Properties of overlapping genes are conserved across microbial genomes (original) (raw)
Abstract
There are numerous examples from the genomes of viruses, mitochondria, and chromosomes that adjacent genes can overlap, sharing at least one nucleotide. Overlaps have been hypothesized to be involved in genome size minimization and as a regulatory mechanism of gene expression. Here we show that overlapping genes are a consistent feature (approximately one-third of all genes) across all microbial genomes sequenced to date, have homologs in more microbes than do non-overlapping genes, and are therefore likely more conserved. In addition, the size, phase (reading frame offset), and distribution, among other characteristics, of overlapping genes are most consistent with the hypothesis that overlaps function in the regulation of gene expression. The upstream sequences and conservation of overlapping orthologs of two model organisms from the genus Prochlorococcus that have significantly different GC-content, and therefore different nucleotide sequences for orthologs, are also consistent with small overlapping sequence regions and programmed shifts in reading frame as a common mechanism in the regulation of microbial gene expression.
Genomes are often compartmentalized into coding regions (genes) and noncoding spacer regions. Although functionally related genes can be closely located, such as in an operon, the fundamental unit of a gene is generally defined as having a unique position within a chromosome. However, early sequencing efforts demonstrated that individual genes can overlap or share one or more nucleotides with adjacent genes (Barrell et al. 1976; Sanger et al. 1977). Originally discovered in viruses, mitochondria, and other extrachromosomal nuclear elements, these overlapping genes, which have been shown to be evolutionarily stable, were originally thought to be the result of evolutionary pressure to conserve sequence length (Miyata and Yasunaga 1978; Krakauer 2000; Scherbakov and Garber 2000). More recently, overlaps have been identified in chromosomal DNA of microbes and higher organisms (Spencer et al. 1986; Williams and Fried 1986; Wellington et al. 1992; Fukuda et al. 1999). Although there is some speculation that they may be rare in nonviral genomes (Krakauer and Plotkin 2002), overlaps have been demonstrated to be potentially important in transcriptional and translational regulators of gene expression and to influence the evolution of genes (Keese and Gibbs 1992; Krakauer 2002). Here we seek to understand the prevalence and properties of overlapping genes across all sequenced microbial genomes, which because of their small size may be influenced by physical size constraints (National Research Council 1999). The characteristics of the overlaps offer insight into the functional role of overlaps in microbes and possibly other organisms.
Results and Discussion
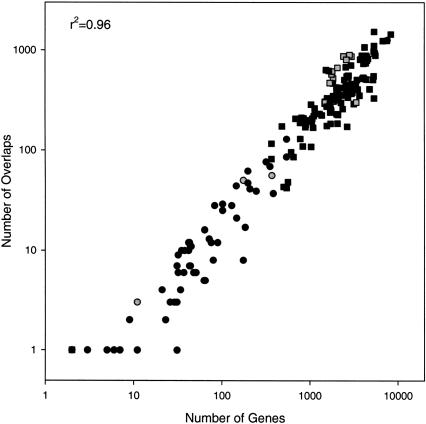
During the genome annotation process, microbial genomes are assigned to coding and noncoding regions by using gene searching models such as Critica, Glimmer, or Generation or by using manual assignment often based on similarity indices such as BLAST (Altschul et al. 1997; Salzberg et al. 1998; Badger and Olsen 1999; http://compbio.ornl.gov/generation). From these defined coding regions, we identified overlapping genes as adjacent genes, located on either DNA strand, that share one or more nucleotides in their coding sequence (CDS). For all publicly available microbial genomes and annotations (ftp://ftp.ncbi.nih.gov/genomes/Bacteria), we find a strong (r2 = 0.96) relationship between the total number of genes and the number of overlapping genes (Fig. 1), which is similar to that found by others for smaller multiorganism data sets (Fukuda et al. 2003) and also consistent with overlap frequencies for single organisms (Fukuda et al. 1999; Scherbakov and Garber 2000; Iwabe and Miyata 2001). As would be expected, there is a similarly robust relationship between the number of overlapping genes and genome size (base pairs) as well (data not shown) because there is a strong relationship between the number of base pairs and number of genes for microbes (Fukuda et al. 2003). The relationship between overlap and gene number is consistent across Eubacteria and Archaebacteria domains as well as for plasmids and chromosomal DNA, suggesting that overlapping genes are a consistent feature across microbial genomes and their extrachromosomal elements for different lineages, occupying diverse environments.
Figure 1.
Number of overlaps versus the total number of genes in the genome for all genomes in the National Center for Biotechnology Information bacteria database (as of May 3, 2003). Plasmids (circles) and chromosomes (squares) are identified by Eubacteria (black) or Archaebacteria (gray) domains. Properties are correlated at r2 = 0.96.
The slope of the linear relationship between the number of genes and overlapping gene pairs (overlaps) is 0.16, which indicates that approximately a third of all genes in the genomes are overlapping because two genes are part of each overlap. It is possible that some of these overlaps could result from misidentification of coding sequences in the annotation process. If this were the case, we would expect a higher percentage of overlaps in genes labeled “hypothetical.” To investigate this, we subanalyzed these hypothetical genes, which canonically connote coding assignments with less statistical confidence. Hypothetical genes account for 53% of all annotated genes and are somewhat less likely to overlap (P = 0.26) than are the remaining genes. Other properties of the overlaps, such as size, phasing, and direction, are similar between hypothetical and nonhypothetical groups. This evidence, as well as other COG analyses, suggests that putative overlapping genes are not the result of misannotation (Fukuda et al. 2003).
We explored whether the relative prevalence of overlapping genes is related to whole genome features. We found that the frequency of overlapping genes is not related to genome compactness, as determined from both the average distance between genes (r2 = 0.02, model 2 least-squares linear regression) and the genome coding percentage (r2 = 0.05, model 2) (Supplemental Fig. 1). The frequency of overlapping genes is also not related to the percent GC content of the genome (r2 = 0.05, model 2), and the overlapping sequences are not statistically different in GC content from non-overlapping sequence within a given genome (P > 0.2). However, based on phylogenetic profiles, genes that overlap have homologs in more organisms (13% increase, P < 0.001) than do non-overlapping genes. Thus, overlaps do not appear to be directly related to GC content or reducing genome size, and it appears that overlapping genes are more likely to be conserved. This tendency toward conservation may be the result of shared information (shared base pairs) resulting in an increased probability of inheritance because each mutation can potentially negatively affect two genes, or the overlap feature itself may have adaptive fitness advantages.
Previous work has shown that overlaps for a given organism can have common characteristics such as strand distribution (Fukuda et al. 1999), phasing (Scherbakov and Garber 2000), codon usage (Fukuda et al. 1999; Kozlov 2000), or size (Merino et al. 1994; Iwabe and Miyata 2001), among others. We investigated these and related properties over all genomes to determine if they are globally conserved. Overlapping genes are distributed irregularly within a genome such that the majority (84%) of overlapping genes occur on the same strand (tandem overlaps, →→) with the remaining (16%) overlaps occurring on opposite DNA strands (antiparallel overlaps, →← or ←→) (Table 1.) This global pattern across microbial genomes is consistent with previous findings for individual organisms and smaller multiple organism data sets showing that antiparallel overlapping genes can be relatively rare (Fukuda et al. 1999, 2003). Tandem and antiparallel overlaps also have markedly different phase (reading frame offset) distributions (Supplemental Fig. 2). Antiparallel overlaps are basically evenly distributed among the three reading frames, whereas tandem overlaps are most common in the +1 (2 + 3_n_ shared bases) and +2 (1 + 3_n_ shared bases) reading frames. In-phase (same reading frame) tandem overlaps, which would require an evolutionarily unstable stop codon read-through (Keese and Gibbs 1992; Krakauer 2000), are exceedingly rare.
Table 1.
Frequency of reading frame offset for the 64,989 overlaps identified in all microbial genomes.
| Phase offset (reading frame shift) | |||
|---|---|---|---|
| Direction | 0 | +1/-1 | +2/-2 |
| Tandem (→→) | 0.001 | 0.259 | 0.578 |
| Antiparallel (→←,←→) | 0.041 | 0.062 | 0.060 |
The unequal distribution among the different directions and reading frame offsets of overlaps across all microbes suggests that there are selective pressures maintaining this pattern. Krakauer (2000) has used an in silico analysis that assigns a probable abundance based on the level of selective independence derived from the plasticity in nucleotide sequence while maintaining a given amino acid sequence. These predictions, which are based on shared information content (IC) alone, suggest the following distribution: antiparallel –2 reading frame shift (1 + 3_n_ shared bases) > tandem +1 (2 + 3n shared bases) and +2 (1 + 3n shared bases) shifts > in-phase antiparallel > antiparallel –1 shift (2 + 3_n_ shared bases) (Krakauer 2000). However, we find that there are significantly more +1 and +2 shift tandem overlaps than for the antiparallel case, suggesting that additional mechanisms must be acting (Table 1). Furthermore, there are similar numbers of antiparallel overlaps in all reading frames, also suggesting that shared IC is probably not the sole mechanism producing the observed distributions.
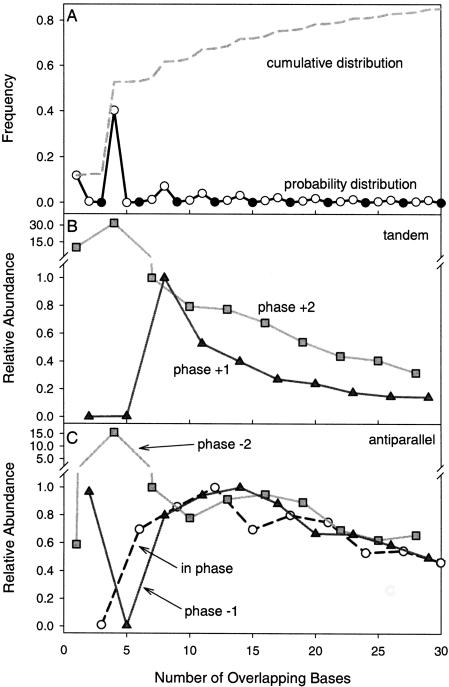
In addition to phasing, the size distribution of overlaps is skewed with most overlaps occurring in smaller size classes. More than 70% of overlaps are <15 bp, and >85% are <30 bp (Fig. 2). However, this distribution is not smooth as the direction and reading frame shift of the overlap affect this distribution dramatically. Notably, there are a few special cases that have a disproportionately large number of members. For example, the majority of the +2/–2 shift (1 + 3_n_ shared bases) overlaps are either single (binary) or four-base overlaps (Fig. 2). The majority of these small +2/–2 shift observations occur for tandem overlaps, but this pattern is also generally true for the less frequent antiparallel genes. Likely because of IC constraints (number of shared bases), longer sizes of +2/–2 phase overlaps are significantly less common and become monotonically rarer as the length of overlap increases. For binary and four-base overlaps, there is a relatively small IC constraint, associated with the overlapping sequence. For example, for tandem binary overlaps the leading stop codon for upstream sequence must either be TAA (TA[A]TG) or TGA (TG[A]TG). This only prohibits the upstream sequence from using TAG as stop codon and therefore represents a relatively small IC constraint. For four-base overlaps, tandem overlaps ([ATGA]) allow the second to last amino acid in the upstream sequence to be one of 12 amino acids (GALKQESPVIRT) and the second amino acid in the downstream sequence to be one of seven different amino acids (MNKITSR) (Kozlov 2000). Both of the groups, which contain hydrophobic, hydrophilic, and other amino acids, impose a small IC constraint on the final amino acid makeup of the two proteins. Similarly, four-base antiparallel overlapping genes, which are restricted to end-on overlaps (→←), have the sequence ([T/CTAA/G]) with 14 amino acids possible. This arrangement also permits a significant degree of flexibility in the second to last amino acid in the sequence with no effect on the secondary sequence (and vice versa). As before, this imposes a relatively small IC constraint and likely accounts for their large numbers. Aside from the unique one and four-base overlaps in the +2/–2 shift group, the number of observations decreases as the length of the overlap increases, likely due to increasing IC constraints (Fig. 2). In short, tandem and antiparallel binary and four-base overlaps (phase +2/–1 overlaps) make up 52% of all overlaps most likely because they have relatively small IC constraints and permit flexibility in the final amino acid composition.
Figure 2.
(A) Size frequency distribution of overlapping genes. Sizes, which are in-frame (i.e., multiples of three), are indicated with filled circles and out-of-phase overlaps have open circles, both with a solid line. The cumulative distribution is plotted as a dashed line. Individual tandem and antiparallel graphs (data not shown) have nearly identical patterns. (B) Distribution of tandem overlaps relative to respective phase maxima above six bases. Tandem in-phase observations were excluded because of few observations. (C) Distribution of antiparallel overlaps relative to respective phase maxima above six bases.
Similar to overlaps with +2/–2 shifts in reading frame, +1/–1 shifted overlaps (2 + 3_n_ shared bases) are generally most prevalent at smaller overlap sizes but also have some special cases (Fig. 2). There are only a few observations of tandem +1 shifted overlaps at 2 or 5 bp because in that orientation and reading frame shift start and stop codon sequence requirements prohibit an evolutionarily stable arrangement. At eight bases and greater, however, there are a significant number of overlaps. The frequency of this tandem +1 shifted overlap decreases sharply with overlap size due to increases in shared IC. Antiparallel –1 shifted overlaps have a similar pattern, again with the 5-bp overlap prohibited. In terms of size distribution, the antiparallel –1 shifted overlaps are relatively constant until they start to decrease monotonically at 17 bases. But overall, as with the other overlaps with shifts in reading frame, +1/–1 shifted overlaps are generally small and are less frequent at longer lengths.
Regardless of precise shift in reading frame, the majority of overlapping genes overlap by only a few base pairs, many of which are the result of unique scenarios that permit a great deal of flexibility in the final amino acid composition of both proteins. The majority of the remaining overlaps are also relatively short, with >80% of the total observations overlapping by <30 bp. The observation that the distribution of overlaps is dramatically skewed toward smaller sizes suggests that a reduction in genome size is not a primary mechanism producing and maintaining overlaps. Alternatively, these short overlapping sequences may be involved in expression regulatory mechanisms (Normark et al. 1983; Krakauer 2000, 2002; Scherbakov and Garber 2000). Short tandem overlapping sequences can use translational coupling via site-specific programmed shifts in reading frame (PSRF) to express both of the overlapping genes in consistent stoichiometries or to allow inhibitory actions (Oppenheim and Yanofsky 1980; Das and Yanofsky 1989; Gesteland and Atkins 1996). Short antiparallel overlaps may also be involved in expression regulation. Compared with tandem overlaps, the relative abundance of antiparallel overlaps decreases less quickly as the length of the overlap increases for the first 20 bases, suggesting that some mechanism is maintaining longer antiparallel overlaps (Fig. 2). These relatively short antiparallel overlaps, which are each complementary to the other overlapping gene, may be involved in expression regulatory mechanisms involving RNA antisense effects such as microRNAs and short interfering RNAs that use small fragments of complementary sequence to regulate expression (Bartel and Bartel 2003; Carrington and Ambros 2003).
We investigated the general properties of overlapping genes further by using two closely related (as determined from 16S/23S/ITS sequences) microbial genomes from the marine cyanobacterium Prochlorococcus spp. (MIT9313 and MED4) that have significantly different %GC content (50.7 and 30.8, respectively) but have 1352 orthologs out of their total 2275 and 1716 respective gene complements as predicted by BLASTP with an e-value threshold set at 1e – 10 (Rocap et al. 2003). Out of the 1352 orthologs, which also are significantly different in %GC content (53.5 and 32.2, respectively), there are 422 and 330 orthologs that are part of an overlapping gene pair in MIT9313 and MED4, respectively. This is generally consistent with the ∼30% of total genes overlapping across all sequenced microbial genomes. Among orthologs that are part of an overlap pair, 292 genes from MIT9313 and 274 from MED4 genes (69% and 83%, respectively), have both members of the overlap pair as orthologs. Thus, the majority of overlapping gene pairs have dual-ortholog overlaps. This suggests that overlapping gene pairs are conserved among organisms for specific genes and function, despite significant differences in the actual base pair composition of the sequences from the differences in GC usage.
Other properties of the Prochlorococcus genomes are consistent with a regulatory role for overlaps. For example, immediately upstream of the stop codon in the leading overlapping gene, both MIT9313 and MED4 each contain multiple, but unique six-base motifs, including strings of adenines adjacent the stop codon in MED4. These strings of adenines, which are found in 19% of the overlapping genes, are twice as likely to be present for overlapping genes as for non-overlapping genes (10%). These conserved motifs and strings of repeating bases near the overlap regions are consistent with the PSRF mechanism and permit cotranslation of two tandem overlap genes (Scherbakov and Garber 2000).
We have shown that overlapping genes are abundant among all microbial genomes, but the number or frequency of overlaps is not related to genome compactness. Overlapping genes have more homologs among microbial genomes than do non-overlapping genes. This property may, in part, be due to their putative regulatory role as the dominant reading frame shifts and length of the overlaps suggests. Overlapping genes in Prochlorococcus spp., which are closely phylogenetically related yet have significantly different nucleotide sequences for orthologs, further support this role.
Because of the small physical size of the organisms, microbial genomes may be under increased pressure to minimize the size of their genome, but this study suggests that overlapping genes are not a direct mechanism to substantially reduce genome size. Nevertheless, regulation of gene expression by overlaps may result in more efficient control and reduce the need for more complex regulatory pathways. This reduction in turn could diminish the number of proteins (and genes) required and thus be indirectly involved in sequence conservation. Other patterns of genomic architecture may also aid in the transcriptional and translational regulation of protein expression and consequently enable microbial genomes to have compact sizes yet enable flexible and efficient physiologies.
Methods
Genomes, which here are defined as contiguous coding regions (plastids and chromosomes from the same organisms are separate “genomes”), were downloaded from the National Institutes of Health National Center for Biotechnology Information ftp server (ftp://ftp.ncbi.nih.gov/genomes/Bacteria) on May 3, 2003. This database included 198 genomes, which are detailed in Supplemental materials.
Only fully assembled genomes were used in analyses. Overlapping genes were defined as adjacent genes, on either strand, that had coding sequences (CDS regions) that shared one or more bases. Conserved genomic motifs among parallel overlapping genes were determined by using the online version of AlignACE 3.0 by searching for motifs within 15 bases of the stop codon of the leading sequence and by allowing seven columns to align and expecting seven sites (http://atlas.med.harvard.edu) (McGuire et al. 2000). Genome compactness was calculated as both the average distance between adjacent genes and the fraction of the genome assigned to genes. Gene conservation was estimated by creating protein–protein phylogenetic profiles by assigning each genome a one or zero based on a minimum BLASTP e-value cutoff of 1e – 5 and summing the ones and zeros for all genomes for each query gene (Altschul et al. 1997; Pellegrini et al. 1999). Other analyses were made by using custom written routines in the MATLAB (version 6.5) programming language.
Acknowledgments
This work was supported by grants from NSF and DOE to S.W.C. The authors would like to thank W. Hess and W.F. Doolittle who commented on an earlier version of the manuscript and the two anonymous reviewers and editors whose comments greatly improved the manuscript.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2433104.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**:** 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16**:** 512-524. [DOI] [PubMed] [Google Scholar]
- Barrell, B.G., Air, G.M., and Hutchison, C.A. 1976. Overlapping genes in bacteriophage-Psix174. Nature 264**:** 34-41. [DOI] [PubMed] [Google Scholar]
- Bartel, B. and Bartel, D.P. 2003. MicroRNAs: At the root of plant development. Plant Physiol. 132**:** 709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. 2003. Role of microRNAs in plant and animal development. Science 301**:** 336-338. [DOI] [PubMed] [Google Scholar]
- Das, A. and Yanofsky, C. 1989. Restoration of a translational stop-start overlap reinstates translational coupling in a mutant Trpb′-Trpa gene pair of the Escherichia-coli tryptophan operon. Nucleic Acids Res. 17**:** 9333-9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, Y., Washio, T., and Tomita, M. 1999. Comparative study of overlapping genes in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae. Nucleic Acids Res. 27**:** 1847-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, Y., Nakayama, Y., and Tomita, M. 2003. On dynamics of overlapping genes in bacterial genomes. Gene 323**:** 181-187. [DOI] [PubMed] [Google Scholar]
- Gesteland, R.F. and Atkins, J.F. 1996. Recoding: Dynamic reprogramming of translation. Ann. Rev. Biochem. 65**:** 741-768. [DOI] [PubMed] [Google Scholar]
- Iwabe, N. and Miyata, T. 2001. Overlapping genes in parasitic protist Giardia lamblia. Gene 280**:** 163-167. [DOI] [PubMed] [Google Scholar]
- Keese, P.K. and Gibbs, A. 1992. Origins of genes: Big-bang or continuous creation. Proc. Natl. Acad. Sci. 89**:** 9489-9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov, N.N. 2000. Analysis of a set of overlapping genes. Doklady Proc. Acad. Sci. USSR 373**:** 119-122. [PubMed] [Google Scholar]
- Krakauer, D.C. 2000. Stability and evolution of overlapping genes. Evolution 54**:** 731-739. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Evolutionary principles of genomic compression. Comments Theor. Biol. 7**:** 215-236. [Google Scholar]
- Krakauer, D.C. and Plotkin, J.B. 2002. Redundancy, antiredundancy, and the robustness of genomes. Proc. Natl. Acad. Sci. 99**:** 1405-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire, A.M., Hughes, J.D., and Church, G.M. 2000. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 10**:** 744-757. [DOI] [PubMed] [Google Scholar]
- Merino, E., Balbás, P., Puente, J.L., and Bolívar, F. 1994. Antisense overlapping open reading frames in genes from bacteria to humans. Nucleic Acids Res. 22**:** 1903-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata, T. and Yasunaga, T. 1978. Evolution of overlapping genes. Nature 272**:** 532-535. [DOI] [PubMed] [Google Scholar]
- National Research Council. 1999. Size limits of very small microorganisms: Proceedings of a workshop. National Academy Press, Washington, DC. [PubMed]
- Normark, S., Bergstrom, S., Edlund, T., Grundstrom, T., Jaurin, B., Lindberg, F.P., and Olsson, O. 1983. Overlapping genes. Ann. Rev. Genet. 17**:** 499-525. [DOI] [PubMed] [Google Scholar]
- Oppenheim, D.S. and Yanofsky, C. 1980. Translational coupling during expression of the tryptophan operon of Escherichia-coli. Genetics 95**:** 785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, M., Marcotte, E.M., Thompson, M.J., Eisenberg, D., and Yeates, T.O. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 96**:** 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap, G., Larimer, F.W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N.A., Arellano, A., Coleman, M., Hauser, L., and Hess, W.R. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424**:** 1042-1047. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L., Delcher, A.L., Kasif, S., and White, O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26**:** 544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger, F., Air, G.M., Barrell, B.G., Brown, N.L., Coulson, A.R., Fiddes, J.C., Hutchison, C.A., Slocombe, P.M., and Smith, M. 1977. Nucleotide-sequence of bacteriophage Phichi174 DNA. Nature 265**:** 687-695. [DOI] [PubMed] [Google Scholar]
- Scherbakov, D.V. and Garber, M.B. 2000. Overlapping genes in bacterial and phage genomes. Mol. Biol. 34**:** 485-495. [PubMed] [Google Scholar]
- Spencer, C.A., Gietz, R.D., and Hodgetts, R.B. 1986. Overlapping transcription units in the Dopa decarboxylase region of Drosophila. Nature 322**:** 279-281. [DOI] [PubMed] [Google Scholar]
- Wellington, C.L., Bauer, C.E., and Beatty, J.T. 1992. Photosynthesis gene superoperons in purple nonsulfur bacteria: The tip of the iceberg. Can. J. Microbiol. 38**:** 20-27. [Google Scholar]
- Williams, T. and Fried, M. 1986. A mouse locus at which transcription from both DNA strands produces messenger-RNAs complementary at their 3′ ends. Nature 322**:** 275-279. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genomes/Bacteria; National Center for Biotechnology Information ftp site for bacteria genomes.
- http://compbio.ornl.gov/generation/; (Generation) Microbial Gene Prediction System Home Page.
- http://atlas.med.harvard.edu; AlignACE motif-finding algorithm home page.