A blood pressure-associated variant of the SLC39A8 gene influences cellular cadmium accumulation and toxicity (original) (raw)
Abstract
Genome-wide association studies have revealed a relationship between inter-individual variation in blood pressure and the single nucleotide polymorphism rs13107325 in the SLC39A8 gene. This gene encodes the ZIP8 protein which co-transports divalent metal cations, including heavy metal cadmium, the accumulation of which has been associated with increased blood pressure. The polymorphism results in two variants of ZIP8 with either an alanine (Ala) or a threonine (Thr) at residue 391. We investigated the functional impact of this variant on protein conformation, cadmium transport, activation of signalling pathways and cell viability in relation to blood pressure regulation. Following incubation with cadmium, higher intracellular cadmium was detected in cultured human embryonic kidney cells (HEK293) expressing heterologous ZIP8-Ala391, compared with HEK293 cells expressing heterologous ZIP8-Thr391. This Ala391-associated cadmium accumulation also increased the phosphorylation of the signal transduction molecule ERK2, activation of the transcription factor NFκB, and reduced cell viability. Similarly, vascular endothelial cells with the Ala/Ala genotype had higher intracellular cadmium concentration and lower cell viability than their Ala/Thr counterpart following cadmium exposure. These results indicate that the ZIP8 Ala391-to-Thr391 substitution has an effect on intracellular cadmium accumulation and cell toxicity, providing a potential mechanistic explanation for the association of this genetic variant with blood pressure.
Introduction
High blood pressure (BP) is a major risk factor for many cardiovascular diseases. It is a multifactorial trait, with an estimated heritability of 0.3–0.5 (1,2). Genome-wide association studies (GWAS) have identified a number of genomic loci at which single nucleotide polymorphisms (SNPs) are associated with inter-individual variation in BP (3). Among them is SNP rs13107325 in the SLC39A8 (solute carrier family 39 member 8, HGNC: 20862) gene on chromosome 4q22, with its major allele associating with increased systolic and diastolic BP (3). However, the molecular and cellular mechanisms underlying its association with BP are unknown.
rs13107325 is a non-synonymous SNP located in exon 8 of SLC39A8, with the major allele having a cytosine (C) and the minor allele having a thymidine (T) at the first position of the codon for amino acid 391 (NC_000004.11:g. 103188709C > T). Thus, the protein, ZIP8, encoded by this gene can be either the Ala391 form (encoded by the C allele) or the Thr391 variant (encoded by the T allele). ZIP8 is a plasma membrane transporter of metal ions including cadmium (Cd2+), zinc, manganese and iron (4,5). It plays a key role in cellular uptake of Cd2+ and in Cd2+-induced cytotoxicity (6,7). Cd2+ exposure has been associated with the development of high BP and other cardiovascular diseases (8–13), which are likely to be the consequence of Cd2+-induced damage to the kidney and vascular endothelial cells (7,14–17), both playing important roles in BP regulation.
In this study, we sought to investigate whether the BP-associated SNP rs13107325 (Ala391Thr) has a functional effect on ZIP8-mediated Cd2+ transport in human embryonic kidney cells (HEK293) and human umbilical vascular endothelial cells (HUVECs) and its subsequent influence on ERK1/2 and NFκB signalling pathways leading to Cd2+-induced cytotoxicity.
Results
In silico structural and bioinformatic analyses
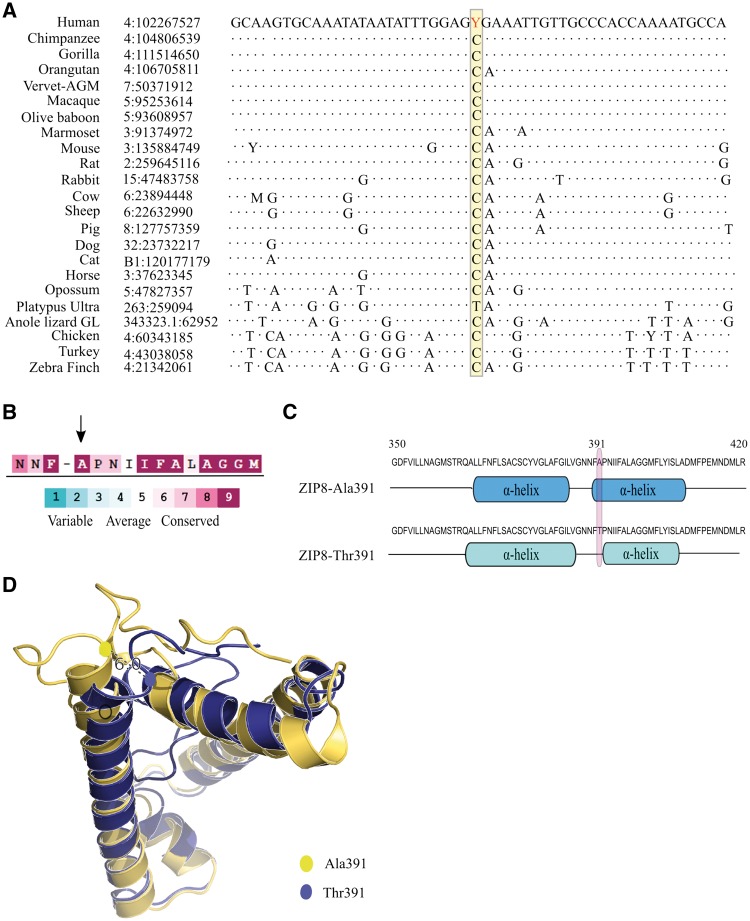
Conservation analysis revealed that the common allele C of rs13107325 had been well conserved in 22 out of the 23 amniote vertebrates as shown in the sequence alignment in Figure 1A, suggesting that the C allele may be important in the preservation of protein function. ConSurf Server analysis also indicated Ala391 residue being highly conserved and crucial for maintaining protein fold predicted by Markov Model based algorithm (normalized conservation score 9, structural residual) when compared with 142 reference sequences (Figure 1B). An analysis with TMHMM prediction suggested that the Ala391 residue was packed inside the α-helical transmembrane domain of the ZIP8 protein, whereas the Thr391 residue was located just outside the α-helix domain in the corresponding protein (Figure 1C).
Figure 1.
Results of computational analyses of ZIP8 Ala391-to-Thr391. (A) A comparison of DNA sequences around the rs13107325 site in 23 mammals. (B) Output of ConSurf Server analysis based on 142 reference sequences, the arrow points the amino acid position of Ala391. (C) Graphical interpretation of the transmembrane helical domains predicted by TMHMM. The number at the top denotes the amino acid position, the Ala391-to-Thr391 substitution is circled in pink, which locates outside the cell membrane. (D) Structural perturbation induced by the Ala391-to-Thr391 substitution as shown by Robetta secondary protein structure prediction. The image shows an overlapped simulation of the two proteins. The Thr391 residue (represented by the blue circle) is at a distance of 6 Å´ from the Ala391 residue (represented by the yellow circle).
This finding prompted further investigation by performing homology-based 3D protein structure modelling using the Robetta approach. Secondary structure analysis was performed to predict in silico the effect of the Ala391-to-Thr391 substitution on the structure and dynamics of ZIP8. This was performed on both the ZIP8-Ala391 and ZIP8-Thr391-containing protein sequences. The analysis revealed a total of 7 α-helical domains for both sequences. A noticeable change was observed at the α-helical structural transition (residues 390-392) in the ZIP8-Thr391 sequence when overlapped with the ZIP8-Ala391 sequence, together with other transmembrane α-helical domain in close proximity shifting their position within the membrane (Figure 1D).
Furthermore, the Sorting Tolerant From Intolerant (SIFT) (18) and Polymorphism Phenotyping v2 (PolyPhen-2) (19) tools predicted that rs13107325 had a possibly damaging effect on protein function.
A research using the UCSC Genome Browser showed that rs13107325 was located in a genomic region without evidence of transcription factor binding, suggesting that it probably does not affect gene transcription. Consistently, an allelic expression imbalance assay of rs13107325 in HUVEC cells showed no difference in SLC39A8 RNA level between the C and T alleles (Supplementary Material, Fig. S1).
Effect of SNP rs13107325 on Cd2+ intracellular accumulation
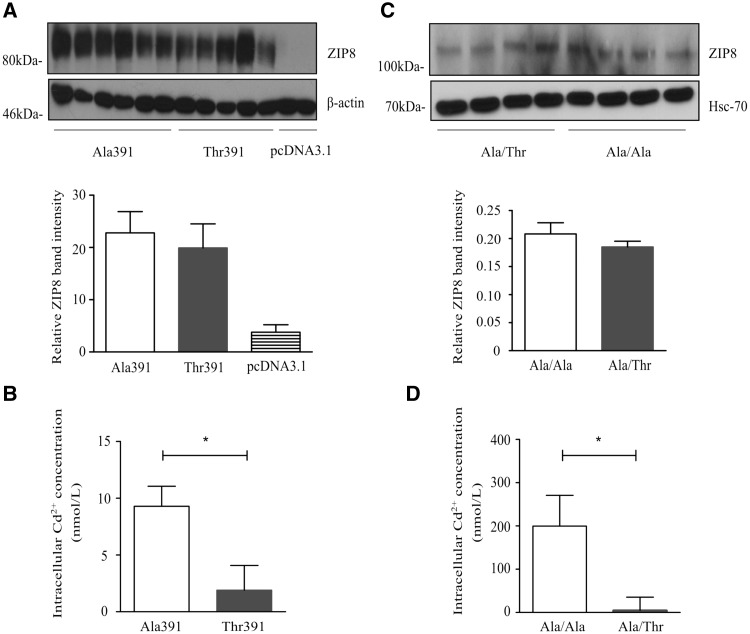
To ascertain an effect of the ZIP8 Ala391-to-Thr391 substitution on Cd2+ transport and intracellular accumulation, HEK293 kidney cells were transfected with either the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid. The efficiency of transfection and specificity of the antibody used for subsequent immunoblotting analysis are shown in Supplementary Material, Fig. S2. Transfected cells were incubated with Cd2+ (1 μmol/l) or vehicle (H2O) for 2 h, followed by measurement of intracellular Cd2+ levels. A range of 1–10 μmol/l of Cd2+ has generally been used in the literature. The concentration of 1 μmol/l used in our study is closer to the _K_m value of 0.62 μmol/l for ZIP8-mediated Cd2+ transport in mouse foetal fibroblasts (4). Immunoblotting analyses demonstrated that HEK293 cells transfected with either the pcDNA-ZIP8-Ala391 or pcDNA-ZIP8-Thr391 plasmid had a substantial increase in total ZIP8 protein level as compared with cells transfected with the empty pcDNA3.1(+) plasmid and that there was no significant difference in ZIP8 level between cells transfected with either the pcDNA-ZIP8-Ala391 and those transfected pcDNA-ZIP8-Thr391 plasmid (Figure 2A). Measurement of Cd2+ in the cell lysates showed that the intracellular Cd2+ concentration was significantly higher in HEK293 cells transfected with the pcDNA-ZIP8-Ala391 plasmid than in those transfected with the pcDNA-ZIP8-Thr391 plasmid (Figure 2B), suggesting that the Ala391-to-Thr391 substitution has an effect on Cd2+ transport and intracellular accumulation.
Figure 2.
Effect of ZIP8 Ala391-to-Thr391 on intracellular Cd2+ accumulation. (A) Immunoblotting analysis of HEK293 cells transfected with the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, with an anti-ZIP8 antibody and an antibody for the loading control protein β-actin. Upper panel: a representative immunoblot image; Lower panel: mean (± SEM) values of ZIP8 band intensities standardized against β-actin band intensities. (B) Intracellular Cd2+ concentrations in HEK293 cells transfected with the pcDNA-ZIP8-Ala391 or pcDNA-ZIP8-Thr391 plasmid, standardized against transfection efficiency and subtracted from intracellular Cd2+ concentrations of HEK293 cells transfected with the empty pcDNA3.1(+) plasmid. Data shown are mean (± SEM) values from three experiments, with duplicate for each plasmid in each experiment. (C) Immunoblotting analysis of HUVECs of the Ala/Ala or Ala/Thr genotype with an anti-ZIP8 antibody and an antibody for the loading control protein Hsc-70. Upper panel: a representative immunoblot image; lower panel: mean (± SEM) values of ZIP8 band intensities standardized against Hsc-70 band intensities. (D) Intracellular Cd2+ concentrations in HUVECs of the Ala/Ala or Ala/Thr genotype, standardized against ZIP8 levels (represented by ZIP8 band intensities with normalization against β-actin band intensities). Data shown are mean (± SEM) values from 3 experiments, with duplicate for each plasmid in each experiment. * Indicates P < 0.05 by paired Student’s _t_-test.
To investigate if the ZIP8 Ala391-to-Thr391 substitution affected intracellular Cd2+ accumulation in vascular endothelial cells, primary cultures of HUVECs of either the Ala/Ala or Ala/Thr genotype were incubated with Cd2+ (1 μmol/l) for 2 h, followed by measurement of Cd2+ accumulation in cell lysates [due to the rarity of minor allele homozygotes, no HUVECs with this genotype (Thr/Thr) were available for our study]. Immunoblotting analyses detected no significant difference in ZIP8 level between HUVECs of the Ala/Ala genotype and HUVECs of the Ala/Thr genotype (Figure 2C). Importantly, Cd2+ assays showed that HUVECs of the Ala/Ala genotype had significantly higher intracellular Cd2+ levels than those of the Ala/Thr genotype (Figure 2D), indicating that SNP rs13107325, leading to the Ala391-to-Thr391 substitution, has an effect on Cd2+ accumulation in vascular endothelial cells.
Effect of SNP rs13107325 on Cd2+-induced cytotoxicity
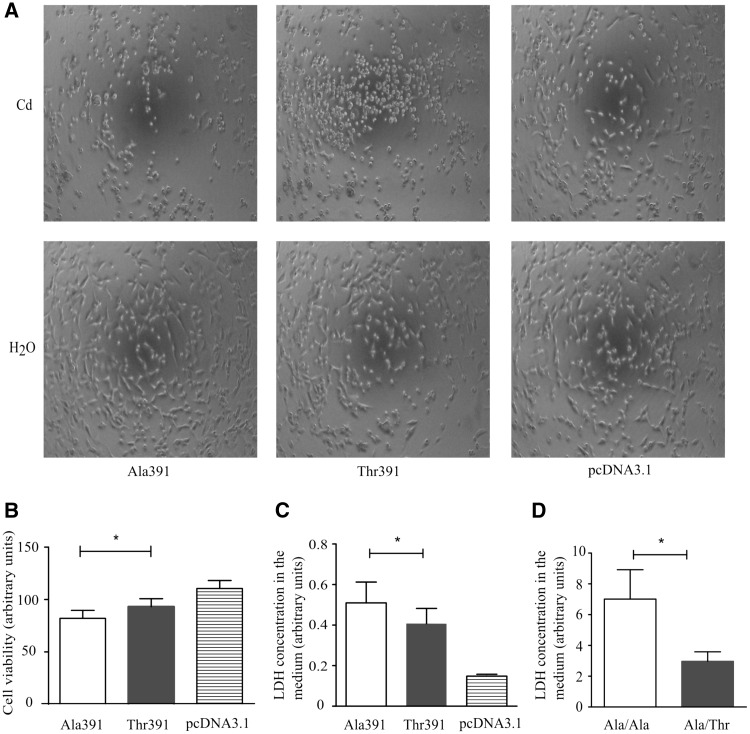
Having found that the ZIP8 Ala391-to-Thr391 substitution affected Cd2+ intracellular accumulation in kidney and endothelial cells, we investigated if it had any influence on Cd2+-induced cytotoxicity. HEK293 cells transfected with either the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid were incubated with 1 μmol/l Cd2+ or vehicle (H2O) for 2 or 24 h, followed by cytotoxicity assays. Assessment of cell morphology revealed that following Cd2+ treatment, HEK293 cells transfected with either the pcDNA-ZIP8-Ala391 or pcDNA-ZIP8-Thr391 plasmid became smaller and changed from spindle-shape to spherical-shape, which were not observed in cells transfected with the pcDNA3.1(+) vector control (Figure 3A). Following Cd2+ treatment for 2 h, the viability of cells expressing the heterologous ZIP8-Ala391 was lower than that of cells expressing the heterologous ZIP8-Thr391, and both had lower viability than cells transfected with the vector control (Figure 3B). Correspondingly, 24 h exposure to 1 μmol/l Cd2+ resulted in larger amounts of LDH released by dead cells in media conditioned by ZIP8-Ala391 expressing cells than in media conditioned by ZIP8-Thr391 expressing cells, and both had significantly higher LDH concentrations compared with media conditioned by cells transfected with the empty plasmid (Figure 3C).
Figure 3.
Effect of ZIP8 Ala391-to-Thr391 on Cd2+-induced cell death. (A) Digital light microscopic images of HEK293 cells transfected with the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, followed by a 24 h incubation with Cd2+ (1 µmol/l). (B) Percentages of viable HEK239 cells transfected with the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, followed by a 2 h incubation with Cd2+ (1 µmol/l), determined by MTS assay. (C) Relative amounts of LDH in media conditioned by HEK239 cells transfected with the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, followed by a 24 h incubation with Cd2+ (1 µmol/l). (D) Relative amounts of LDH in media conditioned by HUVECs of the Ala/Ala or Ala/Thr genotype, followed by a 24 h incubation with Cd2+ (1 µmol/l). Data shown in (B), (C) and (D) are mean (± SEM) values from 3 experiments, with triplicate for each plasmid in each experiment. *Indicates P < 0.05 by Wilcoxon test.
To determine if the effect of the ZIP8 Ala391-to-Thr391 substitution on Cd2+-induced cytotoxicity also occurred in vascular endothelial cells, HUVECs of either the C/C (Ala/Ala) or C/T (Ala/Thr) genotype were incubated with Cd2+ (1 μmol/l) for 24 h, followed by measurement of LDH in culture media. Consistent with the findings in HEK293 cells, media conditioned by HUVECs of the Ala/Ala genotype had significantly higher LDH concentrations than media conditioned by HUVECs of the Ala/Thr genotype (Figure 3D), suggesting a genotype-specific Cd2+ cytotoxic effect.
Effect of SNP rs13107325 on Cd2+-induced ERK1/2 phosphorylation
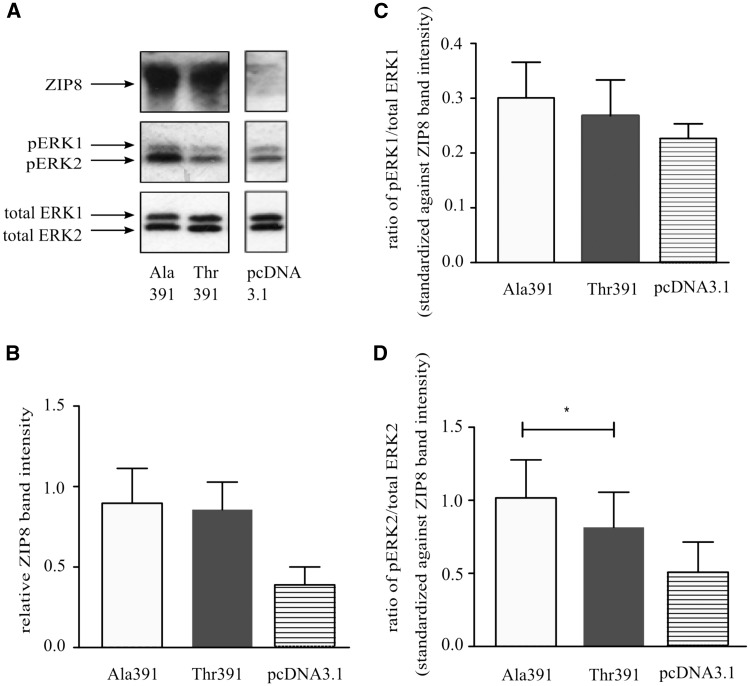
Since MAPK signalling pathway activation has been implicated in Cd2+-induced cytotoxicity (20), we investigated if the ZIP8 Ala391-to-Thr391 substitution had any effect on this pathway in response to Cd2+ exposure. HEK293 cells transfected with either the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid were incubated with Cd2+ (1 μmol/l) for 24 h, followed by immunoblot analyses of the amounts of total and phosphorylated ERK1 and ERK2 in cell lysates. The analyses confirmed increased ZIP8 expression of cells transfected with the pcDNA-ZIP8-Ala391 or pcDNA-ZIP8-Thr391 plasmid as compared with cells transfected with the empty plasmid vector pcDNA3.1(+), and showed that there was no significant difference in ZIP8 level between cells transfected with the pcDNA-ZIP8-Ala391 plasmid and those transfected with the pcDNA-ZIP8-Thr391 plasmid (Figure 4A and B). Following Cd2+ treatment, the levels of phospho-ERK1 and phospho-ERK2 were higher in cells expressing heterologous ZIP8-Ala or ZIP8-Thr391 than in cells transfected with the empty plasmid (Figure 4A,C and D). There was no significant difference in the ratio of phospho-ERK1 versus total ERK1 between pcDNA-ZIP8-Ala391 and pcDNA-ZIP8-Thr391 expressing cells (Figure. 4C). However, the ratio of phospho-ERK2 versus total ERK2 in pcDNA-ZIP8-Ala391 expressing cells was significantly higher than that in pcDNA-ZIP8-Thr391 expressing cells, with standardization for ZIP8 expression levels (Figure. 4D).
Figure 4.
Effect of ZIP8 Ala391-to-Thr391 on ERK phosphorylation in cells incubated with Cd2+. HEK239 cells transfected with the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, were incubated with Cd2+ (1 µmolL) for 24 h, followed by immunoblotting analyses of ZIP8, total ERK1, phosphorylated ERK1, total ERK2, and phosphorylated ERK2. (A) Representative immunoblot images. (B) Mean (± SEM) values of relative intensities of the ZIP8 band. (C) Mean (± SEM) values of the ratio of phosphorylated ERK1 band intensity versus total ERK1 band intensity, standardized against ZIP8 band intensity. (D) Mean (± SEM values of the ratio of phosphorylated ERK2 band intensity versus total ERK2 band intensity, standardized against ZIP8 band intensity. Data shown in (B) to (D) are from three experiments, with triplicate for each plasmid in each experiment. * Indicates P < 0.05 by paired Student’s _t_-test.
Effect of SNP rs13107325 on NFκB activation in response to Cd2+
Since NFĸB has also been implicated in Cd2+-induced cytotoxicity (20), we investigated whether the ZIP8 Ala391-to-Thr391 substitution had an effect on NFĸB activation. HEK293 cells were transfected with either the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, as well as a firefly luciferase reporter plasmid and a renilla luciferase plasmid (the latter to serve as a reference of transfection efficiency). Immunoblot analyses showed no significant difference in ZIP8 level between cells transfected with the pcDNA-ZIP8-Ala391 plasmid and those transfected with the pcDNA-ZIP8-Thr391 plasmid. The transfected cells were incubated with Cd2+ for 2 h, followed by dual-luciferase assays to measure NFκB activity. The assays showed 5-10 fold higher NFκB activity in the ZIP8-expressing cells than in cells transfected with the empty plasmid and a statistically significant 1.2 fold difference between cells transfected with the pcDNA-ZIP8-Ala391 plasmid and those transfected with the pcDNA-ZIP8-Thr391 plasmid (Figure 5A).
Figure 5.
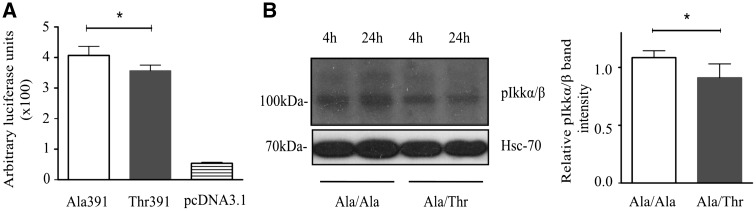
Effect of ZIP8 Ala391-to-Thr391 on NFκB activity in cells incubated with Cd2+. (A) HEK293 cells were transfected with either the pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or pcDNA3.1(+) plasmid, as well as a firefly luciferase reporter plasmid containing copies of the NFκB responsive element upstream of the firefly luciferase reporter gene, and a renilla luciferase plasmid to serve a reference for transfection efficiency. The transfected cells were incubated with Cd2+ (1 μmol/l) for 2 h, and NFκB activity measured by dual-luciferase assays. Data shown are mean (± SEM) luciferase activity from three experiments, with triplicate for each plasmid in each experiment. (B) HUVECs of the Ala/Ala or Ala/Thr genotype were subjected to immunoblotting analyses of the phospho-IKKα/β. Shown on the left is a representative immunoblot image, and on the right are the relative phospho-IKKα/β band intensities standardized against loading control protein (Hsc-70) band intensities in different genotypes. Data shown are mean (±SEM) values from 3 experiments of 4 samples for each genotype, conducted in duplicate. * Indicates P < 0.05 by paired Student’s _t_-test.
To investigate if the effect of the ZIP8 Ala391-to-Thr391 substitution on NFĸB activation also existed in vascular endothelial cells, HUVECs of either the Ala/Ala or Ala/Thr genotype were subjected to immunoblot analysis of the IKK complex (IKKα and IKKβ), a central regulator of NFĸB activation (21). HUVECs of the Ala/Ala genotype had a significantly larger increase in IKKα/β phosphorylation than those of the Ala/Thr genotype, comparing 24 h with 4 h incubation with 1 μmol/l Cd2+ ( Figure 5B and C).
Discussions
In these in vitro experiments, we found that the BP-associated SNP rs13107325 resulting in the ZIP8 Ala391-to-Thr391 substitution exerted an effect on Cd2+ accumulation in kidney and vascular endothelial cells following exposure to extracellular Cd2+, as well as on Cd2+-stimulated intracellular signal pathway activation and cell death. This effect is likely to arise from the amino acid change rather than a difference in expression level, since there was no genotype-dependent difference in the ZIP8 protein level as shown in Figure 2C. This is supported by the results of the computational analysis which indicates that the Ala391-to-Thr391 substitution induces a structural change of the ZIP8 protein. These results demonstrate that rs13107325 is a functional genetic variant which can influence the survival of kidney and vascular endothelial cells, two cell types that play important roles, respectively, in salt-fluid homeostasis and vascular tone regulation, paramount to BP control.
Chronic Cd2+ exposure has been shown to increase BP in animal models (15). Furthermore, some studies in humans have detected an association between Cd2+ exposure and elevated BP (8–10). There is evidence indicating that the effect of Cd2+ on BP is primarily due to its accumulation and toxicity in the kidney (22). It has been reported that greater than one-third of body Cd2+ deposits are found in the kidney, some of which are excreted via the urine (23). The most common form of Cd2+ in the blood circulation is metallothionein (MT)-bound, which is partially filtered by the glomeruli, followed by reabsorption at the proximal tubule (24). A balance of MT and Cd2+ concentration is important for renal protection as non-MT-bound Cd2+ can be toxic (25). Studies of mice with SLC39A8 knock-in have indicated that ZIP8 mediates Cd2+ uptake in the kidney proximal tubule and thereby plays an important role in Cd2+-induced nephrotoxicity and kidney failure (3,26). In line with these literatures, our in vitro study showed that following an incubation with Cd2+, ZIP8-Ala391 expressing HEK293 cells had significantly greater accumulation of intracellular Cd2+, leading to higher Cd2+-induced cell death compared with ZIP8-Thr391 expressing cells. This provides a possible mechanistic explanation for the association between the ZIP8-Ala391 allele and increased BP revealed by GWAS (3).
There is evidence indicating that the effect of Cd2+ on blood pressure is, to some extent, also due to Cd2+-induced vascular endothelial cell damage and dysfunction. Cd2+ can cause vasoconstriction by inhibiting nitric oxide (14), increasing endothelial permeability (16), promoting peroxidation of membrane lipids (27), and inducing endothelial cell death (17). These can promote atherosclerosis, a risk factor for hypertension (28). In agreement, other studies have demonstrated that Cd2+-fed apoE-/- mice have significantly larger atherosclerotic plaques (17,29). Furthermore, epidemiological studies have shown that elevation in blood Cd2+ level is independently associated with early atherosclerotic vessel wall thickening in healthy female individuals and that there is increased frequency of atherosclerosis in a Cd2+ contaminated area in the Netherlands (11,17). In the present study, we found that vascular endothelial cells of the rs13107325 Ala/Ala genotype had higher Cd2+ uptake and were more susceptible to Cd2+-induced cell death than cells of Ala/Thr genotype. This provides another possible mechanistic explanation for the association between the ZIP8-Ala391 allele and increased BP (3).
The cytotoxic effect of Cd2+ has previously been shown to be mediated by the ERK signalling pathway (30). Upon activation, phosphorylated ERK1/2 can enter the nucleus and phosphorylate transcription factors involved in cell-cycle regulation (31). A growing body of evidence suggests that aberrant activation of ERK can promote cell death (30,32). It has been reported that in HEK293 cells, sustained ERK activation can last up to 6 days following Cd2+ exposure (33). In the present study, we found that Cd2+-induced ERK activation was greater in ZIP8-Ala391 expressing cells than in ZIP8-Thr391 expressing cells. This suggests that the greater cytotoxic effect of Cd2+ in ZIP8-Ala391 expressing cells (comparing to ZIP8-Thr391 expressing cells) is in part mediated by a greater ERK activation.
The transcription factor NFκB regulates the expression of many genes with various cellular functions (34,35). Previous studies have demonstrated an association of Cd2+-induced apoptosis with NFκB activation, and trans-activation of anti-apoptotic and/or survival genes, oxidative stress-related genes and inflammatory cytokines (34,36–38). In the present study, we found that NFκB activity was significantly higher in ZIP8-Ala391 expressing cells compared to ZIP8-Thr391 expressing cells following Cd2+ treatment. We also detected higher levels of the NFĸB effector elements phospho-IKKα/β in HUVECs of the Ala/Ala genotype when compared with HUVECs of the Ala/Thr genotype. Therefore, these experiments demonstrate that cells carrying the ZIP8-Ala391 allele had higher levels of NFκB activation in response to Cd2+ exposure, possibly to counteract cell death.
Interestingly, in addition to its relation with BP, rs13107325 has been shown to be associated with a number of other traits and disorders, including blood levels of N-terminal pro-B-type natriuretic peptide levels (39), manganese (40), total cholesterol (41), high density lipoprotein (41–43) and triglyceride levels (41), as well as height (41), body mass index (44), schizophrenia (41,45) and Parkinson’s disease (41), among others (41). Therefore, rs13107325 appears to have pleiotropic effects and it is possible that its influence on BP may involve more than one intermediary pathway. Conversely, the results of our study showing an effect of rs13107325 on Cd2+-induced cell toxicity also provide a possible explanation for the association of this SNP with some other diseases such as schizophrenia and Parkinson’s disease which have been suggestively linked with Cd2+ exposure (46,47).
There are some limitations to our study. First, the study was conducted using human cell models in vitro. Future studies using in vivo models are warranted. It has been reported that global SLC39A8 (-/-) knockout mice were embryonically lethal (48). Therefore, one possibility would be to test the possible effect of conditional knockout, knockdown or knock-in of SLC39A8 on BP in mice with and without Cd2+ exposure (48,49). Second, our study focused on the effect of Cd2+. Further studies to examine possible influences of other metal ions transported by ZIP8, for example Zn2+ and Mn2+, would also be warranted.
In conclusion, the results of this study demonstrate that the BP-associated SNP rs13107325 has a functional effect on ZIP8 which can influence the viability of kidney and vascular endothelial cells. These findings can contribute to a better understanding of the functional roles of genetic variants in BP regulation, which in turn can facilitate the development of new therapeutics for hypertension.
Materials and Methods
Structural prediction
Human ZIP8 protein sequences were retrieved from Ensembl version 78 (51). The longest isoform (ENST00000356736), deemed as canonical, was chosen for further analysis. Conservation analysis was performed on Ensembl using 23 way alignment of high-coverage amniote vertebrate genomes. The ConSurf server was used to predict functional domains based on the evolutionary conservation of amino acid positions, with UniRef protein clusters to infer the phylogenetic relations between homologous sequences (52). The Robetta approach was employed to predict the secondary structure of ZIP8–Ala391 and ZIP8-Thr391 (53). This method segments the protein into domains that are homologous to proteins with known structure, before an optional structure refinement procedure. The domain containing amino acid 391 for both versions of the protein was fully processed and analysed using PyMol to assess the predicted structural effects of the amino acid substitution in the respective functional domain (54). Further to the full structural reconstruction, prediction of Transmembrane Helices in proteins (TMHMM) v2 program was used to predict transmembrane helical domains within the input sequences (55).
Cell culture
HUVECs were isolated from umbilical cords from different individuals (n = 178) using a standard procedure (50). Cords from miscarriages, stillbirths, or HIV positive, group B streptococcus positive or hepatitis B infected mothers were excluded. Cells were maintained in M199 medium (Sigma, M4530) supplemented with 15% foetal bovine serum (Appleton Woods, FB021), l-glutamine, 2.5 μg/ml β-endothelial growth factor (Sigma, E1388), 4.5 μg/ml endothelial cell growth factor (Sigma, E2759), 2.5 μg/ml thymidine (Sigma, 89270), heparin (Sigma, H3393), and 1% penicillin/streptomycin. Cells of passage ≤4 were used for the experiments in this study. HEK293 cells were maintained in DMEM medium (Sigma, D5796) supplemented with 10% foetal bovine serum, 3.34 mM/l l-glutamine, 0.02 units/l penicillin and 20 mg/l streptomycin.
Genotyping
Genomic DNA extracted from HUVECs was genotyped for rs13107325 with the use of TaqMan SNP genotyping assay (Applied Biosystems, c_1827682_10). Genotyping results of a subset of samples were verified by DNA sequencing.
Construction of ZIP8 expression plasmids
Full-length human SLC39A8 cDNA (GenBank Accession No. NM_022154.5) in pCMV6-XL4 plasmid was purchased from OriGene (Origene, SC112817). The cDNA, corresponding to the C allele for SNP rs13107325 and therefore encoding the Ala391 form of ZIP8, was cloned into pcDNA3.1(+) expression plasmid. Using this plasmid (pcDNA-ZIP8-Ala391) as a template, site-directed mutagenesis was carried out to generate a plasmid (pcDNA-ZIP8-Thr391) in which the nucleotide C at the rs13107325 site in the SLC39A8 cDNA was changed to T. The DNA sequences of both plasmids were verified to be correct by sequencing.
Cd2+ transport assay
HEK293 cells were transfected with either pcDNA-ZIP8-Ala391, pcDNA-ZIP8-Thr391, or the pcDNA3.1(+) plasmid, by the calcium phosphate transfection method. Transfected HEK293 cells, and untransfected HUVECs of different genotypes for SNP rs13107325, were subjected to Cd2+ transport assays. In brief, cells were cultured in normal medium to 80–85% confluency and then in serum-free medium containing 1 µM (0.2% v/v in H2O) CdCl2 (Sigma, 439800) for 2 or 24 h. Thereafter, cells were washed with phosphate buffered saline, and then the cell lysates prepared in lysis buffer consisting of 150 mM NaCl, 50 mM Tris 7.5, 1% Nonidet P40, 0.5% sodium deoxycholate, and 1x protease inhibitors. Cd2+ concentrations in the cell lysates were determined using Measure-iTTM cadmium assay kit (Invitrogen, M36353), according to manufacturer’s instructions. The fluorescence intensity indicating Cd2+ content was recorded for each well at 520 nm (λex: 490 nm), with two readings for each sample. The Cd2+ concentration values were extrapolated from a standard curve ranging from 0 to 4.2 mM. In the HEK293 cell assays, Cd2+ values from cells transfected with either the pcDNA-ZIP8-Ala391 or pcDNA-ZIP8-Thr391 plasmid were standardized for ZIP8 protein levels determined by immunoblot analysis and subtracted by those from cells transfected with the pcDNA3.1(+) plasmid.
Cell viability assay
Cytotoxicity induced by Cd2+ exposure was assessed in transfected HEK293 cells and in untransfected HUVECs of different genotypes. Following incubation of cells with 1 μmol/l Cd2+ for 24 h, cell morphological changes were assessed by microscopic examination, and the percentages of viable cells were determined with the use of CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, G3582, or being referred to as MTS) using the following equation:
(Ab490nm in cadmium treated cells)-(Ab490nm in media) (Ab490nm in untreated cells)-(Ab490nm in media)×100%
The lactate dehydrogenase (LDH) released from dead cells into culture media was measured using a lactate dehydrogenase assay kit (Pierce Thermo, 88953) as per the manufacturer’s instructions, and the amounts of LDH in the media were calculated using the following equation:
(LDH activity in media of cadmium treated cells)-(LDH activity in media of untreated cells)(LDH activity in media and lysates of untreated cells)-(LDH activity in media of untreated cells)×100%
Immunoblotting analyses
Total cell lysates were prepared from transfected HEK293 cells and from HUVECs. Protein concentrations in the lysates were measured by the bicinchoninic acid assay (Pierce, 23227). To determine the relative amounts of ZIP8, 15 μg total cellular proteins from each sample were subjected to standard immunoblotting analysis with an anti-ZIP8 antibody (Abcam, ab103182) and subsequently with an anti-β-actin (Abcam, ab8226) or anti-Hsc70 (Santa Cruz, sc7298) antibody as a loading control. To determine the relative quantities of total and phosphorylated ERK1/2, 15 μg proteins from each sample were analysed by immunoblotting with an antibody for phospho-p44/42 MAPK (ERK1/2)(Cell Signalling, 9101) and an antibody for total p44/42 MAPK (Cell Signalling, 9102). The blots were subjected to densitometric analysis with GIMP 2.8.16 software. The relative protein levels were standardized against the levels of the loading control protein.
Luciferase reporter assay
Using X-tremeGENE HP DNA transfection system (Roche, 06366236001), HEK293 cells were transfected with a ZIP8 expression plasmid (pcDNA-ZIP-Ala391 or pcDNA-ZIP-Thr391), a firefly luciferase reporter plasmid containing copies of the NFκB responsive element upstream of the firefly luciferase reporter gene, and a renilla luciferase plasmid to serve as a reference for transfection efficiency. Twenty-four hours after transfection, cells were incubated with 1 μmol/l Cd2+ for a further 2 h before being analysed using a dual luciferase assay kit (Promega, E1910). The NFκB activity was calculated by the ratio of the luminescence intensity yielded by firefly luciferase versus that by renilla luciferase.
Statistical analyses
Paired Student’s t-tests were performed for variables with normal distribution, and Wilcoxon matched-pairs test was used for variables not in normal distribution. Data are presented as mean ± SEM from at least three independent experiments, and all p values are two-tailed.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Prof. Henning Walczak for the generous gift of the NFκB and renilla luciferase plasmids. We are also grateful to Prof. Patricia Munroe and Dr. Qingzhong Xiao for their technical advice to this study.
Conflict of Interest statement. None declared.
Funding
This work is supported by the British Heart Foundation and falls under the portfolio of research conducted within the National Institute for Health Research Leicester Cardiovascular Biomedical Research Unit. It also forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Unit supported and funded by the National Institute of Health Research. Ruoxin Zhang was the recipient of a scholarship (No. 2011831630) from the China Scholarship Council. Claudio Mauro is the recipient of a British Heart Foundation Intermediate Fellowship (FS/12/38/29640). Funding to pay the Open Access publication charges for this article was provided by Queen Mary, University of London.
References
- 1.Havlik R.J., Garrison R.J., Feinleib M., Kannel W.B., Castelli W.P., McNamara P.M. (1979) Blood pressure aggregation in families. Am. J. Epidemiol., 110, 304–312. [DOI] [PubMed] [Google Scholar]
- 2.Miall W.E., Oldham P.D. (1963) The hereditary factor in arterial blood-pressure. Brit. Med. J., 1, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D, Verwoert G.C. International Consortium for Blood Pressure Genome-Wide Association., et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L., Girijashanker K., Dalton T.P., Reed J., Li H., Soleimani M., Nebert D.W. (2006) ZIP8, member of the solute-carrier-39 (SLC39) metal-transporter family: characterization of transporter properties. Mol. Pharmacol, 70, 171–180. [DOI] [PubMed] [Google Scholar]
- 5.Wang C.Y., Jenkitkasemwong S., Duarte S., Sparkman B.K., Shawki A., Mackenzie B., Knutson M.D. (2012) ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J. Biol. Chem., 287, 34032–34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton T.P., He L., Wang B., Miller M.L., Jin L., Stringer K.F., Chang X., Baxter C.S., Nebert D.W. (2005) Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. P. Natl. Acad. Sci. USA, 102, 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B., Schneider S.N., Dragin N., Girijashanker K., Dalton T.P., He L., Miller M.L., Stringer K.F., Soleimani M., Richardson D.D, et al. (2007) Enhanced cadmium-induced testicular necrosis and renal proximal tubule damage caused by gene-dose increase in a Slc39a8-transgenic mouse line. Am. J. Physiol. Cell Physiol., 292, C1523–C1535. [DOI] [PubMed] [Google Scholar]
- 8.Tellez-Plaza M., Navas-Acien A., Crainiceanu C.M., Guallar E. (2008) Cadmium exposure and hypertension in the 1999-2004 National Health and Nutrition Examination Survey (NHANES). Environ. Health Persp., 116, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa H., Nishijo M. (1996) Environmental cadmium exposure, hypertension and cardiovascular risk. J. Cardiovasc. Risk, 3, 11–17. [PubMed] [Google Scholar]
- 10.Staessen J.A., Kuznetsova T., Roels H.A., Emelianov D., Fagard R. (2000) Exposure to cadmium and conventional and ambulatory blood pressures in a prospective population study. Public Health and Environmental Exposure to Cadmium Study Group. Am. J. Hypertens., 13, 146–156. [DOI] [PubMed] [Google Scholar]
- 11.Houtman J.P. (1993) Prolonged low-level cadmium intake and atherosclerosis. Sci. Total Environ., 138, 31–36. [DOI] [PubMed] [Google Scholar]
- 12.Navas-Acien A., Selvin E., Sharrett A.R., Calderon-Aranda E., Silbergeld E., Guallar E. (2004) Lead, cadmium, smoking, and increased risk of peripheral arterial disease. Circulation, 109, 3196–3201. [DOI] [PubMed] [Google Scholar]
- 13.Everett C.J., Frithsen I.L. (2008) Association of urinary cadmium and myocardial infarction. Environ. Res., 106, 284–286. [DOI] [PubMed] [Google Scholar]
- 14.Varoni M.V., Palomba D., Gianorso S., Anania V. (2003) Cadmium as an environmental factor of hypertension in animals: new perspectives on mechanisms. Vet. Res. Commun., 27 Suppl 1, 807–810. [DOI] [PubMed] [Google Scholar]
- 15.Satarug S., Nishijo M., Lasker J.M., Edwards R.J., Moore M.R. (2006) Kidney dysfunction and hypertension: role for cadmium, p450 and heme oxygenases? Tohoku J. Exp. Med., 208, 179–202. [DOI] [PubMed] [Google Scholar]
- 16.Prozialeck W.C., Edwards J.R., Nebert D.W., Woods J.M., Barchowsky A., Atchison W.D. (2008) The vascular system as a target of metal toxicity. Toxicol. Sci., 102, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messner B., Knoflach M., Seubert A., Ritsch A., Pfaller K., Henderson B., Shen Y.H., Zeller I., Willeit J., Laufer G., et al. (2009) Cadmium is a novel and independent risk factor for early atherosclerosis mechanisms and in vivo relevance. Arterioscl. Throm. Vas., 29, 1392–1398. [DOI] [PubMed] [Google Scholar]
- 18.Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protocols, 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 19.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thevenod F. (2009) Cadmium and cellular signaling cascades: to be or not to be? _Toxicol._Appl. Pharmacol, 238, 221–239. [DOI] [PubMed] [Google Scholar]
- 21.Israel A. (2010) The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harbor Perspect. Biol., 2, a000158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajjimaporn A., Botsford T., Garrett S.H., Sens M.A., Zhou X.D., Dunlevy J.R., Sens D.A., Somji S. (2012) ZIP8 expression in human proximal tubule cells, human urothelial cells transformed by Cd + 2 and As + 3 and in specimens of normal human urothelium and urothelial cancer. Cancer Cell Int., 12, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alissa E.M., Ferns G.A. (2011) Heavy metal poisoning and cardiovascular disease. J. Toxicol., 2011, 870125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vestergaard P., Shaikh Z.A. (1994) The nephrotoxicity of intravenously administered cadmium-metallothionein: effect of dose, mode of administration, and preexisting renal cadmium burden. Toxicol. Appl. Pharmacol., 126, 240–247. [DOI] [PubMed] [Google Scholar]
- 25.Friberg L. (1984) Cadmium and the kidney. Environ. Health Persp., 54, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L., Wang B., Hay E.B., Nebert D.W. (2009) Discovery of ZIP transporters that participate in cadmium damage to testis and kidney. Toxicol. Appl. Pharmacol., 238, 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S., Yadav P., Trivedi R., Bansal A.K., Bhatnagar D. (1995) Cadmium-induced lipid peroxidation and the status of the antioxidant system in rat tissues. J. Trace Elem. Med. Biol., 9, 144–149. [DOI] [PubMed] [Google Scholar]
- 28.Bondjers G., Glukhova M., Hansson G.K., Postnov Y.V., Reidy M.A., Schwartz S.M. (1991) Hypertension and atherosclerosis. Cause and effect, or two effects with one unknown cause? Circulation, 84, VI2–V16. [PubMed] [Google Scholar]
- 29.Knoflach M., Messner B., Shen Y.H., Frotschnig S., Liu G., Pfaller K., Wang X., Matosevic B., Willeit J., Kiechl S., et al. (2011) Non-toxic cadmium concentrations induce vascular inflammation and promote atherosclerosis. Circ. J., 75, 2491–2495. [DOI] [PubMed] [Google Scholar]
- 30.Martin P., Pognonec P. (2010) ERK and cell death: cadmium toxicity, sustained ERK activation and cell death. FEBS J., 277, 39–46. [DOI] [PubMed] [Google Scholar]
- 31.Ramos J.W. (2008) The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int. J. Biochem. Cell B., 40, 2707–2719. [DOI] [PubMed] [Google Scholar]
- 32.Cagnol S., Chambard J.C. (2010) ERK and cell death: mechanisms of ERK-induced cell death–apoptosis, autophagy and senescence. FEBS J., 277, 2–21. [DOI] [PubMed] [Google Scholar]
- 33.Martin P., Poggi M.C., Chambard J.C., Boulukos K.E., Pognonec P. (2006) Low dose cadmium poisoning results in sustained ERK phosphorylation and caspase activation. Biochem. Bioph. Res. Co., 350, 803–807. [DOI] [PubMed] [Google Scholar]
- 34.Hart B.A., Lee C.H., Shukla G.S., Shukla A., Osier M., Eneman J.D., Chiu J.F. (1999) Characterization of cadmium-induced apoptosis in rat lung epithelial cells: evidence for the participation of oxidant stress. Toxicology, 133, 43–58. [DOI] [PubMed] [Google Scholar]
- 35.Chen G.G., Liu Z.M., Vlantis A.C., Tse G.M., Leung B.C., van Hasselt C.A. (2004) Heme oxygenase-1 protects against apoptosis induced by tumor necrosis factor-alpha and cycloheximide in papillary thyroid carcinoma cells. J. Cell. Biochem., 92, 1246–1256. [DOI] [PubMed] [Google Scholar]
- 36.Thevenod F., Friedmann J.M., Katsen A.D., Hauser I.A. (2000) Up-regulation of multidrug resistance P-glycoprotein via nuclear factor-kappaB activation protects kidney proximal tubule cells from cadmium- and reactive oxygen species-induced apoptosis. J. Biol. Chem., 275, 1887–1896. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z.M., Chen G.G., Ng E.K., Leung W.K., Sung J.J., Chung S.C. (2004) Upregulation of heme oxygenase-1 and p21 confers resistance to apoptosis in human gastric cancer cells. Oncogene, 23, 503–513. [DOI] [PubMed] [Google Scholar]
- 38.Hyun J.S., Satsu H., Shimizu M. (2007) Cadmium induces interleukin-8 production via NF-kappaB activation in the human intestinal epithelial cell, Caco-2. Cytokine, 37, 26–34. [DOI] [PubMed] [Google Scholar]
- 39.Johansson A., Eriksson N., Lindholm D., Varenhorst C., James S., Syvanen A.C., Axelsson T., Siegbahn A., Barratt B.J., Becker R.C., et al. (2016) Genome-wide association and Mendelian randomization study of NT-proBNP in patients with acute coronary syndrome. Hum. Mol. Genet., 25, 1447–1456. [DOI] [PubMed] [Google Scholar]
- 40.Ng E., Lind P.M., Lindgren C., Ingelsson E., Mahajan A., Morris A., Lind L. (2015) Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum. Mol. Genet., 24, 4739–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickrell J.K., Berisa T., Liu J.Z., Segurel L., Tung J.Y., Hinds D.A. (2016) Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet., 48(7):709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterworth D.M., Ricketts S.L., Song K., Chen L., Zhao J.H., Ripatti S., Aulchenko Y.S., Zhang W., Yuan X., Lim N., et al. (2010) Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscl. Throm. Vas., 30, 2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R., et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet., 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrera N., Arrojo M., Sanjuan J., Ramos-Rios R., Paz E., Suarez-Rama J.J., Paramo M., Agra S., Brenlla J., Martinez S., et al. (2012) Association study of nonsynonymous single nucleotide polymorphisms in schizophrenia. Biol. Psychiat., 71, 169–177. [DOI] [PubMed] [Google Scholar]
- 46.Orisakwe O.E. (2014) The role of lead and cadmium in psychiatry. N. Am. J. Med. Sci., 6, 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komatsu F., Kagawa Y., Kawabata T., Kaneko Y., Chimedregzen U., Purvee B., Otgon J. (2011) A high accumulation of hair minerals in Mongolian people: 2(nd) report; influence of manganese, iron, lead, cadmium and aluminum to oxidative stress, Parkinsonism and arthritis. Curr. Aging. Sci., 4, 42–56. [DOI] [PubMed] [Google Scholar]
- 48.Galvez-Peralta M., He L., Jorge-Nebert L.F., Wang B., Miller M.L., Eppert B.L., Afton S., Nebert D.W. (2012) ZIP8 zinc transporter: indispensable role for both multiple-organ organogenesis and hematopoiesis in utero. PloS One, 7, e36055.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider S.N., Liu Z., Wang B., Miller M.L., Afton S.E., Soleimani M., Nebert D.W. (2014) Oral cadmium in mice carrying 5 versus 2 copies of the Slc39a8 gene: comparison of uptake, distribution, metal content, and toxicity. Int. J. Toxicol., 33,14–20. [DOI] [PubMed] [Google Scholar]
- 50.Jaffe E.A., Nachman R.L., Becker C.G., Minick C.R. (1973) Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest., 52, 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. (2015) Ensembl 2015. Nucleic Acids Res., 43, D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res., 38, W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D.E., Chivian D., Baker D. (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res., 32, W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu MJ, Bao S, Gálvez-Peralta M, Pyle CJ, Rudawsky AC, Pavlovicz RE, Killilea DW, Li C, Nebert DW, Wewers MD, et al. (2013) ZIP8 regulates host defense through zinc-mediated inhibition of NF-Œ∫B. Cell Rep., 3, 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]