Identification of a SUMO-binding motif that recognizes SUMO-modified proteins (original) (raw)
Abstract
Posttranslational modification by the ubiquitin homologue, small ubiquitin-like modifier 1 (SUMO-1), has been established as an important regulatory mechanism. However, in most cases it is not clear how sumoylation regulates various cellular functions. Emerging evidence suggests that sumoylation may play a general role in regulating protein-protein interactions, as shown in RanBP2/Nup358 and RanGAP1 interaction. In this study, we have defined an amino acid sequence motif that binds SUMO. This motif, V/I-X-V/I-V/I, was identified by NMR spectroscopic characterization of interactions among SUMO-1 and peptides derived from proteins that are known to bind SUMO or sumoylated proteins. This motif binds all SUMO paralogues (SUMO-1-3). Using site-directed mutagenesis, we also show that this SUMO-binding motif in RanBP2/Nup358 is responsible for the interaction between RanBP2/Nup358 and sumoylated RanGAP1. The SUMO-binding motif exists in nearly all proteins known to be involved in SUMO-dependent processes, suggesting its general role in sumoylation-dependent cellular functions.
Keywords: Ubc9, posttranslational modification, protein-protein interaction, RanBP2, Nup358
Posttranslational modification by the ubiquitin homologue, small ubiquitin-like modifier 1 (SUMO-1), has been identified as an important mechanism for cellular regulation of transcription, DNA repair, cell cycle progression, protein intracellular trafficking, and nuclear receptor activities (1-5). More than 60 SUMO-1 target proteins and two diseases linked to SUMO-1 modification have been reported (3, 6, 7). Two SUMO paralogues, SUMO-2/3, are closely related and share 97% amino acid sequence identity (8) but are only 46% and 48% identical to SUMO-1, respectively. The in vivo functions of SUMO-2/3 modifications appear to be distinct from that of SUMO-1. All three SUMO paralogues are attached to substrate proteins through a biochemical pathway similar to that of ubiquitination (9). Despite the sequence differences between SUMO-1 and SUMO-2/3, the activation enzyme E1 and conjugation enzyme E2 do not discriminate among the three SUMO molecules (10).
Despite increasing information on the importance of SUMO modification in cellular regulation, the mechanism by which SUMO modification regulates these processes is not well understood. Posttranslational modifications, such as phosphorylation and ubiquitination, and now evidently also sumoylation, modulate protein-protein interactions (11, 12). For example, sumoylation of RanGAP1 results in its interaction with the nuclear pore protein RanBP2/Nup358 (13, 14). Transcription factors P300 and Elk-1, when modified by SUMO-1, recruit histone deacetylase 6 (HDAC6) (15) and HDAC2 (16), respectively.
SUMO modification is likely to provide a new binding site for interactions with other proteins. In principle, SUMO modification could regulate the activity of a protein by altering its conformation. However, this is unlikely to be a general phenomenon, because SUMO modification sites are often located in extended loops, such as in RanGAP1 (17), or in an unstructured terminus, such as in p53 (18). In addition, the modification sites do not appear to require regular secondary structures (19). If SUMO modification provides a site for binding to other proteins, SUMO-binding motifs (SBM) on interacting proteins are likely to be responsible for these sumoylation-dependent protein-protein interactions. However, SBM has not been well defined to date. The previously identified ΦKXE motif binds the E2 enzyme for covalent modification by SUMO but does not bind to SUMO noncovalently (19).
A previous study (20) suggested a putative SBM based on sequence alignment of proteins associated with sumoylated p73α protein in a yeast two-hybrid screen. This putative motif consists of the Ser-X-Ser sequence flanked by three or four acidic residues at the C terminus and a few hydrophobic residues at the N terminus. Interaction between sumoylated targets and this SXS motif, however, was never tested by direct binding experiments. Here, we initially characterized the direct interactions between SUMO-1 and two peptides suggested from the previous study (20) by using NMR spectroscopy. Unexpectedly, these initial NMR studies showed that the SXS motif is not involved in interactions with SUMO-1. We thus undertook investigations to identify the true SBM and to demonstrate interaction between SUMO-modified RanGAP1 and this newly defined SBM within RanBP2/Nup358. This motif exists in nearly all cellular proteins that have been shown to be involved in sumoylation-dependent processes, thereby suggesting that this motif plays an important role in cellular functions modulated by SUMO modification.
Materials and Methods
Peptide Synthesis. Peptides were synthesized by the Peptide Synthesis Core Facility at the City of Hope, purified by HPLC, and verified by mass spectrometry.
Expression and Purification of Recombinant Proteins. 13C/15N-enriched human SUMO-1 (1-97), Ubc9, and SUMO-3 were expressed and purified as described (10). A DNA fragment encoding amino acids 2596-2836 of human RanBP2 was generated by PCR and inserted into pGEX1λ T vector (Amersham Pharmacia Biosciences). Mutants of RanBP2 were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) (see Supporting Text, which is published as supporting information on the PNAS web site), confirmed by sequencing, expressed by the Escherichia coli strain BL21(DE3), and purified by glutathione agarose.
NMR Studies. All NMR samples contained 20 mM phosphate buffer (pH 6.8) and 5 mM DTT in 92% H2O/8% D2O. All NMR spectra were acquired at 17°C on a Bruker (Billerica, MA) 600 MHz NMR spectrometer equipped with four channels, pulsed-field gradient, and pulse-shaping capabilities.
Two-dimensional NOESY, total correlation spectroscopy (TOCSY) (21), and 15N-filtered NOESY and TOCSY spectra were acquired as described (22). Four millimolar unlabeled peptide was titrated into the sample containing 1 mM 13C/15N-labeled SUMO-1 in five steps until SUMO-1 and peptide reached 1:1 molar ratio. At each titration point, 2D 1H-15N heteronuclear sequential quantum correlation (HSQC) spectra were recorded for bound 13C/15N-labeled SUMO-1. The assignments of free SUMO-1 and its complexes with each of the three peptides at pH 6.8 were obtained by using a combination of HNCA, CBCA(CO)NH, and CC(TOCSY-CO)NH spectra (23).
Isothermal Titration Calorimetry (ITC) Measurements. ITC measurements were performed at 30°C by using a Microcal (Amherst, MA) VP-ITC calorimeter. Protein and peptide samples were buffered with 20 mM Tris, pH 7.6/5 mM 2-mercaptoethanol and thoroughly degassed before use. The concentrations were determined by amino acid analysis. The sample cell (1.4 ml) contained either ≈100 μM SUMO-1 or ≈50 μM SUMO-3. A total of 29 injections of 10 μl of peptide solution (≈1 mM) were carried out at 3-min intervals. The heat generated due to dilution of the titrants (peptide) was subtracted for baseline correction. The baseline-corrected data were analyzed with Microcal ORI-GIN Ver. 5.0 software. Experiments were duplicated.
Calculations of Electrostatic Potentials. The surface electrostatic potentials for SUMO-1 were calculated by using the delphi module of insightii (Micron Separations, Westboro, MA) and the NMR structure of SUMO-1 (24), as described (25).
Protein-Binding Assay. The cDNA coding for human RanGAP1 was cloned into pBluescript vector (Stratagene). The in vitro protein-binding assays were performed as described (13). Briefly, _in vitro_-translated RanGAP1 or purified recombinant Ubc9 was used to bind to the RanBP2 domains immobilized on the microtiter plates. Autoradiography or Western blot after SDS/PAGE was used to detect bound RanGAP1 or Ubc9, respectively.
Results
Defining the Region in PIASX That Forms Direct Interactions with SUMO-1. We used NMR spectroscopy to investigate SUMO-1 binding by a synthetic peptide (PIASX-P, see Fig. 1_A_) corresponding to the PIASX sequence containing the SXS motif followed by a few acidic residues. PIASX belongs to a protein family that was originally identified as protein inhibitor for activated STAT transcription factors and is known to possess the E3 ligase activity for SUMO conjugation (26). A member of this family, PIASy, colocalizes with SUMO-1/2 in the promyelocytic leukemia (PML) nuclear bodies (NB) (27) and thus may bind to SUMO-1 and/or -2 or to the SUMO-1 and/or -2 moieties of modified proteins. The synthetic PIASX-P contains a sequence that is conserved among the PIAS family members (20).
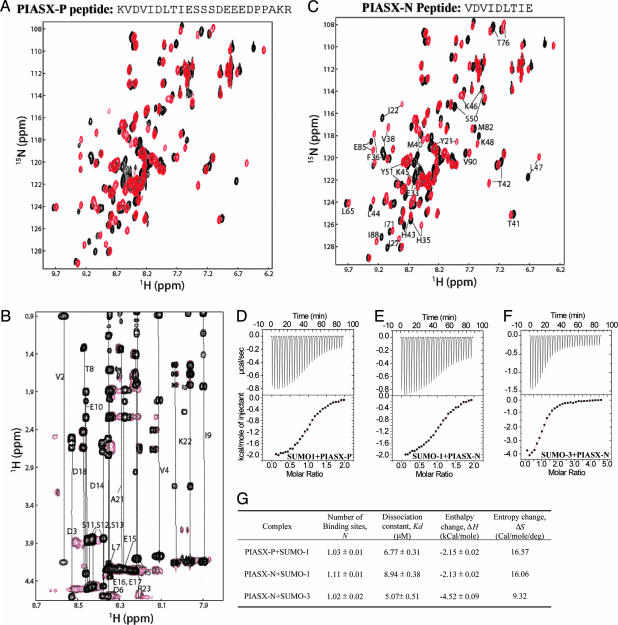
Fig. 1.
Identification of SBM. (A) Superposition of the 1H-15N HSQC spectra of 15N/13C-labeled human SUMO-1, free (black) and in complex (red) with PIASX-P. The sequence of PIASX-P is shown above the spectra. (B) Superposition of the TOCSY spectrum (black) of free PIASX-P and the 15N-filtered TOCSY spectrum (red) of PIASX-P in complex of 1:1 molar ratio with 15N/13C-labeled SUMO-1. Resonance assignments of the free peptide are indicated. (C) Superposition of the 1H-15N HSQC spectra of SUMO-1, free (black) and in complex with PIASX-N (red). The residues in SUMO-1 that are significantly affected by the complex formation are indicated with their assignments. (D-F) ITC measurements for the interaction between PIASX-P or PIASX-N and SUMO-1 or -3. Experimental details are provided in Materials and Methods. (G) Summary of thermodynamic parameters obtained from the ITC measurements shown in D-F.
NMR chemical shift perturbation is extremely sensitive to molecular interactions. Addition of unlabeled PIASX-P to 15N-13C-enriched SUMO-1 resulted in chemical shift changes of specific amino acid residues of SUMO-1, thus confirming that the peptide binds SUMO-1 specifically (Fig. 1 A).
Reciprocally, 15N-filtered TOCSY and NOESY spectra were used to selectively identify residues in PIASX-P that complex with labeled SUMO-1. Superposition of the TOCSY spectrum of free PIASX-P and the 15N-filtered TOCSY spectrum of the peptide complexed with 15N-13C-enriched SUMO-1 (Fig. 1_B_) unexpectedly suggested that the SXS motif within PIASX-P (20), including the central SXS triplet followed by several acidic residues, was not involved in binding SUMO-1. Instead, the segment from Val-2 to Ile-9 of PIASX-P showed the most significant chemical shift changes, indicating that these residues were responsible for binding SUMO-1 (Fig. 1_B_).
To further confirm that this N-terminal region of PIASX-P was responsible for SUMO-1 binding, another peptide (PIASX-N) was synthesized that corresponded to residues Val-2 to Glu-10 of PIASX-P (Fig. 1_C_) with the previously suggested core consensus sequence of the SBM deleted. The interaction between PIASX-N and SUMO-1 was examined by using 15N-1H HSQC spectra (Fig. 1_C_). The PIASX-N peptide induced nearly identical chemical shift changes in SUMO-1 as did PIASX-P, suggesting that the two peptides interacted with SUMO-1 in an identical fashion.
We also used ITC to compare the interactions among SUMO-1 and the peptides PIASX-P and PIASX-N (Fig. 1 D, E, and G). Consistent with the finding from NMR studies, the binding affinity between SUMO-1 and PIASX-N was similar to that between SUMO-1 and PIASX-P, and the small differences may be due to the uncertainties in the estimation of peptide concentrations by amino acid analysis. These data confirm that the N-terminal hydrophobic region of PIASX-P and not the previously suggested SXS motif is responsible for binding SUMO-1. Interestingly, the interaction is both enthalpy and entropy driven, consistent with the hydrophobic nature of the interactions.
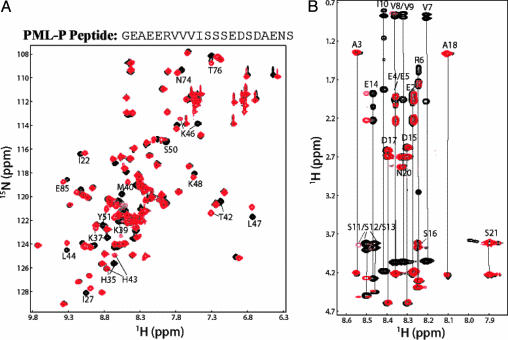
Identification of Residues in the PML Protein Responsible for Binding SUMO-1. PML was initially identified in acute promyelocytic leukemia, in which PML is fused to the retinoic acid receptor, resulting in disaggregation of the PML NBs (7). Sumoylation of PML is an initiating event for PML NB formation (28). Additionally, PML has been reported to associate with sumoylated p73α in a yeast two-hybrid experiment (20). To define the consensus amino acid sequence for binding SUMO-1, we prepared a second synthetic peptide (PML-P; Fig. 2_A_) corresponding to a region of PML that contains the SXS triplet and the C-terminal acidic-amino acid region. NMR experiments similar to those described above were performed to characterize the interaction between PML-P and SUMO-1. The superimposed 15N-1H HSQC spectra of SUMO-1, free and in complex with PML-P, indicated that PML-P complexed SUMO-1 specifically (Fig. 2 A). However, superposition of the TOCSY spectrum of free PML-P and 15N-filtered TOCSY spectrum of PML-P in complex with the 15N-13C-labeled SUMO-1 (Fig. 2_B_) demonstrated that the residues involved in direct association with SUMO-1 are the hydrophobic residues including Val-7, Val-8, Val-9, and Ile-10 but not the residues that constitute SXS motif suggested by Minty et al. (20).
Fig. 2.
Interaction between SUMO-1 and PML. (A) Superposition of the 1H-15N HSQC spectra of SUMO-1, free (black) and in complex with PML-P (red). The sequence of PML-P is shown above the spectra. The residues in SUMO-1 that are significantly affected by the complex formation are indicated with their assignments. (B) Superposition of the TOCSY spectrum of free PML-P (black) and the 15N-filtered TOCSY spectrum of PML-P in complex with 15N-13C-labeled SUMO-1 (red). Resonance assignment of the free peptide is indicated.
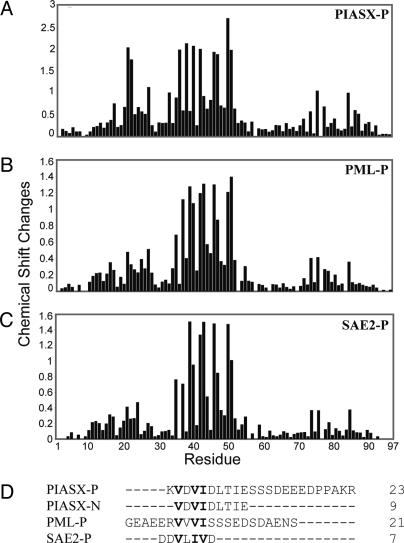
Defining the Consensus Sequence of the SBM. The SUMO-binding regions in PML-P and PIASX-N do not show clear sequence similarity. Therefore, chemical shift perturbation was used to investigate whether PML-P and PIASX-P bind to the same region of SUMO-1. Plots of chemical shift changes in SUMO-1, upon binding to PIASX-P (Fig. 3_A_) or to PML-P (Fig. 3_B_), revealed that both peptides bind to similar regions of SUMO-1. This suggested that both peptides should contain the same amino acid sequence motif responsible for the interaction. Sequence comparison of PIASX-N and PML-P suggested the consensus sequence V-X-V-I for the SBM (Fig. 3_D_).
Fig. 3.
Identification of the consensus sequence. (A-C) Plots of chemical shift changes of SUMO-1 in complex with PIASX-P (A), PML-P (B), and SAE2-P (C) vs. residue number. Chemical shift changes were calculated as square root of (25 × Δδ1H2 + Δδ15N2) to compensate for the difference in chemical shift dispersion between 1H and 15N nuclei. (D) Sequence alignment of the four synthetic peptides (PIASX-P, PIASX-N, PML-P, and SAE2-P) used in the studies to define the sequence requirement for binding SUMO. Sequences are aligned to reveal the conserved motif V-X-V/I-V/I.
Val and Ile are similar amino acids with branched aliphatic side chains. Thus it is possible that Val and Ile are interchangeable for interaction with SUMO-1. Allowing interchange of Val and Ile in the SBM resulted in the identification of potential SUMO-1 interaction sequence V-L-I-V in RanBP2/Nup358 and in SAE2, a subunit of the SUMO activation enzyme (Fig. 3_D_). To test SUMO binding by this sequence, we synthesized a peptide with the sequence D-D-V-L-I-V-D corresponding to the sequence in SAE2 (referred to as SAE2-P), which has the Val and Ile exchanged in the third and fourth positions relative to that in PML and PIASX (Fig. 3_D_). NMR studies demonstrated that this peptide specifically interacts with SUMO-1 (Fig. 6, which is published as supporting information on the PNAS web site) in the same region as does PIASX-P or PML-P (Fig. 3_C_). These results also confirmed that valine and isoleucine are exchangeable in the sequence of the SBM (Fig. 3_D_).
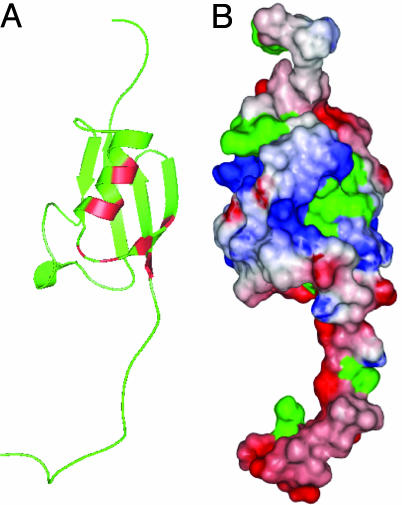
Interaction with SUMO-3 and the Sequence-Dependent Binding Affinities. The SUMO-1 residues that are perturbed by complex formation with the peptides cluster together in the 3D structure (Fig. 4 A and B). These residues at the binding interface of SUMO-1 are relatively conserved among SUMO-1, -2, and -3, suggesting that this SBM can interact with all three SUMO paralogues. Using SUMO-3 as a representative of the nearly identical SUMO-2/3, ITC measurements established that SUMO-3 binds to PIASX-N with a similar affinity as SUMO-1 (Fig. 1 F and G). The differences in binding affinities and thermal dynamic profiles between SUMO-1 and SUMO-3 upon complex formation with PIASX-N may reflect the differences in the sequences between the SUMO paralogues. Thus, this SBM interacts with all three SUMO paralogues but with different affinities and thermal dynamic profiles.
Fig. 4.
The binding site on SUMO-1 for the SBM. (A) Ribbon diagram of the 3D structure of SUMO-1. Residues that show significant chemical shift perturbation upon binding to the peptides are indicated in red, as suggested by the results shown in Fig. 3. (B) Surface representation of the 3D structure of SUMO-1. The orientation of the molecule in B is the same as that in A. The color spectrum of red to blue corresponds to changes from negative to positive potentials over a range of -5to +5 _K_B/electron. Surface hydrophobic residues are indicated in green.
NMR-binding experiments also indicated that the affinity between SUMO-1 and the SBM of a protein depended on the sequence context of the motif. Among the three peptides studied, PIASX-P binds to SUMO-1 with an exchange rate between the free and bound form that is “slow” relative to NMR chemical shift timescale (Fig. 7 A and B, which is published as supporting information on the PNAS web site). Both the PML-P and SAE2-P peptides bind to SUMO-1 in fast exchange relative to the chemical shift timescale (Fig. 7 C and D). Thus their dissociation constants can be estimated directly from NMR chemical shift perturbation, as 93.5 ± 67.1 μM and 68.8 ± 41.8 μM, respectively. The affinities between the SBM-containing peptides and SUMO are similar to those between ubiquitin-binding motifs and ubiquitin (29, 30).
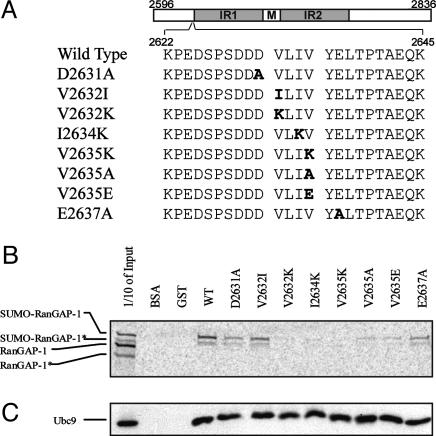
Role of the SBM in RanBP2-RanGAP1 Interaction. The interaction between RanGAP1 and RanBP2 (a component of nuclear pore complexes) is a prototype of sumoylation-mediated protein-protein interaction. In the nuclear import process, binding of sumoylated RanGAP1 to RanBP2 is required for cargo to associate with importin upon hydrolysis of RanGTP (13, 14). The same region of RanBP2 that interacts with sumoylated RanGAP1 also possesses E3 ligase activity for SUMO modification (31). In this report, we have identified residues 2632-2635 of RanBP2 as a SBM (Fig. 5_A_), and this result is consistent with published data. For example, fragments of RanBP2 covering this region bind sumoylated RanGAP1 specifically but not the unmodified RanGAP1 (13, 32). Saitoh et al. (32) also showed that RanBP2 fragments that were truncated in this region (i.e., a fragment encompassing residues 2633-2761) lost the ability to pull down sumoylated RanGAP1.
Fig. 5.
Functional implications of the SBM in RanBP2/Nup358. (A) Design of the mutations within residues 2596-2836 of RanBP2/Nup358. (B) Full length RanGAP1 was translated in vitro by using rabbit reticulocyte transcription/translation extracts in the presence of [35S]methionine. Both RanGAP1 and sumoylated RanGAP1 were produced during the translation process, as shown in the first lane. The asterisks indicate truncated versions of RanGAP1. Translated proteins were assayed for binding to the wild-type or mutant RanBP2/Nup358 fragments, as described in Materials and Methods. (C) Pull down of Ubc9 by the wild-type and mutant RanBP2/Nup358 fragment, as described in Materials and Methods. Western blot with anti-His-tag antibody was used to detect Ubc9.
To confirm our hypothesis that the SBM of RanBP2 is critical for RanBP2's affinity for sumoylated RanGAP1, we individually mutated the three critical hydrophobic residues within RanBP2 (residues 2596-2836; V2632K, V2632I, I2634K, V2635K, V2635A, or V2635E) to test their roles in binding SUMO-modified RanGAP1 (Fig. 5_A_). This region of RanBP2 is not predicted to form any regular secondary structures, and the entire RanBP2 fragment does not form a defined 3D structure, as indicated by the lack of NMR chemical shift dispersion (data not shown). Therefore, the dramatic mutation of hydrophobic V to K (basic) or E (acidic), or the conservative mutation of V2635A, should not affect the structural integrity of RanBP2. Mutation of V2632I investigates whether Val at the first position of the SBM can be replaced by Ile. Additionally, although acidic residues immediately adjacent to the V-L-I-V sequence are not conserved among the proteins containing the SBM (Fig. 3_D_), they could contribute to the affinity of the interaction, because the surface of SUMO-1 that binds to the SBM contains positively charged potentials (Fig. 4_B_). Thus, two additional, individual mutations, D2631A and E2637A, were prepared. Both the wild-type and mutant proteins were expressed and purified as described in Materials and Methods.
We adopted the pull-down assay used previously (13) to investigate binding between sumoylated RanGAP1 and the wild-type and mutant RanBP2 domains. Both RanGAP1 and sumoylated RanGAP1 (labeled with [35S]-methionine) were produced during the coupled in vitro transcription/translation process by using rabbit reticulocyte lysates. Evidently, rabbit reticulocyte extracts contain the necessary factors for SUMO-1 modification (Fig. 5_B_, first lane) (13). Both products appeared as doublets, possibly due to translation initiation from an internal ATG codon in RanGAP1's cDNA. In the pull-down assay, sumoylated RanGAP1, but not the unmodified RanGAP1, complexed with wild-type RanBP2 and with several mutants, and the intensity of sumoylated RanGAP1 pulled down was ordered as: wild type > V2632I > D2631A ≈ E2637A > V2635E ≈ V2635A. The V2632K, I2634K, and V2635K mutants did not interact appreciably with either the modified or unmodified RanGAP1. No or diminished interaction of sumoylated RanGAP1 with the mutant RanBP2 was not likely due to disruption of the structural integrity of RanBP2, because all wild-type and mutant proteins maintained the ability to interact with Ubc9 (Fig. 5_C_). In addition, it is not likely that the mutation in SBM disrupted the sumoylation-independent interaction between RanBP2 and RanGAP1, because the region on RanBP2 that was sensitive to the structural integrity of RanGAP1 is distinct from the region that contains the SBM (13). This result demonstrated that the hydrophobic residues within the SBM of RanBP2 are critical for specific interaction with sumoylated RanGAP1. Negative electrostatic potentials contribute to the affinity of the interaction but are not essential. Thus, the SBM within RanBP2 is functionally significant to mediate the protein-protein interaction of the nuclear pore complex. This finding indicates the functional importance of the SBM identified in this study in sumoylation-dependent cellular processes.
Discussion
Identification of the SBM. Posttranslational modification by the ubiquitin homologues SUMO-1, -2, and -3 plays important roles in diverse cellular processes. Like ubiquitin signaling, SUMO signaling likely occurs through interactions between SUMO moieties of modified target proteins and SBMs within functionally significant protein partners. In this study, we have defined a previously undescribed SBM. Despite the analogous features between SUMO and ubiquitin, this SBM does not resemble ubiquitin-binding motifs nor does it bind to the region of SUMO that is equivalent to the region of ubiquitin interacting with the ubiquitin interaction motifs (Fig. 4). Additionally, this motif binds to all three SUMO paralogues.
Specificity Control in SUMO-Mediated Protein-Protein Interactions. How is specificity controlled in sumoylation-mediated protein-protein interactions by using such a SBM? Previously, the affinity of the interaction between RanBP2 and sumoylated RanGAP1 was shown to depend on the size of the RanBP2 fragments used, and furthermore, SUMO-1 could not compete with sumoylated RanGAP1 for binding to RanBP2 (13, 14). RanBP2 might contain additional site(s) for SUMO-independent interaction with RanGAP1, but such interaction would be too weak to be functionally important by itself and to be detectable by standard biochemical methods. Despite the weak interaction, it contributes synergistically to the association between RanBP2 and sumoylated RanGAP1. This is expected based on a thermodynamic principle: the dissociation constant is related to the free energy changes associated with the interaction by ΔG = -_RT_ln_K_d, where R is a thermal dynamic constant and T is temperature. When the interaction contains two contributions, addition of the free energy changes due to two interactions leads to multiplication of the dissociation constants of the two independent interactions [ln_K_dtotal = ln_K_d1 + ln_K_d2 = ln(_K_d1·_K_d2)]. For example, let's assume that RanGAP1 and RanBP2 bind to each other with a dissociation constant of 1 mM in the absence of SUMO-1 modification. This affinity is physiologically insignificant and too weak to be detected by standard biochemical methods. However, this interaction can significantly decrease the dissociation constant for the association between sumoylated RanGAP1 and RanBP2 by a factor of 1,000. In this way, SUMO modification plays a critical regulatory role in protein-protein interactions, not only in this example, but also in other cellular functions. The dynamic SUMO conjugation and deconjugation system in cells thus provides a rapid and efficient control of cellular protein-protein interactions.
Functional Implication of the SBM in Other Proteins. The V/I-X-V/I-V/I motif exists in nearly all proteins related to SUMO-dependent processes. SUMO modification of PML is a prerequisite for PML NB formation and recruitment of PML NB components, including autoantigen Sp100 (33), transcription factor Daxx (34), and transcriptional coactivator CBP (28, 35). Interestingly, all these known PML NB components contain the SBM. Histone modifications, including acetylation, are important regulatory mechanisms for gene expression (36). The SUMO-modified transcriptional coactivator P300 and transcription factor Elk-1 recruit HDAC6 (15) and HDAC2 (16), respectively, to specific target promoters where they repress transcription. The SBM motif is found in both HDACs. Furthermore, the SBM is found in other proteins that have not been shown to associate with SUMO-dependent processes. It is possible that the SUMO-dependent processes involving these proteins remain to be identified. It is also possible that a given SBM may not be functionally exposed in a protein structure. For example, many proteins harbor one or multiple SUMO consensus modification sites (ψKXE), but only a fraction of these sites are actually modified.
Conclusion
We have identified the consensus sequence, V/I-X-V/I-V/I, as the first SBM. This motif is important for RanBP2-RanGAP1 interaction, and it is likely to play important roles in protein-protein interactions in various cellular functions and to impart control through the dynamic SUMO conjugation and deconjugation system.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. Ronald T. Hay, Hisato Saito, and Michael J. Matunis for helpful discussions and Dr. Weidong Hu for technical assistance with the NMR experiments. This work is supported by National Institutes of Health Grants GM59887 and CA94595 (to Y.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SUMO, small ubiquitin-like modifier; SBM, SUMO-binding motif; HDAC, histone deacetylase; TOCSY, total correlation spectroscopy; HSQC, heteronuclear sequential quantum correlation; ITC, isothermal titration calorimetry; NB, nuclear body; PML, promyelocytic leukemia.
References
- 1.Poukka, H., Aarnisalo, P., Karvonen, U., Palvimo, J. J. & Janne, O. A. (1999) J. Biol. Chem. 274**,** 19441-19446. [DOI] [PubMed] [Google Scholar]
- 2.Muller, S., Ledl, A. & Schmidt, D. (2004) Oncogene 23**,** 1998-2008. [DOI] [PubMed] [Google Scholar]
- 3.Seeler, J. S. & Dejean, A. (2003) Nat. Rev. Mol. Cell. Biol. 4**,** 690-699. [DOI] [PubMed] [Google Scholar]
- 4.Kotaja, N., Karvonen, U., Janne, O. A. & Palvimo, J. J. (2002) J. Biol. Chem. 277**,** 30283-30288. [DOI] [PubMed] [Google Scholar]
- 5.Chauchereau, A., Amazit, L., Quesne, M., Guiochon-Mantel, A. & Milgrom, E. (2003) J. Biol. Chem. 278**,** 12335-12343. [DOI] [PubMed] [Google Scholar]
- 6.Steffan, J. S., Agrawal, N., Pallos, J., Rockabrand, E., Trotman, L. C., Slepko, N., Illes, K., Lukacsovich, T., Zhu, Y. Z., Cattaneo, E., et al. (2004) Science 304**,** 100-104. [DOI] [PubMed] [Google Scholar]
- 7.Kakizuka, A., Miller, W. H., Jr., Umesono, K., Warrell, R. P., Jr., Frankel, S. R., Murty, V. V., Dmitrovsky, E. & Evans, R. M. (1991) Cell 66**,** 663-674. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh, H. & Hinchey, J. (2000) J. Biol. Chem. 275**,** 6252-6258. [DOI] [PubMed] [Google Scholar]
- 9.Hay, R. T. (2001) Trends Biochem. Sci. 26**,** 332-333. [DOI] [PubMed] [Google Scholar]
- 10.Tatham, M. H., Kim, S., Yu, B., Jaffray, E., Song, J., Zheng, J., Rodriguez, M. S., Hay, R. T. & Chen, Y. (2003) Biochemistry 42**,** 9959-9969. [DOI] [PubMed] [Google Scholar]
- 11.Farrow, N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., Forman-Kay, J. D. & Kay, L. E. (1994) Biochemistry 33**,** 5984-6003. [DOI] [PubMed] [Google Scholar]
- 12.Swanson, K. A., Kang, R. S., Stamenova, S. D., Hicke, L. & Radhakrishnan, I. (2003) EMBO J. 22**,** 4597-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matunis, M. J., Wu, J. & Blobel, G. (1998) J. Cell Biol. 140**,** 499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. (1997) Cell 88**,** 97-107. [DOI] [PubMed] [Google Scholar]
- 15.Girdwood, D., Bumpass, D., Vaughan, O. A., Thain, A., Anderson, L. A., Snowden, A. W., Garcia-Wilson, E., Perkins, N. D. & Hay, R. T. (2003) Mol. Cell 11**,** 1043-1054. [DOI] [PubMed] [Google Scholar]
- 16.Yang, S. H. & Sharrocks, A. D. (2004) Mol. Cell 13**,** 611-617. [DOI] [PubMed] [Google Scholar]
- 17.Bernier-Villamor, V., Sampson, D. A., Matunis, M. J. & Lima, C. D. (2002) Cell 108**,** 345-356. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez, M. S., Desterro, J. M., Lain, S., Midgley, C. A., Lane, D. P. & Hay, R. T. (1999) EMBO J. 18**,** 6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, D., Tatham, M. H., Yu, B., Kim, S., Hay, R. T. & Chen, Y. (2002) J. Biol. Chem. 277**,** 21740-21748. [DOI] [PubMed] [Google Scholar]
- 20.Minty, A., Dumont, X., Kaghad, M. & Caput, D. (2000) J. Biol. Chem. 275**,** 36316-36323. [DOI] [PubMed] [Google Scholar]
- 21.Wuthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 22.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenblatt, J., Konrat, R. & Kay, L. E. (1997) J. Am. Chem. Soc. 119**,** 6711-6721. [Google Scholar]
- 23.Clore, G. M. & Gronenborn, A. M. (1994) Methods Enzymol. 239**,** 349-363. [DOI] [PubMed] [Google Scholar]
- 24.Bayer, P., Arndt, A., Metzger, S., Mahajan, R., Melchior, F., Jaenicke, R. & Becker, J. (1998) J. Mol. Biol. 280**,** 275-286. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Q., Jin, C., Liao, X., Shen, Z., Chen, D. J. & Chen, Y. (1999) J. Biol. Chem. 274**,** 16979-16987. [DOI] [PubMed] [Google Scholar]
- 26.Jackson, P. K. (2001) Genes Dev. 15**,** 3053-3058. [DOI] [PubMed] [Google Scholar]
- 27.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15**,** 3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong, S., Muller, S., Ronchetti, S., Freemont, P. S., Dejean, A. & Pandolfi, P. P. (2000) Blood 95**,** 2748-2752. [PubMed] [Google Scholar]
- 29.Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D. & Hicke, L. (2002) Nat. Cell Biol. 4**,** 389-393. [DOI] [PubMed] [Google Scholar]
- 30.Kang, R. S., Daniels, C. M., Francis, S. A., Shih, S. C., Salerno, W. J., Hicke, L. & Radhakrishnan, I. (2003) Cell 113**,** 621-630. [DOI] [PubMed] [Google Scholar]
- 31.Pichler, A., Gast, A., Seeler, J. S., Dejean, A. & Melchior, F. (2002) Cell 108**,** 109-120. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh, H., Pizzi, M. D. & Wang, J. (2002) J. Biol. Chem. 277**,** 4755-4763. [DOI] [PubMed] [Google Scholar]
- 33.Szostecki, C., Guldner, H. H., Netter, H. J. & Will, H. (1990) J. Immunol. 145**,** 4338-4347. [PubMed] [Google Scholar]
- 34.Yang, X., Khosravi-Far, R., Chang, H. Y. & Baltimore, D. (1997) Cell 89**,** 1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bannister, A. J. & Kouzarides, T. (1996) Nature 384**,** 641-643. [DOI] [PubMed] [Google Scholar]
- 36.Richards, E. J. & Elgin, S. C. (2002) Cell 108**,** 489-500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information