Phylogenomic analysis of the family Peptostreptococcaceae (Clostridium cluster XI) and proposal for reclassification of Clostridium litorale (Fendrich et al. 1991) and Eubacterium acidaminophilum (Zindel et al. 1989) as Peptoclostridium litorale gen. nov. comb. nov. and Peptoclostridium acidaminophilum comb. nov (original) (raw)
Abstract
In 1994, analyses of clostridial 16S rRNA gene sequences led to the assignment of 18 species to Clostridium_ cluster XI, separating them from _Clostridium sensu stricto(Clostridium_ cluster I). Subsequently, most cluster XI species have been assigned to the family Peptostreptococcaceae_ with some species being reassigned to new genera. However, several misclassified _Clostridium species remained, creating a taxonomic conundrum and confusion regarding their status. Here, we have re-examined the phylogeny of cluster XI species by comparing the 16S rRNA gene-based trees with protein- and genome-based trees, where available. The resulting phylogeny of thePeptostreptococcaceae_ was consistent with the recent proposals on creating seven new genera within this family. This analysis also revealed a tight clustering of Clostridium litorale andEubacterium acidaminophilum. Based on these data, we propose reassigning these two organisms to the new genus__Peptoclostridium_ as__Peptoclostridium litorale_ gen. nov. comb. nov. (the type species of the genus) and Peptoclostridium acidaminophilum comb. nov., respectively. As correctly noted in the original publications, the generaAcetoanaerobium and__Proteocatella_ also fall within cluster XI, and can be assigned to the _Peptostreptococcaceae.Clostridium sticklandii, which falls within radiation of genus _Acetoanaerobium, is proposed to be reclassified as _Acetoanaerobium sticklandii comb. nov. The remaining misnamed members of the _Peptostreptococcaceae, [__Clostridium_]hiranonis, [__Clostridium_]paradoxum and [__Clostridium_]thermoalcaliphilum, still remain to be properly classified.
Keywords: Taxonomy, Gram-positive bacteria, anaerobes, genome analysis, 16S rRNA
In the past, obligately anaerobic spore-forming bacteria that stained Gram-positive were often assigned to the genus Clostridium_, which resulted in a single genus with more than 200 validly named species with vastly different properties, including proteolytic and cellulosolytic bacteria, some pathogenic and some benign (Parte, 2014). In 1994, Collins and colleagues used 16S rRNA gene sequences to divide clostridial species into 19 clusters; each cluster included several proposed genera and corresponded to a family-level taxon (Collins et al., 1994). In subsequent studies, most of those groupings have been confirmed and the taxonomy of many former Clostridium_ species has been streamlined by reassigning them to new genera. It has become clear that only _Clostridium cluster I species (Clostridium sensu stricto_) are sufficiently close to the type species Clostridium butyricum_ to qualify as members of the same genus. This view has been reinforced by the recent work byLawson & Rainey (2016), who removed from the genus _Clostridium even cluster II species,C lostridium histolyticum_,Clostridium limosum_ and Clostridium proteolyticum, which are the closest relatives of cluster I, and reassigned them to the new genus _Hathewaya. In the clostridial classification update in the latest edition of Bergey’s, many former Clostridium species have been moved to families Lachnospiraceae, Peptostreptococcaceae and_Ruminococcaceae in the orderClostridiales_ and to the familyErysipelotrichaceae_ in the class__Erysipelotrichia_ (Ludwig et al., 2009a). However, most of these organisms still retained the ‘__Clostridium’ name (Rainey et al., 2009; Parte, 2014), resulting in a taxonomic and nomenclature conundrum, where the Clostridium species designation did not necessarily indicate a close relationship to the type speciesClostridium butyricum_ and such Clostridium_ sensu stricto organisms as _Clostridium botulinum andClostridium tetani_. To deal with this conundrum, the NCBI Taxonomy Database and SILVA database display such misnamed organisms as [__Clostridium_] species (Federhen, 2012, 2015; Yilmaz et al., 2014); for clarity, this approach is also used in this work. This designation helps in highlighting the problem but obviously does not resolve it.
In 2013, Yutin and Galperin proposed to resolve the inconsistency of having 15 validly described Clostridium species in the familyPeptostreptococcaceae by tentatively assigning them, along with Eubacterium tenue andEubacterium yurii, to the new genus Peptoclostridium_ (Yutin & Galperin, 2013). They have argued that, although imperfect and tentative, such a solution was still better than either listing these organisms as [__Clostridium_] species or preserving theClostridium_ species name for the bacteria that clearly do not belong to the genus _Clostridium or even the family _Clostridiaceae. Unfortunately, the proposal to include all diverse species into a single genus proved to be unsatisfactory. In addition, this proposal did not comply with the Bacteriological Code (Parker et al., 2015), and its partial adoption by some databases only increased the confusion in clostridial nomenclature.
Recently, this nomenclature problem was partly resolved by three papers that reassigned 10 of those 15 [__Clostridium_] species to the new genera. First, Gerritsen and colleagues proposed creating four new genera _Romboutsia, _Intestinibacter, _Terrisporobacter and _Asaccharospora, which accommodated five former [__Clostridium_] species: _Clostridium bartlettii, _Clostridium glycolicum, _Clostridium irregulare, _Clostridium lituseburense and _Clostridium mayombei (Gerritsen et al., 2014). Earlier this year, Sasi Jyothsna and colleagues proposed creation of two more genera, _Paraclostridium and _Paeniclostridium, to accommodate three more such species: _Clostridium bifermentans, _Clostridium ghonii and _Clostridium sordellii (Sasi Jyothsna et al., 2016). Finally, Lawson and colleagues proposed reassigning [__Clostridium_]difficile and _Clostridium mangenotii to the new genus _Clostridioides (Lawson et al., 2016). Although these studies substantially streamlined the nomenclature of the family _Peptostreptococcaceae, several members of this family remain listed as [_Clostridium] or [__Eubacterium] species. Here, we report a re-analysis of the available sequence data for the members of the familyPeptostreptococcaceae, confirm the groupings proposed by Gerritsen et al.(2014), Sasi Jyothsna et al.(2016) and Lawson et al.(2016) and propose re-assigning [__Clostridium]litorale and [__Eubacterium]acidaminophilum to the new genus _Peptoclostridium. We additionally propose transfer of the genera Acetoanaerobium and_Proteocatella to the Peptostreptococcaceae_ and reclassification of [__Clostridium_] sticklandii asAcetoanaerobium sticklandii.
The original description of the clostridial cluster XI (Collins et al., 1994) included 19 validly described species assigned to eight proposed genera with an additional genus reserved for ‘__Clostridium aminobutyricum_’ (not validly described, currently listed as Clostridium sp. DSM 2634=ATCC 13726). Subsequent analyses of 16S rRNA gene sequences showed that members of three proposed genera (listed as genera 6, 7 and 8 in Collins et al., 1994), namely ‘_C. aminobutyricum’, Clostridium felsineum_,Clostridium formicaceticum_, and _Clostridium halophilum, along with the more recently described Clostridium caminithermale, form a relatively compact separate group that also includes Clostridium aceticum and_Anaerovirgula multivorans (Brisbarre et al., 2003; Chen_et al._, 2006; Pikuta_et al._, 2006; Gerritsen_et al._, 2014; Poehlein_et al._, 2015; Lawson_et al._, 2016). In the 2009 edition of Bergey’s, these species were assigned to Clostridiaceae 2, a sister group to Peptostreptococcaceae_ (Ludwig et al., 2009b), and are listed the same way in the SILVA database (Yilmaz et al., 2014). In our hands, 16S rRNA gene sequences from these organisms also formed a separate group. Also, aside from the genome of _C. aceticum (Poehlein et al., 2015), there was almost no sequence data to verify the 16S rRNA gene-based tree. Therefore, these organisms were excluded from further analysis. The phylogeny of the remaining members of cluster XI and thePeptostreptococcaceae was analysed using 16S rRNA gene-based, protein-based and whole genome-based phylogenetic trees.
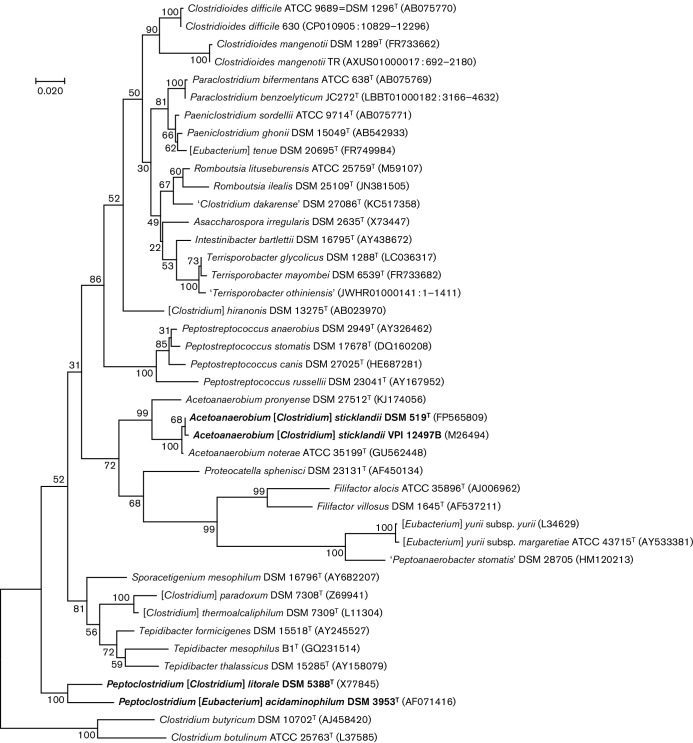
The 16S rRNA gene trees reconstructed using neighbour-joining and maximum likelihood methods (Figs 1 and S1, available in the online Supplementary Material) were similar to those reported previously (Collins et al., 1994; Chen, 2006; Gerritsen et al., 2014; Lawson et al., 2016; Sasi Jyothsna et al., 2016). They showed clear groupings of the species within the genera__Filifactor, _Peptostreptococcus and _Tepidibacter. These trees also confirmed grouping of the former [_Clostridium]glycolicus and _Clostridium mayombei, which had been reassigned to the genus Terrisporobacter (Gerritsen et al., 2014), of the former_C . ghonii with C. sordellii, reassigned to the genus Paeniclostridium (Sasi Jyothsna et al., 2016), and ofC_ . difficile and _C. mangenotii, recently reassigned to the genus_Clostridioides_ (Lawson et al., 2016). Given the reclassification of the former C. bartlettii as Intestinibacter bartlettii_;C. irregulare as _Asaccharospora irregularis; _C. lituseburense as _Romboutsia lituseburensis (Gerritsen et al., 2014); and of C. bifermentans_ as _Paraclostridium bifermentans (Sasi Jyothsna et al., 2016), only five validly described members of the family Peptostreptococcaceae_(four from the original cluster XI and Clostridium hiranonis) remained listed as [__Clostridium] species. Finally, in accordance with the original publications (Pikuta et al., 2009; Bes_et al._, 2015), the 16S rRNA gene tree showed that members of the genera Acetoanaerobium (Acetoanaerobium noterae and _Acetoanaerobium pronyense) and_Proteocatella (Proteocatella sphenisci) also fall within cluster XI and family _Peptostreptococcaceae. We therefore formally propose reassigning genera Acetoanaerobium and_Proteocatella to the familyPeptostreptococcaceae_.
Fig. 1.
16S rRNA gene-based phylogenetic tree of the family Peptostreptococcaceae._ The sequences from type strains (indicated with superscript T) were used and listed under their DSM accession numbers, where available. GenBank accession numbers are listed in parentheses. For Clostridioides difficile_ 630, _Clostridioides mangenotii TR,Paraclostridium benzoelyticum_JC272T, and ‘Terrisporobacter othiniensis_’, 16S rRNA gene sequences were taken from the respective genomic entries. Quotation marks indicate the organisms whose names have not yet been validly published. The organisms that this work proposes to be renamed are indicated in boldtype. The sequences were aligned using muscle (Edgar, 2004) and the tree was inferred using the maximum-likelihood method based on the Tamura–Nei model (Tamura & Nei, 1993) as implemented in mega6 (Tamura et al., 2013); for the initial neighbour-joining tree, see Fig. S1. The tree was rooted using sequences from C. butyricum and C. botulinum_, members ofClostridium sensu stricto.
The phylogenetic trees generated using 16S rRNA gene sequences also showed that__C. litorale_ and _E. acidaminophilum formed a well-supported separate branch (Fig. 1), which has also been seen in previous reports (Baena et al., 1999; Pikuta et al., 2009;Gerritsen et al., 2014; Rainey et al., 2015; Lawson et al., 2016), see e.g. Fig. 143 in Wade (2009). This association suggested that these two species might qualify for inclusion into the same genus. Indeed, 16S rRNA genes of _C. litorale and _E. acidaminophilum share 94 % sequence identity over 1500 bases, which is close to the threshold for a single genus suggested byYarza et al. (2008, 2014) and supported by Tindall et al. (2010). Previously, Collins et al. (1994) assigned _C. litorale to a separate genus in cluster XI. Later, Baena et al. (1999)determined the 16S rRNA gene sequence of _E. acidaminophilum, showed its tight clustering with _C. litorale, and recommended creating a new genus to accommodate these two species. We wanted an independent means of verifying the close relation between C. litorale and _E. acidaminophilum and checking the consistency of other groupings seen on the 16S rRNA gene tree. To this end, we reconstructed a concatenated alignment of 50 widespread ribosomal proteins and also alignments for DNA-directed RNA polymerase beta subunit (RpoB) and DNA gyrase subunit B (GyrB) from various members of Peptostreptococcaceae. These alignments were sufficiently long to allow fine mapping of phylogenetic relations and analyse deep phylogenetic lineages (Yutin et al., 2012; Yutin & Galperin, 2013). However, the use of such trees in clostridial phylogeny is limited by the fact that protein sequences are currently available only for a limited number of strains (Table S1). Fortunately, genome sequences of C. litorale and_E. acidaminophilum have already been made available by Poehlein et al.(2014a, b).
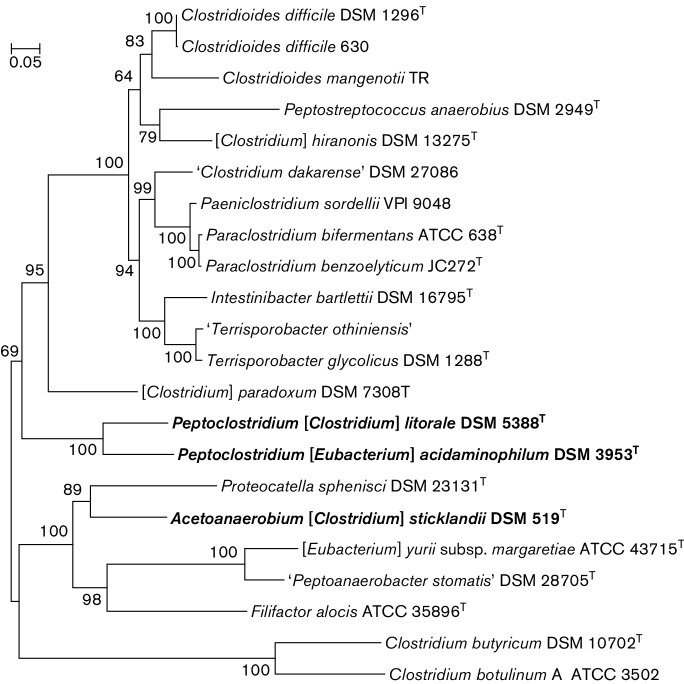
The ribosomal proteins-based tree (Fig. 2) confirmed the close relationship between C. litorale andE. acidaminophilum, as well as the groupings proposed previously by Gerritsen et al.(2014), Sasi Jyothsna et al.(2016) and Lawson et al.(2016). The RpoB and GyrB trees (Fig. S2) had similar topologies and also showed__C. litorale_ and E. acidaminophilum_ branching together. In addition, grouping of _C. litorale andE. acidaminophilum_ could be seen on the tree built from whole-genome alignments of members of Peptostreptococcaceae_ (Fig. S3a), using the approaches developed by Goris et al.(2007), Deloger et al.(2009) and Varghese et al.(2015). Visualization of this tree on a two-dimensional plot using principal component analysis (Fig. S3b) also showed _C. litorale andE. acidaminophilum_ clustering separately from other species.
Fig. 2.
Ribosomal protein-based tree of the members of _Peptostreptococcaceae. A maximum-likelihood tree was built using the PhyML program (Guindon et al., 2010) from a concatenated alignment of 50 ribosomal proteins (L1–L7, L9–L11, L13–L24, L27–L29, L31–L36 and S2–S20), with a total of 6269 aligned positions, as described previously (Yutin et al., 2012; Yutin & Galperin, 2013). The organisms proposed for renaming are indicated in boldtype. The names of the organisms that have not been validly published are in quotation marks. The tree was rooted using sequences from _C. butyricum and _C. botulinum.
In addition to sequence information, C. litorale andE. acidaminophilum share several important physiological properties, including the ability to grow on betaine and such amino acids as glycine and serine, metabolizing them via Stickland reaction (Dietrichs et al., 1991), and the lack of utilization of carbohydrates. These traits are usually not seen in other representatives of__Peptostreptococcaceae_ (Table S2). The most conspicuous difference between the two species is that C. litorale is a spore former, whereasE. acidaminophilum is not (Zindel et al., 1988; Fendrich et al., 1990). However, E. acidaminophilum_ encodes almost as many proteins as _C. litorale (Table S1), including some very similar core sporulation proteins (Galperin et al., 2012), see Table S3. Therefore, the lack of sporulation inE. acidaminophilum_ is likely due to a relatively recent loss of certain sporulation genes in that particular lineage. Based on these data, we propose reclassification of C. litorale andE. acidaminophilum into the new genus__Peptoclostridium_ as _Peptoclostridium litorale gen. nov. comb. nov. and _Peptoclostridium acidaminophilum comb. nov., respectively.
It must be noted that the current proposal of genus Peptoclostridium_ is substantially different from the one put forward previously by Yutin & Galperin (2013), which included all 15 validly described [__Clostridium_] members of thePeptostreptococcaceae. As noted above, 10 out of those 15 [__Clostridium] species have already been re-assigned to new genera within Peptostreptococcaceae(Gerritsen et al., 2014; Lawson et al., 2016; Sasi Jyothsna et al., 2016). The proposed renaming of [__Clostridium_]sticklandii is discussed below. Two other species,__Clostridium paradoxum and _Clostridium thermoalcaliphilum, consistently cluster together and are weakly linked to Tepidibacter_ species (seeFigs 1 and 2 and Gerritsen et al. (2014); Sasi Jyothsna et al., 2016); they might have to be reassigned to a separate genus. C. hiranonis_ does not fit into any of the current genera and will probably have to be elevated to the genus level as well. Of the other [__Eubacterium_] members of thePeptostreptococcaceae_, _E. tenue is closely related toPaeniclostridium ghonii_ andPaeniclostridium sordellii_ (Wade, 2009; Sasi Jyothsna et al., 2016) and clearly falls within the genus__Paeniclostridium. The misnamedE. yurii fits into the recently proposed genus__Peptoanaerobacter_ (Sizova et al., 2015). ‘__Clostridium dakarense’ (Lo et al., 2013) is related to the members of genusRomboutsia and could be assigned to that genus (Fig. 1).
Among the [Clostridium] species that have not been validly described or deposited in microbial culture databases, ‘Clostridium venationis_’ (16S rRNA gene GenBank accession number EU089966, 99 % identical to that of the type strain ofC. mangenotii) is a candidate for inclusion into the genus Clostridioides (Fig. 1). According to the GenBank entry, this organism was a psychrotolerant, spore-forming, proteolytic bacterium isolated from a meat processing facility. A similar organism has been reportedly isolated from an anaerobically enriched uranium-contaminated soil sediment (Yang et al., 2012). These two isolates substantially expand the ecological range of Clostridioides_ species. Of other [__Clostridium_] species known only by their 16S rRNA gene entries, ‘Clostridiummaritimum_’ (EU089965) and ‘Clostridium ruminantium_’ (EU089964 andKJ722512) fall within the genus _Romboutsia, while ‘_Clostridium metallolevans_’ (DQ133569,Meyer et al., 2007) belongs to__Terrisporobacter_ (Fig. 1).
It has been previously noted that [Clostridium]sticklandii strain DSM 519T is closely related toC. difficile (Stadtman & McClung, 1957; Fonknechten_et al._, 2010). However, on the 16S rRNA gene-based tree, this strain falls within the radiation of the genus _Acetoanaerobium and next to the _Proteocatella and Filifactor branches (Fig. 1 and Bes et al., 2015; Scaria et al., 2015). Accordingly, on the ribosomal protein tree that did not have any representatives of Acetoanaerobium, it clustered with Proteocatella sphenisci_ (Fig. 2). Therefore, we formally propose reclassification of [__Clostridium_] sticklandii asAcetoanaerobium sticklandii comb. nov. It should be noted that the assignment of the genus _Acetoanaerobium to familyPeptostreptococcaceae eliminates the need for Family XIX Incertae Sedis, created in the second edition of Bergey’s (Ludwig et al., 2009a). A summary of the proposed name changes and an updated nomenclature of the family _Peptostreptococcaceae are provided in Tables S4 and S5.
Description of Peptoclostridium gen. nov.
Peptoclostridium[Pep.to.clos.tri′di.um. Gr. v. _peptô_ digest; N.L. neut. dim. n. __Clostridium a bacterial genus name (from Gr. n. klôstêr a spindle); N.L. neut. dim. n.Peptoclostridium_ the digesting clostridium].
Obligately anaerobic, motile, straight or slightly curved rods, 0.5–1.5×2–8 µm, metabolizing amino acids and oligopeptides but not carbohydrates. Gram-staining variable, Gram-positive-type cell wall contains_meso_-diaminopimelate. Cells grow at 15–40 °C, no growth at 42 °C; pH range is from 6.5 to 8.4, optimum pH is 7.1–7.4, can grow in defined media containing biotin using glycine or serine as sole carbon and energy source. Betaine and sarcosine (N_-methylglycine) can be used as carbon and energy sources in the presence of electron donors, such as H2 or amino acids alanine, leucine, isoleucine, valine or phenylalanine. Utilization of glycine is via Stickland reaction catalysed by the glycine reductase complex and requires selenium (Dietrichs et al., 1991). Glycine is metabolized to acetate, CO2 and NH3; serine is metabolized to acetate, ethanol, CO2, H2 and NH3. In complex media, acetate and butyrate are the major fermentation products. Growth is stimulated by low amounts of NaCl but completely inhibited by 6 % NaCl. Oxidase and catalase negative. Sulfate, thiosulfate and nitrate are not reduced. Cultured representatives of Peptoclostridium_ have been isolated from anaerobic mud in wastewater and marine sediments. Metagenomics analyses detected 16S rRNA genes from potential members of this genus in microbial consortia performing anaerobic dechlorination of hexachlorobenzene and polychlorinated biphenyls and dibenzofurans (Yoshida et al., 2005; Ho & Liu, 2011; Zhou et al., 2015), an anaerobic cellulolytic microbial consortium from mangrove soil (Gao et al., 2014), as well as in a methanogenic bioreactor degrading terephthalate (Nobu et al., 2015). The G+C content of genomic DNA ranges from 41.3 to 44.0 mol%. The type species is _Peptoclostridium litorale (basonymClostridium litorale Fendrich _et al._1991).
Description of Peptoclostridium litorale comb. nov.
_Peptoclostridium litorale(li.to.ra′le. L. neut. adj. litorale coastal, referring to the source of the organism).
Basonym: _Clostridium litorale Fendrich _et al._1991.
The description of Peptoclostridium litorale is identical to that provided forClostridium litorale (Fendrich et al., 1990; Rainey et al., 2009). In addition to those described for the genus, has the following distinguishing properties. Cells stain Gram-negative. Form ovoid subterminal spores 1.5–2.0 µm in diameter. Colonies are brown, circular, with irregular margins.
The type strain W6T=ATCC 49638T=DSM 5388T was isolated from anoxic marine sediment near the coast of the Jade Bay (Jadebusen) of the North Sea in Germany (Fendrich et al., 1990). Sequence data from a whole-genome sequencing project (Poehlein et al., 2014a) are available in GenBank accession no. JJMM01000000. The G+C content of the type strain chromosomal DNA was originally reported as 26.1 mol% (Fendrich et al., 1990) but genome sequencing gave a much higher figure of 41.3 mol% (Poehlein et al., 2014a).
Description of Peptoclostridium acidaminophilum comb. nov.
_Peptoclostridium acidaminophilum(a.cid.a.mi.no′phi.lum. N.L. neut. adj. acidaminophilum loving amino acids).
Basonym: _Eubacterium acidaminophilum Zindel et al. 1989.
The description of Peptoclostridium acidaminophilum is identical to that forEubacterium acidaminophilum (Zindel et al., 1988; Rainey et al., 2009). In addition to those described for the genus, has the following distinguishing properties. Cells stain Gram-positive but behave Gram-negative in the KOH test. Spores are not observed. At least 1–2 mM NaCl are required for growth. Can utilize glycylglycine, glycylglycylglycine, glutathione and hydantoic acid.
The type strain al-2T=ATCC 49065T=DSM 3953T was isolated from anaerobic black mud from a waste water ditch near Konstanz, Germany (Zindel et al., 1988). The complete genomic sequence of the type strain has been determined (Poehlein et al., 2014b) and is available in GenBank accession no. CP007452. The G+C content of the type strain chromosomal DNA is 44.0 mol%.
Description of Acetoanaerobium sticklandii comb. nov.
_Acetoanaerobium sticklandii(stick.lan′di.i N.L. gen. masc. n. sticklandii named after British biochemist Leonard Hubert Stickland, who described a key reaction in clostridial amino acid metabolism).
Basonym: _Clostridium sticklandii Stadtman & McClung (1957) (Approved Lists 1980).
The description of Acetoanaerobium sticklandii_ is identical to that provided earlier for _Clostridium sticklandii (Stadtman & McClung, 1957; Rainey_et al._, 2009). The type strain is ATCC 12662T=DSM 519T=JCM 1433T, isolated from black mud from the east shore of San Francisco Bay (Stadtman & Barker, 1951). A detailed comparison of this strain withAcetoanaerobium noterae_ NOT-3T(=ATCC 35199T) and _Acetoanaerobium pronyense ST07-YET (=DSM 27512T) has been published (Bes et al., 2015). The complete genome sequence of the type strain is available (Fonknechten et al., 2010); its G+C content is 33.3 mol%.
Acknowledgements
We thank Drs Aharon Oren (The Hebrew University of Jerusalem), Boris Belitsky (Tufts University) and Sean Turner (NCBI) for helpful comments and Drs Yuri Wolf and Eugene Koonin (NCBI) for advice on phylogenetic trees. This work was supported by the NIH Intramural Research Program at the U.S. National Library of Medicine.
Supplementary Data
Supplementary File 1
References
- Baena S., Fardeau M. L., Woo T. H., Ollivier B., Labat M., Patel B. K.(1999). Phylogenetic relationships of three amino-acid-utilizing anaerobes, Selenomonas acidaminovorans, ‘Selenomonas acidaminophila_’ and Eubacterium acidaminophilum, as inferred from partial 16S rDNA nucleotide sequences and proposal of_Thermanaerovibrio acidaminovorans gen. nov., comb. nov. and_Anaeromusa acidaminophila_ gen. nov., comb. nov. Int J Syst Bacteriol 49969–974. 10.1099/00207713-49-3-969 [DOI] [PubMed] [Google Scholar]
- Bes M., Merrouch M., Joseph M., Quéméneur M., Payri C., Pelletier B., Ollivier B., Fardeau M. L., Erauso G., Postec A.(2015). Acetoanaerobium pronyense sp. nov., an anaerobic alkaliphilic bacterium isolated from a carbonate chimney of the Prony Hydrothermal Field (New Caledonia). Int J Syst Evol Microbiol 652574–2580. 10.1099/ijs.0.000307 [DOI] [PubMed] [Google Scholar]
- Brisbarre N., Fardeau M. L., Cueff V., Cayol J. L., Barbier G., Cilia V., Ravot G., Thomas P., Garcia J. L., Ollivier B.(2003). Clostridium caminithermale sp. nov., a slightly halophilic and moderately thermophilic bacterium isolated from an Atlantic deep-sea hydrothermal chimney. Int J Syst Evol Microbiol 531043–1049. 10.1099/ijs.0.02471-0 [DOI] [PubMed] [Google Scholar]
- Chen S., Song L., Dong X.(2006). Sporacetigenium mesophilum gen. nov., sp. nov., isolated from an anaerobic digester treating municipal solid waste and sewage. Int J Syst Evol Microbiol 56721–725. 10.1099/ijs.0.63686-0 [DOI] [PubMed] [Google Scholar]
- Collins M. D., Lawson P. A., Willems A., Cordoba J. J., Fernandez-Garayzabal J., Garcia P., Cai J., Hippe H., Farrow J. A.(1994). The phylogeny of the genus_Clostridium_: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44812–826. 10.1099/00207713-44-4-812 [DOI] [PubMed] [Google Scholar]
- Deloger M., El Karoui M., Petit M. A.(2009). A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol 19191–99. 10.1128/JB.01202-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrichs D., Meyer M., Rieth M., Andreesen J. R.(1991). Interaction of selenoprotein PA and the thioredoxin system, components of the NADPH-dependent reduction of glycine in Eubacterium acidaminophilum and Clostridium litorale. J Bacteriol 1735983–5991. 10.1128/jb.173.19.5983-5991.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C.(2004). muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 321792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S.(2012). The NCBI Taxonomy database. Nucleic Acids Res 40D136–D143. 10.1093/nar/gkr1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S.(2015). Type material in the NCBI Taxonomy Database. Nucleic Acids Res 43D1086–D1098. 10.1093/nar/gku1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrich C., Hippe H., Gottschalk G.(1990). Clostridium halophilium sp. nov. and C. litorale sp. nov., an obligate halophilic and a marine species degrading betaine in the Stickland reaction. Arch Microbiol 154127–132. 10.1007/BF00423321 [DOI] [Google Scholar]
- Fonknechten N., Chaussonnerie S., Tricot S., Lajus A., Andreesen J. R., Perchat N., Pelletier E., Gouyvenoux M., Barbe V., et al. (2010). Clostridium sticklandii, a specialist in amino acid degradation:revisiting its metabolism through its genome sequence. BMC Genomics 11555. 10.1186/1471-2164-11-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Mekhedov S. L., Puigbo P., Smirnov S., Wolf Y. I., Rigden D. J.(2012). Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 142870–2890. 10.1111/j.1462-2920.2012.02841.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z. M., Xu X., Ruan L. W.(2014). Enrichment and characterization of an anaerobic cellulolytic microbial consortium SQD-1.1 from mangrove soil. Appl Microbiol Biotechnol 98465–474. 10.1007/s00253-013-4857-2 [DOI] [PubMed] [Google Scholar]
- Gerritsen J., Fuentes S., Grievink W., van Niftrik L., Tindall B. J., Timmerman H. M., Rijkers G. T., Smidt H.(2014). Characterization of_Romboutsia ilealis_ gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera_Romboutsia_ gen. nov., Intestinibacter gen. nov., Terrisporobacter gen. nov. and_Asaccharospora_ gen. nov. Int J Syst Evol Microbiol 641600–1616. 10.1099/ijs.0.059543-0 [DOI] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M.(2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 5781–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O.(2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Ho C. H., Liu S. M.(2011). Effect of coplanar PCB concentration on dechlorinating microbial communities and dechlorination in estuarine sediments. Chemosphere 8248–55. 10.1016/j.chemosphere.2010.10.012 [DOI] [PubMed] [Google Scholar]
- Lawson P. A., Rainey F. A.(2016). Proposal to restrict the genus_Clostridium_ (Prazmowski) to Clostridium butyricum and related species. Int J Syst Evol Microbiol 661009–1016. 10.1099/ijsem.0.000824 [DOI] [PubMed] [Google Scholar]
- Lawson P. A., Citron D. M., Tyrrell K. L., Finegold S. M.(2016). Reclassification of_Clostridium difficile_ as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 4095–99. 10.1016/j.anaerobe.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Lo C. I., Mishra A. K., Padhmanabhan R., Samb B., Sow A. G., Robert C., Couderc C., Faye N., Raoult D., et al. (2013). Non-contiguous finished genome sequence and description of Clostridium dakarense sp. nov. Stand Genomic Sci 914–27. 10.4056/sigs.4097825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Schleifer K.-H., Whitman W. B.(2009a). Taxonomic outline of the phylum Firmicutes. Bergey's Manual of Systematic Bacteriology, 2nd edn,Vol 3: The Firmicutes 15–17. Edited by De Vos P., Garrity G. M., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H., Whitman W. B.New York: Springer. [Google Scholar]
- Ludwig W., Schleifer K.-H., Whitman W. B.(2009b). Revised road map to the phylum Firmicutes. Bergey's Manual of Systematic Bacteriology, 2nd edn,Vol 3: The Firmicutes 1–16. Edited by De Vos P., Garrity G. M., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H., Whitman W. B.New York: Springer. [Google Scholar]
- Meyer J., Schmidt A., Michalke K., Hensel R.(2007). Volatilisation of metals and metalloids by the microbial population of an alluvial soil. Syst Appl Microbiol 30229–238. 10.1016/j.syapm.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Nobu M. K., Narihiro T., Rinke C., Kamagata Y., Tringe S. G., Woyke T., Liu W. T.(2015). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 91710–1722. 10.1038/ismej.2014.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. T., Garrity G. M., Tindall B. J.(2015). International code of nomenclature of prokaryotes. Int J Syst Evol Microbiol 20 10.1099/ijsem.0.000778 [DOI] [Google Scholar]
- Parte A. C.(2014). LPSN–list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42D613–D616. 10.1093/nar/gkt1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuta E. V., Itoh T., Krader P., Tang J., Whitman W. B., Hoover R. B.(2006). Anaerovirgula multivorans gen. nov., sp. nov., a novel spore-forming, alkaliphilic anaerobe isolated from Owens Lake, California, USA. Int J Syst Evol Microbiol 562623–2629. 10.1099/ijs.0.64198-0 [DOI] [PubMed] [Google Scholar]
- Pikuta E. V., Hoover R. B., Marsic D., Whitman W. B., Lupa B., Tang J., Krader P.(2009). Proteocatella sphenisci gen. nov., sp. nov., a psychrotolerant, spore-forming anaerobe isolated from penguin guano. Int J Syst Evol Microbiol 592302–2307. 10.1099/ijs.0.002816-0 [DOI] [PubMed] [Google Scholar]
- Poehlein A., Alghaithi H. S., Chandran L., Chibani C. M., Davydova E., Dhamotharan K., Ge W., Gutierrez-Gutierrez D. A., Jagirdar A., et al. (2014a). First insights into the genome of the amino acid-metabolizing bacterium Clostridium litorale DSM 5388. Genome Announc 2e00754-14. 10.1128/genomeA.00754-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlein A., Andreesen J. R., Daniel R.(2014b). Complete genome sequence of amino acid-utilizing Eubacterium acidaminophilum al-2 (DSM 3953). Genome Announc 2e00573-14. 10.1128/genomeA.00573-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlein A., Cebulla M., Ilg M. M., Bengelsdorf F. R., Schiel-Bengelsdorf B., Whited G., Andreesen J. R., Gottschalk G., Daniel R., Dürre P.(2015). The complete genome sequence of_Clostridium aceticum_: a missing link between Rnf- and cytochrome-containing autotrophic acetogens. MBio 6e01168-15. 10.1128/mBio.01168-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey F. A., Hollen B. J., Small A.(2009). Genus I.Clostridium. Bergey's Manual of Systematic Bacteriology, 2nd edn,Vol. 3: The Firmicutes 738–828. Edited by De Vos P., Garrity G. M., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H., Whitman W. B.New York: Springer. [Google Scholar]
- Rainey F. A., Hollen B. J., Small A. M.(2015). Clostridium. Bergey's Manual of Systematics of Archaea and Bacteria, 1–122. Edited by DeVos P., Chun J., Dedysh S., Hedlund B., Kämpfer P., Rainey F. A., Trujillo M., Whitman W. B.John Wiley & Sons, Inc. [Google Scholar]
- Sasi Jyothsna T. S., Tushar L., Sasikala C., Ramana C. V.(2016). Paraclostridium benzoelyticum gen. nov., sp. nov., isolated from marine sediment and reclassification of Clostridium bifermentans as_Paraclostridium bifermentans_ comb. nov. Proposal of a new genus Paeniclostridium gen. nov. to accommodate_Clostridium sordellii_ and Clostridium ghonii. Int J Syst Evol Microbiol 661268–1274. 10.1099/ijsem.0.000874 [DOI] [PubMed] [Google Scholar]
- Scaria J., Suzuki H., Ptak C. P., Chen J. W., Zhu Y., Guo X. K., Chang Y. F.(2015). Comparative genomic and phenomic analysis of Clostridium difficile and Clostridium sordellii, two related pathogens with differing host tissue preference. BMC Genomics 16448. 10.1186/s12864-015-1663-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova M. V., Chilaka A., Earl A. M., Doerfert S. N., Muller P. A., Torralba M., McCorrison J. M., Durkin A. S., Nelson K. E., Epstein S. S.(2015). High-quality draft genome sequences of five anaerobic oral bacteria and description of_Peptoanaerobacter_ stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae. Stand Genomic Sci 1037. 10.1186/s40793-015-0027-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman T. C., Barker H. A.(1951). Studies on the methane fermentation. X. A new formate-decomposing bacterium, Methanococcus vannielii. J Bacteriol 62269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman T. C., McClung L. S.(1957). Clostridium sticklandii nov. spec. J Bacteriol 73218–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M.(1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall B. J., Rosselló-Móra R., Busse H. J., Ludwig W., Kämpfer P.(2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60249–266. 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- Varghese N. J., Mukherjee S., Ivanova N., Konstantinidis K. T., Mavrommatis K., Kyrpides N. C., Pati A.(2015). Microbial species delineation using whole genome sequences. Nucleic Acids Res 436761–6771. 10.1093/nar/gkv657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade W. G.(2009). The Firmicutes. Bergey’s Manual of Systematic Bacteriology, 2nd edn,Vol. 3 865–891. Edited by De Vos P., Garrity G. M., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H., Whitman W. B.New York: Springer. [Google Scholar]
- Yang F., Tiedje J., Zhou J., Marsh T. L.(2012). Firmicutes and their roles in uranium immobilization. US Department of Energy Subsurface Biogeochemical Research Annual Meeting University-Led Research, 64 Washington, DC: U. S. Department of Energy Office of Science. [Google Scholar]
- Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K. H., Ludwig W., Glöckner F. O., Rosselló-Móra R.(2008). The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31241–250. 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K. H., Whitman W. B., Euzéby J., Amann R., Rosselló-Móra R.(2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12635–645. 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
- Yilmaz P., Parfrey L. W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F. O.(2014). The SILVA and ‘All-species Living Tree Project (LTP)’ taxonomic frameworks. Nucleic Acids Res 42D643–D648. 10.1093/nar/gkt1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Takahashi N., Hiraishi A.(2005). Phylogenetic characterization of a polychlorinated-dioxin- dechlorinating microbial community by use of microcosm studies. Appl Environ Microbiol 714325–4334. 10.1128/AEM.71.8.4325-4334.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Puigbò P., Koonin E. V., Wolf Y. I.(2012). Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7e36972. 10.1371/journal.pone.0036972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Galperin M. Y.(2013). A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 152631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhang C., Zhang D., Awata T., Xiao Z., Yang Q., Katayama A.(2015). Polyphasic characterization of an anaerobic hexachlorobenzene-dechlorinating microbial consortium with a wide dechlorination spectrum for chlorobenzenes. J Biosci Bioeng 12062–68. 10.1016/j.jbiosc.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Zindel U., Freudenberg W., Rieth M., Andreesen J. R., Schnell J., Widdel F.(1988). Eubacterium acidaminophilum sp. nov., a versatile amino acid-degrading anaerobe producing or utilizing H2 or formate. Description and enzymatic studies. Arch Microbiol 150254–266. 10.1007/BF00407789 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1