Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution (original) (raw)
Abstract
The platypus (2_n_ = 52) has a complex karyotype that has been controversial over the last three decades. The presence of unpaired chromosomes and an unknown sex-determining system especially has defied attempts at conventional analysis. This article reports on the preparation of chromosome-specific probes from flow-sorted chromosomes and their application in the identification and classification of all platypus chromosomes. This work reveals that the male karyotype has 21 pairs of chromosomes and 10 unpaired chromosomes (E1-E10), which are linked by short regions of homology to form a multivalent chain in meiosis. The female karyotype differs in that five of these unpaired elements (E1, E3, E5, E7, and E9) are each present in duplicate, whereas the remaining five unpaired elements (E2, E4, E6, E8, and E10) are absent. This finding indicates that sex is determined by the alternate segregation of the chain of 10 during spermatogenesis so that equal numbers of sperm bear either one of the two groups of five elements, i.e., five X and five Y chromosomes. Chromosome painting reveals that these X and Y chromosomes contain pairing (XY shared) and differential (X- or Y-specific) segments. Y differential regions must contain male-determining genes, and X differential regions should be dosage-compensated in the female. Two models for the evolution of the sex-determining system are presented. The resolution of the longstanding debate over the platypus karyotype is an important step toward the understanding of mechanisms of sex determination, dosage compensation, and karyotype evolution.
Keywords: monotremes, multivalent chain, X-inactivation, sex determination
The duck-billed platypus (Ornithorhynchus anatinus) is one of three extant species of the egg-laying monotremes whose ancestors are believed to have diverged from the mammalian lineage ≈210 million years ago and some 30 million years before the divergence of marsupials and placental mammals (1). Its unique mixture of mammalian, avian, and reptilian features (2) has excited biologists for more than two centuries. For geneticists, the platypus undoubtedly provides a special outgroup species because it represents the earliest offshoots of the mammalian lineage. Studies in monotremes can answer questions about sex determination, genomic imprinting, and dosage compensation in mammals. Accurate chromosome identification is crucial for these studies. The correct chromosome number of 2_n_ = 52 was determined for the platypus in 1975 (3). The karyotype was first considered to be reptilian (4), but this idea was subsequently refuted (5). However, difficulty was encountered in the analysis of the karyotype, as a number of chromosomes, variably assessed as between four and eight, appeared to be unpaired, a feature not seen in such numbers in any other mammal except the other two monotremes, the short- and long-beaked echidnas (6, 7). These unpaired chromosomes were believed to form a multivalent chain at meiosis. However, it remained controversial as to whether unpaired chromosomes, and therefore formation of a meiotic chain, occur in both sexes (3, 7, 8) or males only (9). The largest of the unpaired chromosomes was presumed to be the X as it was present in two copies in the female. Subsequently, the X was shown to carry several genes that are X-linked in mammals (10-12), although human Xp genes were mapped to platypus autosomes 1 and 2 (13), and the mammalian SRY testis-determining gene could not be found (14). The X chromosome clearly lay at one end of the chain (7, 8) and a tiny “parachute-like” element was at the other end (9). It was not possible to identify a Y chromosome unambiguously. The mechanism of sex determination in monotremes was a mystery.
Whether dosage compensation for the platypus X chromosome occurs has also been debated for decades. Semiquantitative PCR for three genes on the X shows that transcription from these loci is equivalent in both sexes (2), but could not distinguish between X inactivation and differential/reduced expression from both X chromosomes. Replication asynchrony between the two X chromosomes was described in echidna lymphocytes (7), but it appeared to be confined to the short arm (8), which is paired and should require no dosage compensation. No asynchrony was found in fibroblasts.
This study reports the sorting of male platypus chromosomes and production of chromosome-specific paint probes for all but the smallest unpaired chromosome. These paint probes identified 10 unpaired chromosomes in the male and regions of homology between them that explain their involvement in a multivalent chain. In the female five of these unpaired chromosomes are absent and the remaining five are present in duplicate. Thus the female karyotype does not contain any unpaired elements. The sex chromosome complement in the male is therefore heterozygous and interpreted as X1Y1X2Y2X3Y3X4Y4X5Y5. This remarkable finding has implications for sex chromosome evolution and X chromosome dosage compensation.
Materials and Methods
Primary fibroblast cultures from the toe web of the Australian platypus O. anatinus (2_n_ = 52) were established routinely in standard medium at 32°C (Animal Experimentation Ethics Committee Permit RCG.07.03 to F.G. and Environment Australian Capital Territory Permit LI 2002 270 to J.A.M.G.). Flow sorting, chromosome paint production, and fluorescence in situ hybridization were performed according to the protocol described (15). Chromosomes were prepared for sorting as described (16), with slight modification. Colcemid (0.1 μg/ml) was added to flasks of male platypus fibroblasts and incubated for only 30 min instead of the usual 4-6 h. Longer incubation times caused interphase cells to detach from the flasks. Mitotic cells were collected by carefully tapping the culture flasks and were centrifuged at 400 × g for 5 min. The cell pellet was resuspended in 10 ml of hypotonic KCl (75 mM KCl/0.2 mM spermine/0.5 mM spermidine) and allowed to swell for 1 h instead of the usual 15 min at room temperature. From this point onward the standard protocol was followed (15, 16).
The paints produced were hybridized to male (three individuals) and female (one individual) metaphase preparations. Images were captured by using qfish software (Leica Microsystems, Cambridge, U.K.) and a cooled charge-coupled device camera (Photometrics Sensys, Tucson, AZ) mounted on a Leica DMRXA microscope equipped with a ×63, 1.3 numerical aperture objective. Cy3, FITC, and 4′,6-diamidino-2-phenylindole (DAPI) signals were captured separately as 16-bit black and white images and merged to a color image. The DAPI image was enhanced with a spatial filter to obtain enhanced chromosome bands. GTG banding was performed by using standard protocols.
All image processing was performed with Leica CW4000 software.
Results
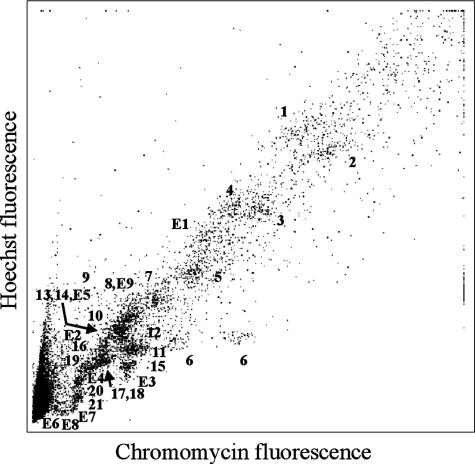
The aim of the analysis was the identification of all platypus chromosomes to resolve the controversial karyotype, in particular to identify the unpaired chromosomes and determine their homology relationships by using chromosome-specific paint probes produced from flow-sorted chromosomes. The modification of the normal sort preparation protocol achieved separation of almost all chromosomes in the flow karyotype (Fig. 1), and degenerate oligonucleotide primed-PCR provided chromosome-specific DNA free of contamination from other chromosomes. Only chromosomes 8 and E9, and chromosomes 13, 14, and E5, sorted together in two peaks and were combined in two paint probes. However, a unique Eq paint probe was prepared from a single sorted chromosome. Chromosomes 1-7, plus E1, are the larger chromosomes, and the nucleolar chromosome 6 is represented by two peaks caused by polymorphism of the “stalk” of this chromosome (see Fig. 2). Numbers 8-21 represent the medium and small chromosomes that cluster together at the lower end of the flow karyotype.
Fig. 1.
Flow karyotype of male O. anatinus. Almost all chromosomes are represented by individual peaks. Only chromosomes 8 and E9, and chromosomes 13, 14, and E5, sort together.
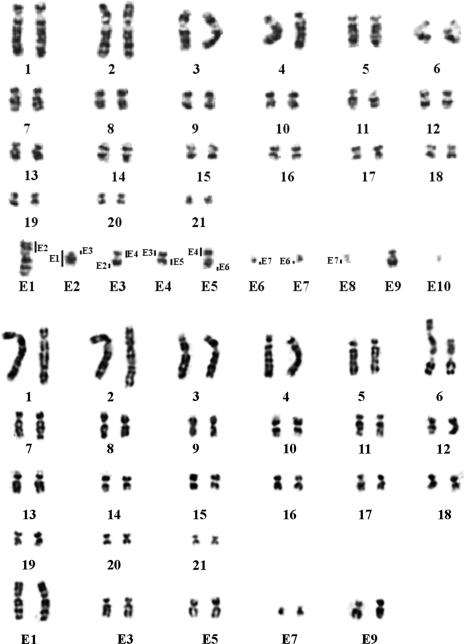
Fig. 2.
G-banded karyotypes of O. anatinus. (Upper) The male platypus (2_n_ = 52) has 21 chromosome pairs and 10 unpaired chromosomes labeled E1-E10. The order of these chromosomes determined by their pairing regions is indicated by vertical bars. (Lower) G-banded female karyotype of O. anatinus. There are no unpaired chromosomes.
Fig. 2 Upper is the G-banded karyotype of a male platypus, in which each chromosome has been identified by using the chromosome-specific paints (see Fig. 3 for examples). The upper part of the karyotype shows the 21 paired chromosomes that form bivalents at meiosis. Chromosomes 1-7 can be identified easily by their size, centromere position, and G bands. Chromosomes 1 and 4 have large light-stained centromeric regions corresponding to heterochromatin; chromosomes 2 and 3 have dark-stained centromeres. Chromosome 6 has long satellite arms, which are heteromorphic, and carry the nucleolus organizer regions (8). Chromosomes 8-21 are much harder to distinguish by morphology and G banding and remained unclassified in previous reports. Chromosome painting classified these chromosomes and established their characteristic patterns of G banding (or reverse 4′,6-diamidino-2-phenylindole banding) and centromere position.
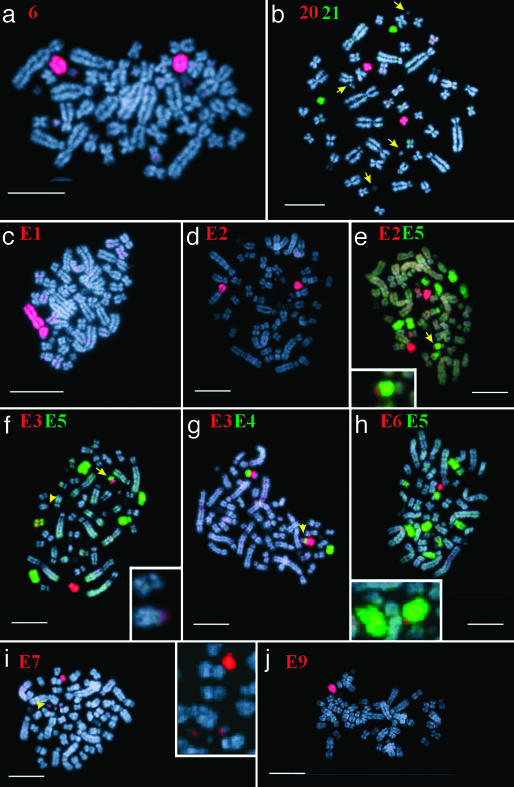
Fig. 3.
Examples of chromosome painting on male metaphases of the platypus. Chromosome paints used are indicated in each image, and unpaired chromosomes E1-E9 are present in single copy. (a) Chromosome 6 painted in red. (b) Chromosome 20 (red) and 21 (green), arrows point to tiny unpaired chromosomes. (_c_-i) Shown is the region of homology, which links chromosomes E1-E8 (see text). (Bars represent 10 μm.)
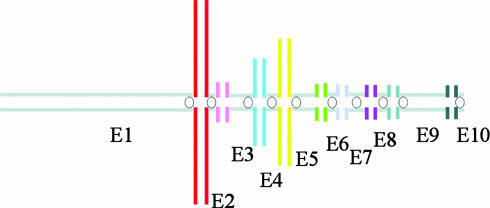
The 10 unpaired chromosomes identified by painting and designated E1-E10 are arranged in the lower part of the karyotype (Fig. 2 Upper). The order of all the chromosomes in the multivalent was established, and the parts (some of them relatively small) shared by each unpaired chromosome were determined by using single- and dual-color fluorescence in situ hybridization. Chromosome E1 can easily be recognized by its specific size and G bands. Chromosome E2 is acrocentric. Chromosomes E3 and E5 are submetacentric and share a heterochromatic region proximal on the long arm. Chromosome E4 is smaller then E3 and E5 and is metacentric. Chromosomes E6, E7, E8, and E10 are tiny chromosomes. E7 is the biggest of these four and is metacentric, as is E8. E6 is submetacentric. E10 is the smallest, which makes its centromere position difficult to determine (see also Fig. 3_b_). E9 is submetacentric and much larger than the tiny E6, E7, E8, and E10. Fig. 2 Lower is a G-banded karyotype of a female platypus in which E1, E3, E5, E7, and E9 are paired and E2, E4, E6, E8, and E10 are absent; there are no unpaired chromosomes.
Fig. 3 gives examples of the fluorescence in situ hybridization results on male metaphases. Fig. 3_a_ shows chromosome 6 identified by its specific paint. The p-arm of this chromosome consists of satellite DNA and the moderately repetitive ribosomal genes. Only the tip of 6p is painted, as the rest of the short arm is composed of repetitive DNA that is blocked by competition in the hybridization procedure. Fig. 3_b_ shows the hybridization of chromosome paint 20 (red) and 21 (green) onto the two smallest paired chromosomes with arrows showing four smaller unpaired chromosomes, each of different morphology. Fig. 3 c and d shows paint E1 and E2, respectively; E1 paints the largest unpaired chromosome plus another unpaired element in the chain; the homology between them implies that it must be E2. Paint E2 paints this second element plus the short arm of E1 and a region on the long arm of the next element in the chain, E3. The latter region was difficult to detect but can be seen in Fig. 3_e_ Inset. Fig. 3_e_ also shows the hybridization of paint E5 not only to E5 and the heterochromatic region of E3, but also to chromosomes pairs 13 and 14, which sort together with E5 in the flow karyotype (Fig. 1). In Fig. 3_f_, paint E3 (red) paints chromosome E3 plus the short arm of E2 (see arrowhead and Inset), and the short arm of E4 (see arrow). It also paints a band (yellow) on E5, as E3 and E5 share a common region of heterochromatin. E5 (green) paints E5 (the chromosome with the red heterochromatic region) and the long arm of E4 (arrow). As in Fig. 3_e_, paint E5 also paints chromosomes pairs 13 and 14 that sort together with E5 in the flow karyotype (Fig. 1). Fig. 3_g_ shows paint E3 in red and paint E4 in green. Paint E4 paints the short arms of E3 and E5 in addition to the entire E4. Again, paint E3 results in a red heterochromatic band on E5. Fig. 3_h_ shows the tiny element E6 in red and E5 in green. Fig. 3_h_ Inset shows that E6 paints the distal end of E5q. Fig. 3_i_ presents E7 in red, and the Inset shows that this probe also paints the tiny elements E6 and E8 (confirmed by dual-color painting, image not shown), thus linking E6-E8; the unlabeled E10 is also visible in this Inset. Fig. 3 _b_-i thus links E1 to E8. Fig. 3_j_ shows the larger element E9 without visual hybridization with E8 or E10.
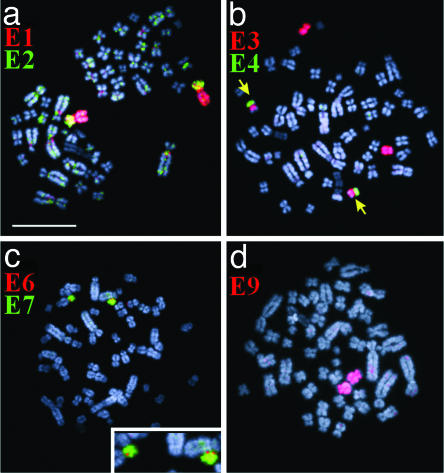
Fig. 4 shows the results of chromosome painting onto female metaphases. Fig. 4_a_ presents paint E1 in red and paint E2 in green. Element E2 itself is not present in female metaphases, instead element E1 is a paired chromosome in the female platypus; its short arm is in yellow as it is painted both by paints E1 and E2. Fig. 4_b_ shows the hybridization of paint E3 in red and E4 in green. E3 paints the entire E3 and the heterochromatic region on E5; E3 and E5 are present in pairs. E4 is not present, but the E4 paint probe hybridizes to its homologous region on E5p (green) and E3p (masked by the red E3 paint in this merged image). Fig. 4_c_ shows paint E6 in red and E7 in green: E7 forms a pair, but E6 is not present in the female platypus and the E6 paint reveals a tiny E6 pairing region on both homologues of E7. In 70% (n = 20) of the cells, the E7 pair is heteromorphic in size. Fig. 4_d_ shows that E9 also forms a pair in the female platypus. The results of these hybridization experiments demonstrate that the female platypus lacks the even-numbered elements and that the odd-numbered elements are all present in pairs. Although it is possible that other platypus populations may have different karyotypes, we believe that the findings in the one female and three males that we studied are representative of the species. Furthermore, the diploid number of chromosomes was always 2_n_ = 52 in three different reports in specimens from a total of 20 individuals (3, 7, 8).
Fig. 4.
Examples of chromosome painting on female metaphases of the platypus. Chromosome paints used are indicated in each image. Chromosomes E1, E3, E7, and E9 are present in two copies, and pairing regions are indicated for E2 (a), E4 (b), and E6 (c) (see text). (d) Two copies of Eq in red. (Bar represents 10 μm.)
In conclusion, successive links have been demonstrated between elements E1 and E8 of the chain. We could not detect homology between E8 and E9 or between E9 and E10 (for which no chromosome-specific probe could be made). However, Grützner et al. (17) have now analyzed the chain in male meiotic preparations, using our paint probes and telomeric probes, and have shown that E8 is linked to E9 and E10 is linked to E9 at the end of the chain.
Discussion
Chromosome-specific paint probes from the male platypus were used to identify and classify the platypus karyotype and resolve the decades-long debate about the nature of the unpaired chromosomes and their participation in the multivalent chain in male meiosis. We show that the male has 21 pairs of homologous chromosomes and 10 unpaired chromosomes, designated E1-E10 (Figs. 1 and 2). In the female, the odd-numbered elements (E1, E3, E5, E7, and E9) are present in two copies, and the even-numbered elements (E2, E4, E6, E8, and E10) are absent. Thus females have no unpaired chromosomes and are not expected to produce a multivalent chain in meiosis. As sex in platypus is associated with particular combinations of these elements, either homozygous E1, E3, E5, E7, and E9 in the female or heterozygous E1, E3, E5, E7, E9/E2, E4, E6, E8, and E10 in the male, by convention the odd elements are considered as X chromosomes and the even elements as Y chromosomes.
The X1Y1X2Y2X3Y3X4Y4X5Y5 sex chromosome constitution that we describe in the male platypus seems unique among mammals in the number of chromosomes involved in the multivalent chain at meiosis. Meiotic chains have been described in certain plants (18, 19), termites, and spiders (20-23). In mammals a meiotic chain has been reported in a primate with an X1X2Y1Y2 quadrivalent at male meiosis (24). It is unclear how the complex platypus sex chromosome system can achieve alternate segregation of X and Y chromosomes into individual sperm with high probability (17) and apparently without reproductive wastage.
Homologous regions of variable size, demonstrated here by chromosome painting, link the 10 unpaired elements in the male platypus karyotype. These homologous regions allow chiasmata to form during prophase of male meiosis I, thus forming a multivalent chain as shown in Fig. 5. The differential (unpaired) regions in the multivalent cannot form chiasmata and the horizontal lines in Fig. 5 represent these regions. In somatic cells, the female has a double dose of genes carried by these differential segments of the five X chromosomes.
Fig. 5.
Diagram of the multivalent chain in male meiotic cells. Horizontal and vertical lines represent differential and paired regions, respectively.
Because the E1 short arm pairs almost entirely with E2, it follows that genes known to map to the short arm of E1 [the human Xq genes _GLA, PLP, F8, F9_, and _RCP_ and the human Xp genes _ALAS2_ and _GATA1_ (11, 12)] all are present in the platypus in double doses in both males and females. This region therefore has no need for dosage compensation. However, four human Xq genes (AR, GDX, P3, and G6PD) (11, 25) that map to E1q, a region that we now confirm to be the differential region of the platypus X, should require dosage compensation. The other four X chromosomes (E3, E5, E7, and E9) also have regions that are not represented on the Y chromosomes and are thus present in single dose in the male. It will be of interest to discover whether dosage compensation occurs for these X chromosomes as it does apparently for the differential region of E1 (2). Ohno (26) explained the stability of the X chromosome across mammalian species by the inactivation mechanism. This stability would be disrupted if parts of the X chromosome were translocated to autosomes. As discussed below, exchanges between the ancestral X and autosomes may have occurred during the evolution of the multivalent in platypus, which would imply that a mechanism for dosage compensation based on X inactivation must accommodate multiple X chromosomes. On the other hand, dosage compensation may have been achieved in this species without X inactivation.
The five Y chromosomes share homologies with the X chromosomes only at their pairing segments. The differential segments of E2, E4, E6, E8, and E10, although not detectable by our methods, are likely to contain male-determining and other male-specific sequences. In the absence of a detectable homologue of SRY (14), the molecular basis for testis determination in the platypus is unknown. It will be important to explore male-specific sequences on the Y chromosomes for a novel sex-determining gene. Nevertheless, our results show that sex in the platypus is determined by alternate segregation of X and Y chromosomes during male meiosis. This finding is confirmed by Grützner et al. (17) using the chromosome-specific paint probes described here on platypus sperm, which reveals that male haploid germ cells contain either X or Y chromosomes but not both.
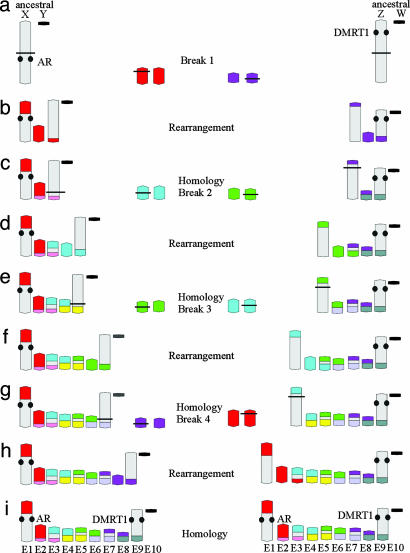
Our findings have consequences for the postulated model (8) of evolution >210 million years of the platypus sex chromosome system. Most models of sex-chromosome evolution assume an origin from an ancestral pair of sex chromosome in which one member acquires a male-determining role in a differential segment. It is possible that the system in platypus originated from such an ancestral pair. We propose a model (alternative to ref. 8) in which part of an original sex chromosome is repeatedly involved in exchanges with autosomes. In the absence of information on the gene content of any element except E1 and E9, it cannot be certain which end of the chain represents the initial sex pair. One possibility (Fig. 6 Left) is that the ancestral therian X and Y initiated the chain from a large X and an attenuated Y chromosome with small pairing regions and large differential regions (Fig. 6_a_). A reciprocal exchange between the X and an autosome leads to a meiotic chain of four (Fig. 6_b_). Element 2 in this chain is homologous with the short arm of E1 (Fig. 6_c_). The next event is an exchange between the derivative X and another autosome (Fig. 6_c_). The new element E3 has a region derived from the original X, whereas the new element E4 does not. Two similar events lead to the final 10 elements in which the derivative X is involved on each occasion. Note that in the diagram after each rearrangement we have given one of the exchanged segments and its homologous segment a different color to emphasize the regions that subsequently pair with one another in the multivalent. It is evident that E1, E3, E5, E7, and E9 have X regions and the other elements do not. Four successive X-autosome exchanges can provide a explanation for the homologies within the 10 elements of the chain. An alternative possibility (Fig. 6 Right) is that E9 and E10 originated from an ancestral sex chromosome pair and that E1 and E2, with homology to the eutherian X chromosome, represent the latest addition to the chain. Either pathway might be an early stage in the evolution of sex-determination systems from birds to mammals. The recent observation that DMRT1, the putative sex-determining gene carried by the Z chromosome in birds, maps to chromosome E9 in platypus (17) supports the second possibility. In this model the ZW pair would be the ancestors of chromosomes E9 and E10 and the starting point in the evolution of the monotreme multivalent. Comparative mapping studies with birds and reptile species may provide answers to the most likely mechanism of evolution of the sex chromosome system in monotremes and may help to identify the key sex-determining genes.
Fig. 6.
A model for the evolution of the platypus sex chromosome complement. Evolution starts with an ancestral pair of differential chromosomes, one of them is repeatedly involved in exchanges with an autosome. Two pathways are shown. (Left) The ancestral pair has homology to the mammalian X chromosome (indicated by the androgen receptor gene mapped to E1). (Right) The homology is to the avian Z based on the mapping of DMRT1 on E9. The model can be explained step by step. On the left there are two differential chromosomes (ancestral X-Y chromosomes). A break (indicated by a horizontal line) occurs within one of these and within a homolog of an autosomal pair (a). These breaks lead to a rearrangement and a chain of four elements. Element E1 and E3 are the derivative chromosomes, and E2 is the nonderivative homolog of the original autosomal pair (b). The color change from red to pink indicates the pairing regions of the element E2 with its “old” autosomal homolog within the chain of four elements (c). A new break and rearrangement with another autosomal pair (blue) result in a chain of six elements (c and d). The derivative blue homolog is rearranged and can be found on elements E3 and E5, whereas the nonderivative blue homolog forms element E4. Similarly, a color change from blue to yellow indicates the pairing regions with its old autosomal homolog (e). Note that the new element E3 has a region derived from the original X, whereas the new element E4 does not. Two additional rearrangements result in the chain of 10 elements as detected by chromosome painting. The right pathway, which starts with ancestral Z-W chromosomes, can be explained similarly.
Acknowledgments
This work was performed at the Cambridge Resource Centre for Comparative Genomics at the University of Cambridge, which was supported by a generous grant from the Wellcome Trust (to M.A.F.-S.). The Australian Research Council supports F.G. and J.A.M.G.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Woodburne, M. O., Rich, T. H. & Springer, M. S. (2003) Mol. Phylogenet. Evol. 28**,** 360-385. [DOI] [PubMed] [Google Scholar]
- 2.Grützner, F., Deakin, J., Rens, W., El-Mogharbel, N. & Graves, J. A. M. (2003) Comp. Biochem. Phys. 136**,** 867-881. [DOI] [PubMed] [Google Scholar]
- 3.Bick, Y. A. & Sharman, G. B. (1975) Cytobios 14**,** 17-28. [PubMed] [Google Scholar]
- 4.Matthey, R. (1949) Les Chromosomes des Vertebrés (Rouge, Lausanne, Switzerland).
- 5.VanBrink, J. (1959) Chromosoma 10**,** 1-72. [DOI] [PubMed] [Google Scholar]
- 6.Bick, Y. A., Murtagh, C. & Sharman, G. B. (1973) Cytobios 7**,** 233-243. [PubMed] [Google Scholar]
- 7.Murtagh, C. E. (1977) Chromosoma 65**,** 37-57. [Google Scholar]
- 8.Wrigley, J. M. & Graves, J. A. M. (1988) Chromosoma 96**,** 231-147. [DOI] [PubMed] [Google Scholar]
- 9.Bick, Y. A. (1992) in Platypus and Echidnas, ed. Augee, M. L. (Royal Zoological Society of New South Wales, Sydney), pp. 64-68.
- 10.Mitchell, J. M., Wilcox, S. A., Watson, J. M., Lerner, J. L., Woods, D. R., Schefler, J., Hearn, J. P., Bishop, C. E. & Graves, J. A. M. (1998) Hum. Mol. Genet. 7**,** 429-434. [DOI] [PubMed] [Google Scholar]
- 11.Watson, J. M., Spencer, J. A., Riggs, A. D. & Graves, J. A. M. (1990) Proc. Natl. Acad. Sci. USA 87**,** 7125-7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcox, S. A., Watson, J. M., Spencer, J. A. & Graves, J. A. M. (1996) Genomics 35**,** 66-70. [DOI] [PubMed] [Google Scholar]
- 13.Watson, J. M., Spencer, J. A., Riggs, A. D. & Graves, J. A. M. (1991) Proc. Natl. Acad. Sci. USA 88**,** 11256-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves, J. A. M. (2002) Trends Genet. 18**,** 259-264. [DOI] [PubMed] [Google Scholar]
- 15.Rens, W., O'Brien, P. C. M., Yang, F., Graves, J. A. M. & Ferguson-Smith, M. A. (1999) Chromosome Res. 7**,** 461-474. [DOI] [PubMed] [Google Scholar]
- 16.Yang, F., Carter, N. P., Shi, L. & Ferguson-Smith, M. A. (1995) Chromosoma 103**,** 642-652. [DOI] [PubMed] [Google Scholar]
- 17.Grützner, F., Rens, W., Tsend-Ayush, E., El-Mogharbel, N., O'Brien, P. C. M., Jones, R. C., Ferguson-Smith, M. A. & Graves, J. A. M. (2004) Nature, in press. [DOI] [PubMed]
- 18.Cleland, R. E. (1962) The Cytogenetics of Oenothera (Academic, New York).
- 19.Wiens, D. & Barlow, B. A. (1975) Science 187**,** 1208-1209. [DOI] [PubMed] [Google Scholar]
- 20.Syren, R. M. & Luykx, P. (1977) Nature 266**,** 167-168. [DOI] [PubMed] [Google Scholar]
- 21.Syren, R. M. & Luykx, P. (1981) Chromosoma 82**,** 65-88. [Google Scholar]
- 22.Luykx, P. & Syren, R. M. (1981) Experientia 37**,** 819-820. [Google Scholar]
- 23.Rowell, D. M. (1985) Chromosoma 93**,** 169-176. [Google Scholar]
- 24.Mudry, M. D., Rahn, I. M. & Solari, A. J. (2001) Am. J. Prim. 54**,** 65-78. [DOI] [PubMed] [Google Scholar]
- 25.Spencer, J. A., Watson, J. M., Lubahn, D. B., Joseph, D. R., French, F. S., Wilson, E. M. & Graves, J. A. M. (1991) J. Hered. 82**,** 134-139. [DOI] [PubMed] [Google Scholar]
- 26.Ohno, S. (1967) Chromosomes and Sex-Linked Genes (Springer, Berlin).