Outbreak of Klebsiella pneumoniae Producing a New Carbapenem-Hydrolyzing Class A β-Lactamase, KPC-3, in a New York Medical Center (original) (raw)
Abstract
From April 2000 to April 2001, 24 patients in intensive care units at Tisch Hospital, New York, N.Y., were infected or colonized by carbapenem-resistant Klebsiella pneumoniae. Pulsed-field gel electrophoresis identified a predominant outbreak strain, but other resistant strains were also recovered. Three representatives of the outbreak strain from separate patients were studied in detail. All were resistant or had reduced susceptibility to imipenem, meropenem, ceftazidime, piperacillin-tazobactam, and gentamicin but remained fully susceptible to tetracycline. PCR amplified a _bla_KPC allele encoding a novel variant, KPC-3, with a His(272)→Tyr substitution not found in KPC-2; other carbapenemase genes were absent. In the outbreak strain, KPC-3 was encoded by a 75-kb plasmid, which was transferred in vitro by electroporation and conjugation. The isolates lacked the OmpK35 porin but expressed OmpK36, implying reduced permeability as a cofactor in resistance. This is the third KPC carbapenem-hydrolyzing β-lactamase variant to have been reported in members of the Enterobacteriaceae, with others reported from the East Coast of the United States. Although producers of these enzymes remain rare, the progress of this enzyme group merits monitoring.
Carbapenems have the broadest activity spectra of any β-lactam antibiotic and are often the most appropriate agents for use in the treatment of infections caused by multiresistant gram-negative bacteria. The emergence and spread of acquired carbapenem resistance are therefore a major concern and have been dubbed a “global sentinel event” (31). Resistance occasionally arises from the combination of impermeability with an AmpC-type or extended-spectrum β-lactamase (ESBL) (4, 17); these enzymes have only very weak carbapenemase activity, and a greater potential concern is the slow emergence of resistance mediated by β-lactamases that hydrolyze carbapenems strongly (15). These β-lactamases include acquired metallo-β-lactamases that belong to molecular class B and that are encoded by alleles of _bla_IMP, _bla_VIM, _bla_SPM, or _bla_GIM and carbapenem-hydrolyzing class D β-lactamases encoded by various _bla_OXA alleles. Although still infrequent, the Acinetobacter and Pseudomonas spp. producing these enzymes have a wide and growing geographic distribution and have been associated with hospital outbreaks (2, 5, 15, 35).
There are fewer reports of acquired metallo-carbapenemases in members of the Enterobacteriaceae, but detection is complicated by the fact that the carriage of the corresponding genes does not always confer obvious resistance (15). Nevertheless, _bla_IMP alleles have been detected in Klebsiella, Citrobacter, and Serratia spp. in Japan (25), Singapore (13), the People's Republic of China (7), and Taiwan (GenBank accession number AF322577), while _bla_VIM alleles have been found in an isolate of Serratia marcescens in Korea (AY030343) and an Escherichia coli isolate from Greece (AY339625). Two klebsiellae producing IMP or VIM enzymes have also been detected in the United Kingdom (N. Woodford and D. M. Livermore, unpublished data). Finally, there are a few reports of Enterobacteriaceae with class A β-lactamases able to hydrolyze carbapenems. These β-lactamase enzymes, not yet found in nonfermenters, include the near-identical IMI-1 (29) and NMC-A (23) types, both from Enterobacter cloacae, and SME-1 (21, 37) and SME-2 (27), both from Serratia marcescens. IMI, NMC, and SME types have been found rarely, sometimes in isolates collected even before imipenem was launched, and show no evidence of spread or transfer in vitro. Potentially of more significance are recent reports of KPC carbapenemases. The first of these, KPC-1, was found in a Klebsiella pneumoniae isolate from North Carolina (38); KPC-2 was then found in isolates of K. pneumoniae from Baltimore, Maryland (20), and Salmonella enterica serotype Cubana, also in Maryland (18), as well as in Klebsiella oxytoca and K. pneumoniae isolates from New York (3, 39). These recent discoveries raise the disturbing possibility that the dissemination of KPC enzymes may have already begun in the eastern United States. In the present report, we describe an outbreak of infections in New York caused by carbapenem-resistant K. pneumoniae strains that produced another KPC variant, designated KPC-3.
(This work was presented in part previously [K. Young et al., 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-50, 2003].)
MATERIALS AND METHODS
The outbreak.
Between April 2000 and April 2001, 24 patients in intensive care units (ICUs) at the Tisch Hospital, NYU Medical Center, were infected by carbapenem-resistant K. pneumoniae. The Tisch Hospital has eight ICUs, and the outbreak affected the medical, surgical, cardiovascular, and coronary care units, all housed on the same floor of the hospital. Medical and surgical staff often moved among the four units, providing an opportunity for nosocomial spread.
Bacterial isolates and reference strains.
Three clinical isolates, CL 5761 to CL 5763, obtained from separate patients at the NYU Medical Center and identified as K. pneumoniae by standard laboratory methods, were studied in detail. Each was reported to be resistant to imipenem (meropenem was not tested) and multiple other antimicrobials by the clinical laboratory, where Vitek and Kirby-Bauer methods were used. The isolates were sent to the Merck Clinical Laboratory, where isolate CL 5762 was noted to contain two colony variants on MacConkey and blood agar plates; these were tested separately as CL 5762A and CL 5762B. Isolates CL 5761, CL 5762A, CL 5762B, and CL 5763 subsequently were referred to the Health Protection Agency's Antibiotic Resistance Monitoring and Reference Laboratory in London, United Kingdom.
Antimicrobial susceptibility testing.
MICs were determined by a standard NCCLS broth microdilution method or by Etest (Cambridge Diagnostic Services Ltd., Cambridge, United Kingdom). Susceptibilities to β-lactams were also determined by microdilution in the presence of clavulanic acid at a fixed concentration of 4 μg/ml; a ≥8-fold reduction in the MICs of oxyimino-β-lactams in the presence of clavulanic acid was taken to imply ESBL production (28). A susceptible K. pneumoniae strain, CL 15245, was included for comparison.
Pulsed-field gel electrophoresis (PFGE).
Overnight growth from sheep blood agar was suspended in 2 ml of sterile saline to an optical density at 550 nm of 0.5, and 500 μl of the suspension was transferred to a 1.5-ml microcentrifuge tube and spun for 5 min at 4°C at 20,200 × g. The cell pellet was washed once in 1.0 ml of Carl's buffer (50 mM EDTA, 20 mM NaCl, 10 mM Tris [pH 7.6]) and then resuspended in 100 μl of Carl's buffer with an equal volume of molten plug agarose (1.6% agarose in Carl's buffer). Two agarose plugs were made by transferring 100 μl of this mixture to two plug molds. These plugs were solidified on ice for 15 min, transferred into 2-ml microcentrifuge tubes containing 1.0 ml of ES buffer (0.5 M EDTA, 1% Sarkosyl) and 20 μl of proteinase K (20 mg/ml), and incubated overnight at 55°C before being washed in at least five changes of TE (10 mM Tris, 1.0 mM EDTA). Half of one plug was soaked in 1.0 ml of TE for 30 min and then in 150 μl of 1× XbaI restriction buffer with 15 U of XbaI and then incubated for at least 3 h at 37°C. Half of this digested half-plug was loaded on a 1% agarose gel made in 0.5× Tris-borate-EDTA (TBE). Electrophoresis was carried out in 0.5× TBE buffer at 14°C with an initial switch time of 1 s and a final switch time of 40 s for 16 h at 6 V/cm. The gel was stained with ethidium bromide (1 μg/ml) for 20 min, destained with distilled water for 20 min, and viewed under UV transillumination. Gel images were captured by using a Gel Doc 1000 system (Bio-Rad, Hercules, Calif.).
Hydrolysis of imipenem and isoelectric focusing.
Hydrolysis of 0.1 mM imipenem (Merck, Hoddesdon, United Kingdom) was monitored by UV spectrophotometry at 297 nm and 37°C in 10 mM phosphate buffer (pH 7.0). Activity was standardized relative to protein concentration, which was determined by the Bio-Rad protein microassay, which is based on the Bradford method (2a). Bovine serum albumin was used as the standard. Uninduced β-lactamase activity was assayed by growing bacterial cultures overnight in nutrient broth (NB), diluting them 10-fold into fresh broth, incubating them for 4 h, harvesting the cells, and disrupting the cells by sonication. Cultures of K. pneumoniae were grown overnight at 37°C on nutrient agar, with or without 0.125 times the MIC of imipenem, potentially to induce β-lactamase. The cells were harvested into 2-ml volumes of 10 mM phosphate buffer (pH 7.0) and disrupted by four cycles of freezing and thawing. Debris was removed by centrifugation at 15,000 × g for 15 min, and the supernatants were retained at −20°C and then assayed against imipenem as described above.
Inhibition of carbapenemase activity by EDTA was investigated by incubating 50 μl of crude enzyme extract with 900 μl of 0.1 mM EDTA (pH 8.0) at 37°C for 1 h and then subjecting the mixture to an assay with 0.1 mM imipenem as described above. Extracts of Pseudomonas aeruginosa 101/1477 with an IMP-1 enzyme (14) and a derivative of E. coli JM109 with an NMC-A enzyme (23) were used as controls.
Samples of the crude enzyme extracts were analyzed by isoelectric focusing as described previously (36).
PCR for resistance genes.
Isolates were examined by PCR using Taq polymerase (Invitrogen, Paisley, United Kingdom) and previously reported conditions with primers specific for _bla_IMP (32), _bla_VIM (35), _bla_SME (27), _bla_KPC (38), and _bla_IMI or _bla_NMC (Table 1).
TABLE 1.
PCR primers used in this study
| Gene | Primer sequences (5′-3′) | Product size (bp) | Reference |
|---|---|---|---|
| _bla_IMP | For, CTACCGCAGCAGAGTCTTTG | 587 | 32 |
| Rev, AACCAGTTTTGCCTTACCAT | |||
| _bla_VIM | For, AGTGGTGAGTATCCGACAG | 261 | 35 |
| Rev, ATGAAAGTGCGTGGAGAC | |||
| _bla_SME | For, AACGGCTTCATTTTTGTTTAG | 830 | 27 |
| Rev, GCTTCCGCAATAGTTTTATCA | |||
| _bla_KPC | For, TGTCACTGTATCGCCGTC | ca. 1,000 | 38 |
| Rev, CTCAGTGCTCTACAGAAAACC | |||
| _bla_IMI/_bla_NMC | For, CCA TTC ACC CAT CAC AAC | 440 | This study (based on GenBank accession no. U50278) |
| Rev, CTA CCG CAT AAT CAT TTG C | |||
| waaE | For, CTG TCG CGT CTG CTA CAG TT | 851 | This study (based on GenBank accession no. AF146532) |
| Rev, CCG GAT AGA TCG CTT TTG TG | |||
| qnr | For, GAT AAA GTT TTT CAG CAA GAG G | 593 | 10 |
| Rev, ATC CAG ATC GGC AAA GGT TA |
The plasmid-carried quinolone resistance gene qnr was also sought (10) (Table 1). PCR conditions were denaturation at 95°C for 1 min, annealing at 47, 52, or 57°C for 30 s, and extension at 72°C for 1 min, for 30 cycles on a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany). Multiple annealing temperatures were used with the gradient thermocycler in an attempt to amplify related, but nonidentical, sequences.
The product of waaE is a glucosyl transferase involved in core lipopolysaccharide biosynthesis (30), and its disruption was associated with a 16-fold decrease in imipenem susceptibility of a laboratory mutant of K. pneumoniae (J. Tomas, M. C. Conejo, S. Merino, M. Regue, V. J. Benedi, A. Pascual, and L. Martinez-Martinez, Proc. Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1517, 2001). Primers were designed to amplify an 851-bp fragment containing the entire waaE gene (Table 1). This fragment was amplified from isolate CL 5761 and sequenced as detailed below.
Sequencing of a _bla_KPC PCR product.
A ca. 1-kb PCR product obtained from strain CL 5761 by two independent PCRs with _bla_KPC primers was purified with a Recovery DNA purification kit II (Hybaid, Ashford, United Kingdom) and sequenced in both directions with a dye-labeled ddNTP Terminator cycle sequencing kit (Beckman Coulter UK Ltd., High Wycombe, United Kingdom). Samples were analyzed on a CEQ 2000 automated sequencer (Beckman Coulter). Initially, primers 5 and 10 (38) were used, and the data were confirmed with four internal primers designed on the basis of the obtained sequence. A contig was assembled with the GeneBuilder component of BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) and translated with BioEdit (6). The signal sequence was predicted with SignalP version 1.1 (http://www.cbs.dtu.dk/services/SignalP/), and the molecular weight and pI of the mature peptide were predicted by using the ExPASy Compute pI/M w tool (http://www.expasy.ch/tools/pi_tool.html).
Location and in vitro transfer of _bla_KPC.
Plasmids were isolated from K. pneumoniae isolates with a QIAfilter midi kit (QIAGEN, Valencia, Calif.) used in accordance with the manufacturer's instructions and were separated by electrophoresis through a 0.6% agarose gel in the presence of 1× TBE buffer at a constant voltage of 90 V for 16 h at 4°C. Supercoiled DNA of plasmid R100.1 was used as a size standard (24, 33), as were plasmids extracted from E. coli strains 39R861 (34) and V517 (16). Extracted plasmids were transformed by electroporation into competent cells prepared from susceptible K. pneumoniae strain CL 15245, which had its full complement of porins, and also into competent cells of E. coli strains DH5α, LS279, and LS584 (41). Electroporation (Gene Pulser; Bio-Rad) was performed at 2.48 kV, 25 μF, and 200 Ω in 0.2-cm-gap cuvettes by using 50 μl of cells and 1 μl of DNA. Immediately after the pulse was applied, the cells were placed on ice, suspended in 1 ml of SOC medium (Invitrogen, Carlsbad, Calif.), and incubated, with shaking, for 1 h at 37°C. Transformants were selected by plating 100 μl of cells onto Luria-Bertani agar containing ceftazidime (2 μg/ml) or imipenem (2 μg/ml).
DNA from clinical isolates and transformants was transferred from the agarose gel to a Duralon-UV nylon membrane (Stratagene, La Jolla, Calif.) by capillary action overnight by use of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) transfer buffer and was fixed by using a Stratalinker UV cross-linker (Stratagene) on the “autocrosslink” setting. The DNA on the membrane was hybridized with a fluorescein-labeled KPC-3 DNA probe created with the Prime-It Fluor fluorescence labeling kit (Stratagene) with primers 5 and 10 (31) in accordance with the manufacturer's instructions. Hybridization was performed using the Illuminator chemiluminescent detection system (Stratagene) in accordance with the manufacturer's protocol. Pictures of the agarose gel and Southern blot were taken with an Image Station 440CF (Kodak, Rochester, N.Y.).
Conjugal transfer of _bla_KPC-3 to E. coli J62-2 (Rifr) was attempted by a filter-mating protocol. Transconjugants were selected on Mueller-Hinton agar containing rifampin (100 μg/ml) and either ceftazidime (10 μg/ml) or meropenem (1 μg/ml). The plasmid content of transformants was determined as described above, while plasmids were extracted from transconjugants as described previously (11). The presence of a _bla_KPC allele in transconjugants and transformants was confirmed by PCR (38).
Outer membrane protein (OMP) profiles.
Isolates of K. pneumoniae were grown in Luria-Bertani broth and NB as high- and low-osmolarity media, respectively. Outer membranes were isolated and prepared for electrophoresis by a procedure adapted from published methods (9, 40). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (8) were run with 11% acrylamide and 0.21% bisacrylamide, with 5 μg of protein loaded per lane.
Nucleotide sequence accession numbers.
The sequences of _bla_KPC-3 and waaE from K. pneumoniae strain CL 5761 reported in the present paper have been deposited in GenBank under accession numbers AF395881 and AF482468, respectively.
RESULTS
History and control of the outbreak.
Twenty-four patients in ICUs at the Tisch Hospital, NYU Medical Center, were colonized or infected with carbapenem-resistant K. pneumoniae between April 2000 and April 2001 (Table 2). Klebsiellae with this phenotype had not been detected in the hospital previously. All infections were nosocomially acquired, with the patients having been hospitalized from 9 to 374 days prior to isolation of the organism. Risk factors for acquisition included prolonged hospitalization, an ICU stay, and ventilator usage. Carbapenem-resistant organisms were isolated predominantly from respiratory secretions but also from urine and blood. Fourteen of the 24 patients were infected, and 8 of these died, with the Klebsiella infection considered causative or contributory. The isolates were also broadly resistant to many antibiotic classes, and various therapies were used with little effect. Owing to their resistance, infections caused by these klebsiellae proved extremely difficult to eradicate, and physical isolation of colonized or infected patients was necessary in order to confine the outbreak. The importance of an aseptic technique was reemphasized to the house staff by providing an infection control on-call service to all ICU staff on all shifts. As the sites of the infections changed from respiratory secretions to urine and blood, contamination of Foley catheters was suspected. Indeed, it was discovered that Foley catheters were a common source of contamination due to an improper technique being used to empty the collection bags. The outbreak was eventually controlled and eliminated by April 2001 by a combination of intensive infection control measures and rigorous local surveillance, these safeguards remain in place.
TABLE 2.
Details of patients from whom carbapenem-resistant K. pneumoniae was isolated
| Patient | Date isolated (mo/yr) | Site(s) | ICU type(s)a | Status | Outcome |
|---|---|---|---|---|---|
| 1 | 4/00 | Sputum | CCU | Infection | Died |
| 2 | 5/00 | Sputum | MICU | Infection | Died |
| 3 | 5/00 | Sputum | MICU | Colonization | Recovered |
| 4 | 5/00 | Sputum and blood | SICU | Infection | Died |
| 5 | 5/00 | Sputum and urine | CCU | Colonization | Recovered |
| 6 | 5/00 | Sputum | MICU | Infection | Recovered |
| 7 | 6/00 | Sputum and urine | SICU | Infection | Recovered |
| 8 | 6/00 | Urine | SICU | Infection | Recovered |
| 9 | 7/00 | Wound | SICU and MICU | Colonization | Recovered |
| 10 | 7/00 | Sputum | MICU | Colonization | Recovered |
| 11 | 7/00 | Blood and urine | MICU | Infection | Recovered |
| 12 | 7/00 | Blood and urine | CCU and MICU | Infection | Died |
| 13 | 8/00 | Blood | SICU | Infection | Died |
| 14 | 8/00 | Blood | SICU | Infection | Died |
| 15 | 8/00 | Sputum | SICU | Colonization | Recovered |
| 16 | 9/00 | Sputum | CVCU | Infection | Recovered |
| 17 | 9/00 | Wound | SICU | Infection | Died |
| 18 | 9/00 | Sputum | MICU | Colonization | Recovered |
| 19 | 10/00 | Sputum | MICU | Infection | Died |
| 20 | 12/00 | Wound | SICU | Colonization | Recovered |
| 21 | 1/01 | Sputum | MICU | Colonization | Recovered |
| 22 | 2/01 | Sputum | SICU | Colonization | Recovered |
| 23 | 3/01 | Wound | SICU | Colonization | Recovered |
| 24 | 4/01 | Catheter | SICU | Infection | Recovered |
PFGE analysis.
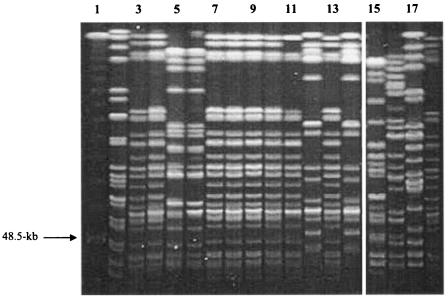
PFGE analysis of K. pneumoniae isolates recovered during the outbreak identified a predominant strain (isolated from 19 patients), but isolates representing two unrelated strains were also recovered (Fig. 1). Isolates CL 5761, CL 5762, and CL 5763, which were indistinguishable from each other and belonged to the outbreak strain, were examined in greater detail at the Merck Clinical Laboratory and the Antibiotic Resistance Monitoring and Reference Laboratory. Two colony variants of CL 5762 were noted and tested separately as CL 5762A and CL 5762B.
FIG. 1.
PFGE analysis of XbaI-digested genomic DNA from carbapenem-resistant isolates of K. pneumoniae. Lane 1 contains a concatemer of phage λ DNA used as a molecular size marker. Isolates representing the predominant outbreak strain are shown in lanes 3, 4, 7 to 11, 13, and 18. Other lanes show isolates unrelated to the outbreak strain; the types represented in lanes 5 and 6 and in lanes 12 and 14 were related organisms, each isolated from two or more patients.
Antibiotic susceptibilities.
Isolates CL 5761, CL 5762B, and CL 5763 were nonsusceptible to imipenem and meropenem based on MICs of >4 μg/ml (22) and more resistant to ertapenem (MICs, >32 μg/ml) (Table 3). Isolate CL 5762A appeared susceptible to carbapenems at NCCLS breakpoints, although the MICs were higher for this isolate than for the control strain and rose further after storage for 1 year. The MICs of several β-lactams were reduced by the presence of clavulanate at 4 μg/ml; this synergy was most obvious with isolate CL 5762A (the least-carbapenem-resistant organism). All of the isolates were resistant to oxyimino cephalosporins with MICs of ≥256 mg/liter (Table 3), but only isolate CL 5762A was inferred to produce an ESBL based on the criterion of a ≥8-fold reduction in the MICs of oxyimino-β-lactams by clavulanate (28); however, even this isolate did not show the great susceptibility to ceftazidime or clavulanate typical of ESBL producers. Synergy between carbapenem and clavulanate was weak or absent (Table 3). All of the isolates were resistant to other cephalosporins, piperacillin-tazobactam, amikacin, and gentamicin and were at least intermediately resistant to ciprofloxacin; isolate CL 5762B was resistant to polymyxin B, but the others were susceptible. All were susceptible to tetracycline.
TABLE 3.
Antibiotic susceptibilitiesa and carbapenemase activities of clinical isolates of K. pneumoniae
| Antibioticb | Inhibitor | MIC (μg/ml) | ||||
|---|---|---|---|---|---|---|
| CL 5761 | CL 5762A | CL 5762B | CL 5763 | CL 15245 | ||
| Imipenem | − | 16 | 2 | 16 | 16 | ≤0.125 |
| + CLAc | 8 | 0.5 | 8 | 8 | ≤0.25 | |
| Meropenem | − | 8 | 1 | 16 | 16 | ≤0.125 |
| + CLA | 8 | 2 | 32 | 16 | ≤0.125 | |
| Ertapenemd | − | >32 | 0.25 | >32 | >32 | NTe |
| Aztreonam | − | >256 | 256 | >256 | >256 | ≤0.25 |
| + CLA | 256 | 64 | >256 | >256 | ≤0.25 | |
| Ceftazidime | − | 256 | 256 | >256 | >256 | ≤0.25 |
| + CLA | 128 | 32 | 256 | >256 | ≤0.25 | |
| Ceftriaxone | − | 256 | >256 | >256 | >256 | ≤0.25 |
| + CLA | 64 | 32 | 128 | 256 | ≤0.25 | |
| Cefepime | − | 32 | 128 | 128 | 128 | ≤0.25 |
| + CLA | 32 | 8 | 64 | 64 | ≤0.25 | |
| Cefoxitin | − | 128 | 32 | 256 | 256 | 2 |
| + CLA | 256 | 32 | 256 | 256 | <1 | |
| Piperacillind | + Tazo- bactam | >256 | >256 | >256 | >256 | NT |
| Amikacin | − | 16 | 32 | 32 | 32 | 0.5 |
| Gentamicin | − | 8 | 8 | 16 | 16 | ≤0.25 |
| Chloramphenicol | − | 128 | 64 | 64 | 128 | 4 |
| Ciprofloxacin | − | 4 | 2 | 4 | 16 | ≤0.06 |
| Polymyxin B | − | 0.25 | 0.25 | 64 | 0.125 | 0.25 |
| Tetracycline | − | 4 | 1 | 4 | 4 | 1 |
Detection of a carbapenem-hydrolyzing β-lactamase.
Cell extracts prepared from each of the four selected K. pneumoniae isolates hydrolyzed imipenem in the spectrophotometric assay, and the extracts had similar specific activities (Table 3) despite the susceptibility of CL 5762A to carbapenems. The ratios of β-lactamase activity determined in the presence or absence of induction ranged from 0.5 to 1.2, indicating no significant inducibility. Imipenem hydrolysis was not inhibited by preincubation of extracts with 0.1 mM EDTA for 1 h, whereas the activity of the IMP-1 enzyme from P. aeruginosa 101/1477 was inhibited by this treatment. These data suggest that the K. pneumoniae isolates produced a nonmetallo-β-lactamase, and this conclusion was supported by the absence of PCR products with primers for _bla_IMP and _bla_VIM alleles. Likewise, no products were obtained with primers for _bla_IMI or _bla_SME, whereas a product of ca. 1 kb was obtained from the four isolates with primers for _bla_KPC.
Molecular characterization of _bla_KPC-3.
Both strands of the PCR product from strain CL 5761 were sequenced by direct cycle sequencing. The product contained an open reading frame of 882 bp encoding 293 amino acids, with two nucleotide changes not present in _bla_KPC-1 (GenBank accession number AF297554). These mutations were predicted to result in amino acid substitutions of Ser(174)→Gly and His(272)→Tyr (Fig. 2). The KPC-2 enzyme (AY034847) also has the Ser(174)→Gly substitution. The novel enzyme and corresponding allele were designated KPC-3 and _bla_KPC-3, respectively. The first 24 residues of the _bla_KPC-3 translation product were predicted to be a cleavable signal peptide; hence, the mature KPC-3 enzyme had a predicted molecular weight of 28,503 and a predicted pI of 5.97. The latter value was comparable to the pI of ca. 6.5 determined by isoelectric focusing (data not shown).
FIG. 2.
Comparison of KPC enzyme sequences. The predicted 24-residue cleavable signal peptide is shaded.
Analysis and transfer of plasmid DNA.
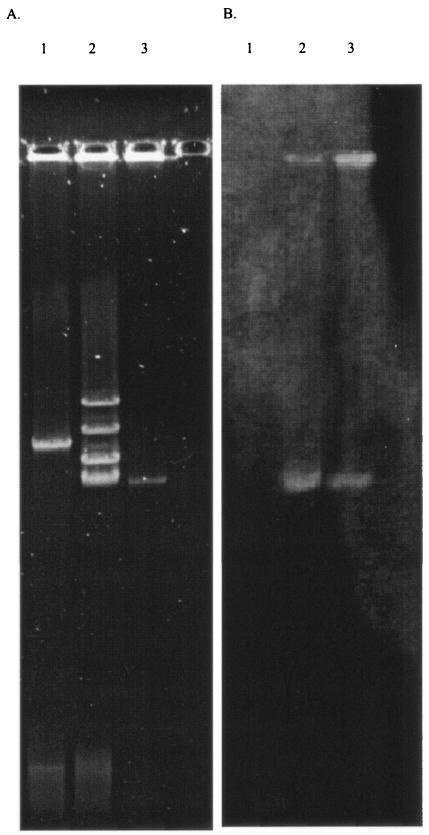
Isolates CL 5761 (Fig. 3), CL 5762A, CL 5762B, and CL 5763 each contained four large plasmids; isolates CL 5762A and CL 5762B contained a fifth, slightly smaller plasmid. Plasmid extracts prepared from all strains were transformed into E. coli DH5α, LS279, and LS584 by electroporation. Transformants of all three E. coli strains were obtained during selection with ceftazidime at 2 μg/ml but not with imipenem at 2 μg/ml. The _bla_KPC-3 allele was detected in all the transformants by PCR, and plasmid analysis indicated that they had acquired a single plasmid of ca. 75 kb, corresponding to the smallest of the four plasmids present in all the isolates (Fig. 3a); this plasmid hybridized with a _bla_KPC probe (Fig. 3b) and was transferable also by conjugation from isolates CL 5761 and CL 5763 to E. coli strain J62-2 at frequencies of 10−4 to 10−6 per input donor cell. No transformants were obtained with K. pneumoniae CL 15245 as the recipient.
FIG. 3.
Plasmid profiles (A) and a Southern blot hybridized with a _bla_KPC probe (B) of K. pneumoniae CL 5761 (lanes 2) and a representative _bla_KPC-3-containing E. coli electrotransformant (lanes 3). Lanes 1 show the 94-kb plasmid R100.1 (22, 31) as a guide to molecular size.
The MICs of carbapenems, extended-spectrum cephalosporins, and monobactams, but not cefoxitin, increased for transformants of E. coli DH5α. The MICs of β-lactams for them were consistently lower than those for the parent strain, CL 5761 (Table 4). Resistance to amikacin was transferred, but only a fourfold increase in gentamicin resistance was noted. Resistance to ciprofloxacin was not transferred, and qnr was not detected in either the transformants or the clinical isolates.
TABLE 4.
Antibiotic susceptibilities of E. coli transformants with the 75-kb KPC-3-encoding plasmid
| Antibiotic | MIC (μg/ml) for: | ||
|---|---|---|---|
| DH5α | DH5α (KPC-3)a | CL 5761 | |
| Imipenem | 0.12 | 1 | 8 |
| Meropenem | ≤0.03 | 0.25 | 16 |
| Ceftazidime | 0.12 | 16 | >32 |
| Ceftriaxone | ≤0.25 | 4 | 128 |
| Aztreonam | ≤0.25 | 16 | >256 |
| Cefoxitin | 4 | 8 | 256 |
| Amikacin | 0.5 | 16 | 16 |
| Gentamicin | 0.25 | 1 | 16 |
| Streptomycin | 2 | 2 | 2 |
| Chloramphenicol | 8 | 8 | 128 |
| Ciprofloxacin | 0.06 | 0.06 | 4 |
| Polymyxin B | 0.5 | 0.5 | 1 |
Analysis of OMPs.
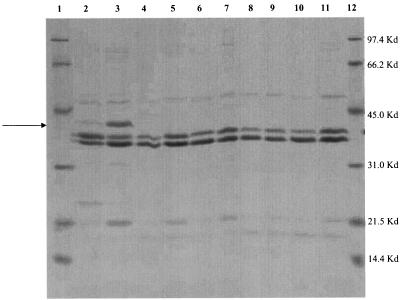
Two major OMPs, migrating between the 45.0- and 31.0-kDa standards, were observed in all the K. pneumoniae isolates (Fig. 4). A third, slightly larger band was induced in the susceptible control strain, CL 15245, after growth in a low-osmolarity medium (NB) (Fig. 4, lane 3); this protein corresponds to OmpK35, a homolog of E. coli porin OmpF (8). The three clinical isolates (including variants CL 5762A and CL 5672B) lacked the OmpK35 porin even after growth under inducing conditions of low osmolarity.
FIG. 4.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of OMPs extracted from carbapenem-resistant K. pneumoniae isolates. Lanes 1 and 12 show protein molecular size markers as indicated in kilodaltons to the right of the gel (Bio-Rad). Other lanes contain control strain CL 15245 (lanes 2 and 3), CL 5761 (lanes 4 and 5), CL 5762A (lanes 6 and 7), CL 5762B (lanes 8 and 9), and CL 5763 (lanes 10 and 11). Extracts in lanes 2, 4, 6, 8, and 10 were prepared from cells grown in Luria-Bertani broth (high osmolarity); those in lanes 3, 5, 7, 9, and 11 were prepared from cells grown in NB (low osmolarity). The arrow indicates the band corresponding to the inducible porin OmpK35.
Amplification and sequencing of waaE.
Disruption of the waaE gene has been implicated in decreased imipenem susceptibility in K. pneumoniae mutants in vitro (Tomas et al., 41st ICAAC). To determine whether changes in waaE contributed to the resistance phenotype of clinical isolates harboring _bla_KPC-3, this gene was sequenced from isolate CL 5761. In comparison with a reference sequence (GenBank accession number AF146532), the waaE sequence of K. pneumoniae CL 5761 contained six nucleotide changes; five were silent, but one was predicted to cause an Ile(31)→Val substitution. It seems unlikely that this conservative change would have a major effect on structure and function.
DISCUSSION
Nosocomially acquired, carbapenem-resistant K. pneumoniae isolates were first seen at the Tisch Hospital in April 2000, being recovered from the respiratory secretions of patients in ICUs. K. pneumoniae ranks fourth as a cause of hospital-acquired pneumonia in the United States (1), with its principal nosocomial reservoirs reported to be contaminated medical equipment, the hands of hospital staff, and the gastrointestinal tracts of patients (26). PFGE of isolates from 24 patients identified one predominant strain from 19 patients, although other resistant strains were also identified. The isolates were obtained from debilitated patients who had received multiple intensive antibiotic treatments, and antibiotic pressure may have been a factor selecting for initial colonization and the development of further resistance.
Three representatives of the outbreak strain were studied and shown to produce a carbapenem-hydrolyzing enzyme; sequencing revealed it to be a novel member of the KPC family, designated KPC-3. These isolates also lacked OmpK35, which may have reduced permeability and have potentiated their resistance. We also found an Ile(31)→Val substitution in waaE, a gene associated previously with imipenem resistance in a laboratory mutant of K. pneumoniae (Tomas et al., 41st ICAAC), but this substitution was considered unlikely to contribute to the carbapenem resistance of the outbreak strain.
In addition to their carbapenem resistance, these isolates were mostly resistant to cephalosporins, penicillins, and aminoglycosides. Variant CL 5762A showed greater susceptibility to carbapenems and other β-lactams, but no explanation for this behavior was found; the organism had a similar carbapenemase-specific activity and a similar OMP profile to the other three isolates.
Using transformation, conjugation, and hybridization studies, we located the _bla_KPC allele on a ca. 75-kb plasmid and showed that this allele also carried an aminoglycoside resistance determinant conferring resistance to amikacin and low-level resistance to gentamicin—a resistance profile consistent with an AAC(6′) variant. MICs for KPC-3-producing transformants of porin-proficient E. coli DH5α tended to be lower than those reported for KPC-1-producing transformants of the same strain (38). This finding may reflect either alterations in the substrate specificity of KPC-3 contingent on amino acid differences or the quantity of enzyme produced. The transformant was also much less resistant than the original isolate, probably owing to the lack of any permeability lesion.
Carbapenem-resistant klebsiellae remain rare. No such isolates were reported to The Surveillance Network from 1998 to 2001 (12), and a 24-hospital U.S. survey of K. pneumoniae from 1996 to 2000 revealed only four resistant isolates (from a single center) among 1,123 isolates examined; these isolates produced the KPC-2 enzyme (19). Nevertheless, since 2000, KPC family enzymes have been detected in the New England and Mid-Atlantic regions of the United States, predominantly in Klebsiella spp. but also in a variety of enteric organisms. Recent reports of KPC-2 include a Salmonella isolate in Maryland (18), K. oxytoca and K. pneumoniae isolates in New York (3, 39), and an E. cloacae isolate in Massachusetts (A. Hossain, M. J. Ferraro, R. M. Pino, R. B. Dew III, E. S. Moland, T. J. Lockhart, K. S. Thomson, R. V. Goering, and N. D. Hanson, Proc. Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-664, 2003). The KPC-3 enzyme, the variant described here, has also been identified in E. coli in New Jersey (T. Hong, E. S. Moland, B. Abdalhamid, N. D. Hanson, J. Wang, C. Sloan, D. Fabian, A. Farrajallah, J. Levine, and K. S. Thomson, Proc. Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-665, 2003). Transfer of these enzymes has been demonstrated in vitro both here and in one other study (20), and it may be that plasmids encoding them are beginning to spread in the clinical setting. If so, this would be a disturbing development, owing to the multiresistance of the resulting strains. Clearly, surveillance of these and other carbapenemases is warranted. The main selective factors remain uncertain; however, all members of the KPC family identified thus far (18, 38) confer greater resistance to cephalosporins than to carbapenems. Cephalosporin use, therefore, may be as selective for their spread as carbapenem use. Furthermore, whereas the presence of this enzyme alone is sufficient to result in cephalosporin resistance, resistance to carbapenems appears to require the presence of an additional mechanism, such as decreased permeability.
Acknowledgments
The work of N.W. and D.M.L. on emerging β-lactamases is supported, in part, by the EU/FP6-funded COBRA project (6-PCRD LSHM-CT-2003-503-335).
REFERENCES
- 1.Anonymous. 2000. National Nosocomial Infection Surveillance (NNIS) System Report, data summary from January 1992-April 2000, issued June 2000. Am. J. Infect. Control 28**:**429-448. [DOI] [PubMed] [Google Scholar]
- 2.Bou, G., G. Cerveró, M. A. Domínguez, C. Quereda, and J. Martínez-Beltrán. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38**:**3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72**:**248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39**:**55-60. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41**:**563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, Y.-W., M. Afzal-Shah, E. T. S. Houang, M.-F. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45**:**710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41**:**95-98. [Google Scholar]
- 7.Hawkey, P. M., J. Xiong, H. Ye, H. Li, and F. H. M'Zali. 2001. Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China. FEMS Microbiol. Lett. 194**:**53-57. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Alles, S., S. Alberti, D. Alvarez, A. Domenech-Sanchez, L. Martinez-Martinez, J. Gil, J. M. Tomas, and V. J. Benedi. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145**:**673-679. [DOI] [PubMed] [Google Scholar]
- 9.Hosaka, M., N. Gotoh, and T. Nishino. 1995. Purification of a 54-kilodalton protein (OprJ) produced in NfxB mutants of Pseudomonas aeruginosa and production of a monoclonal antibody specific to OprJ. Antimicrob. Agents Chemother. 39**:**1731-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47**:**559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kado, C. I., and S.-T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145**:**1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlowsky, J. A., M. E. Jones, C. Thornsberry, I. R. Friedland, and D. F. Sahm. 2003. Trends in antimicrobial susceptibilities among Enterobacteriaceae isolated from hospitalized patients in the United States from 1998 to 2001. Antimicrob. Agents Chemother. 47**:**1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh, T. H., L.-H. Sng, G. S. Babini, N. Woodford, D. M. Livermore, and L. M. C. Hall. 2001. Carbapenem-resistant Klebsiella pneumoniae in Singapore producing IMP-1 β-lactamase and lacking an outer membrane protein. Antimicrob. Agents Chemother. 45**:**1939-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J.-M. Frère, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43**:**902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3**:**489-495. [DOI] [PubMed] [Google Scholar]
- 16.Macrina, F. L., D. J. Kopecko, K. R. Jones, D. J. Ayers, and S. M. McCowen. 1978. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1**:**417-420. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Martínez, L., A. Pascual, S. Hernández-Allés, D. Alvarez-Díaz, A. I. Suárez, J. Tran, V. J. Benedí, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43**:**1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miriagou, V., L. S. Tzouvelekis, S. Rossiter, E. Tzelepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47**:**1297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moland, E. S., J. A. Black, J. Ourada, M. D. Reisbig, N. D. Hanson, and K. S. Thomson. 2002. Occurrence of newer β-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob. Agents Chemother. 46**:**3837-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moland, E. S., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51**:**711-714. [DOI] [PubMed] [Google Scholar]
- 21.Naas, T., L. Vandel, W. Sougakoff, D. M. Livermore, and P. Nordmann. 1994. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38**:**1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.Nordmann, P., S. Mariotte, T. Naas, R. Labia, and M.-H. Nicolas. 1993. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 37**:**939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohtsubo, H., and E. Ohtsubo. 1976. Isolation of inverted repeat sequences, including IS1, IS2, and IS3, in Escherichia coli plasmids. Proc. Natl. Acad. Sci. USA 73**:**2316-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38**:**71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11**:**589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Queenan, A. M., C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos, R. C. J. Moellering, Jr., J. P. Quinn, J. Hindler, A. A. Medeiros, and K. Bush. 2000. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44**:**3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasheed, J. K., G. J. Anderson, H. Yigit, A. M. Queenan, A. Doménech-Sánchez, J. M. Swenson, J. W. Biddle, M. J. Ferraro, G. A. Jacoby, and F. C. Tenover. 2000. Characterization of the extended-spectrum β-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob. Agents Chemother. 44**:**2382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen, B. A., K. Bush, D. Keeney, Y. Yang, R. Hare, C. O'Gara, and A. A. Medeiros. 1996. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother 40:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regué, M., N. Climent, N. Abitiu, N. Coderch, S. Merino, L. Izquierdo, M. Altarriba, and J. M. Tomás. 2001. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J. Bacteriol. 183**:**3564-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richet, H. M., J. Mohammed, L. C. McDonald, and W. R. Jarvis. 2001. Building communication networks: international network for the study and prevention of emerging antimicrobial resistance. Emerg. Infect. Dis. 7**:**319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (_bla_imp) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34**:**2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silver, L., M. Chandler, E. B. de la Tour, and L. Caro. 1977. Origin and direction of replication of the drug resistance plasmid R100.1 and of a resistance transfer factor derivative in synchronized cultures. J. Bacteriol. 131**:**929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Threlfall, E. J., B. Rowe, J. L. Ferguson, and L. R. Ward. 1986. Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J. Hyg. 97**:**419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsakris, A., S. Pournaras, N. Woodford, M.-F. I. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38**:**1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodford, N., M.-F. I. Palepou, G. S. Babini, B. Holmes, and D. M. Livermore. 2000. Carbapenemases of Chryseobacterium (Flavobacterium) meningosepticum: distribution of blaB and characterization of a novel metallo-β-lactamase gene_, blaB3_, in the type strain, NCTC 10016. Antimicrob. Agents Chemother. 44**:**1448-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, Y. J., P. Wu, and D. M. Livermore. 1990. Biochemical characterization of a β-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob. Agents Chemother. 34**:**755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45**:**1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yigit, H., A. M. Queenan, J. K. Rasheed, J. W. Biddle, A. Domenech-Sanchez, S. Alberti, K. Bush, and F. C. Tenover. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob. Agents Chemother. 47**:**3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshihara, E., N. Gotoh, T. Nishino, and T. Nakae. 1996. Protein D2 porin of the Pseudomonas aeruginosa outer membrane bears the protease activity. FEBS Lett. 394**:**179-182. [DOI] [PubMed] [Google Scholar]
- 41.Young, K., and L. L. Silver. 1991. Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J. Bacteriol. 173**:**3609-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]