter, R. Fundele, and T. Haaf. 2000. Demethylation of the zygotic paternal genome. Nature 403**:**501-502. [DOI] [PubMed] [Google Scholar]
- 16.Nan, X., H.-H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393**:**386-389. [DOI] [PubMed] [Google Scholar]
- 17.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12**:**2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108**:**193-199. [DOI] [PubMed] [Google Scholar]
- 19.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99**:**247-257. [DOI] [PubMed] [Google Scholar]
- 20.Panning, B., and R. Jaenisch. 1996. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 10**:**1991-2002. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer, G. P., S. D. Steigerwald, R. S. Hansen, S. M. Gartler, and A. D. Riggs. 1990. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc. Natl. Acad. Sci. USA 87**:**8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsahoye, B. H. 2002. Nearest-neighbor analysis. Methods Mol. Biol. 200**:**9-15. [DOI] [PubMed] [Google Scholar]
- 23.Santos, F., B. Hendrich, W. Reik, and W. Dean. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241**:**172-182. [DOI] [PubMed] [Google Scholar]
- 24.Smith, A. 1991. Culture and differentiation of embryonic stem cells. J. Tissue Cult. Methods 13**:**89-94. [Google Scholar]
- 25.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414**:**277-283. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka, S., T. Kunath, A. K. Hadjantonakis, A. Nagy, and J. Rossant. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282**:**2072-2075. [DOI] [PubMed] [Google Scholar]
- 27.Tucker, K. L., C. Beard, J. Dausmann, L. Jackson-Grusby, P. W. Laird, H. Lei, E. Li, and R. Jaenisch. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10**:**1008-1020. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 8**:**1184-1197. [DOI] [PubMed] [Google Scholar]
- 29.Wewer, U. M., D. Tichy, A. Damjanov, M. Paulsson, and I. Damjanov. 1987. Distinct antigenic characteristics of murine parietal yolk sac laminin. Dev. Biol. 121**:**397-407. [DOI] [PubMed] [Google Scholar]
- 30.Wiles, M. V., and G. Keller. 1991. Multiple hematopoietic lineages develop from embryonic stem (ES) cells in culture. Development 111**:**259-267. [DOI] [PubMed] [Google Scholar]