Bystander gene activation by a locus control region (original) (raw)
Abstract
Random assortment of genes within mammalian genomes establishes the potential for interference between neighboring genes with distinct transcriptional specificities. Long-range transcriptional controls further increase this potential. Exploring this problem is of fundamental importance to understanding gene regulation. In the human genome, the _Ig_β (CD79b) gene is situated between the pituitary-specific human growth hormone (hGH) gene and its locus control region (hGH LCR). Igβ protein is considered B-cell specific; its only known role is in B-cell receptor signaling. Unexpectedly, we found that _hIg_β is transcribed at high levels in the pituitary. This _Ig_β transcription is dependent on pituitary-specific epigenetic modifications generated by the hGH LCR. In contrast, expression of _Ig_β at its native site in B cells is independent of hGH LCR activity. These studies demonstrated that a gene with tissue-restricted transcriptional determinants (B cell) can be robustly activated in an unrelated tissue (pituitary) due to fortuitous positioning within an active chromatin domain. This ‘bystander' gene activation pathway impacts on current concepts of tissue specificity and models of active chromatin domains.
Keywords: CD79b, chromatin, _Ig_β, locus control region, hGH
Introduction
Transcriptional controls in higher eukaryotes can be exerted over extended distances (Ptashne, 1986; Bresnick and Tze, 1997). Remote regulatory elements can function by establishing extensive ‘activated' chromatin domains that encompass target promoters (Vyas et al, 1992; Hebbes et al, 1994; Bulger and Groudine, 1999; Elefant et al, 2000; Forsberg and Bresnick, 2001; Ho et al, 2002). Alternatively, remote regulatory elements can be brought into close juxtaposition with target promoters via long-range ‘looping' or ‘tracking' mechanisms (Mueller-Storm et al, 1989; Tewari et al, 1996; McDowell and Dean, 1999; Carter et al, 2002; Tolhuis et al, 2002). Sets of transcriptional regulatory determinants responsible for long-range gene activation pathways are generally referred to as locus control regions (LCRs) (Forrester et al, 1987; Grosveld et al, 1987). The identification of LCRs in a wide array of gene systems suggests that long-range transcriptional controls constitute common pathways for gene activation in higher eukaryotes.
The extensive distances over which long-range controls can be exerted, and the close packing and random arrangement of many loci in the mammalian genome, establishes the potential for mutual interference among neighboring genes with distinct developmental or tissue specificities. Understanding how this significant problem in gene regulation is overcome is of fundamental importance. Insulator and boundary elements have been identified that can block a subset of such unwanted interactions (Chung et al, 1993; Hebbes et al, 1994; Bell and Felsenfeld, 1999). However, simple linear models of insulator/barrier function would be incompatible with effective insulation between loci with overlapping transcriptional control determinants. This suggests that more complex models of gene ‘insulation' need to be formulated. Alternatively, certain loci may in fact not be fully insulated from the influence of unrelated transcriptional determinants and chromatin domains in their local environment. In this case, genes may be expressed in ‘inappropriate' tissues or developmental settings. Such lack of precision in gene expression may be tolerated by the cell and/or might be compensated by post-transcriptional regulatory pathways.
The human growth hormone (hGH) cluster contains five genes expressed specifically in the pituitary (hGH-N) or in the placenta (hCS-L, hCS-A, hGH-V, and hCS-B). The hGH LCR in these two tissues is composed of partially overlapping sets of HS. HSI and HSII are specific to pituitary chromatin, HSIV is specific to placental chromatin, while HSIII and HSV are formed in the chromatin of both tissues (Jones et al, 1995). The mechanism of long-range activation by the hGH LCR has been best defined in the pituitary. The two closely linked pituitary-specific HS, HSI and HSII, are situated 14.5–15.0 kb 5′ to the hGH-N promoter. These HS appear to recruit HAT activity with subsequent bidirectional spreading of histone acetylation (Elefant et al, 2000; Ho et al, 2002). These reactions establish a continuous 32 kb domain of acetylated chromatin that extends to encompass the hGH-N promoter. Epigenetic alterations within this domain are required for trans factor binding at the hGH-N promoter and for effective hGH-N transcriptional activation (Ho et al, 2002). Targeted inactivation of HSI results in loss of the acetylated histone domain in pituitary chromatin and is accompanied by 90–95% loss of hGH-N transgene expression (Ho et al, 2002). Thus hGH-N transcriptional activation and the establishment of the extensively modified hGH LCR chromatin domain are intimately linked processes and both are dependent on the function of HSI.
The human _CD79b/Ig_β gene (referred to throughout as _hIg_β) functions specifically in B lymphocytes (Hermanson et al, 1988). Igβ proteins form heterodimers with Igα to create the signal transduction subunit of the B-cell receptor (Campbell et al, 1991; Clark et al, 1992). As such, Igβ plays a critical and nonredundant role in B-cell differentiation and function (Papavasiliou et al, 1995). There are no additional functions defined for this protein. Considering this B-cell specificity, it is interesting and potentially problematic that the _hIg_β gene is situated between HSI of the hGH LCR and the hGH-N promoter (Bennani-Baïti et al, 1998a, 1998b). This organization places the _hIg_β locus within a hyperacetylated hGH LCR chromatin domain in somatotrope nuclei. The close packing of the hGH-N, _hIg_β, and the hGH LCR thus presents a complex and potentially informative model to study mechanisms by which genes are shielded from each other's transcriptional control elements.
In the present study, we report the unexpected observation that _hIg_β is actively transcribed in the pituitary. Interestingly, activation of the _hIg_β locus in the pituitary occurs in the absence of B-cell-specific transcription factors required for _hIg_β expression in B cells. This activation of _hIg_β in the pituitary reflects its fortuitous positioning within the activated chromatin domain generated by the hGH LCR. Thus, _hIg_β is not shielded from the hGH LCR as might be expected, but instead is robustly activated by it. This ‘bystander' gene activation may represent a relatively common phenomenon. As such it would have important implications for models of developmental and tissue-specific gene regulation within the mammalian genome.
Results
An hIg_β_ transgene with extensive flanking regions is appropriately expressed in mouse B cells
The five genes within the hGH cluster are activated in either pituitary or placenta. In both situations, this activation is dependent on the action of a set of remote 5′ hGH LCR elements (Introduction). The B-cell-specific _hIg_β gene is situated between the hGH LCR and the hGH cluster. We sought to identify a separate set of chromatin determinants in this region that were responsible for activation of _hIg_β in B cells. Defining such determinants would allow us to model inter-relationships of transcriptional control elements within this tightly packed region of the genome. As an initial step, we assessed the expression of _hIg_β from the 87 kb hGH/P1 transgene (Figure 1A). This previously described transgene (Su et al, 2000) contains _hIg_β along with extensive flanking sequences. The flanking sequences include 31 kb of 5′-flanking sequences encompassing the entire hGH LCR and extending into the striated muscle-specific SCN4A gene and 51 kb of 3′-flanking sequences that contain hGH-N and three of the four placentally expressed genes of the hGH gene cluster. The expression of _hIg_β from the hGH/P1 transgene was compared to that of the endogenous mouse (m) _Ig_β (Figure 1B). _hIg_β was robustly expressed in the spleens of four hGH/P1 transgenic mouse lines. Expression per transgene copy was maintained within a two-fold range, varying from 25 to 42% of endogenous _mIg_β. Immunofluorescent analysis of transgenic spleens confirmed that hIgβ protein expression was restricted to the B-cell compartment (Figure 1C). B-cell specificity was further substantiated by flow sorting of splenocytes (data not shown). These results indicated that the hGH/P1 transgene contains determinants sufficient to establish an autonomous, site-of-integration-independent chromatin environment that supports robust expression of _hIg_β in mouse B cells.
Figure 1.
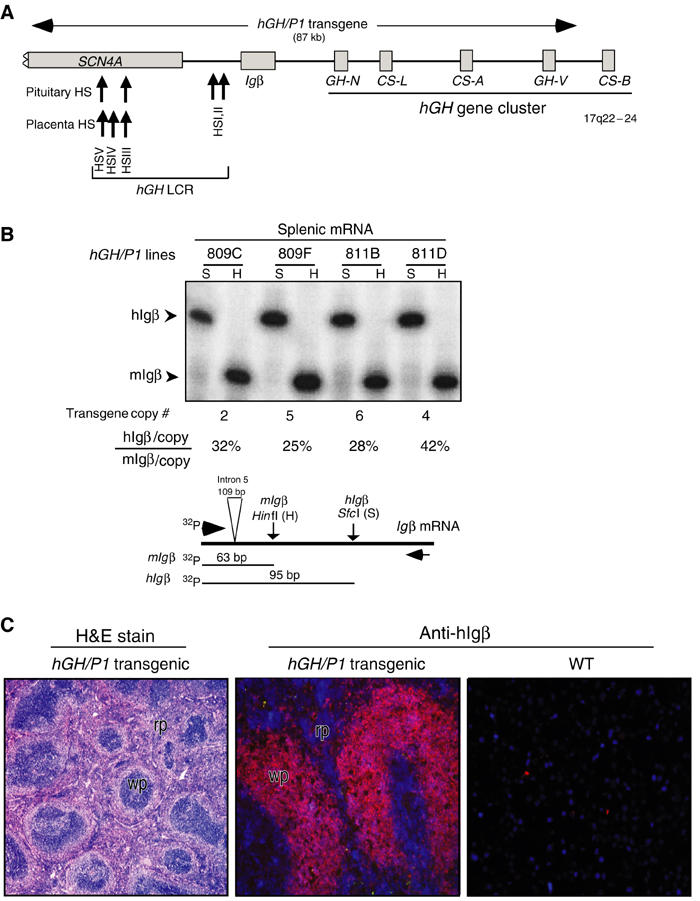
hIg_β expression in the spleens of hGH/P1 transgenic mice. (A) Map of the hIg_β/hGH locus and the hGH/P1 transgene. Each gene is represented by a labeled box. SCN4A, _hIg_β, and the five genes in the hGH cluster are represented. The horizontal arrow above the locus indicates the hGH/P1 transgene. DNaseI HS of the hGH LCR are indicated by upward arrows along with their respective tissue specificities. (B) Splenic expression of _hIg_β from the hGH/P1 transgene is copy number dependent. _hIg_β mRNA and endogenous _mIg_β mRNA were coamplified (RT/PCR) and the cDNAs were distinguished by restriction analysis. A representative RT/PCR analysis is presented in the autoradiograph and a diagram of the assay is shown below. The 5′ primer was 32P-end labeled and the PCR products were digested with _Hin_fI (H) that exclusively digests the _mIg_β PCR product and _Sfc_I (S) that exclusively digests the _hIg_β PCR product. The 5′-labeled fragments generated from mouse (63 bp) and human _Ig_β (95 bp) cDNAs are indicated (arrowheads). These signals were quantified from four unique hGH/P1 transgenic lines. The transgene copy number for each line is noted below each lane. Transgene expression per transgene copy was normalized to endogenous _mIg_β and values are indicated as percentages below each respective lane. (C) _hIg_β transgene is selectively expressed in the white pulp of the mouse spleen. (Left) Hematoxylin and eosin staining of an hGH/P1 transgenic spleen showing normal architecture of the spleen and indicating the defined areas of red pulp (rp) and white pulp (wp). (Middle) hGH/P1 transgenic spleen stained with anti-hIgβ (Texas red). The intense hIgβ staining is restricted to white pulp (wp) consistent with the local abundance of B lymphocytes. (Right) Nontransgenic, wild-type (WT) spleen stained with anti-hIgβ. The absence of a signal confirms the species specificity of the anti-hIgβ antibody.
hIg_β_ transgene is expressed in the mouse pituitary
The fidelity of _hIg_β expression from the hGH/P1 transgene was evaluated by a tissue survey (Figure 2A). As expected, the spleen contained high levels of _hIg_β and _mIg_β mRNAs. In contrast, these mRNAs were at or below trace levels in the brain, heart, kidney, and liver, and at a slightly higher level in the intestine. Surprisingly, a strong _hIg_β mRNA signal was detected in the pituitary. While an exact quantitative comparison of _hIg_β mRNA levels in B cells and somatotropes is complicated by the presence of mixed cell populations in the spleen and pituitary, it was clear that the _hIg_β mRNA level in the pituitary was robust, approximating that in the spleen.
Figure 2.
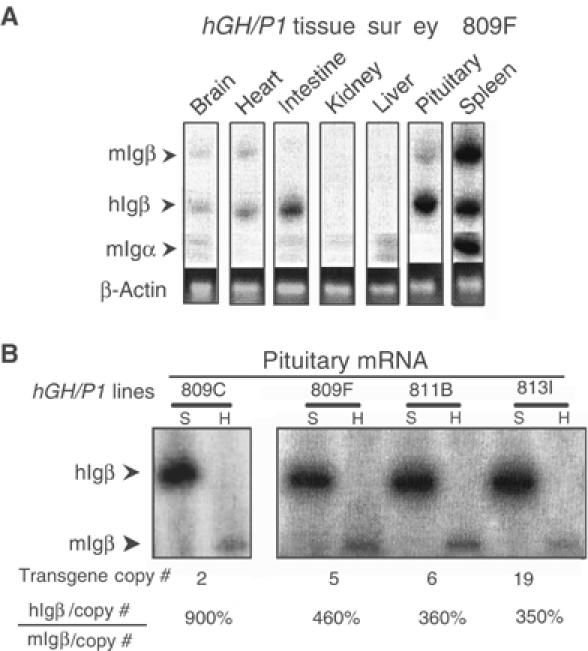
_Ig_β is transcribed from the hGH/P1 transgene in the mouse pituitary. (A) Tissue survey of hGH/P1 transgenic mice revealed abundant _hIg_β mRNA in the pituitary. A co-RT/PCR endonuclease cleavage assay was used to analyze the tissue specificity of _hIg_β expression in a representative hGH/P1 transgenic mouse line (line 809F). The RT/PCR assay was identical to Figure 1B with the exception that the 3′ primer is 32P-end labeled. The figure shows the PCR products digested with _Sfc_I; the larger band (336 bp) corresponds to _mIg_β and the smaller (250 bp) to _hIg_β (labeled arrows). A separate RT/PCR of _mIg_α mRNA was carried out to monitor B-lymphocyte contamination in each tissue. The levels of _hIg_β, _mIg_β, and mIg_α are shown relative to a β_-actin mRNA loading control (ethidium bromide-stained gel). (B) Pituitary expression of _hIg_β mRNA from the hGH/P1 transgene was copy number dependent. Pituitaries from four hGH/P1 transgenic mouse lines were assayed by RT/PCR for _hIg_β mRNA. The level of _hIg_β mRNA was normalized to endogenous mouse pituitary _Ig_β mRNA and this value is shown as a percentage below each pair of lanes after correction for gene copy number.
Analysis of all four hGH/P1 transgenic lines confirmed the robust expression of _hIg_β mRNA in the pituitary. These studies further revealed that the pituitary expression was copy number dependent, varying by less than three-fold (Figure 2B and real-time RT/PCR analysis, data not shown). Consistent with the expression of the _hIg_β transgene in the pituitary, we also detected the endogenous _mIg_β mRNA in this tissue, although at significantly lower levels.
To address the possibility that _hIg_β mRNA in transgenic mouse pituitary might reflect B-cell contamination, we assayed for _mIg_α mRNA (Figure 2A). _Ig_β and _Ig_α are encoded on separate chromosomes in both human and mouse genomes (Campbell et al, 1991; Clark et al, 1992). As expected, _mIg_α mRNA was easily detected in transgenic mouse splenic RNA. However, _mIg_α mRNA was absent in the pituitary RNA samples, and could not be detected in any of the other surveyed transgenic tissues (Figure 2A). We concluded that the _hIg_β gene is robustly transcribed in the transgenic mouse pituitary.
The overall structure of hIg_β_ mRNA in transgenic pituitary is very similar to that in B cells
The structure of the _hIg_β transcripts in the pituitary was assessed. We considered two possible origins for the pituitary _hIg_β transcripts that would differentiate them from authentic (B cell) _Ig_β mRNAs. First, these transcripts could represent noncoding (intergenic) RNAs randomly generated from the active hGH LCR chromatin domain. Intergenic transcription has been described throughout the human β-globin LCR (Ashe et al, 1997; Routledge and Proudfoot, 2002) and it has been implicated in chromatin modification (Gribnau et al, 2000). Second, _hIg_β may be transcribed in the pituitary from an alternative promoter distinct from that used in B cells. To address these possibilities, _hIg_β mRNA size was analyzed by Northern blotting. These data revealed that _hIg_β mRNA in transgenic mouse pituitaries was of the same size as in transgenic mouse purified lymphocytes and human B cells (Figure 3A). The normal size of the pituitary _hIg_β mRNA was incompatible with its generation by random transcription within the LCR domain.
Figure 3.
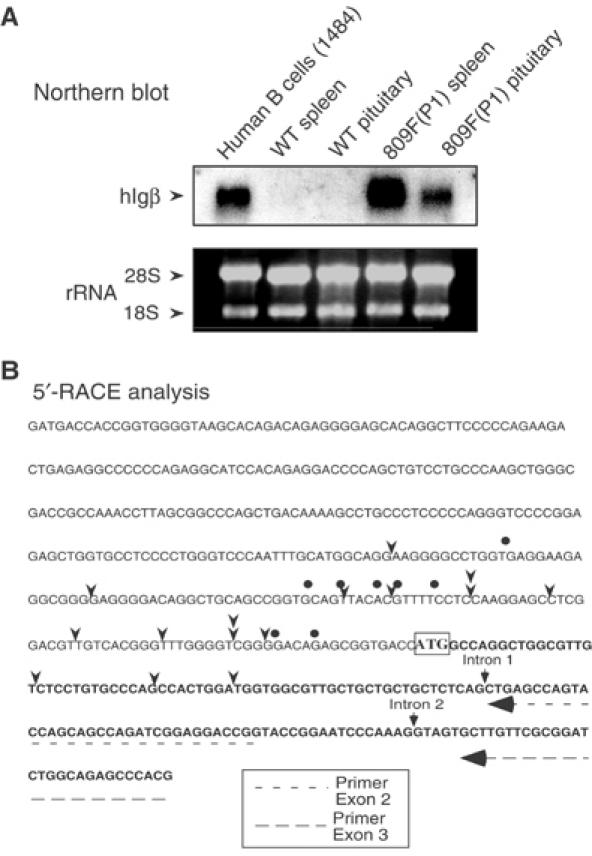
Structural comparison of _hIg_β mRNAs in the transgenic pituitary and B cells. (A) Northern blot analysis. Total RNAs from a human B-cell line (1484), from wild-type (WT) mouse lymphocytes (spleen) and pituitary, and lymphocytes (spleen) and pituitary of hGH/P1 transgenic mouse line 809F were hybridized with a [32P] labeled _hIg_β cDNA probe. The gel was stained with ethidium bromide to visualize 18S and 28S rRNAs as loading control. The _hIg_β mRNA hybridizing bands comigrated in all tissues. No additional bands were detected. The absence of signal in the lanes containing WT lymphocytes and pituitary RNA confirmed the species specificity of the _hIg_β probe. (B) Mapping the 5′ terminus of pituitary and B-cell _Ig_β mRNAs. The sequence of the _hIg_β 5′-flanking region and 5′-terminal transcribed region are shown; the translation start site, ATG, is boxed. The 5′ termini of the transcribed mRNAs in B cells and transgenic pituitary were determined by 5′ RACE. The arrowheads indicate transcription start sites of _hIg_β in the hGH/P1 transgenic mouse pituitary mRNA population. The dots indicate the _hIg_β start sites in human B-cell line 1484. The figure also indicates the locations of the primers used for the 5′-RACE assay (dashed arrows) and the positions of introns 1 and 2.
To determine whether the pituitary and B-cell _hIg_β mRNAs begin at different promoters, the structures of their respective 5′ termini were determined by 5′ rapid amplification of cDNA ends (5′ RACE). _hIg_β mRNAs isolated from the hGH/P1 transgenic pituitary and human B-cell line revealed multiple transcription initiation sites distributed across an overlapping 160-nucleotide window (Figure 3B). This scattering of 5′ termini is consistent with a previous study (Thompson et al, 1996) and with the TATA-less/GC-rich structure of the _hIg_β promoter (Geng and Johnson, 1993; Donohoe and Blomberg, 1997; Dong et al, 2000). Of particular note, the 5′-terminal sequences in the 14 randomly selected pituitary _hIg_β cDNAs were continuous with the genomic sequence encompassing exon 1 of the _hIg_β gene. These data argued against the initiation of transcription from an alternative promoter/exon. The 5′-RACE sequence data also showed that pituitary _hIg_β is normally spliced across intron 1. The correct size of the _hIg_β RT–PCR product extending between exons 5 and 6 (see Figure 1B) confirmed that _hIg_β pituitary mRNA was also correctly spliced across intron 5. Thus, on the basis of a number of structural parameters, we concluded that the _hIg_β mRNA in the pituitary is closely related, if not identical, to the mature, authentic B-cell _hIg_β mRNA.
Ig_β_ mRNA is expressed in the human pituitary
We next considered whether the unexpected expression of _hIg_β mRNA in the mouse pituitary reflected a peculiarity of the transgenic model. PolyA-primed cDNA was generated from normal human pituitary and from two human pituitary somatotrope adenomas (Figure 4). Controls for this study included mRNA samples from a transgenic pituitary, from human peripheral blood leukocytes (PBLs), and from a human erythroid cell line (K562). The identity of the pituitary samples was confirmed by the presence of hGH mRNA. _hIg_β mRNA was detected in all three human pituitary samples in the absence of _hIg_α mRNA. In contrast, the control PBLs contained both _hIg_β and _hIg_α mRNAs but neither was detected in K562 cells. The relative levels of _hIg_β mRNA and hGH mRNA were similar in a comparison of the normal pituitary and pituitary adenoma #361. This comparison, however, revealed a relatively higher level of hGH mRNA in adenoma #373. This variation between the two adenomas may reflect the enrichment for somatotropes within the surgical samples or be a peculiarity of the adenoma pathology. This was not further explored. From these data we concluded that the transcription of _hIg_β from the hGH/P1 transgene in the mouse pituitary faithfully recapitulated the expression of _hIg_β in primary human pituitary tissues.
Figure 4.
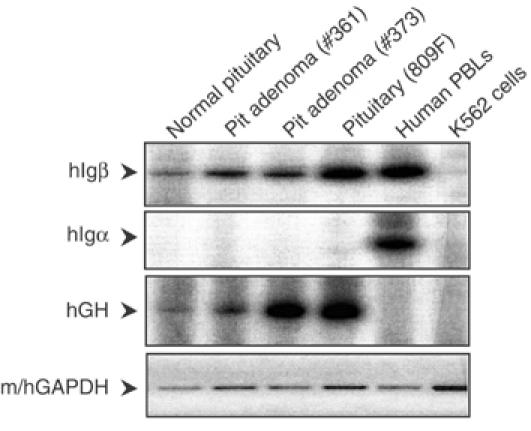
_hIg_β mRNA is expressed in human pituitaries. (Top panel) A normal human pituitary and two human pituitary adenomas (#361 and #373) were evaluated for _hIg_β mRNA by RT/PCR using 32P-labeled RT–PCR primers. Positive controls were human peripheral blood lymphocytes (PBLs), hGH/P1 transgenic pituitary (809F line), and the negative control was the human erythroid cell line K562. (Second panel) _Ig_α was monitored by RT/PCR with labeled primers. (Third panel) hGH RT/PCR with labeled primers identified the human and transgenic mouse pituitaries. (Bottom panel) Detection of m/hGAPDH by RT–PCR represented the loading control (ethidium bromide-stained products). All RT/PCR reactions were assayed in the linear range of amplification.
hIg_β_ transcription in the pituitary is activated by the hGH LCR
_Ig_β expression in B cells has been shown to reflect the activities of B-cell-specific transcription factors (Omori and Wall, 1993; Thompson et al, 1996; Akerblad et al, 1999). Therefore, the expression of _hIg_β in the pituitary is likely to reflect an alternative pathway of gene activation. We speculated that this alternative mode of activation might reflect the localization of the _hIg_β gene within the chromatin domain established in the pituitary by the hGH LCR. HSI of the hGH LCR has a unique and necessary role in establishing this activated chromatin domain (Introduction). We generated a series of transgenic lines carrying the _hIg_β gene flanked by native sequences that either included (−_8.0Ig_β transgene) or excluded HSI (−_1.3Ig_β transgene and −_0.2Ig_β transgene) (Figure 5A). A semiquantitative co-RT/PCR assay was used to assess _hIg_β mRNA levels in the pituitaries from mice representing three or more independent lines carrying each of the three _hIg_β transgenes. _hIg_β mRNA was robustly expressed from the transgenes containing HSI (−_8.0Ig_β). In contrast, _hIg_β mRNA expression was dramatically lower in mice carrying transgenes truncated 3′ to HSI (−_1.3Ig_β and −_0.2Ig_β) (Figure 5B, left). _hIg_β mRNA levels in representative lines were separately assessed by real-time PCR and normalized to endogenous mouse GAPDH mRNA (data not shown, see Materials and methods). The two sets of mRNA assays were in full agreement. These data support the central role of HSI in activating _hIg_β transcription in the pituitary.
Figure 5.
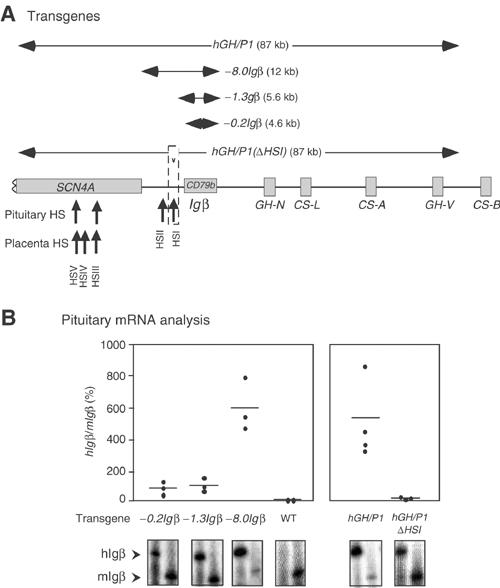
_hIg_β expression in the transgenic pituitary is dependent on the hGH LCR. (A) Transgene constructs. A map of the hGH gene cluster is shown. Horizontal arrows above the map indicate the extent of each transgene. The sizes of the constructs are indicated in parentheses and the designation of each construct indicates the extent of sequences 5′ from the _hIg_β gene promoter. Note that hGH/P1 and −_8.0Ig_β transgenes include HSI whereas this determinant is excluded from the −_0.2Ig_β and −_1.3Ig_β transgenes. In _hGH/P1(Δ_HSI), a 99 bp segment (dashed box), corresponding to the critical core elements of HSI, has been deleted from the hGH/P1 transgene (Ho 2002). (B) HSI of the hGH LCR is a critical determinant of pituitary _hIg_β expression. The ratios of pituitary _hIg_β to _mIg_β mRNAs in the series of _hIg_β transgenic mouse lines (A) are shown. Mice from F1 or later generations and from at least three independent lines (dots) were analyzed (_X_-axis). The _hIg_β to _mIg_β mRNA ratios in each transgenic pituitary were plotted on the _Y_-axis as a percentage. The horizontal lines represent the mean values for each construct in the three or more lines analyzed. Representative co-RT/PCR endonuclease cleavage assays corresponding to each construct are shown below the graph. Wild-type mouse (WT) pituitary RNA was used as negative control. Expression ratios were corrected for transgene copy number.
The involvement of HSI in the activation of _hIg_β in the pituitary was further tested by a second approach. Deletion of a core 99 bp segment of HSI (hGH/P1_Δ_HSI transgene; see Figure 5A) destroys HSI formation, dramatically decreases acetylation throughout the hGH LCR, and results in a 20- to 40-fold decrease in hGH-N transgene expression (Ho et al, 2002). The impact of HSI on _hIg_β transcription was tested by comparing levels of _hIg_β mRNA in the pituitaries of lines carrying the intact hGH/P1 transgene to lines carrying the inactivated HSI (hGH/P1_Δ_HSI). This comparison revealed a mean 47-fold decrease in _hIg_β mRNA in the lines lacking a functional HSI (Figure 5B, right). The data were confirmed by independent real-time PCR analysis of _hIg_β mRNAs in representative lines (data not shown). Thus, the activation of pituitary _hIg_β transcription was dependent on HSI, the major hGH LCR determinant in the pituitary.
Expression of hIg_β_ in B cells is independent of hGH LCR action
The formation of HSI of the hGH LCR is specific to the pituitary (Jones et al, 1995) and hGH-N mRNA is not found in the spleen (Figure 6A). Despite this tissue specificity of hGH LCR function, the pronounced effect of HSI on _hIg_β expression in the pituitary prompted us to formally test whether HSI enhances _hIg_β expression in B cells. Comparison of _hIg_β mRNA levels in the spleens of hGH/P1 and _hGH/P1(Δ_HSI) mice revealed equally robust and copy number-dependent levels from both transgenes (Figure 6B; compare with Figure 1B). These data are consistent with the tissue specificity of hGH LCR function and support the conclusion that HSI activates _hIg_β transcription in the pituitary but has no appreciable function in the B-cell chromatin context.
Figure 6.
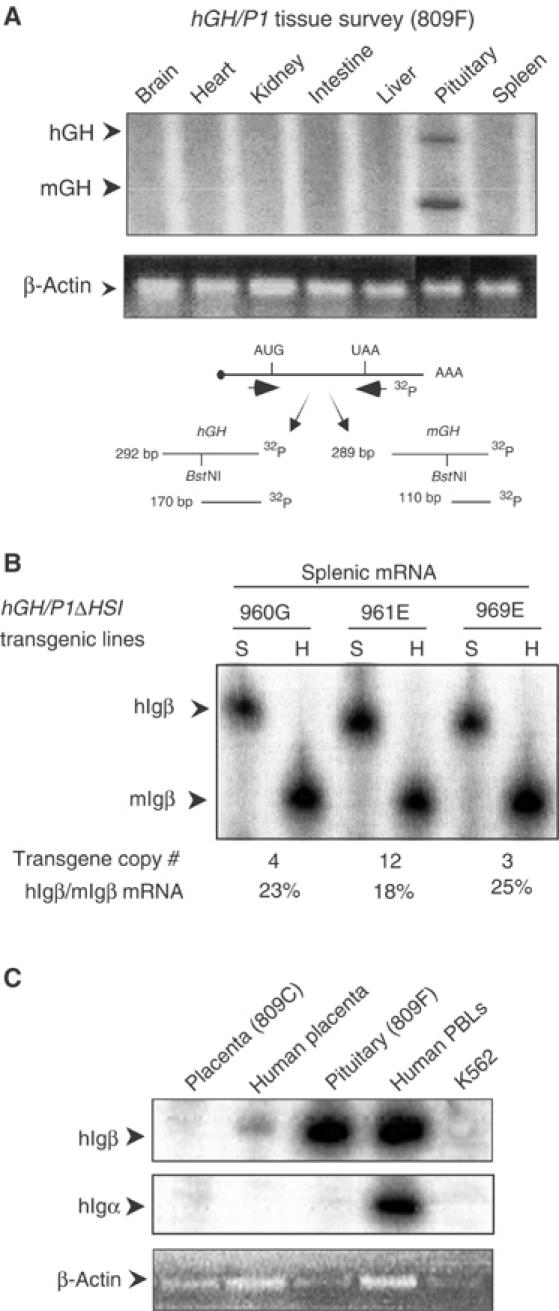
Activation of hIg_β transcription by the hGH LCR is limited to the pituitary. (A) hGH-N mRNA is restricted to the pituitary. mGH and hGH mRNAs were detected by an RT/PCR endonuclease cleavage assay. This assay, shown below the autoradiograph, was applied to tissues from an hGH/P1 transgenic mouse. Arrows to the left of the autoradiograph (upper panel) indicate the expected positions of the mGH and hGH RT/PCR products. In the lower panel, β_-actin PCR products in each lane are visualized by ethidium bromide staining. (B) _hIg_β transgene expression in the spleen is not linked to HSI activity. Splenic RNA samples from three independent lines of mice carrying the hGH/P1_Δ_HSI construct (lines 960G, 961E, and 969E) were analyzed for _hIg_β expression by the co-RT/PCR endonuclease cleavage assay (Figure 1B). The level of transgene expression per gene copy was determined. The percentage of _hIg_β to endogenous _mIg_β expression is indicated below the autoradiograph. (C) hIgβ is not expressed in the placenta. RNA samples from an hGH/P1 placenta (line 809C), a human term placenta, a transgenic pituitary, human PBLs, and an erythroid cell line (K562) were each analyzed for _hIg_β expression by RT/PCR. Ig_α mRNA was assessed to detect potential B-cell contamination of the tissue samples. The lower panel visualizes the products of a β_-actin RT/PCR assay by ethidium bromide staining shown as a loading control.
hIg_β_ is not activated by the hGH LCR in the placenta
The hGH LCR activates expression of pituitary and placental genes from the hGH cluster via distinct epigenetic pathways. In the pituitary, the LCR establishes a continuous 32 kb domain of acetylated chromatin (Elefant et al, 2000; Ho et al, 2002). In contrast, acetylation of the LCR and the GH genes in placental chromatin is highly localized to two discontinuous regions, HSV–HSIII region and the region encompassing the four closely linked placental genes within the cluster (Kimura et al, 2004). Of particular note, levels of histone H3 and H4 acetylation in the region encompassing _hIg_β (between HSIII and the hGH gene cluster) are quite low in placental chromatin when compared to the pituitary. The dependence of _hIg_β gene activation in the pituitary on its positioning within a highly acetylated chromatin domain could therefore be further addressed by analyzing _hIg_β expression in the placenta. In comparison to the robust expression of _hIg_β mRNA in the transgenic pituitary and human B cells, _hIg_β mRNA in the placental samples could only be detected at trace levels (Figure 6C and data not shown). These data support the conclusion that activation of _hIg_β is directly related to its location within the highly acetylated LCR chromatin domain formed in the pituitary.
hIg_β_ mRNA in the pituitary is not effectively expressed at the protein level
The presence of _hIg_β mRNA in the pituitary suggested that hIgβ protein might be synthesized at this site. hIgβ protein could theoretically contribute to a novel somatotrope signaling complex. hIgβ protein expression was therefore assessed by Western analysis of transgenic pituitary extracts. A strong hIgβ signal was detected in the transgenic spleen extracts and human B-cell extracts. In contrast, the Western blots failed to reveal convincing evidence for hIgβ in the pituitary (Figure 7). Immunofluorescent microscopy was carried out using hGH/P1 transgenic pituitaries. These studies were essentially negative with only a rare cell showing evidence of positive hIgβ staining (data not shown). We concluded that the abundant _hIg_β mRNA in the pituitary is either poorly translated or hIgβ protein, once synthesized, is rapidly catabolized due to the lack of a partnering Igα.
Figure 7.
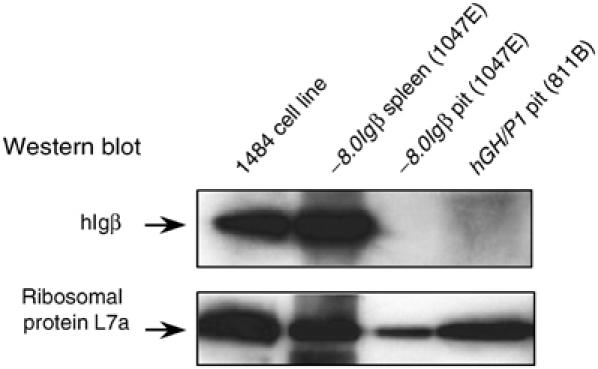
hIgβ protein does not accumulate in _hIg_β transgenic mouse pituitaries. Protein extracts from −_8.0Ig_β and hGH/P1 transgenic pituitaries (lines 1047E and 811B, respectively), human B-cell line 1484, and a −_8.0Ig_β transgenic spleen (line 1047E) were studied by Western analysis. The top panel was incubated with an antibody to hIgβ and the bottom panel with an antibody to ribosomal protein L7a. Both panels were then developed with secondary antibodies and signals detected by enhanced chemiluminescence. The positions of hIgβ and the ribosomal protein L7 are indicated.
Discussion
Regulation of eukaryotic gene transcription reflects complex interactions among transcription factors, cofactor complexes, and epigenetically modified chromatin structures. The present study revealed that the activated chromatin environment established in pituitary somatotropes by the hGH LCR triggers expression of the adjacent B lymphocyte-specific _hIg_β gene. This ectopic activation of _hIg_β reflects its fortuitous positioning within the hGH LCR chromatin domain. Activation of the ‘bystander' _hIg_β gene by the hGH LCR occurs in the absence of B-cell-specific trans factors. The _cis_-acting selectivity of this effect is underscored by the observation that _Ig_α, the B-cell-specific partner of _Ig_β, remains unexpressed in the mouse and human pituitaries. Thus, _hIg_β can be activated in two tissues by two distinct mechanisms: it can be driven in the spleen by a pathway controlled by B-cell-specific transcription factors and it can be driven in the pituitary by the epigenetic influence of an encompassing somatotrope-specific chromatin domain. The latter ‘bystander' gene activation pathway, which is unlikely to be unique, adds to the complexity of gene regulation and tissue specification models.
The mechanistic basis for _hIg_β expression in pituitary was initially explored by defining the structure of the _hIg_β transcripts. Northern blot analysis, RT/PCR across splice junctions, and 5′ RACE indicated that pituitary _hIg_β transcripts corresponded to full-length, polyadenylated _hIg_β mRNAs (Figures 1B, 3A and B). 5′-RACE analysis further revealed a heterogeneity of the 5′ ends of _hIg_β in both B cells and pituitary that is consistent with the lack of a TATA motif and the GC-rich structure of the _hIg_β promoter (Geng and Johnson, 1993; Donohoe and Blomberg, 1997; Dong et al, 2000). Thus, the _hIg_β gene, although considered B-cell specific, appears to be robustly expressed as a normally structured mRNA in the pituitary.
Although the structures of the _Ig_β mRNAs in B cells and in the pituitary are remarkably similar, subtle differences were noted. Northern analyses (Figure 3A and data not shown) revealed a more sharply focused _Ig_β mRNA band in B cells than in the pituitary and the 5′-RACE analysis revealed that the 5′ termini of the _Ig_β mRNA in B cells were more tightly grouped than in the pituitary. These observations may reflect qualitative differences in the accuracy of transcriptional initiation in B cells and by the ‘bystander' pathway of _Ig_β activation in the pituitary.
It is of note that endogenous _mIg_β mRNA is also detected in mouse pituitaries. However its levels, when corrected for gene copy number, are substantially lower than _hIg_β mRNA expression from the hGH/P1 transgene (Figure 2A and B). This could reflect a difference in the chromatin environment and transcriptional control of the human and murine GH loci. Whereas primates have a five-gene GH cluster, rodents have a single, isolated GH gene (Barsh et al, 1983; Chen et al, 1989). Whether the single mGH gene is under LCR control has not been determined. Computer searches reveal a conserved noncoding region in the mouse genome 5′ to the _mIg_β gene that can be aligned with HSI of the hGH LCR. However, it is not known if this sequence represents a functional correlate of the hGH LCR. A definitive comparison should emerge once detailed epigenetic and functional analyses of the mGH locus are established.
The ‘bystander' effect exerted by the hGH LCR on _hIg_β in the pituitary raised the question of whether _hIg_β exerts a reciprocal activation of hGH in B lymphocytes. Analysis of hGH/P1 transgenic mice failed to reveal hGH-N mRNA in spleen and analysis of human B cells was similarly negative for hGH-N mRNA (Figure 6A). These results suggest that the _hIg_β gene is activated by localized chromatin modifications that do not extend downstream to encompass the hGH-N promoter. This model can now be tested by defining the epigenetic modifications linked to _hIg_β expression in B cells.
The observation that genes with distinct expression patterns can be located in close proximity has led to the concept that these genes must be functionally shielded from one another (Dillon and Sabbattini, 2000). Such shielding may be mediated by boundary or insulator elements that protect adjacent genes from mutual positive or negative interference. This model has been most actively supported in studies of the chicken (c) β_-globin_ locus (Chung et al, 1993, 1997; Hebbes et al, 1994). A constitutive HS located at the 5′ border of the hyperacetylated c_β_-globin cluster defines the active domain and separates it from an adjacent heterchromatic region (Prioleau et al, 1999). Although such boundaries may be present in certain genomic regions, their function(s) have yet to be extensively tested in their native contexts. The present finding suggests that this model is not universal and that gene expression in higher eukaryotes may not be so neatly controlled in all cases.
Activation of the _hIg_β gene in the pituitary may be difficult to explain solely on the basis of its presence within an active chromatin domain. For example, the _h_α-globin genes are situated within a region of constitutively open chromatin and yet their expression is erythroid specific (Vyas et al, 1992). Therefore, an open chromatin environment does not guarantee transcription activation. While the activated chromatin context is an essential prerequisite for activation, transcription factor association is certainly involved in this process. There are interesting relationships between B lymphocyte and somatotrope transcription factors that may be relevant to _hIg_β activation in the pituitary. Motifs involved in _Ig_β expression bind factors such as early B-cell factor (EBF), Oct-2, LyF1/μB, and PU.1 (Omori and Wall, 1993; Thompson et al, 1996; Akerblad et al, 1999). The octamer motif, bound by Oct-2 and the ubiquitous Oct-1, is a major determinant of _Ig_β promoter activity. Oct-1, Oct-2, and Pit-1 belong to the family of POU homeodomain transcription factors. Pit-1 is abundantly expressed in pituitary cells (Ingraham et al, 1988; Asa et al, 1993) and is critical to hGH-N activation (Shewchuk et al, 1999, 2002). Crossbinding of Pit-1 and the Oct factors to corresponding cis elements has been demonstrated in vitro, although it has not been possible to confirm actual crossactivation using cell transfection models (Elsholtz et al, 1990). It is possible that the _hIg_β promoter, when in the activated hGH LCR chromatin domain, is permissive to binding by abundant and closely related factors such as Pit-1. Alternatively, the active chromatin conformation within the hGH LCR may promote direct association of the basal transcriptional complex. The latter scenario might predict a slightly less constrained site of transcription initiation of _hIg_β in the pituitary relative to B cells.
‘Bystander' activation of hIg_β, while secondary to the effects of the hGH LCR, may have a subsequent impact on hGH expression itself. Studies in the β_-globin locus have demonstrated that two genes, when under the control of a common LCR, can alter each other's expression via transcriptional competition (Giglioni et al, 1984; Dillon et al, 1997). Prior studies of the hGH cluster revealed that direct linkage of HSI,II to the hGH-N transgene results in overexpression of serum hGH and a giant phenotype in transgenic mice (Jones et al, 1995). This superinduction of hGH could reflect the artificial proximity of HSI,II to the hGH promoter in the transgene construct, or alternatively, it might reflect the exclusion of a competing _hIg_β transcriptional unit. Weighing against this second possibility is the observation that mice homozygous for deletion of the _mIg_β gene promoter lack evidence of growth abnormalities (Gong and Nussenzweig, 1996; Nussensweig, personal communication). Thus transcriptional inactivation of _Ig_β, at least in the mouse, fails to significantly impact on mGH expression. Whether bystander activation of a gene can have a secondary impact on its local environment can now be explored in greater detail.
The present data suggest that genes and their _cis_-acting sequences are not necessarily organized into discrete and functionally insulated chromosomal domains. The prevalence of long-range transcriptional controls in higher eukaryotes and the presence of extensive domains of modified chromatin make it likely that transcriptional interactions among closely packed genes may be the rule rather than the exception. Such ‘bystander' effects impact on current concepts regarding tissue specificity, developmental controls, and widen the considerations of how gene control circuits are organized. Together, these observations add another layer of complexity to our understanding of gene activation mechanisms.
Materials and methods
Cell lines and primary cells
Human K562 erythroid cell line and human lymphoblastoid cell line CRL-1484 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin. Peripheral blood lymphocytes (PBLs) from a normal donor were purified from fresh, heparinized whole-blood samples on Ficoll–Paque gradients (Amersham Biosciences). Studies of human samples were approved by the University of Pennsylvania Institutional Review Board.
Human pituitary and placenta samples
A preparation of polyA-primed cDNA from normal human pituitaries was a gift of Dr Roman Perez-Fernandez (Compostela University). Pituitary GH-secreting adenomas were donated by Dr Peter J Snyder (University of Pennsylvania). Portions of human full-term placentas were obtained from the Obstetrics service at the University of Pennsylvania.
RT/PCR analyses
In all cases, 0.5 μg of total RNA extracted from tissues or cell lines was reverse transcribed with an oligo-dT primer in the presence of AMV reverse transcriptase, then coamplified using a primer set corresponding to regions conserved between human and mouse (Table I). Either the 5′ or 3′ primer was end-labeled with [γ-32P]ATP by T4 polynucleotide kinase. This primer set spans intron 5 in order to distinguish cDNA from amplified genomic DNA by fragment sizes. PCR products were digested with a restriction enzyme; _mIg_β cDNA is specifically cleaved by _Hin_fI, _hIg_β cDNA by _Sfc_I, and hGH-N by Bst_NI. Fragments were separated on 6% polyacrylamide/denaturing gels, and bands were quantified on a Storm 840 PhosphorImager (Molecular Dynamics, Sunnyvale, CA). The ratios of digested cDNA fragments were normalized to transgene copy number. β_-actin and GAPDH cDNAs were amplified for 24 cycles with unlabeled primers and analyzed on agarose gels. All PCR results were determined to be within the linear range of amplification by analysis of serial cycles.
Table 1.
Primers and probes used in the experiments
| | Sequences (5′ to 3′) | | | ---------------------------- | -------------------------------------- | | PCR primers | | | mhIgβ5′ | 5′GGAGGAAGATCACACCT3′ | | mhIgβ3′ | 5′ATCCCCAGAGAACTCC3′ | | mhIgα5′ | 5′GTTCAGGAAACGATGGCAGA3′ | | mhIgα3′ | 5′TCACTAAGTGGCCCTGACAGA3′ | | mhGH5′ | 5′GCCTGCTCTGCCTGC3′ | | mhGH3′ | 5′GACTGGATGAGCAGCAG3′ | | mh β-Actin 5′ | 5′TGTGATGGTGGGAATGGGTCAG3′ | | mh β-Actin 3′ | 5′TCGGTGAGCAAGCACAGGGTG3′ | | mhGAPDH 5′ | 5′GCCAAAAGGGTCATCATCTC3′ | | mhGAPDH 3′ | 5′CTGCTTCACCACCTTCTTGA3′ | | | | | | 5′-RACE primers | | | Exon 6 hIgβ 3′ | 5′TCATGGGGCGACCTGGCTC3′ | | Oligo-dT-adaptor primer 5′ | 5′GACTCGAGTCGACATCGAT(17)3′ | | Exon 3 hIgβ 3′ | 5′CGTGGGCTCTGCCAGATCCGCGAACAAGC3′ | | Exon 2 hIgβ 3′ | 5′CGGTCCTCCGATCTGGCTGCTGGTACTGGCTCAG3′ | | | | | | Probes | | | 5′ primer for dot blot probe | 5′TCATCCTCTTCATCATCGTGCC3′ | | 3′ primer for dot blot probe | 5′TGCTGCCCTTGTCCTTCTAC3′ |
RNA quantification by real-time RT/PCR
Pituitary RNA samples were reverse transcribed with random hexamers in the presence of MMLV reverse transcriptase. The cDNA was diluted to 10 ng/μl and was used for real-time PCR using TaqMan reagents on an ABI Prism 7700 Sequence Detector (Applied Biosystems). All samples were run in duplicate. Amplification of the _hIg_β mRNA was normalized with a reference mGAPDH mRNA probe. All primers and probes for real-time analysis of _hIg_β and mGAPDH mRNAs were standardized reagents purchased from Applied Biosystems.
Northern analyses
Total RNA was extracted from lymphocytes (spleen), pituitaries, and cell lines with RNA-Bee (Tel-Test, TX). A 20 μg portion of each RNA sample was separated on a 1.2% agarose gel containing 2.2 M formaldehyde in 1 × MOPS, and transferred to a Zeta-Probe membrane (Bio-Rad). The membrane was probed with 32P-labeled _hIg_β cDNA probe at 65°C in PerfectHyb buffer (SigmaAldrich). The membrane was then washed (1 × SSC, 0.1% SDS, 0.5 × SSC, 0.1% SDS, and finally 0.1 × SSC, 0.1% SDS) at room temperature to 65°C and signals were detected by exposure to a phosphorimager screen.
5′ rapid amplification of cDNA ends
mRNAs isolated from B-cell line 1484 and hGH/P1 transgenic pituitaries were reverse transcribed using an antisense _hIg_β-specific primer corresponding to a site in exon 6 of hIgβ (5′TCATGGGGCGACCTGGCTC3′). The RT product was ethanol precipitated in the presence of carrier (glycoblue) and resuspended in water. A polyA tail was added to the 3′ end of the transcribed cDNA using terminal dinucleotidyl transferase (Promega). An initial PCR was run using the oligo-dT-adaptor primer (Table I) and an _hIg_β-specific antisense primer on exon 3 (Table I). A subsequent nested PCR was run using the same oligo-dT-adaptor primer and _hIg_β-specific antisense primer corresponding to a region of exon 2 (Table I). The products were cloned into the pGEMT vector system I (Promega) and sequenced by chain termination.
FACS analysis of mouse spleen B cells for hIg_β_ protein
Spleen cells were harvested, suspended in PBS, and lymphocytes were isolated on Ficoll gradients. Cells were stained for 30 min using a panel of monoclonal antibodies conjugated to fluorescent dyes: anti-mouse B220-APC (B-cell surface marker) (Pharmingen, catalog number 01129A), anti-mouse Igβ FITC (Southern Biotechnology Associates, catalog number 1830–02), and anti-human Igβ PE (Pharmingen, catalog number 555679). Isotype-matched monoclonal controls were labeled with FITC, PE, and APC. Using a FACSCalibur flow cytometer (BD Immunocytometry systems) 10 000–20 000 cell events were acquired. The lymphocyte population to be analyzed was selected by side scatter and forward scatter. Analyses of the stained cells were performed using Cell Quest Software.
Protein extraction and Western blotting
Proteins from tissues and cell lines were purified in proteinase inhibitor buffer (1 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol (DTT), 10 mM Tris–HCl (pH 7.4), 100 mg of phenylmethylsulfonyl fluoride (PMSF)/ml, 2 mg of aprotinin/ml, 2 mg of pepstatin A/ml) by repeated pipetting. After clarification, the supernatants were separated and transferred using NuPage Bis–Tris gels system (Invitrogen). Signals were detected using monoclonal mouse anti-human Igβ (Pharmingen) and polyclonal anti-L7a rabbit serum (Ji et al, 2003). Sheep anti-mouse IgG-HRP (Santa Cruz) and donkey anti-rabbit IgG-HRP (Amersham) were used as secondary antibodies.
Immunofluorescent microscopy
Polyclonal goat anti-hIgβ antibodies (Santa Cruz Biotechnologies) and anti-hGH monoclonal antibodies (mAb 9) (Bennani-Baïti et al, 1998a) were used at a 1:1000 dilution. As secondary antibodies, donkey anti-goat Cy3 (Vector) and donkey anti-mouse Cy2 (Vector) were used at 1:200. Immunohistochemistry was performed on fresh frozen mouse spleen and pituitary sections. Defrosted slides were placed in buffered neutralized formalin for 5 min, transferred to distilled water, blocked with protein blocking agent (Immunotech) for 10 min and incubated overnight with the primary antibody at 4°C. The next day the slides were washed and incubated for 30 min at 37°C with the secondary antibodies. Finally, the slides were incubated for 10 s with 4′,6-diamidino-2 phenylindole, dihydrochloride (DAPI), covered with mounting medium and observed under a fluorescence microscope.
Transgenic constructs and generation of transgenic mouse lines
hGH/P1 wild-type and _hGH/P1(Δ_HSI) deletion lines have been previously reported (Su et al, 2000; Ho et al, 2002) (Table II). −_8.0Ig_β contains a 12 kb genomic fragment derived by _Eco_RI digestion of P1 6057 that includes 8.0 kb 5′ to the _hIg_β gene and 975 bp 3′ to the _hIg_β polyA addition site. −_1.3Ig_β contains a 5.6 kb genomic fragment generated by _Bgl_II and _Eco_RI digestions with 1.3 kb of 5′-flanking sequences. −_0.2Ig_β contains a 4.6 kb genomic fragment generated by _Bsi_EI and _Eco_RI digestions that contains 200 bp of 5′-flanking region. Each construct was microinjected into fertilized mouse oocytes (C57BL/6 × SJL) to generate the transgenic lines (Transgenic & Chimeric Mouse Core, University of Pennsylvania). Founders were identified by dot-blot analyses of tail DNA using a PCR-generated, 740 bp probe, corresponding to a region between exons 4 and 6 of the _hIg_β gene (Table I). Transgenic lines used in the present study are listed in Table II.
Table 2.
Transgenes, line designations, construct sizes, and copy numbers
| Transgene | Construct size (kb) | Line designation | Copy number |
|---|---|---|---|
| −_0.2Ig_β | 4.6 | 1008A | 2 |
| 1007A | 2 | ||
| 1096C | 4 | ||
| −_1.3Ig_β | 5.6 | 1014A | 3 |
| 1014F | 2 | ||
| 1092Fa | 3 | ||
| −_8.0Ig_β | 12 | 989D | 7 |
| 1002D | 10 | ||
| 1047C | 3 | ||
| hGH/P1 | 87 | 809C | 2 |
| 809F | 5 | ||
| 811B | 6 | ||
| 811D | 4 | ||
| 813I | 19 | ||
| _hGH/P1(Δ_HSI) | 87 | 960G | 4 |
| 961E | 12 | ||
| 969E | 3 |
Acknowledgments
We thank Dr Roman Perez-Fernandez (Compostela University) and Dr Peter J Snyder (University of Pennsylvania) for gifts of human pituitary mRNA and pituitary adenoma tissue samples, respectively, and Dr Yugong Ho for the _hGH/P1(Δ_HSI) transgenic lines. We acknowledge support from the University of Pennsylvania Transgenic Mouse Core (P30DK50306, P30DK19525, P30CA16520), the Morphology Core (P30DK50306), and the Flow Cytometry and Cell Sorting Cores (P30CA16520). We thank Drs Marisa Bartolomei, Thomas Kadesch, and Gerd Blobel and members of the Liebhaber and Cooke laboratories for critical reading of the manuscript. This work was supported by NIH grant HD25147 (NEC and SAL) and by a Leukemia and Lymphoma Society postdoctoral fellowship 5013-03 (IC).
References
- Akerblad P, Rosberg M, Leanderson T, Sigvardsson M (1999) The B29 (immunoglobulin beta-chain) gene is a genetic target for early B-cell factor. Mol Cell Biol 19: 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asa SL, Puy LA, Lew AM, Sundmark VC, Elsholtz HP (1993) Cell type-specific expression of the pituitary transcription activator pit-1 in the human pituitary and pituitary adenomas. J Clin Endocr Metab 77: 1275–1280 [DOI] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ (1997) Intergenic transcription and transinduction of the human beta-globin locus. Gene Dev 11: 2494–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh GS, Seeburg PH, Gelinas RE (1983) The human growth hormone gene family: structure and evolution of the chromosomal locus. Nucleic Acids Res 11: 3939–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (1999) Stopped at the border: boundaries insulators. Curr Opin Genet Dev 9: 191–198 [DOI] [PubMed] [Google Scholar]
- Bennani-Baïti IM, Asa SL, Song D, Iratni R, Liebhaber SA, Cooke NE (1998a) DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. Proc Natl Acad Sci USA 95: 10655–10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennani-Baïti IM, Cooke NE, Liebhaber SA (1998b) Physical linkage of the human growth hormone gene cluster and the CD79b (Ig beta/B29) gene. Genomics 48: 258–264 [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Tze L (1997) Synergism between hypersensitive sites confers long-range gene activation by the beta-globin locus control region. Proc Natl Acad Sci USA 94: 4566–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M (1999) Looping versus linking: toward a model for long-distance gene activation. Gene Dev 13: 2465–2477 [DOI] [PubMed] [Google Scholar]
- Campbell KS, Hager EJ, Friedrich RJ, Cambier JC (1991) IgM antigen receptor complex contains phosphoprotein products of B29 and mb-1 genes. Proc Natl Acad Sci USA 88: 3982–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P (2002) Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626 [DOI] [PubMed] [Google Scholar]
- Chen EY, Liao YC, Smith DH, Barrera-Saldaña HA, Gelinas RE, Seeburg PH (1989) The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics 4: 479–497 [DOI] [PubMed] [Google Scholar]
- Chung JH, Bell AC, Felsenfeld G (1997) Characterization of the chicken beta-globin insulator. Proc Natl Acad Sci USA 94: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Clark MR, Friedrich RJ, Campbell KS, Cambier JC (1992) Human pre-B and B cell membrane mu-chains are noncovalently associated with a disulfide-linked complex containing a product of the B29 gene. J Immunol 149: 2857–2863 [PubMed] [Google Scholar]
- Dillon N, Sabbattini P (2000) Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. BioEssays 22: 657–665 [DOI] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F (1997) The effect of distance on long-range chromatin interactions. Mol Cell 1: 131–139 [DOI] [PubMed] [Google Scholar]
- Dong S, Lester L, Johnson LF (2000) Transcriptional control elements and complex initiation pattern of the TATA-less bidirectional human thymidylate synthase promoter. J Cell Biochem 77: 50–64 [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Blomberg BB (1997) The 14.1 surrogate light chain promoter has lineage- and stage-restricted activity. J Immunol 158: 1681–1691 [PubMed] [Google Scholar]
- Elefant F, Cooke NE, Liebhaber SA (2000) Targeted recruitment of histone acetyltransferase activity to a locus control region. J Biol Chem 275: 13827–13834 [DOI] [PubMed] [Google Scholar]
- Elsholtz HP, Albert VR, Treacy MN, Rosenfeld MG (1990) A two-base change in a POU factor-binding site switches pituitary-specific to lymphoid-specific gene expression. Gene Dev 4: 43–51 [DOI] [PubMed] [Google Scholar]
- Forrester WC, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M (1987) Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res 15: 10159–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg EC, Bresnick EH (2001) Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. BioEssays 23: 820–830 [DOI] [PubMed] [Google Scholar]
- Geng Y, Johnson LF (1993) Lack of an initiator element is responsible for multiple transcriptional initiation sites of the TATA-less mouse thymidylate synthase promoter. Mol Cell Biol 13: 4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglioni B, Casini C, Mantovani R, Merli S, Comi P, Ottolenghi S, Saglio G, Camaschella C, Mazza U (1984) A molecular study of a family with Greek hereditary persistence of fetal hemoglobin and beta-thalassemia. EMBO J 3: 2641–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Nussenzweig MC (1996) Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science 272: 411–414 [DOI] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell 5: 377–386 [DOI] [PubMed] [Google Scholar]
- Grosveld F, Van Assendelft GB, Greaves DR, Kollias G (1987) Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51: 975–985 [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J 13: 1823–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson GG, Eisenberg D, Kincade PW, Wall R (1988) B29: a member of the immunoglobulin gene superfamily exclusively expressed on beta-lineage cells. Proc Natl Acad Sci USA 85: 6890–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Cooke NE, Liebhaber SA (2002) A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol Cell 9: 291–302 [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Chen R, Mangalam HJ, Elsholtz HP, Flynn SE, Lin CR, Simmons DM, Swanson L, Rosenfeld MG (1988) A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell 55: 519–529 [DOI] [PubMed] [Google Scholar]
- Ji X, Kong J, Liebhaber SA (2003) In vivo association of the stability control protein αCP with actively translating mRNAs. Mol Cell Biol 23: 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BK, Monks BR, Liebhaber SA, Cooke NE (1995) The human growth hormone gene is regulated by a multicomponent locus control region. Mol Cell Biol 15: 7010–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura AP, Liebhaber SA, Cooke NE (2004) Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol Endocrinol 18: 1018–1032 [DOI] [PubMed] [Google Scholar]
- McDowell JC, Dean A (1999) Structural and functional cross-talk between a distant enhancer and the epsilon-globin gene promoter shows interdependence of the two elements in chromatin. Mol Cell Biol 19: 7600–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Storm HP, Sogo JM, Schaffner W (1989) An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge [erratum appears in Cell 1989 Oct 20;59(2):405]. Cell 58: 767–777 [DOI] [PubMed] [Google Scholar]
- Omori SA, Wall R (1993) Multiple motifs regulate the B cell-specific promoter of the B29 gene. Proc Natl Acad Sci USA 90: 11723–11727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavasiliou F, Misulovin Z, Suh H, Nussenzweig MC (1995) The role of Ig beta in precursor B cell transition and allelic exclusion. Science 268: 408–411 [DOI] [PubMed] [Google Scholar]
- Prioleau MN, Nony P, Simpson M, Felsenfeld G (1999) An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J 18: 4035–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M (1986) Gene regulation by proteins acting nearby and at a distance. Nature 322: 697–701 [DOI] [PubMed] [Google Scholar]
- Routledge SJ, Proudfoot NJ (2002) Definition of transcriptional promoters in the human beta globin locus control region. J Mol Biol 323: 601–611 [DOI] [PubMed] [Google Scholar]
- Shewchuk BM, Asa SL, Cooke NE, Liebhaber SA (1999) Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J Biol Chem 274: 35725–35733 [DOI] [PubMed] [Google Scholar]
- Shewchuk BM, Liebhaber SA, Cooke NE (2002) Specification of unique Pit-1 activity in the hGH locus control region. Proc Natl Acad Sci USA 99: 11784–11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Liebhaber SA, Cooke NE (2000) The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J Biol Chem 275: 7902–7909 [DOI] [PubMed] [Google Scholar]
- Tewari R, Gillemans N, Harper A, Wijgerde M, Zafarana G, Drabek D, Grosveld F, Philipsen S (1996) The human beta-globin locus control region confers an early embryonic erythroid-specific expression pattern to a basic promoter driving the bacterial lacZ gene. Development 122: 3991–3999 [DOI] [PubMed] [Google Scholar]
- Thompson AA, Wood WJ, JrGilly MJ, Damore MA, Omori SA, Wall R (1996) The promoter and 5′ flanking sequences controlling human B29 gene expression. Blood 87: 666–673 [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, De Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1456 [DOI] [PubMed] [Google Scholar]
- Vyas P, Vickers MA, Simmons DL, Ayyub H, Craddock CF, Higgs DR (1992) _Cis_-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell 69: 781–793 [DOI] [PubMed] [Google Scholar]