The polarity-inducing kinase Par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC (original) (raw)
Abstract
Par (partitioning-defective) genes were originally identified in Caenorhabditis elegans as determinants of anterior/posterior polarity. However, neither their function in vertebrate development nor their action mechanism has been fully addressed. Here we show that two members of Par proteins, 14-3-3 (Par-5) and atypical PKC (aPKC), regulate the serine/threonine kinase Par-1 to control Xenopus gastrulation. We find first that Xenopus Par-1 (xPar-1) is essential for gastrulation but not for cell fate specification during early embryonic development. We then find that xPar-1 binds to 14-3-3 in an aPKC-dependent manner. Our analyses identify two aPKC phosphorylation sites in xPar-1, which are essential for 14-3-3 binding and for proper gastrulation movements. The aPKC phosphorylation-dependent binding of xPar-1 to 14-3-3 does not markedly affect the kinase activity of xPar-1, but induces relocation of xPar-1 from the plasma membranes to the cytoplasm. Finally, we show that Xenopus aPKC and its binding partner Xenopus Par-6 are also essential for gastrulation. Thus, our results identify a requirement of Par proteins for Xenopus gastrulation and reveal a novel interrelationship within Par proteins that may provide a general mechanism for spatial control of Par-1.
Keywords: 14-3-3, aPKC, gastrulation, Par-1, Xenopus
Introduction
Cell polarity is a crucial factor for a variety of cellular and developmental processes. Recent studies have revealed that several evolutionarily conserved proteins are involved in establishing and maintaining cell polarity. Par genes are known as determinants of asymmetric cell division in one-cell embryos of Caenorhabditis elegans (Kemphues et al, 1988). During this asymmetric cell division, Par proteins are localized asymmetrically. Par-1, a serine/threonine kinase, is restricted at the posterior cortex (Guo and Kemphues, 1995), whereas Par-3, Par-6 and atypical protein kinase C (aPKC) localize at the anterior cortex (Etemad-Moghadam et al, 1995; Tabuse et al, 1998; Hung and Kemphues, 1999). Par-3 contains three PDZ (PSD-95/Dlg/ZO-1) domains, and Par-6 contains a single PDZ domain and a CRIB (Cdc42/Rac interactive binding)-like motif. Par-5, a 14-3-3 protein, is symmetrically distributed throughout the cytoplasm (Morton et al, 2002).
After the initial characterization in C. elegans, orthologs of Par genes have been identified in higher eukaryotes (Ohno, 2001; Etienne-Manneville and Hall, 2003; Macara, 2004). The vertebrate homologs of Par-3, Par-6 and aPKC directly bind to one another and form a multiprotein complex, which regulates the organization of epithelial tight junctions in Madin-Darby canine kidney (MDCK) cells, and axon specification in cultured hippocampal neurons (Ohno, 2001; Shi et al, 2003). Mammalian homologs of Par-1 were also identified as a kinase activity that phosphorylates microtubule-associated proteins (Drewes et al, 1997). However, roles of Par proteins during vertebrate embryonic development have not been determined. Moreover, the detailed molecular mechanisms of their action have not been fully elucidated. Particularly, although the action mechanisms and physiological significance of the Par-3/Par-6/aPKC complex have been relatively well characterized (Ohno, 2001; Etienne-Manneville and Hall, 2003), little is known about regulatory mechanisms and function of Par-1.
To address these issues in Xenopus, we isolated a Xenopus homolog of Par-1 and have shown that Par-1 regulates morphogenetic movements during gastrulation. We have then shown that Par-1 binds to 14-3-3/Par-5 in an aPKC-dependent manner and that aPKC phosphorylates two conserved residues in Par-1 to create the binding sites for 14-3-3. The binding of Par-1 to 14-3-3 has an important role in gastrulation and is essential for relocation of Par-1 from plasma membranes to the cytoplasm. Furthermore, we show that aPKC is also essential for gastrulation. Thus, our results identify a novel interrelationship among Par proteins, spatial control of Par-1 by 14-3-3 and aPKC, that is essential for gastrulation. This mechanism may provide a general molecular basis for control of cell polarity in various biological processes.
Results and discussion
Par-1 is required for proper gastrulation movements
We first isolated a Xenopus Par-1 (xPar-1) cDNA from a Xenopus gastrula cDNA library. xPar-1 is expressed ubiquitously throughout embryogenesis, both maternally and zygotically (data not shown). To examine whether xPar-1 is involved in gastrulation, which is crucial for the vertebrate body plan and involves coordinated morphogenetic movements with polarized cell behaviors (Keller et al, 1992; Wallingford et al, 2002), we performed animal cap assays. It is known that activin-treated animal caps exhibit morphogenetic elongation characteristic of gastrulation and express various molecular markers (Symes and Smith, 1987) (see Figure 1A and B). The noncanonical Wnt pathway regulates convergent extension movements during gastrulation (Wallingford et al, 2002), and xWnt5 or xWnt11, which activates this pathway, inhibits activin-induced elongation of animal caps (Moon et al, 1993; Du et al, 1995) (see also Figure 1A). Injection of wild-type xPar-1 mRNA (xPar-1 wt) severely inhibited activin-induced elongation in animal caps (Figure 1A), whereas it did not inhibit the induction of expression of various molecular markers (Figure 1B). A previous report suggests that Par-1 is a positive regulator of the canonical Wnt pathway, which is mediated by β-catenin (Sun et al, 2001). β-Catenin did not inhibit elongation of caps (Figure 1A), suggesting that the inhibition of gastrulation by xPar-1 is not through excessive signaling of the canonical Wnt pathway. Moreover, expression of either of two dominant-negative forms of xPar-1, xPar-1 KR (a kinase-negative form) and xPar-1 TASA (a nonactivatable form; Drewes et al, 1997), also inhibited activin-induced elongation without altering cell differentiation (Figure 1A and B). Then, we examined the effect of expression of xPar-1 wt, xPar-1 KR or xPar-1 TASA in whole embryos. Expression of any of the three forms of xPar-1 in the dorsal marginal zone of four-cell stage embryos caused severe morphogenetic defects; the tadpoles showed dorsal flexure with a shortened body axis (data not shown and see Figure 1E). This phenotype is indistinguishable from that seen in the tadpoles in which gastrulation movements are specifically impaired (Sokol, 1996; Deardorff et al, 1998; Djiane et al, 2000; Tada and Smith, 2000; Winklbauer et al, 2001). Essentially the same results were obtained with rat Par-1C/MARK1 (rPar-1C) (Drewes et al, 1997) in animal caps and whole embryos (data not shown). Thus, both gain-of-function and dominant-negative manipulations of Par-1 produced apparently identical gastrulation defects. We therefore conclude that xPar-1 is involved in gastrulation movements without affecting cell fate specification and that the regulated, appropriate activity of xPar-1 is necessary.
Figure 1.
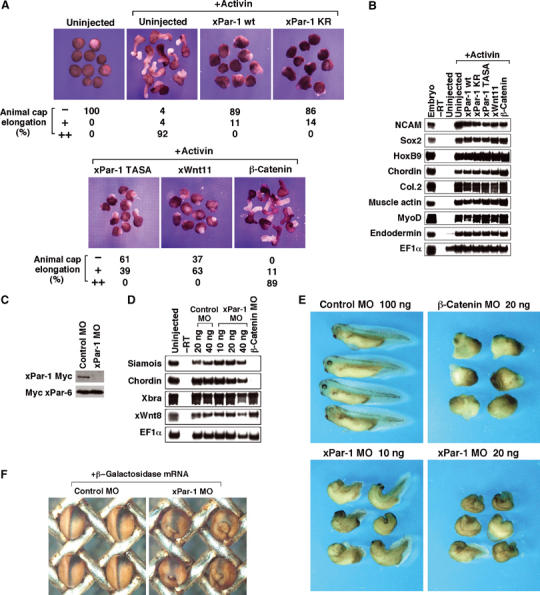
Par-1 is essential for gastrulation movements. (A) xPar-1 wt, xPar-1 KR and xPar-1 TASA inhibit elongation of animal cap explants in response to activin. Four-cell embryos were injected with mRNAs encoding xPar-1 wt (400 pg; _n_=37), xPar-1 KR (400 pg; _n_=49), xPar-1 TASA (400 pg; _n_=44), xWnt11 (500 pg; _n_=19) or β-catenin (500 pg; _n_=37). Animal caps derived from uninjected or injected embryos were left untreated or treated with 10 ng/ml activin and cultured to neurula stage. Control caps remained round. The percentages of elongated animal caps are shown below the photograph. −, no elongation; +, partial elongation; ++, strong elongation. (B) The animal caps from (A) were analyzed by RT–PCR. NCAM and Sox2 (neural marker), HoxB9 (spinal cord marker), chordin and Col.2 (notochord marker), muscle actin and MyoD (muscle marker) and endodermin (endoderm marker) were examined. _EF-1_α serves as a loading control. No signal was observed in the absence of reverse transcriptase (−RT). (C) xPar-1 morpholino antisense oligonucleotide (MO) blocks translation of injected xPar-1 mRNA. xPar-1-Myc mRNA or Myc-xPar-6 mRNA was coinjected into embryos with xPar-1 MO or control MO and embryo lysates were analyzed by Western blotting with anti-Myc antibody. (D) xPar-1 MO does not change cell fate specification in whole embryos. Embryos were injected into both dorsal blastomeres of the four-cell stage with MOs. Whole embryos were harvested at stage 11 for RT–PCR. (E) Effects of xPar-1 MO on whole embryos. Control MO, β-catenin MO or xPar-1 MO was injected into both dorsal blastomeres at the four-stage and cultured until stage 35. Upper left panel, embryos injected with 100 ng of control MO (91% with complete normal structures; _n_=85); upper right, embryos injected with 20 ng of β-catenin MO (96% with ventralization; _n_=48); lower left, embryos injected with 10 ng of xPar-1 MO (82% with dorsal bending and a shortened body axis; _n_=100); lower right, embryos injected with 20 ng of xPar-1 MO (100% with dorsal bending and a shortened body axis; _n_=18). (F) Lineage tracer analysis of gastrulation movements in xPar-1 MO-injected embryos. Indicated combinations of mRNA or MO were injected into one right dorsoanimal blastomere at eight-cell stage and subjected to β-gal staining at stage 15. Dorsal views of injected embryos are shown with the anterior side at the top. β-gal-stained cells are blue. Left panel, embryos injected with 20 ng of control MO and 20 pg of β-galactosidase mRNA; right panel, embryos injected with 20 ng of xPar-1 MO and 20 pg of β-galactosidase mRNA.
Par-1 morpholino blocks gastrulation
To further show requirement of xPar-1 function for gastrulation in vivo, we used an antisense morpholino oligo (MO) against xPar-1. Coinjection of xPar-1 MO with xPar-1-Myc or Myc-Xenopus Par-6 (xPar-6) mRNA resulted in almost complete inhibition of xPar-1-Myc mRNA translation (Figure 1C), indicating that xPar-1 MO is specific and efficient. Then xPar-1 MO or control MO was injected into the dorsal marginal zones of four-cell stage embryos. At the tadpole stage, the xPar-1 MO-injected embryos showed a dorsal bending phenotype with a shortened body axis, indicating that gastrulation movements are severely inhibited (Figure 1E). Next, xPar-1 MO or control MO was coinjected with a cell lineage tracer into one right dorsoanimal blastomere of eight-cell stage embryos. At neurula stages, xPar-1 MO-injected embryos failed to close the neural plate and exhibited externally visible endoderm on the dorsal side (Figure 1F, right). In control MO-injected embryos at neurula stages, cells expressing a cell lineage tracer nuclear β-galactosidase (nβ-gal) were restricted along the midline region as a result of normal gastrulation movements (Figure 1F, left). By contrast, cells expressing nβ-gal were expanded laterally in xPar-1 MO-injected embryos (Figure 1F, right), indicating that cells were not properly converging during gastrulation. For reference, we injected β-catenin MO. β-Catenin MO-injected embryos showed a severe ventralized phenotype with no axial structures, as previously reported (Heasman et al, 2000) (Figure 1E). In contrast, in xPar-1 MO-injected embryos, axial structures remained although the morphology was abnormal (Figure 1E), suggesting that xPar-1 function is independent of the β-catenin-dependent primary axis induction. To examine cell fate specification in these embryos, we performed RT–PCR analyses. In β-catenin MO-injected embryos at stage 11, the expression levels of a dorsal endoderm marker siamois and a dorsal mesoderm marker chordin were markedly reduced, while the expression levels of a pan-mesodermal marker Xbra and a ventral mesoderm marker xWnt8 were unaffected (Figure 1D). However, in xPar-1 MO-injected embryos, the expression levels of siamois, chordin, Xbra and xWnt8 were not reduced (Figure 1D). These results taken together indicate that xPar-1 is essential for morphogenetic movements during gastrulation, but not for cell fate specification.
Par-1 binds to 14-3-3 in an aPKC-dependent manner
To elucidate the action mechanism of Par-1, physical interactions of Par-1 with other Par proteins were examined. In recent reports, the binding of 14-3-3/Par-5 to Par-1 or Par-3 was observed (Benton et al, 2002; Brajenovic et al, 2004). Our co-immunoprecipitation assay detected the binding of xPar-1 or rat Par-3 (rPar-3), but not xPar-6 or a Xenopus aPKC isotype PKCλ (xPKCλ), to mouse 14-3-3ɛ (Figure 2A). In Drosophila, the binding of 14-3-3ɛ to Par-1 was reported to be independent of a phosphoserine binding pocket of 14-3-3ɛ (Benton et al, 2002). We then examined the region of 14-3-3ɛ that interacts with xPar-1. Several mutations of 14-3-3 are known to impair its function (Chang and Rubin, 1997). One is a charge reversal mutation (E183K) in the phosphoserine binding pocket, and two other mutations, F199Y and Y214F, are located in a hydrophobic region of unknown function. In our co-precipitation assay, the binding of 14-3-3ɛ to xPar-1 was severely impaired in any of the E183K, F199Y and Y214F mutations (Figure 2B), suggesting that both the phosphoserine binding pocket and the hydrophobic region of 14-3-3ɛ are important for the interaction of 14-3-3ɛ with vertebrate Par-1. Because a member of the mammalian Par-1 family, c-TAK1/Par-1A, is known to be able to phosphorylate Cdc25C and KSR to create their binding site for 14-3-3 (Peng et al, 1998; Muller et al, 2001), it seemed possible that xPar-1 autophosphorylation might create its own 14-3-3 binding sites. However, xPar-1 KR, a kinase-dead form of xPar-1, was found to bind to 14-3-3ɛ as well as xPar-1 wt (Figure 2C), suggesting that another kinase phosphorylates xPar-1 to create a 14-3-3 binding site. To identify a protein kinase that stimulates the binding of xPar-1 to 14-3-3ɛ, we tested a wide range of kinase inhibitors. Of the inhibitors tested, a broad inhibitor of protein kinases, staurosporine, and a PKC inhibitor bisindolylmaleimide I strongly inhibited the binding of xPar-1 to 14-3-3ɛ (Figure 2D). In contrast, the binding of Par-3 to 14-3-3ɛ, although sensitive to staurosporine, was not inhibited by bisindolylmaleimide I (Figure 2E). Therefore, PKC activity is specifically required for the binding of Par-1 to 14-3-3ɛ. We then hypothesized the involvement of aPKC, a member of Par proteins, in the binding of Par-1 to 14-3-3ɛ. To test this hypothesis, we examined the effect of expression of various forms of Xenopus PKCλ (xPKCλ) on the binding between Par-1 and 14-3-3ɛ. Expression of wild-type xPKCλ or an active form of xPKCλ (xPKCλ AE; Uberall et al, 1997), but not a kinase-negative form of xPKCλ (xPKCλ KE; Nakaya et al, 2000), markedly enhanced the binding of xPar-1 or rPar-1c to 14-3-3ɛ (Figure 2F and G, and also see Figure 2B and C). These results suggest that Par-1 binds to 14-3-3ɛ in an aPKC-dependent manner.
Figure 2.
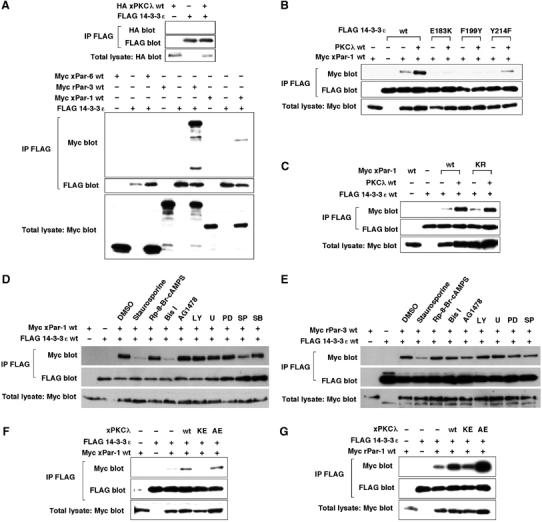
Par-1 binds to 14-3-3 in an aPKC-dependent manner. (A) The binding of xPar-1 or rPar-3, but not xPar-6 or xPKCλ, to 14-3-3ɛ. FLAG-tagged 14-3-3ɛ was coexpressed with HA-tagged xPKCλ, Myc-tagged xPar-6, rPar-3 or xPar-1 in HEK293 cells. Lysates of cells were immunoprecipitated (IP) with anti-FLAG antibody and co-immunoprecipitated proteins were detected by immunoblotting with anti-Myc or HA antibody. Comparable amounts of 14-3-3ɛ were immunoprecipitated in each lane. The expression level of xPKCλ, xPar-6, rPar-3 or xPar-1 in total lysates was similar. (B) Both the phosphoserine binding pocket and the hydrophobic region of 14-3-3ɛ are important for the interaction of 14-3-3ɛ with vertebrate Par-1. (C) xPar-1 KR binds to 14-3-3ɛ as well as xPar-1 wt. (D) PKC activity is required for the binding of Par-1 to 14-3-3ɛ. Cells were transfected and treated with a broad inhibitor of protein kinases staurosporine, a PKA inhibitor Rp-8-Br-cAMPS, a PKC inhibitor bisindolylmaleimide I (Bis I), a receptor tyrosine kinase inhibitor AG1478, a PI3K inhibitor LY294002 (LY), an MEK inhibitor U0126 (U) or PD98059 (PD), a JNK inhibitor SP600125 (SP) or a p38 inhibitor SB203580 (SB) for 2 h. Cell lysates were subjected to immunoprecipitation. (E) PKC activity is not required for the binding of Par-3 to 14-3-3ɛ. (F, G) xPKCλ markedly enhances the binding of xPar-1 or rPar-1c to 14-3-3ɛ.
Two conserved residues in Par-1 are phosphorylated by aPKC
To test direct phosphorylation of xPar-1 by xPKCλ, we isolated FLAG-tagged xPKCλ and Myc-tagged xPar-1 KR. Our kinase assay has shown that xPKCλ is able to phosphorylate xPar-1 in vitro (Figure 3A). To identify an aPKC phosphorylation site (s) in xPar-1, we constructed a series of truncated mutants of xPar-1 (see Figure 3B) and tested their ability to serve as a substrate for xPKCλ. The spacer region, but not the kinase domain or the C-tail domain, was phosphorylated by xPKCλ (Figure 3C). Furthermore, a fragment corresponding to residues 493–684, but not the region corresponding to residues 309–492 or residues 400–581, was phosphorylated by xPKCλ (Figure 3D). Thus, an xPKCλ phosphorylation site (s) in xPar-1 may be present in the region corresponding to residues 582–684. It is known that basic amino acids near a phosphorylation site are required for PKC phosphorylation of substrates. Within this region (residues 582–684) of xPar-1, there are 14 serine/threonine residues that locate near basic amino acids. Out of the 14 residues, 11 are evolutionarily conserved. Then, we constructed more than 10 mutants of the fragment (residues 493–684) in which one or several of the 11 residues were mutated to alanine (s), and tested their ability to serve as a substrate for xPKCλ. Part of the results is shown in Figure 3E. A mutant (S589A/S590A/S592A/T593A) was partially defective in serving as a substrate (lane 8), whereas a mutant (T629A/S631A/S634A/T637A/S638A) was not (lane 7). A mutant (S646A) was partially defective (lane 4), whereas a mutant (S665A) was not (data not shown). To identify a site in the four residues (Ser589, Ser590, Ser592 and Thr593), we made the four mutants, S589A/S590A/S592A, S589A/S590A/T593A, S589A/S592A/T593A and S590A/S592A/T593A. Among them, only S589A/S590A/S592A was phosphorylated to the same extent as the wild-type fragment (lane 6 and data not shown), suggesting that Thr593 is a phosphorylation site. In fact, a mutant (T593A) was phosphorylated to a lesser extent (lane 3). Thus, both Thr593 and Ser646 are candidate sites for phosphorylation. Consistent with this, a mutant fragment (T593A/S646A) was not phosphorylated at all (lane 5). In the full-length constructs of xPar-1, a single mutation at Thr593 or Ser646 reduced the extent of phosphorylation by xPKCλ by about 30–40%, whereas double mutation reduced it dramatically (Figure 3F). These results strongly suggest that xPKCλ phosphorylates xPar-1 on Thr593 and Ser646.
Figure 3.
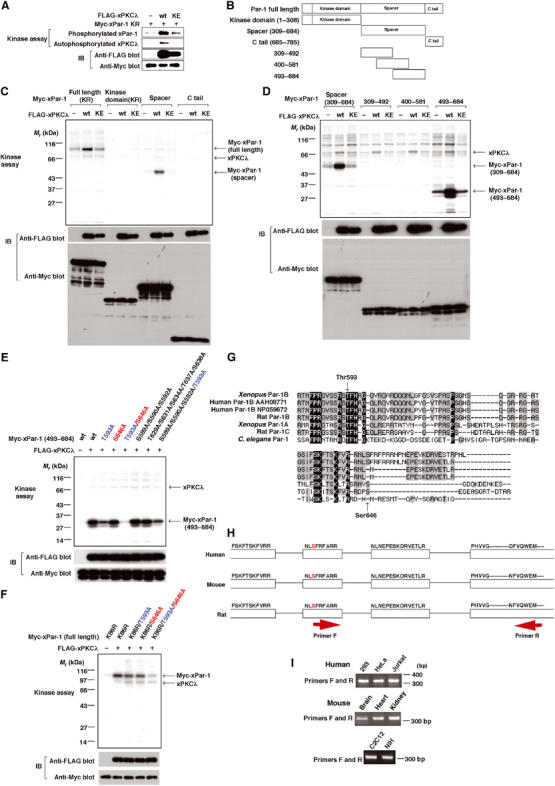
Two conserved residues in Par-1 are phosphorylated by aPKC. (A) xPKCλ phosphorylates xPar-1 in vitro. FLAG-tagged xPKCλ protein was produced in HEK293 cells and purified. Immunoprecipitated Myc-tagged xPar-1 KR (a kinase-dead form of xPar-1) was used as a substrate for kinase assay to prevent autophosphorylation of xPar-1. Phosphorylated xPar-1 and autophosphorylated xPKCλ were detected by autoradiography (upper two panels). Immunoblotting data (IB) confirmed that equal amounts of Myc-tagged xPar-1 KR protein were immunoprecipitated, and that FLAG-xPKCλ was purified (lower two panels). (B) Schematic representation of a series of truncated mutants of xPar-1. (C) xPKCλ phosphorylates the spacer region of xPar-1. (D) xPKCλ phosphorylates the region (residues 493–684) of xPar-1. (E) xPKCλ phosphorylates the region (residues 493–684) of xPar-1 on Thr593 and Ser646. (F) xPKCλ phosphorylates xPar-1 (full-length) on Thr593 and Ser646. (G) Sequence alignment of residues 581–670 of xPar-1 (Xenopus Par-1B) with the corresponding regions of human Par-1B, rat Par-1B, Xenopus Par-1A, rat Par-1C and C. elegans Par-1. (H) Schematic diagram of partial genomic DNA sequences for mammalian Par-1B genes. Exon and intron regions are represented by white boxes and black lines, respectively. Amino-acid sequences are shown above the white boxes. The serine residues that are corresponding to Ser646 in xPar-1 are red. (I) Par-1B mRNA, which has a region encoding an NLSFRFARR sequence, is present in mammal cells and tissues. Total RNAs were isolated from the respective cells or tissues and reverse-transcribed with random primers, followed by amplification with the primers that were designed as shown in (H). The sequences of the primer pairs used were as follows: primer F (human and mouse), 5′-tctttcaggtttgccagaagg; primer R (human), 5′-catctcccactgcacgaagtc; primer R (mouse), 5′-catctcccactgcacaaagtt.
We aligned the residues 581–670 of xPar-1 (noted as Xenopus Par-1B in Figure 3G) with the corresponding regions of other Par-1 orthologs reported. Thr593 in xPar-1 is completely conserved, while Ser646 is apparently not (Figure 3G). However, rat Par-1B that we isolated by RT–PCR contained a serine residue that corresponds to Ser646 in xPar-1 (data not shown). Thus, we searched genomic DNA sequences for mammalian Par-1B genes in the database and found a potential exon, which encodes a sequence NLSFRFARR that contains a serine residue that corresponds to Ser646 in xPar-1 (Figure 3H, red). This insert starts with a consensus splice acceptor ‘ag' and ends with a donor sequence ‘gt'. By RT–PCR, mammalian Par-1B mRNA that contains the region encoding an NLSFRFARR sequence was detected in various cell lines and tissues (Figure 3H and I). Therefore, Ser646 in xPar-1 may be evolutionarily conserved at least in vertebrates.
aPKC-dependent binding of Par-1 to 14-3-3 regulates subcellular localization of Par-1 and plays an essential role in gastrulation
Next we further addressed functional significance of aPKC phosphorylation of xPar-1. A single alanine mutation at either Thr593 or Ser646 completely abolished the binding of xPar-1 to 14-3-3ɛ (Figure 4A). Therefore, aPKC phosphorylation of xPar-1 is required for xPar-1 to bind to 14-3-3ɛ. Neither the sequence around Thr593 nor that around Ser646 matches the canonical 14-3-3 binding motifs, RSXpS/TXP or RXXXpS/TXP (Tzivion and Avruch, 2002) (see Figure 3G). Several 14-3-3 binding proteins that lack a canonical high-affinity 14-3-3 binding motif have been shown to require more than one phosphorylation site for stable binding (Tzivion and Avruch, 2002). Therefore, we can speculate that phosphorylation of both Thr593 and Ser646 is required for the stable binding of xPar-1 to 14-3-3ɛ. We then examined the role of aPKC-mediated phosphorylation of xPar-1 in Xenopus embryos. While injection of a low dose of xPar-1 wt mRNA caused weak gastrulation defects (Figure 4B, middle), injection of the same dose of xPar-1 T593A/S646A mRNA caused much stronger defects in gastrulation (Figure 4B, right). Moreover, while injection of a low dose of xPar-1 wt mRNA partially inhibited activin-induced elongation in animal caps, injection of the same dose of xPar-1 T593A/S646A mRNA strongly inhibited elongation (Figure 4C). These results taken together suggest that aPKC-dependent binding of xPar-1 to 14-3-3 is important for regulated gastrulation movements in Xenopus embryos. To find out functional consequences of xPar-1 binding to 14-3-3, we first tested the kinase activity of xPar-1. Neither aPKC nor 14-3-3ɛ affected markedly the ability of xPar-1 to phosphorylate an exogenous substrate, microtubule-associated protein Tau (Figure 4D). Moreover, those mutants of xPar-1 that are defective in 14-3-3 binding had essentially the same kinase activity as wild-type xPar-1 (Figure 4E). These data suggest that aPKC-dependent binding of xPar-1 to 14-3-3ɛ does not markedly affect the kinase activity of xPar-1. Next, we investigated whether phosphorylation of xPar-1 affects its subcellular localization. Cells expressing xPar-1 wt or xPar-1 T593A/S646A were lysed and the cell extracts were fractionated as Nonidet P-40-soluble supernatants and Nonidet P-40-insoluble fractions. Both xPar-1 wt and xPar-1 T593A/S646A were mainly recovered in insoluble membrane fractions (Figure 4F). Coexpression of 14-3-3ɛ or xPKCλ released xPar-1 wt, but not xPar-1 T593A/S646A, from insoluble membrane fractions to soluble cytosolic fractions (Figure 4F). Moreover, relocation of xPar-1 from the plasma membranes to the cytoplasm in Xenopus explants was induced by expression of 14-3-3ɛ and xPKCλ (Figure 4G). In contrast to wild-type xPar-1, xPar-1 T593A/S646A stayed almost exclusively in the plasma membrane regions (Figure 4G). Thus, aPKC-dependent binding of xPar-1 to 14-3-3 induces relocation of xPar-1 from plasma membranes to the cytoplasm, which is likely to be important for xPar-1 function during gastrulation.
Figure 4.
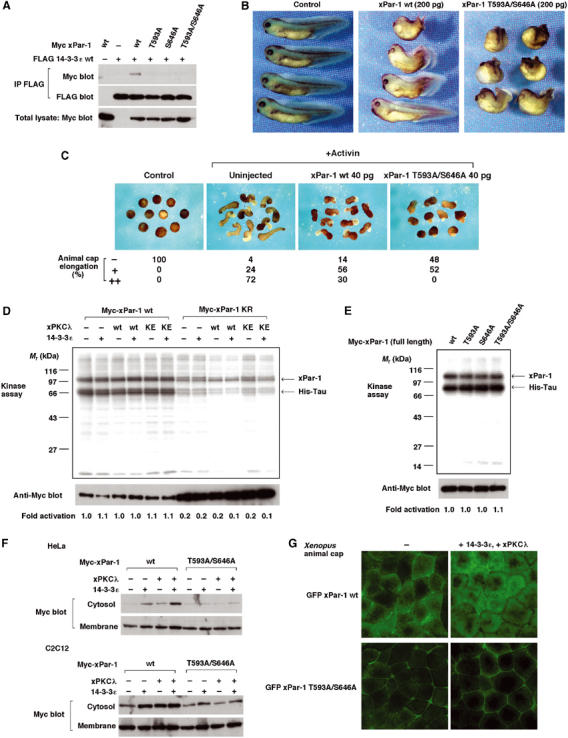
aPKC-dependent binding of Par-1 to 14-3-3 regulates subcellular localization of Par-1 and plays an essential role in gastrulation. (A) aPKC phosphorylation of xPar-1 is required for xPar-1 to bind to 14-3-3ɛ. (B) aPKC-dependent binding of xPar-1 to 14-3-3ɛ is important for regulated gastrulation movements in Xenopus embryos. Embryos were injected dorsally at the four-cell stage, and were cultured until stage 35 and photographed. All embryos are oriented with the anterior toward the left. The phenotypes were scored as previously described (Habas et al, 2003). Embryos with exposed endodermal tissues and dramatically reduced anterior–posterior axis were scored as severe gastrulation defects. Embryos with a partially shortened axis and a slightly bent body were scored as mild gastrulation defects. Left panel, control embryos; middle panel, embryos injected with 200 pg of xPar-1 wt mRNA (22.8% with severe gastrulation defects and 25.3% with mild gastrulation defects; _n_=79); right panel, embryos injected with 200 pg of xPar-1 T593A/S646A mRNA (75.0% with severe gastrulation defects and 12.5% with mild gastrulation defects; _n_=80). (C) xPar-1 T593A/S646A inhibits activin-induced elongation more severely than wild-type xPar-1 in animal caps. Four-cell stage embryos were injected with mRNAs encoding xPar-1 wt (40 pg; _n_=50) or xPar-1 T593A/S646A (40 pg; _n_=52) and animal cap assay was performed. (D) Neither aPKC nor 14-3-3 affects markedly the ability of xPar-1 to phosphorylate an exogenous substrate. Cells were transfected with the indicated combinations of expression vectors. Cells were lysed and Myc-tagged xPar-1 was immunoprecipitated by anti-Myc antibody. Kinase assay for immunoprecipitated xPar-1 was performed using His-tagged Tau as an exogenous substrate. Phosphorylation of Tau and autophosphorylation of xPar-1 were detected. Numbers below the panels represent average relative kinase activities of xPar-1. To determine relative kinase activity, radioactivity incorporated into His-Tau was normalized to Myc-xPar-1 levels in each reaction. The relative activity of the control (lane 1) was set as 1. The average relative kinase activities were calculated from three independent experiments (lanes 1–6) or two independent experiments (lanes 7–12). (E) Those mutants of xPar-1 that are defective in 14-3-3 binding have essentially the same kinase activity as wild-type xPar-1. Numbers below the panels represent average relative kinase activities of xPar-1. (F) Coexpression of 14-3-3ɛ or xPKCλ releases xPar-1 wt, but not xPar-1 T593A/S646A, from insoluble membrane fractions to soluble cytosolic fractions. Cells were transfected with the indicated combinations of expression vectors. Detergent-free lysis and then Western blot analysis of soluble (includes the cytoplasm) and insoluble (includes the plasma membranes) fractions were performed. (G) aPKC-dependent binding of xPar-1 to 14-3-3ɛ induces relocation of xPar-1 from plasma membranes to the cytoplasm in Xenopus explants. GFP-xPar-1 wt or GFP-xPar-1 T593A/S646A mRNA was injected into four-cell stage embryos and animal caps were dissected. Subcellular localization of xPar-1 proteins was observed by fluorescence microscopy.
aPKC and its binding partner Par-6 are essential for gastrulation
Finally, we examined whether aPKC is also essential for gastrulation in Xenopus embryos. xPKCλ is ubiquitously expressed throughout embryogenesis (data not shown). Activin-induced elongation of animal caps was strongly inhibited by expressing xPKCλ KE but not xPKCλ wt (Figure 5A). The cell fate specification was not changed (Figure 5B). Injection of xPKCλ KE mRNA, but not xPKCλ wt, in the dorsal marginal zone of four-cell stage embryos caused severe morphogenetic defects; the embryos showed a dorsal bending phenotype with a shortened body axis (Figure 5C). The gastrulation defects caused by injection of xPKCλ KE mRNA were rescued by coinjection of xPKCλ wt mRNA in animal caps and whole embryos (Figure 5C and D). These results indicate that xPKCλ is required for gastrulation movements. We next extended our analysis to the binding partners of aPKC, Par-3 and Par-6. Overexpression of rPar-3 or xPar-6 in animal caps markedly inhibited elongation without affecting cell fate specification (Figure 5A and B). xPar-6 4A, a mutant form of xPar-6 in which four crucial residues within the PDZ domain were replaced by alanines (Joberty et al, 2000), did not inhibit elongation significantly (Figure 5A), indicating that the interaction of xPar-6 with target proteins through the PDZ domain is important. Injection of rPar-3 or xPar-6 mRNA in the dorsal marginal zone of four-cell stage embryos caused gastrulation defects, while xPar-6 4A had a much weaker effect (data not shown). We then examined the requirement of xPar-6 function by using an antisense MO against xPar-6, which was specific and efficient (Figure 5E). At neurula and tadpole stages, the xPar-6 MO-injected embryos showed a phenotype characteristic of incomplete gastrulation movements without a change in the expression profiles of molecular markers (Figure 5F–H). These results collectively indicate that the aPKC/Par-3/Par-6 complex regulates gastrulation movements. This is consistent with our idea that aPKC phosphorylation of Par-1 is important for gastrulation movements.
Figure 5.
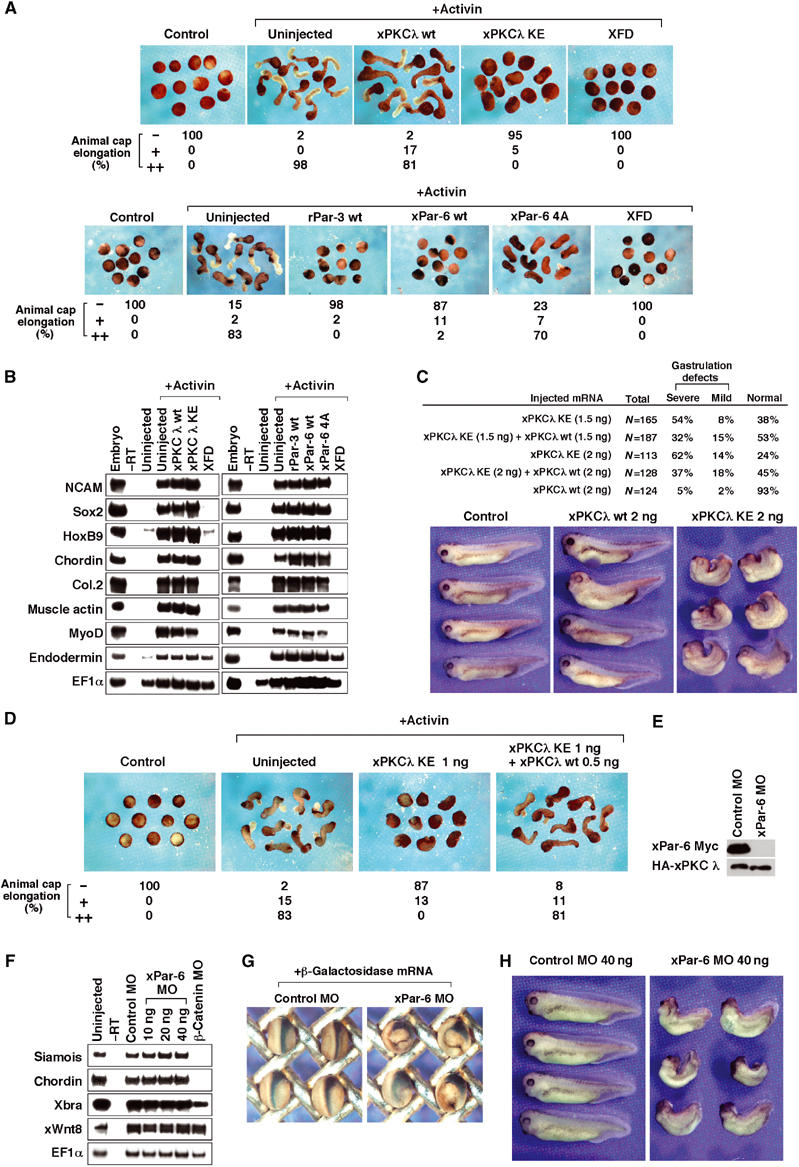
aPKC and its binding partner Par-6 are essential for gastrulation. (A) xPKCλ KE inhibits elongation of animal cap explants in response to activin (upper panels). Four-cell embryos were injected with mRNAs encoding xPKCλ wt (1 ng; _n_=41), xPKCλ KE (1 ng; _n_=58) or dominant-negative FGF receptor (XFD) (Amaya et al, 1991) (1 ng; _n_=49) and animal cap assay was performed. XFD, which is known to inhibit gastrulation movements, was used as a control. Par-3 and Par-6 inhibit elongation of animal cap explants in response to activin (lower panels). Four-cell embryos were injected with mRNAs encoding rPar-3 wt (1 ng; _n_=43), xPar-6 wt (1 ng; _n_=46), xPar-6 4A mutant (1 ng; _n_=47) or XFD (1 ng; _n_=46). Animal caps are cultured and photographed. (B) The cell fate specification is not affected by aPKC, Par-3 or Par-6. The animal caps from (A) were analyzed by RT–PCR. XFD, which is known to inhibit cell fate specification, was used as a control. (C) xPKCλ is required for gastrulation movements. Indicated sets of mRNA were injected into both dorsal blastomeres at the four-cell stage, and the embryos were cultured until stage 35 and the phenotypes were scored as previously described (Habas et al, 2003). All embryos are oriented with the anterior toward the left. (D) The gastrulation defects caused by injection of xPKCλ KE mRNA were rescued by coinjection of xPKCλ wt mRNA in animal caps. (E) xPar-6 morpholino antisense oligonucleotide (MO) effectively blocks translation of injected xPar-6 mRNA. xPar-6-Myc or HA-xPKCλ mRNA was injected into embryos with xPar-6 MO or control MO and embryo lysates were analyzed by Western blotting with anti-Myc antibody. xPar-6-Myc mRNA used in this experiment contains the 5′ UTR, which is recognized by xPar-6 MO. (F) xPar-6 MO does not change cell fate specification in whole embryos. The indicated amounts of MO were injected into both dorsal blastomeres at the four-cell stage and whole embryos were harvested at stage 11 for RT–PCR. (G) Lineage tracer analysis of gastrulation movements in xPar-6 MO-injected embryos. Left panel, embryos injected with 80 ng of control MO and 20 pg of β-galactosidase mRNA; right panel, embryos injected with 80 ng of xPar-6 MO and 20 pg of β-galactosidase mRNA. (H) Effects of xPar-6 MO on whole embryos. Control MO or xPar-6 MO was injected into both dorsal blastomeres of the four-stage embryos and cultured until stage 35. Left panel, embryos injected with 40 ng of control MO (99% with complete normal structures; _n_=82); right panel, embryos injected with 40 ng of xPar-6 MO (81% with dorsal bending and a shortened body axis; _n_=41).
14-3-3 and aPKC regulate Par-1 to control vertebrate gastrulation
Our results have demonstrated that Par-1 is essential for gastrulation movements, but is dispensable for cell fate specification during Xenopus early embryonic development. Although one member of the Par-1 family, Emk, has been knocked out in mice with no apparent effect on gastrulation (Bessone et al, 1999; Hurov et al, 2001), it is possible that multiple knockouts of the Par-1 family members lead to gastrulation defects in mice. Moreover, our results have identified a novel interrelationship within Par proteins: regulation of Par-1 by 14-3-3/Par-5 and aPKC, which is important for proper gastrulation movements. Because gain-of-function and loss-of-function manipulations of Par-1 lead to indistinguishable gastrulation defects, a precisely regulated, appropriate activity of Par-1 is required. Part of this regulation would be attributed to the controlled subcellular localization of Par-1 by 14-3-3 and aPKC. After completion of this study, a paper by Hurov et al (2004) appeared showing aPKC phosphorylation of human Par-1 (hPar-1). They have identified one of the two phosphorylation sites we identified in this study, and have shown that the phosphorylation negatively regulates the plasma membrane localization of hPar-1 by an unknown mechanism. They reported that aPKC phosphorylation inhibits the kinase activity of hPar-1. In our experiments, aPKC phosphorylation does not affect the ability of xPar-1 to phosphorylate an exogenous substrate Tau (see Figure 4D and E). This apparent discrepancy might be caused by differences in the method of the kinase assay (e.g. substrates used) and/or differences in the species of Par-1 used, hPar-1 or xPar-1. A previous study in Drosophila has shown that Par-1 binds to 14-3-3 in a manner that is independent of phosphorylation (Benton et al, 2002), in contrast with our results showing that aPKC phosphorylation of xPar-1 is required for 14-3-3 binding. This discrepancy might also be caused by the species difference. Benton and St Johnston (2003) have recently shown in Drosophila that Par-3 also binds to 14-3-3 in a Par-1-dependent manner, and that Par-3 is excluded from a specific region of the plasma membrane by 14-3-3 in oocytes and epithelial cells. These 14-3-3-dependent mechanisms for Par-3 and Par-1 would thus ensure a proper activity of the polarity proteins. Kinoshita et al (2003) have shown that during Xenopus gastrulation, Dishevelled and Rac are localized around the tips of the elongated cells, which are polarized and become narrow along the mediolateral direction. We might speculate that Par proteins also show a polarized distribution in these cells during Xenopus gastrulation movements. Finally, it should be pointed out that the present identification of the role of Par proteins in gastrulation movements may open a new way to address the functional significance of new observations in functions and regulations of Par proteins.
Materials and methods
Molecular cloning and plasmid construction
The Xenopus gastrula cDNA library was screened using the 0.35-kb fragment in the EST sequence (GenBank accession number BG409740) as a probe, and a full-length cDNA clone of xPar-1 was obtained. While this work was in progress, Nakajo et al isolated a cDNA (unpublished, GenBank accession number AB071963) that is identical to the cDNA clone we isolated and used in this study. Molecular cloning of Xenopus Par-1 homologs that have different sequences from our cDNA clone was also reported (Ossipova et al, 2002). The entire coding region of xPar-1 was amplified by PCR and cloned into the expression vector. The cDNAs of rPar-1C, xPar-6, rPar-3 and xPKCλ were isolated by RT–PCR and cloned into the expression vector. To obtain a kinase-negative form of xPar-1 (xPar-1 KR), Lys86 in xPar-1 was replaced by arginine. To obtain a nonactivatable form of xPar-1 (xPar-1 TASA), Thr212 and Ser216 were replaced by alanines. To obtain a kinase-negative form of xPKCλ, Lys275 in xPKCλ was replaced by glutamate. To obtain an active form of xPKCλ, Ala122 was replaced by glutamate. To obtain xPar-6 4A that contains mutations in the PDZ domain, Lys168, Pro169, Leu170 and Gly171 in xPar-6 were replaced by alanines. These mutations were made by site-directed mutagenesis (Stratagene) and confirmed by DNA sequencing.
Embryo manipulations
Xenopus embryos were obtained by in vitro fertilization of eggs with testes homogenates. Fertilized embryos were dejellied by treatment with 2% cysteine hydrochloride and cultured in 0.1 × MBS (1.5 mM Hepes pH 7.4, 8.8 mM NaCl, 0.1 mM KCl, 0.24 mM NaHCO3, 0.082 mM MgSO4, 0.03 mM Ca(NO3)2 and 0.041 mM CaCl2). Injection of embryos was performed in 5% Ficoll in 0.1 × MBS. The amounts of injected mRNAs are described in the text and figure legends. In vitro synthesis of capped mRNA was performed using mMESSAGE mMACHINE (Ambion) according to the manufacturer's instructions. For the whole embryo phenotypes, mRNAs were injected into two dorsal blastomeres at the four-cell stage, and then photographed at the tadpole stage. For animal cap assay, mRNAs were injected into animal poles of four-cell stage embryos. Animal caps were dissected at stage 8 and cultured in 1 × Steinberg's solution (10 mM Hepes pH 7.4, 60 mM NaCl, 0.67 mM KCl, 0.83 mM MgSO4 and 0.34 mM Ca(NO3)2) containing 1 mg/ml BSA (Sigma) and 10 ng/ml recombinant human Activin A (Genzyne-techne). The caps were photographed at stage 18–20 and harvested for RT–PCR. The sequences of primer pairs for RT–PCR are described in Dr Robertis' web site (http://www.hhmi.ucla.edu/derobertis/) or elsewhere (Djiane et al, 2000; Heasman et al, 2000; Kusakabe et al, 2001).
Morpholino oligos
The anti-xPar-1 morpholino (xPar-1 MO; 5′-TCGGCAGCGGTGTCCTGGTGGTCAT-3′), the anti-xPar-6 morpholino (xPar-6 MO; 5′-GTCCCCGGTTCATGTTGCCAGTGCA-3′), the anti-β-catenin morpholino (Heasman et al, 2000) and the standard control morpholino (control MO) were purchased from Gene Tools, LLC. Sequences complementary to the predicted start codon are underlined. The target sequence for xPar-1 MO does not contain the 5′ untranslated region (5′ UTR) of the xPar-1 cDNA. The target sequence for xPar-6 MO contains the 5′ UTR of the xPar-6 cDNA (GenBank accession number BJ072852). These MOs were resuspended in sterile water and injected in doses that are described in the text and figure legends. In the experiments shown in Figures 1C and 5E, the embryos injected with the indicated combinations of mRNAs and MOs were cultured until stage 9 and crushed in a buffer containing 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 2 mM EGTA, 25 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1% NP-40, 10 mM NaF, 1 mM vanadate, 2 mM DTT, 1 mM PMSF and 0.5% aprotinin. The supernatants were used for immunoblotting with anti-Myc antibody (A-14; Santa Cruz Biotechnology) and anti-HA antibody (Y-11; Santa Cruz Biotechnology).
Detection of _β_-galactosidase
mRNA encoding nuclear β-galactosidase was coinjected as a lineage tracer. Injected embryos were fixed in 2% formaldehyde, 100 mM PIPES (pH 7.4), 2 mM MgCl2 and 1.25 mM EGTA. After washing three times with PBS containing 2 mM MgCl2, embryos were transferred to a staining solution (PBS buffer containing 20 mM K3Fe(CN)6, 20 mM K4Fe(CN)6, 2 mM MgCl2, 0.01% deoxycholate, 0.02% NP-40 and 1 mg/ml X-gal) and incubated at 37°C overnight.
Cell culture, transfection and co-immunoprecipitation
HEK293 cells, C2C12 cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), 15% FCS and 10% calf serum, respectively. HEK293 cells were cultured on collagen-coated plates. Cells were transfected by using Lipofectamine 2000 (Invitrogen) for HEK293 cells, Lipofectamine Plus reagent (Invitrogen) for C2C12 cells and FuGENE6 (Roche) for HeLa cells according to the manufacturer's protocol. Transfected cells were lysed in a buffer consisting of 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 2 mM EGTA, 25 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1% NP-40, 10 mM NaF, 1 mM vanadate, 2 mM DTT, 1 mM PMSF and 0.5% aprotinin. Anti-FLAG M2 antibody (Sigma) was used for immunoprecipitation. Precipitates were washed three times with the same buffer containing no detergent.
Kinase assay
In kinase assay for xPKCλ, FLAG-tagged xPKCλ was expressed in HEK293 cells, precipitated by anti-FLAG M2 affinity gel (Sigma) and eluted by FLAG (DYKDDDDK) peptide. Purified xPKCλ was mixed with immunoprecipitated substrates in a kinase reaction buffer (50 mM Tris pH 7.5, 12.5 mM MgCl2, 1 mM EGTA and 1 mM DTT) and incubated for 10 min at 30°C in the presence of [γ-32P]ATP. The reactions were stopped by addition of Laemmli's sample buffer and boiled. The reaction products were analyzed by SDS–PAGE, followed by autoradiography. For kinase assay for xPar-1, immunoprecipitated xPar-1 was incubated with His-tagged Tau in a kinase reaction buffer for 10 min at 30°C.
Materials
Staurosporine was purchased from Sigma; Rp-8-Br-cAMPS, bisindolylmaleimide I, AG1478, LY294002, SP600125 and SB203580 were from Calbiochem; U0126 and PD98059 were from Promega.
Acknowledgments
We thank S Torii and H Yamanaka for technical advice and helpful discussion, and H Hanafusa for a Xenopus gastrula cDNA library. We also thank members of our laboratory for their help. This work was supported by grants-in-aid from the Ministry of Education, Science and Culture of Japan (to EN).
References
- Amaya E, Musci TJ, Kirschner MW (1991) Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 66: 257–270 [DOI] [PubMed] [Google Scholar]
- Benton R, Palacios IM, St Johnston D (2002) Drosophila 14-3-3/PAR-5 is an essential mediator of PAR-1 function in axis formation. Dev Cell 5: 659–671 [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D (2003) Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115: 691–704 [DOI] [PubMed] [Google Scholar]
- Bessone S, Vidal F, Le Bouc Y, Epelbaum J, Bluet-Pajot MT, Darmon M (1999) EMK protein kinase-null mice: dwarfism and hypofertility associated with alterations in the somatotrope and prolactin pathways. Dev Biol 214: 87–101 [DOI] [PubMed] [Google Scholar]
- Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G (2004) Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem 279: 12804–12811 [DOI] [PubMed] [Google Scholar]
- Chang HC, Rubin GM (1997) 14-3-3 epsilon positively regulate Ras-mediated signaling in Drosophila. Genes Dev 11: 1132–1139 [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Tan C, Conrad LJ, Klein PS (1998) Frizzled-8 is expressed in the Spemann organizer and plays a role in early morphogenesis. Development 125: 2687–2700 [DOI] [PubMed] [Google Scholar]
- Djiane A, Riou J, Umbhauer M, Boucaut J, Shi D (2000) Role of frizzled 7 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development 127: 3091–3100 [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89: 297–308 [DOI] [PubMed] [Google Scholar]
- Du SJ, Purcell SM, Christian JL, McGrew LL, Moon RT (1995) Identification of distinct classes and functional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol Cell Biol 15: 2625–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ (1995) Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83: 743–752 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2003) Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol 15: 67–72 [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ (1995) par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620 [DOI] [PubMed] [Google Scholar]
- Habas R, Dawid IB, He X (2003) Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev 17: 295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C (2000) Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol 222: 124–134 [DOI] [PubMed] [Google Scholar]
- Hung TJ, Kemphues KJ (1999) PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126: 127–135 [DOI] [PubMed] [Google Scholar]
- Hurov JB, Stappenbeck TS, Zmasek CM, White LS, Ranganath SH, Russell JH, Chan AC, Murphy KM, Piwnica-Worms H (2001) Immune system dysfunction and autoimmune disease in mice lacking Emk (Par-1) protein kinase. Mol Cell Biol 21: 3206–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H (2004) Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol 14: 736–741 [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG (2000) The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2: 531–539 [DOI] [PubMed] [Google Scholar]
- Keller R, Shih J, Domingo C (1992) The patterning and functioning of protrusive activity during convergence and extension of the Xenopus organiser. Development, (Suppl): 81–91 [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS (1988) Identification of genes required for cytoplasmic localization in early C.elegans embryos. Cell 52: 311–320 [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Iioka H, Miyakoshi A, Ueno N (2003) PKC delta is essential for Dishevelled function in a noncanonical Wnt pathway that regulates Xenopus convergent extension movements. Genes Dev 17: 1663–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe M, Masuyama N, Hanafusa H, Nishida E (2001) Xenopus FRS2 is involved in early embryogenesis in cooperation with the Src family kinase Laloo. EMBO Rep 2: 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG (2004) Parsing the polarity code. Nat Rev Mol Cell Biol 5: 220–231 [DOI] [PubMed] [Google Scholar]
- Moon RT, Campbell RM, Christian JL, McGrew LL, Shih J, Fraser S (1993) Xwnt-5A: a maternal Wnt that affects morphogenetic movements after overexpression in embryos of Xenopus laevis. Development 119: 97–111 [DOI] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ (2002) The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo. Dev Biol 241: 47–58 [DOI] [PubMed] [Google Scholar]
- Muller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK (2001) C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell 8: 983–993 [DOI] [PubMed] [Google Scholar]
- Nakaya M, Fukui A, Izumi Y, Akimoto K, Asashima M, Ohno S (2000) Meiotic maturation induces animal–vegetal asymmetric distribution of aPKC and ASIP/PAR-3 in Xenopus oocytes. Development 127: 5021–5031 [DOI] [PubMed] [Google Scholar]
- Ohno S (2001) Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol 13: 641–648 [DOI] [PubMed] [Google Scholar]
- Ossipova O, He X, Green J (2002) Molecular cloning and developmental expression of Par-1/MARK homologues XPar-1A and XPar-1B from Xenopus laevis. Gene Expr Patterns 2: 145–150 [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ III, Wu Z, Stephenson MT, Piwnica-Worms H (1998) C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ 9: 197–208 [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN (2003) Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112: 63–75 [DOI] [PubMed] [Google Scholar]
- Sokol SY (1996) Analysis of Dishevelled signalling pathways during Xenopus development. Curr Biol 6: 1456–1467 [DOI] [PubMed] [Google Scholar]
- Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT (2001) PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol 3: 628–636 [DOI] [PubMed] [Google Scholar]
- Symes K, Smith JC (1987) Gastrulation movements provide an early marker of mesoderm induction in Xenopus. Development 101: 339–349 [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S (1998) Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125: 3607–3614 [DOI] [PubMed] [Google Scholar]
- Tada M, Smith JC (2000) Xwnt11 is a target of Xenopus Brachyury: regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development 127: 2227–2238 [DOI] [PubMed] [Google Scholar]
- Tzivion G, Avruch J (2002) 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem 277: 3061–3064 [DOI] [PubMed] [Google Scholar]
- Uberall F, Giselbrecht S, Hellbert K, Fresser F, Bauer B, Gschwendt M, Grunicke HH, Baier G (1997) Conventional PKC-alpha, novel PKC-epsilon and PKC-theta, but not atypical PKC-lambda are MARCKS kinases in intact NIH 3T3 fibroblasts. J Biol Chem 272: 4072–4078 [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Fraser SE, Harland RM (2002) Convergent extension. The molecular control of polarized cell movement during embryonic development. Dev Cell 2: 695–706 [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Medina A, Swain RK, Steinbeisser H (2001) Frizzled-7 signalling controls tissue separation during Xenopus gastrulation. Nature 413: 856–860 [DOI] [PubMed] [Google Scholar]