Evolution of dnmt-2 and mbd-2-like genes in the free-living nematodes Pristionchus pacificus, Caenorhabditis elegans and Caenorhabditis briggsae (original) (raw)
Abstract
Whole genome sequencing of several metazoan model organisms provides a platform for studying genome evolution. How representative are the genomes of these model organisms for their respective phyla? Within nematodes, for example, the free-living soil nematode Caenorhabditis elegans is a highly derived species with unusual genomic characters, such as a reduced Hox cluster (Curr. Biol., 13, 37–40) and the absence of a Hedgehog signaling system. Here, we describe the recent loss of a DNA methyltransferase-2 gene (dnmt-2) in C.elegans. A _dnmt-2_-like gene is present in the satellite model organism Pristionchus pacificus, another free-living nematode that diverged from C.elegans 200–300 million years ago. In contrast, C.elegans, Caenorhabditis briggsae and P.pacificus all contain an _mbd-2_-like gene, which encodes another essential component of the methylation system of higher animals and fungi. Cel-mbd-2 is expressed throughout development and RNA interference (RNAi) experiments result in variable phenotypes. In contrast, Cbr-mbd-2 RNAi results in paralyzed larval or adult worms suggesting recent changes of gene function within the genus Caenorhabditis. We speculate that both genes were part of an ancestral DNA methylation system in nematodes and that gene loss and sequence divergence have abolished DNA methylation in C.elegans.
INTRODUCTION
Whole genome sequencing of several metazoan model organisms, such as Caenorhabditis elegans, Drosophila melanogaster, Ciona intestinalis, mouse and human, offers many opportunities for studying the evolution of genes, gene families and genome organization. In the past, such comparisons have provided important insight into the evolution of genes controlling development, immunity and human health [for review see (1)]. From an evolutionary perspective, future genome research can address two important problems. On the one hand, the sequencing of representatives of additional phyla can reveal how genomic alterations influence major evolutionary transitions (2). On the other hand, the study of distantly related species of the same phylum can indicate the degree of intraphyletic genomic variation. How representative are the genomes of the model organisms, C.elegans, Drosophila and mouse for nematodes, arthropods and vertebrates in general? Is the absence of certain genes or signaling pathways, such as Hedgehog signaling in C.elegans, the result of recent gene loss or does it represent the basic pattern of most nematode species?
To address this type of questions, one has to study the genome of additional representatives of these phyla in more detail. Pristionchus pacificus is a free-living nematode of the Diplogastridae family and has been established as a satellite model organism in evolutionary developmental biology (3–5). P.pacificus propagates as self-fertilizing hermaphrodites and has a 4 day life cycle under laboratory conditions. C.elegans and P.pacificus evolutionarily diverged 200–300 million years ago and, therefore, represent distinct members of the free-living nematodes (Figure 1) (6). One biological process that has been studied in great detail in P.pacificus is the development of the vulva, the egg-laying structure of nematode females and hermaphrodites [for reviews see (5,7)]. In recent years, a genomic approach has complemented developmental studies by generating a physical and a genetic map and performing three sequencing projects [expressed sequence tag (EST), bacterial artificial chromosome (BAC) end and fosmid end sequencing] (8–10).
Figure 1.
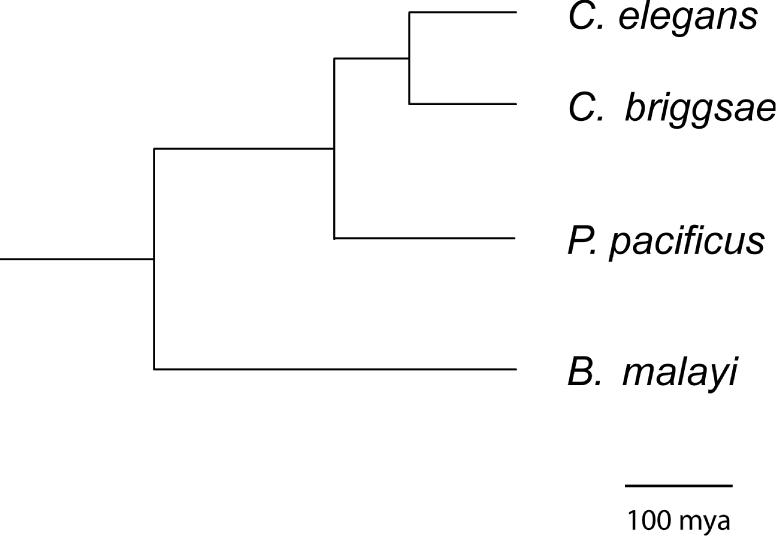
Phylogeny of the three nematodes P.pacificus, C.briggsae and C.elegans [modified after (39)]. C.elegans and C.briggsae are both members of the Rhabditidae family and belong to the genus Caenorhabditis. Recent molecular studies suggest that both species to be separated for ∼100–120 million years (13). Note that the genus Caenorhabditis contains at least 18 species and that C.elegans and C.briggsae belong to the highly derived ‘Elegans’ subgroup (31). P.pacificus is a member of the Diplogastridae family and has been suggested to be separated from C.elegans for 200–300 million years (6). For a recent phylogenetic analysis of the Diplogastridae see (40). Scale bar is 100 million years.
Here, we describe the differences in the gene repertoire of P.pacificus, C.elegans and Caenorhabditis briggsae with regard to two essential components of the methylation system of higher animals and fungi, dnmt-2 and mbd-2, respectively. Whereas P.pacificus contains both genes, dnmt-2 is completely absent from C.elegans and C.briggsae. mbd-2 is present in all three nematodes, but shows differences in sequence and gene structure, and RNA interference (RNAi) experiments in C.elegans and C.briggsae suggest recent changes in gene function.
MATERIALS AND METHODS
Strains
The strains used in this study were P.pacificus PS312 (Pasadena, CA), PS1843 (Port Angeles, WA) and RS106 (Schierenberg, Poland) (11); C.elegans N2 (12); and C.briggsae AF16 (13). Worms were grown on an Escherichia coli OP50 bacterial lawn at 20°C as described elsewhere (11).
Ppa-dnmt-2 cloning
Ppa-dnmt-2 was identified using BLAST search of the EST and BAC end sequences (http://appadb.eb.tuebingen.mpg.de/) with the Drosophila dnmt-2 gene (accession no. NM_164992). We identified the EST clone rt12b04.y1 and the BAC clone PPBAC50-E23z to contain sequences with similarity to Drosophila dnmt-2. To clone the 5′ and 3′ regions of Ppa-dnmt-2, we used the GeneRacer Kit (Invitrogen) following the manufacturer's instructions. As gene specific primers (GSP) we used the following primers:
AG5008: 5′-GAGTGAAATGCCGATACATT-3′ and AG5007: 5′-AAGGGAGAGACATTGTCCACAACTCAG-3′ for 5′-RACE; and AG5505: 5′-TCTCAATCGTCCCCTCTTGTTGTTAAG-3′ and AG5506: 5′-TCCCCTCTTGTTGTTAAGAGCCTTTTG-3′ for 3′-RACE experiments. To define the genomic structure, a PCR reaction mixture containing of 10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTP, 1 U Taq polymerase and 1 μM of primers AG5355: 5′-ATGCATCCTATGCATGAAGGAG-3′ and AG5356: 5′-5′-GCACACTTTCCGAACACATAAC-3′ was used. Thermocycling was performed in a Perkin–Elmer (Norwalk, CT) Gene Amp 9700 PCR machine under conditions consisting of an initial denaturation step (94°C for 3 min), followed by 30 cycles of 94°C for 10 s, 55°C for 30 s, and 72°C for 2 min, and a final incubation at 72°C for 5 min. The GenBank accession number for the Ppa-dnmt-2 is AY766101.
Cloning mdb-2 genes from P.pacificus, C.elegans and C.briggsae
We found an EST similar to the Drosophila mbd-2/3 gene CG8208-PB (accession no. NM_141650) in P.pacificus and performed RACE experiments, using the following GSP: AG5111: 5′-ATCTCACGGTGAAGTGAAGAAGTGC-3′ and AG5110: 5′-ATAACGACCATTCCAGCAATCAATG-3′ for 5′-RACE; and AG5108: 5′-ATTGATTGCTGGAATGGTCGTTATG-3′ and AG5109: 5′-TCAGGAAGCACTTCTTCACTTCACC-3′ for 3′-RACE; and AG5435: 5′-ATGG-GTCGAAGTCGATCTG-3′ and AG5436: 5′-CGGACAAGACTATTGGGTAG-3′ to obtain the genomic structure of the gene. Using the Ppa-mbd-2 sequence, we identified similar sequences of C.briggsae and C.elegans in WORMBASE. To confirm the expression of these genes, we used the following GSP: AG6298: 5′-AGAACTCGTTTTTCTTGTGC-3′ and AG6299: 5′-AGAGAAGCGAATATGACACG-3′ for 5′-RACE; AG6296: 5′-AAGCCGATGCTCTATTCC-3′ and AG6297: 5′-ACGTAAAAACTGGATTCGAC-3′ for 3′-RACE of C.briggsae; AG6521: 5′-TCATCCAAAGTACTTCATAAC-3′ and AG6522: 5′-GTAGTCATTGCCTCTTCTGG-3′ for 5′-RACE; and AG6519: 5′-ATGGCAAAAGCAGATGCTCTG-3′ and AG6520: 5′-ATC AAAACTGGATTCGATGG-3′ for 3′-RACE of C.elegans. The GenBank accession number for the Ppa-mbd-2 is AY766102.
Amplified fragment length polymorphism (AFLP) analysis
We used starved cultures to avoid contamination of nematode DNA with bacterial DNA. Worms were washed at least five times at hourly intervals, centrifuged at 2000 r.p.m. for 2 min and subsequently frozen overnight. Frozen pellets were ground using a pestle and mortar, and genomic DNA was isolated using an AquaPure Genomic DNA Isolation Kit (BioRad) according to the manufacturer's instructions. AFLP fingerprinting was performed according to Cervera et al. (14), using a Perkin–Elmer 9600 thermal cycler. Genomic DNA (250 ng) was digested with MspI or HpaII and EcoRI restriction enzymes and ligated with the adapters. Pre-selective amplification (20 cycles) was performed with an MspI/HpaII primer containing one selective nucleotide (GATGAGTCTCGATCGGC) and an EcoRI primer containing no selective nucleotide (GACTGCGTACCAATTC). The amplification cycle profile was 30 s at 94°C, 60 s at 56°C and 60 s at 72°C. Selective primers had two additional nucleotides for EcoRI (E+2) at their 3′ end and three additional nucleotides for MspI/HpaII (M+3). EcoRI primers were labeled with 33P. The selective amplification was performed as follows: 30 s at 94°C, 30 s annealing step (temperature see below) and 30 s at 72°C for a total of 36 cycles. The annealing temperature in the first cycle was 65°C and was subsequently reduced by 0.7°C for 12 cycles, and continued for the remaining 23 cycles at 56°C. The resulting PCR products were separated on a 5% denaturing polyacrylamide gel using a SequiGen 38 × 50 cm gel apparatus (BioRad Laboratories). Electrophoresis was performed at constant power, 80 W, for ∼3 h. After electrophoresis, gels were dried on a 3 mm Whatman paper and exposed to X-ray films for 18 h at −70°C. The following primer combinations were used in the analysis: E-TG/MH-TG, E-TG/MH-AT, E-TG/MH-GC, E-TG/MH-CG, E-TG/MH-TA, E-AT/MH-AT, E-AT/MH-AT, E-AT/MH-GC, E-AT/MH-CG and E-AT/MH-TA. We performed additional AFLP experiments using the restriction enzymes Sau3AI or NdeII and obtained similar results.
RT–PCR experiments
C.elegans eggs were harvested from gravid worms by bleaching. Hatched L1 worms were allowed to grow at 20°C in OP50-seeded NGM plates and harvested at specific time points to obtain synchronized cultures of L1, L2, L3, L4 and young adult worms. Total RNA was isolated using Trizol (Invitrogen); cDNA was synthesized using SuperScriptII Reverse Transcriptase (Invitrogen). Non-linear RT–PCR was performed using the primers AG6519 and AG6521 for the first round and AG6520 and AG6522 (see above). Cycling parameters were as for Ppa-dnmt-2 cloning.
Gene mapping
Ppa-dnmt-2 and Ppa-mbd-2 were mapped using a meiotic mapping panel prepared from 42 randomly picked F2 animals from a cross between phenotypically marked hermaphrodites of the California strain and males of the Washington strain as described previously (8).
Morpholino and RNAi
The morpholino oligonulceotide (GeneTools, OR) 5′-TAGTCTTCAAAAGGCTCTTAACAAC-3′ was used to knockdown Ppa-dnmt-2 activity. Concentrations ranging from 100 μM to 1 mM were injected into P.pacificus adult hermaphrodites. For C.briggsae and C.elegans mbd-2 downregulation, T7 promoter sequence-tagged primers were used to amplify the entire open reading frame. Double-stranded RNA was synthesized using the MegaScript T7 kit (Ambion, TX) following the manufacturer's instructions. An aliquot of 500 ng/μl were injected into adult worms. In all the cases, injected worms were singled out to fresh plates, kept at 20°C until they laid eggs and were transferred again. The phenotype of the progeny of Cel-mbd-2 and Cbr-mbd-2 RNAi treated animals were scored under the dissecting scope. The progeny of several injected animals from independent rounds of injection was scored.
RESULTS
A _dnmt-2_-like gene in P.pacificus
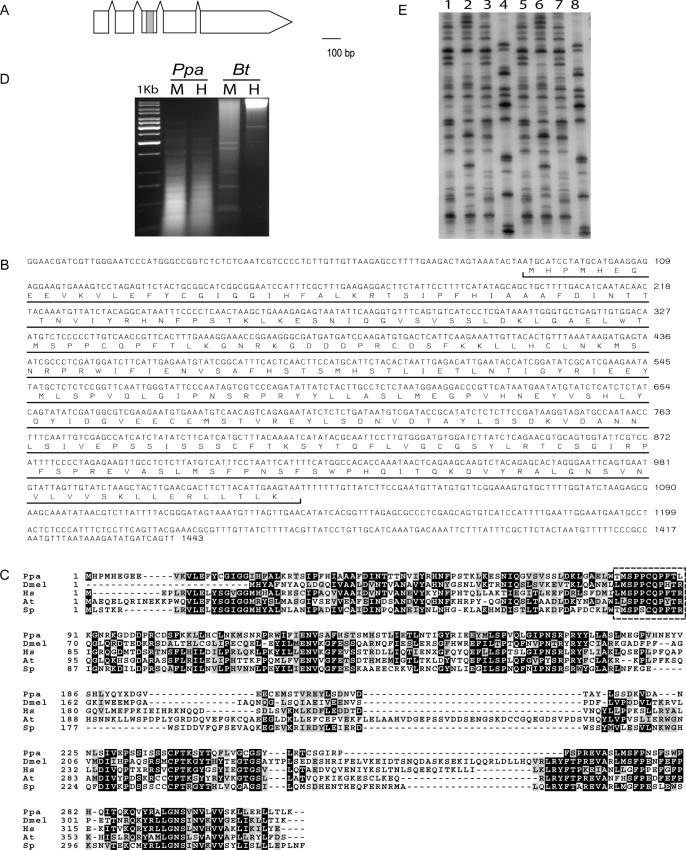
Given that the C.elegans genome has been completely sequenced, one obvious question is whether the genome of P.pacificus contains genes that are absent in C.elegans, but present in other metazoans, such as Drosophila and mouse. Indeed, we were able to identify remnants of one such gene in the BAC end sequence of PPBAC50-E23. This gene is almost similar to the DNA methyltranferase-2 (dnmt-2) genes of other animals (Figure 2) (15,16). In general, animal DNA methyltransferases can be subdivided into three subfamilies, dnmt-1, dnmt-2 and dnmt-3, based on sequence similarity (16). dnmt-1 has been regarded as a ‘maintenance’ methyltransferase, whereas dnmt-3 functions as ‘_de novo_’ methyltransferase. Recent evidence suggests that dnmt-2 enzymes are active cytosine-5′-methyltransferases (17–19).
Figure 2.
Structure, sequence alignment and analysis of Ppa-dnmt-2. (A) Gene structure of Ppa-dnmt-2. The gene contains four introns and is not _trans_-spliced to SL1. The gray box indicates the proposed transferase domain. (B) cDNA sequence of Ppa-dnmt-2 as obtained by 5′-RACE and 3′-RACE experiments. (C) Sequence alignment of _Ppa_-DNMT-2 with the DNMT-2 proteins of D.melanogaster (Dmel), Homo sapiens (Hs), Arabidopsis thaliana (At) and Schizosaccharomyces pombe (Sp). Note that the proposed active site in the domain IV, as determined in fungi and mammals, is conserved in _Ppa_-DNMT-2 (dotted box) (41). (D) Restriction digest of genomic DNA of P.pacificus (Ppa) and Bos taurus (Bt, positive control) with the methylation-sensitive restriction enzyme, HpaII (H) and methylation-insensitive isoschizomer MspI (M). No differences in the restriction patterns have been observed in P.pacificus. (E) Comparison of the AFLP patterns of three strains of P.pacificus from California (lanes 1 and 5), Washington (lanes 2 and 6) and Poland (lanes 3 and 7) and C.elegans N2 wild-type animals (lanes 4 and 8). In lanes 1–4, DNA has been digested with the methylation-sensitive restriction enzyme HpaII and in lanes 5–8 with the methylation-insensitive isoschizomer MspI. No difference in the AFLP pattern was observed in P.pacificus.
Next, we examined whether the obtained BAC end sequence corresponds to a real gene. We amplified a full-length cDNA clone and could show that the gene is expressed throughout the development of the organism (Figure 2B and data not shown). The _Ppa_-DNMT-2 protein is highly conserved relative to other DNMT-2 proteins with amino acid identities of 34% between P.pacificus and Drosophila and 40% between P.pacificus and mouse (Figure 2C). Taken together, we have identified the first dnmt-2 gene of a nematode and can show that the gene encodes for a protein with high sequence similarity to other DNMT-2 proteins.
DNA methylation is not a genome-wide phenomenon in P.pacificus
DNA methylation differs among metazoans from the apparent absence of methylation (as in C.elegans), to methylation of small fractions of the genome (as in Drosophila) to genome-wide methylation in mammals (15,16). In C.elegans, the absence of DNA methylation is consistent with the absence of a _dnmt-2_-like gene, which encodes one of the essential components of the methylation machinery (20). Nippostrongylus brasiliensis is the only other nematode that has been previously tested for DNA methylation, but as for C.elegans, no signs of DNA methylation were detected (21).
We sought to determine whether DNA methylation is a genome-wide phenomenon in P.pacificus. To this end, total genomic DNA was digested with methylation-sensitive restriction enzymes, i.e. HpaII and methylation-insensitive isoschizomers, i.e. MspI. No differences in the restriction patterns have been observed indicating that DNA methylation is not a genome-wide phenomenon in P.pacificus (Figure 2D). Similarly, no differences were observed after AFLP analysis of HpaII and MspI digested animals (Figure 2E).
We tried to assess the function of Ppa-dnmt-2 by reducing gene activity using the morpholino technology (MO) (6). Morpholinos are synthetic DNA analogs that have been shown to efficiently block translation of their target RNAs in a variety of animal species (22). A Ppa-dnmt2(MO) containing the translation start site of Ppa-dnmt2 has no obvious phenotype under laboratory conditions. The progeny of Ppa-dnmt2(MO) injected animals grow and live normally and have a normal brood size (data not shown). Thus, silencing of Ppa-dnmt2(MO) has no obvious phenotype under standard laboratory conditions. This result parallels the mouse knockout of DNMT2, which also has no obvious phenotype in ES cells (23). Similarly, depletion of Dnmt2 by RNAi in Drosophila has no apparent phenotype in embryonic development (18).
Rapid sequence evolution of the MBD2 proteins in nematodes
Besides DNA methyltransferases, methyl-CpG binding proteins (MBD) are essential components of the DNA methylation system [for review see (16)]. Vertebrate genomes usually encode four MBD proteins, whereas invertebrates contain only one gene. The invertebrate genes have been named mbd-2/3 because they have the highest sequence similarity to mbd-2 and mbd-3 of vertebrates. To determine whether nematodes contain similar genes, we used an in silico approach.
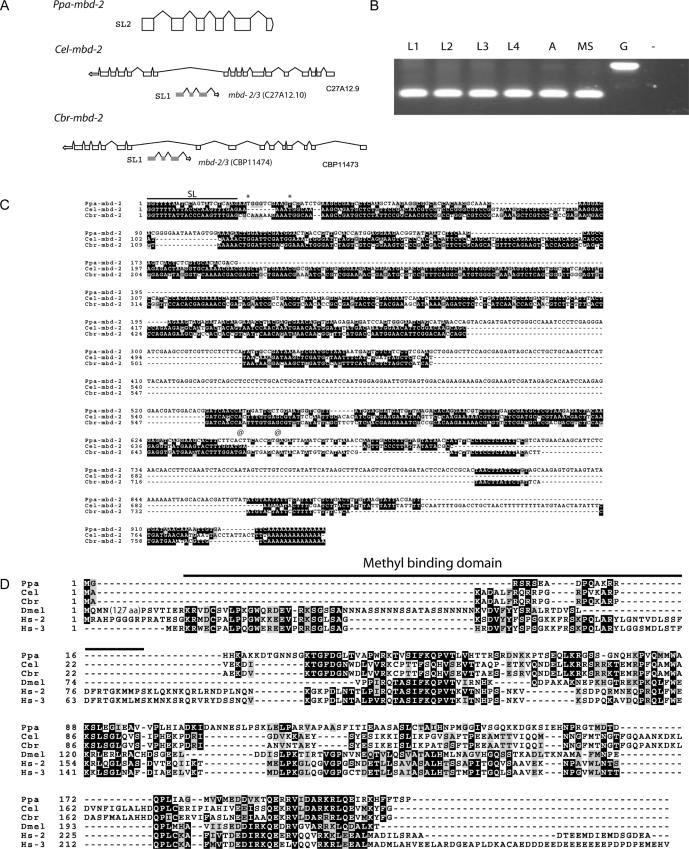
In P.pacificus, we identified an _mbd-2/3_-like gene in the BAC clone PPBAC26-H09. We cloned a full-length cDNA and showed that the gene is SL2 spliced and therefore most likely a member of an operon (Figure 3A and B). By genome walking, we could show that the neighboring genes to Ppa-mbd-2/3 are similar to the C.elegans genes R05D11.5 and D2030.5. Interestingly, R05D11.5 is SL2 spliced and is located in the operon CE0P1484 (24). The existence of operons in P.pacificus had recently been demonstrated, but it remains unknown whether operon organization is generally conserved between P.pacificus and C.elegans (25). As the gene name ‘_mbd-2/3_’ would be inconsistent with the nomenclature rules in nematodes, we refer to the _mbd_-like genes as ‘_mbd-2_’. The _Ppa_-MBD2 protein has an overall amino acid identity of 27 and 25% to the Drosophila and mouse proteins, respectively (Figure 3D).
Figure 3.
Structure and sequence alignment of nematode mbd-2 genes. (A) Ppa-mbd-2 is _trans_-spliced to an SL2 leader sequence and contains five introns. Cel-mbd-2 and Cbr-mbd-2 are located in the intron of C27A12.9 and CBP11473, respectively and are _trans_-spliced to SL1. (B) Stage specific expression of Cel-mbd-2 by RT–PCR analysis. Cel-mbd-2 is expressed throughout development. MS, mixed stages; L1–L4, first to fourth larval stage; A, adult; G, amplification from genomic DNA; −, negative control. (C) cDNA sequences of Ppa-mbd-2, Cel-mbd-2 and Cbr-mbd-2 as obtained by 5′-RACE and 3′-RACE experiments. The SL1 and SL2 splice leader sequences are indicated by a black line. Conserved nucleotides are shown in black. *, conceptual translational start site; @, stop codon. (D) Sequence alignment of the MBD-2 proteins of the three nematodes P.pacificus (Ppa), C.elegans (Cel), C.briggsae (Cbr) and D.melanogaster (Dmel) and the two human proteins MBD2 (Hs2) and MBD3 (Hs3). Note that the methyl-binding domain (black line) is nearly completely absent in the proteins of all three nematodes. Conserved amino acids are shown in black, similar amino acids in gray.
The annotation of the C.elegans genome did not contain a gene encoding an MBD-2-like protein (WormBase release WS120). However, we identified an _mbd-2_-like gene in the recently sequenced genome of C.briggsae (Figure 3A, C and D). C.briggsae is one of the closest known relatives of C.elegans and both nematodes diverged from a common ancestor roughly 100 million years ago (Figure 1) (13). Cbr-mbd-2 is located in an intron of the gene CBP11473 on chromosome I (Figure 3A). With this information in hand, we re-analyzed the C.elegans genome and identified a Cel-mbd-2 gene that shows the same organization as Cbr-mbd-2 (Figure 3A). Cel-mbd-2 is located in an intron of the orthologous gene C27A12.9, but had not been annotated previously. In contrast to Ppa-mbd-2, Cel-mbd-2 and Cbr-mbd-2 are SL1 spliced and there is no indication that the two genes would be members of an operon (Figure 3A and C).
The sequence comparison of the mbd-2 genes from C.elegans, C.briggsae, P.pacificus and Drosophila revealed a striking pattern of sequence conservation. In particular, the nematode sequences lack a typical methyl-binding domain, which was shown to be crucial for the function of MBD-2 proteins for DNA methylation (Figure 3C and D) (16). The absence of a methyl-binding domain provides additional evidence for the absence of DNA methylation in these nematodes.
mbd-2 RNAi results in distinct phenotypes in C.elegans and C.briggsae
RT–PCR experiments indicate that Cel-mbd-2 is expressed throughout development (Figure 3B). To investigate a potential function of the mbd-2 genes in C.elegans and C.briggsae, we used the RNAi technology (26). mbd-2(RNAi) results in distinct phenotypes in these two closely related nematodes. Cbr-mbd-2(RNAi) results in a strong Uncoordinated movement (Unc) phenotype. Animals become paralyzed and eventually die at various stages during post-embryonic development (Figure 4A). Specifically, 35% of RNAi treated animals die during larval development and 17% as adults. The remaining animals had no visible phenotype and produced normal progeny. In contrast, Cel-mbd-2(RNAi) results in animals with a variety of phenotypes. In total, 18% of the progeny of RNAi treated animals had an Unc phenotype (Figure 4C). However, these animals did not show signs of complete paralyzation as was seen in more than 50% of C.briggsae animals after Cbr-mbd-2(RNAi). A total of 11% of the Cel-mbd-2(RNAi) treated animals undergo a rupture in the region of the gonad (Figure 4B). In addition, a few animals with a multivulva phenotype have been observed. However, the frequency of this phenotype was below 1% and, therefore, more detailed vulval cell lineage analysis of such animals could not be performed. Embryonic lethality has not been observed in RNAi treated animals of both species. Taken together, Cel-mbd-2 and Cbr-mbd-2 RNAi treatment result in strong morphological defects, which suggest a function of both genes during post-embryonic development.
Figure 4.
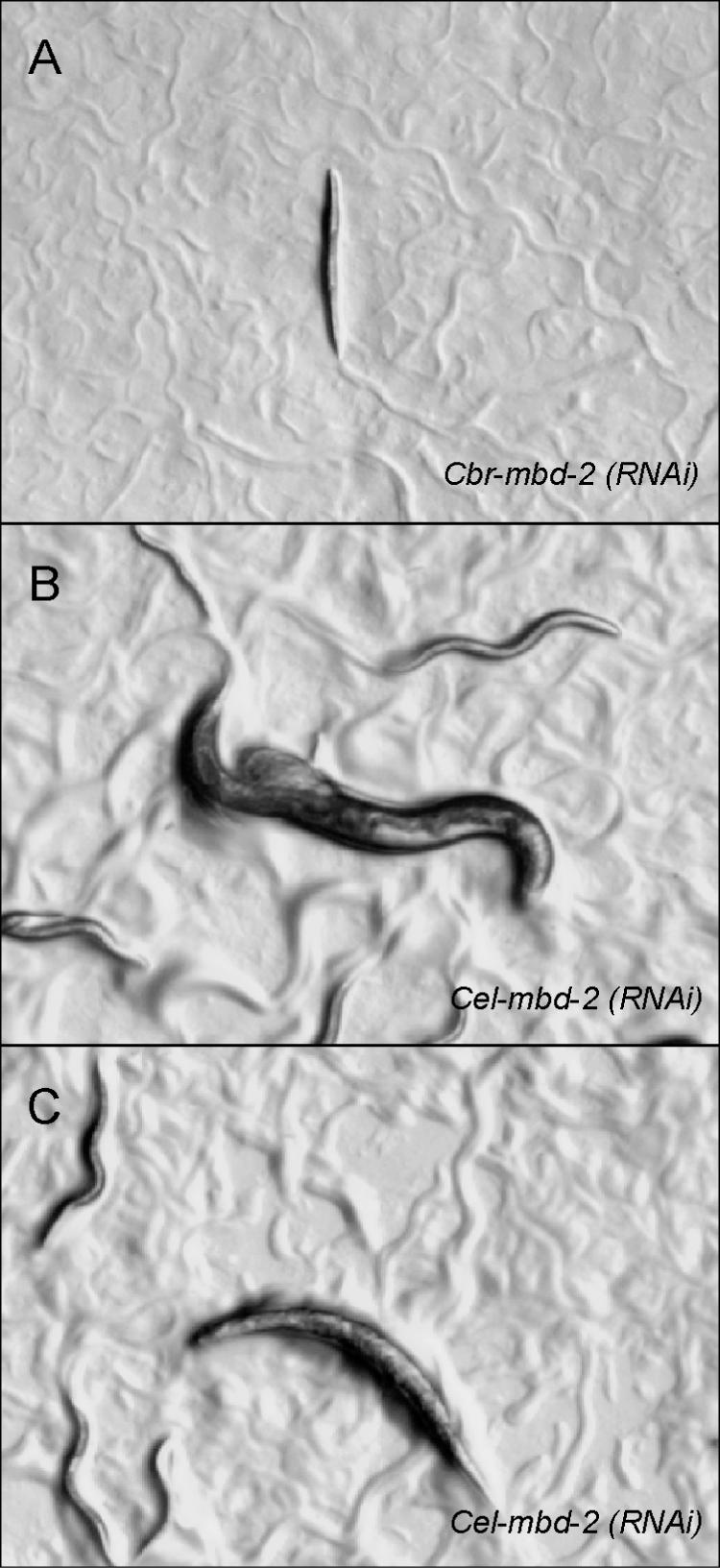
Phenotype of the progeny of RNAi treated animals. (A) _Cbr-mbd-2_RNAi results in progeny with a strong paralyzation. (B and C) _Cel-mbd-2_RNAi treated animals result in progeny that rupture at the gonad (B) or have an Unc phenotype (C).
Ppa-dnmt-2 is located on chromosome IV
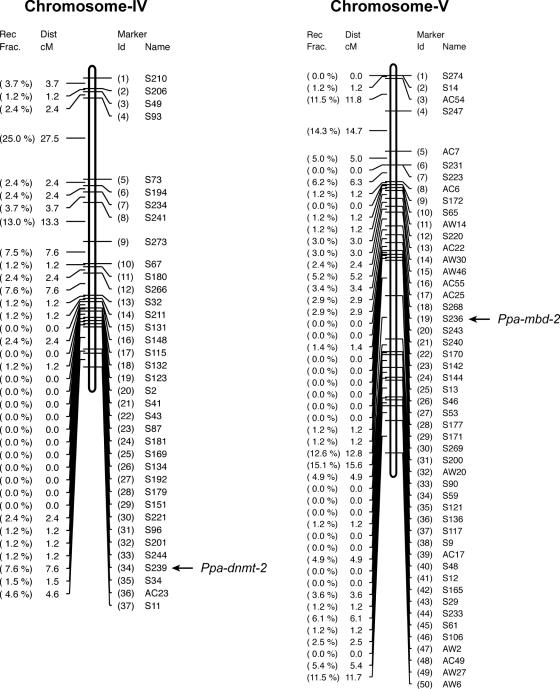
Given the gene loss of dnmt-2 in Caenorhabditis, it is of interest to determine the chromosomal location of Ppa-dnmt-2. Recent work has identified that the chromosomes of C.elegans and P.pacificus are in large parts homologous to one another (9,27). Chromosomal rearrangements between both species are common, but are nearly exclusively intrachromosomal (27). We generated single-stranded conformational polymorphism (SSCP) markers in Ppa-dnmt-2 and Ppa-mbd-2 and placed these SSCP markers on the genetic linkage map (8). Ppa-mbd-2 is located on chromosome V of P.pacificus, which has been identified to correspond to C.elegans chromosome I (Figure 5) (M. Zheng and R. J. Sommer, unpublished observation). Cel-mbd-2 and Cbr-mbd-2 are both located on chromosome I, indicating that the chromosomal location of this gene has been evolutionarily conserved. The SSCP marker S239 in Ppa-dnmt-2 is located at the tip of chromosome IV of P.pacificus between the markers S244 and S34 (Figure 5).
Figure 5.
Chromosomal localization of Ppa-dnmt-2 and Ppa-mbd-2.
DISCUSSION
We show differences in the gene repertoire of the three soil nematodes P.pacificus, C.briggsae and C.elegans. Although _dnmt2_-like genes cannot be identified in C.briggsae and C.elegans, a clear 1:1 ortholog to insect and vertebrates genes does exist in P.pacificus. Ppa-dnmt-2 is the first _dnmt-2_-like gene to be described from a nematode species and this finding suggests a relatively recent loss of dnmt-2 in the evolutionary lineage that gave rise to Caenorhabditis. Whole genome sequencing projects currently ongoing might provide additional information with regard to the exact time of the gene loss: Caenorhabditis remanei, Caenorhabditis japonica and Caenorhabditis sp. CB5161 are currently being sequenced, and information about the presence or absence of _dnmt-2_-like genes in these genomes will indicate whether the gene loss occurred before or after the divergence of these Caenorhabditis species.
Although the P.pacificus genome contains a dnmt-2 and an _mbd-2_-like gene, we were so far unable to provide any evidence for DNA methylation in this nematode species. Unfortunately, little is known about DNA methylation in nematodes in general. Besides C.elegans, only Nippostrongyloides has been investigated and no signs of DNA methylation have been observed in either of the two species (21). However, as for the data provided for P.pacificus in this report, the existing data from C.elegans and Nippostrongyloides do not completely rule out the existence of low-level methylation at particular developmental stages. For example, the genome of D.melanogaster was considered for a long time to be completely unmethylated. Recent reports, however, clearly demonstrated methylation of specific regions of the genome during embryogenesis (28,29). In addition, DNA methylation might have evolved substantially among nematodes. Considering the deep phylogenetic branches within the phylum (30), only the direct analysis of more distantly related nematodes can address this question further.
The evolution of mbd-2, although not as extreme as that of dnmt-2, also provides some indications for recent evolutionary changes in Caenorhabditis. First, sequence comparison of the _mbd-2_-like genes of C.briggsae, C.elegans and P.pacificus indicates some deviations from the consensus sequence that is not observed in any known plant, fungi, insect or vertebrate sequences. The methyl-binding domain is partially absent from the genes of the three nematodes (Figure 3B). However, these modified enzymes could perform other biochemical functions that are completely unrelated to DNA methylation (see below). Second, reduction of mbd-2 function by RNAi results in distinct phenotypes in C.briggsae and C.elegans, suggesting recent functional diversification of Cel-mbd-2 or Cbr-mbd-2 or both. Normally, functional differences between C.briggsae and C.elegans are rare and have been reported for only a small amount of genes and processes (13,31–34). Morphologically both species are indistinguishable (35), but molecular studies indicate substantial differentiation of the genome (13).
The Unc phenotypes seen for RNAi treated animals of mbd-2 genes in Caenorhabditis, suggest that MBD proteins may have an ancient neuronal function dating back to early metazoa and points toward a possible medical significance. It has been known for some time that the methylcytosine-binding protein MeCP2 is required in mammalian neurons for normal brain function, and MeCP2 mutations cause the progressive autism Rett syndrome (RTT) in humans and an RTT-like phenotype in mice (36,37). More evidence that methyl-binding proteins can evolve to have functions other than the binding of methylated DNA is available from studies of the mouse orthologs of mbd-2, MBD2 versus MBD3. Although closely related, these two proteins have qualitatively distinct functions (38). The RNAi phenotypes in C.elegans and C.briggsae demonstrate a vital function of MBD2 proteins that is probably unrelated to methylation. The independence of mammalian MBD3 from methylation may reflect such a function that has been conserved in metazoa, and such methylation-independent processes may at least account for the autistic phenotypes seen in humans with RTT. Therefore, future studies could use Cel-mbd-2 as a potential model for studying these methylation-independent but disease-related functions of MBD proteins in more detail.
This study provides further evidence for the absence of DNA methylation in free-living nematodes. One might speculate that the absence of DNA methylation in C.elegans and other related nematode species provides evolutionary plasticity of ‘DNA methylation’ genes to adopt new functions. One approach to study this further would be a more detailed RNAi analysis of _mbd-2_-like genes in additional Caenorhabditis species, once the genome sequence of these species is available. In addition, whole genome sequencing of more nematode genomes provides a platform to study gene loss (and maybe gain) of additional genes related to DNA methylation. The compactness of many nematode genomes makes them an attractive group of organisms for studies on the genetic repertoire of eukaryotic genomes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr R. Hong for discussions and critically reading the manuscript.
DDBJ/EMBL/GenBank accession nos+ AY766101 and AY766102
REFERENCES
- 1.Brown T.A. (2002) Genomes, 2nd edn. Wiley-Liss, NY. [PubMed] [Google Scholar]
- 2.Collins F.S., Green,E.D., Guttmacher,A.E. and Guyer,M.S. (2003) A vision for the future of genomics research. Nature, 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 3.Sommer R.J. (2000) Comparative genetics: a third model nematode species. Curr. Biol., 10, R879–R881. [DOI] [PubMed] [Google Scholar]
- 4.Simpson P. (2002) Evolution of development in closely related species of flies and worms. Nature Rev. Genet., 3, 907–917. [DOI] [PubMed] [Google Scholar]
- 5.Rudel D. and Sommer,R.J. (2003) The evolution of developmental mechanisms. Dev. Biol., 264, 15–37. [DOI] [PubMed] [Google Scholar]
- 6.Pires-daSilva A. and Sommer,R.J. (2004) Conservation of the global sex determination gene tra-1 in distantly related nematodes. Genes Dev., 18, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sommer R. (2001) As good as they get: cells in nematode vulva development and evolution. Curr. Opin. Cell Biol., 13, 715–720. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan J., Sinz,W., Lanz,C., Brand,A., Nandakumar,R., Raddatz,G., Witte,H., Keller,H., Kipping,I., Pires-daSilva,A. et al. (2002) A bacterial artificial chromosome-based genetic linkage map of the nematode Pristionchus pacificus. Genetics, 162, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan J., Sinz,W., Jesse,T., Wiggers-Perebolte,L., Jansen,K., Buntjer,J., van der Meulen,M. and Sommer,R.J. (2003) An integrated physical and genetic map of the nematode Pristionchus pacificus. Mol. Genet. Genomics, 269, 715–722. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasan J., Otto,G.W., Kahlow,U., Geisler,R. and Sommer,R.J. (2004) AppaDB: an AcedB database for the nematode satellite organism Pristionchus pacificus. Nucleic Acids Res., 32 (Database issue), D421–D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommer R.J. and Sternberg,P.W. (1996) Evolution of nematode vulval fate patterning. Dev. Biol., 173, 396–407. [DOI] [PubMed] [Google Scholar]
- 12.Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein L.D., Bao,Z., Blasiar,D., Blumenthal,T., Brent,M.R., Chen,N., Chinwalla,A., Clarke,L., Clee,C., Coghlan,A. et al. (2003) The Genome Sequence of Caenorhabditis briggsae: A Platform for Comparative Genomics. PLoS Biol., 1, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cervera M.T., Ruiz-Garcia,L. and Martinez-Zapater,J.M. (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol. Genet. Genomics, 268, 543–552. [DOI] [PubMed] [Google Scholar]
- 15.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 16.Hendrich B. and Tweedie,S. (2003) The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet., 19, 269–277. [DOI] [PubMed] [Google Scholar]
- 17.Hermann A., Schmitt,S. and Jeltsch,A. (2003) The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem., 278, 31717–31721. [DOI] [PubMed] [Google Scholar]
- 18.Kunert N., Marhold,J., Stanke,J., Stach,D. and Lyko,F. (2003) A Dnmt2-like protein mediates DNA methylation in Drosophila. Development, 130, 5083–5090. [DOI] [PubMed] [Google Scholar]
- 19.Tang L.Y., Reddy,M.N., Rasheva,V., Lee,T.L., Lin,M.J., Hung,M.S. and Shen,C.K. (2003) The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J. Biol. Chem., 278, 33613–33616. [DOI] [PubMed] [Google Scholar]
- 20.The C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C.elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 21.Tweedie S., Charlton,J., Clark,V. and Bird,A. (1997) Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol. Cell. Biol., 17, 1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heasman J. (2002) Morpholino oligos: making sense of antisense?. Dev. Biol., 243, 209–214. [DOI] [PubMed] [Google Scholar]
- 23.Okano M., Xie,S. and Li,E. (1998) Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res., 26, 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blumenthal T., Evans,D., Link,C.D., Guffanti,A., Lawson,D., Thierry-Mieg,J., Thierry-Mieg,D., Chiu,W.L., Duke,K., Kiraly,M. et al. (2002) A global analysis of Caenorhabditis elegans operons. Nature, 417, 851–854. [DOI] [PubMed] [Google Scholar]
- 25.Lee K.Z. and Sommer,R.J. (2003) Operon structure and _trans_-splicing in the nematode Pristionchus pacificus. Mol. Biol. Evol., 20, 2097–2103. [DOI] [PubMed] [Google Scholar]
- 26.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 27.Lee K.Z., Eizinger,A., Nandakumar,R., Schuster,S.C. and Sommer,R.J. (2003) Limited microsynteny between the genomes of Pristionchus pacificus and Caenorhabditis elegans. Nucleic Acids Res., 31, 2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tweedie S., Ng,H.H., Barlow,A.L., Turner,B.M., Hendrich,B. and Bird,A. (1999) Vestiges of a DNA methylation system in Drosophila melanogaster?. Nature Genet., 23, 389–390. [DOI] [PubMed] [Google Scholar]
- 29.Lyko F., Ramsahoye,B.H. and Jaenisch,R. (2000) DNA methylation in Drosophila melanogaster. Nature, 408, 538–540. [DOI] [PubMed] [Google Scholar]
- 30.De Ley P. and Blaxter,M. (2002) Systematic Position and Phylogeny. In Lee,D.L. (ed.), The Biology of nematodes. Taylor & Francis, London, NY, 1–30. [Google Scholar]
- 31.Kirouac M. and Sternberg,P.W. (2003) _cis_-Regulatory control of three cell fate-specific genes in vulval organogenesis of Caenorhabditis elegans and C.briggsae. Dev. Biol., 257, 85–103. [DOI] [PubMed] [Google Scholar]
- 32.Wang X. and Chamberlin,H.M. (2002) Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev., 16, 2345–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudel D. and Kimble,J. (2002) Evolution of discrete Notch-like receptors from a distant gene duplication in Caenorhabditis. Evol. Dev., 4, 319–333. [DOI] [PubMed] [Google Scholar]
- 34.Fitch D.H., Bugaj-Gaweda,B. and Emmons,S.W. (1995) 18S ribosomal RNA gene phylogeny for some Rhabditidae related to Caenorhabditis. Mol. Biol. Evol., 12, 346–358. [DOI] [PubMed] [Google Scholar]
- 35.Sudhaus W. and Kiontke,K. (1996) Phylogeny of Rhabditis subgenus Caenorhabditis (Rhabditidae, Nematoda). J. Zoolog. Syst. Evol. Res., 34, 217–233. [Google Scholar]
- 36.Kriaucionis S. and Bird,A. (2003) DNA methylation and Rett syndrome. Hum. Mol. Genet., 12, R221–R227. [DOI] [PubMed] [Google Scholar]
- 37.Zoghbi H.Y. (2003) Postnatal neurodevelopmental disorders: meeting at the synapse? Science, 302, 826–830. [DOI] [PubMed] [Google Scholar]
- 38.Hendrich B., Guy,J., Ramsahoye,B., Wilson,V.A. and Bird,A. (2001) Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev., 15, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaxter M.L., De Ley,P., Garey,J.R., Liu,L.X., Scheldeman,P., Vierstraete,A., Vanfleteren,J.R., Mackey,L.Y., Dorris,M., Frisse,L.M. et al. (1998) A molecular evolutionary framework for the phylum Nematoda. Nature, 392, 71–75. [DOI] [PubMed] [Google Scholar]
- 40.Fürst von Lieven A. and Sudhaus,W. (2000) Comparative and functional morphology of the buccal cavity of Diplogastrina (Nematoda) and a first outline of the phylogeny of this taxon. J. Zoolog. Syst. Evol. Res., 38, 37–63. [Google Scholar]
- 41.Dong A., Yoder,J.A., Zhang,X., Zhou,L., Bestor,T.H. and Cheng,X. (2001) Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res., 29, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]